Abstract
Cellular and biochemical studies support a role for all five human RecQ helicases in DNA replication; however, their specific functions during this process are unclear. Here we investigate the in vivo association of the five human RecQ helicases with three well-characterized human replication origins. We show that only RECQ1 (also called RECQL or RECQL1) and RECQ4 (also called RECQL4) associate with replication origins in a cell cycle-regulated fashion in unperturbed cells. RECQ4 is recruited to origins at late G1, after ORC and MCM complex assembly, while RECQ1 and additional RECQ4 are loaded at origins at the onset of S phase, when licensed origins begin firing. Both proteins are lost from origins after DNA replication initiation, indicating either disassembly or tracking with the newly formed replisome. Nascent-origin DNA synthesis and the frequency of origin firing are reduced after RECQ1 depletion and, to a greater extent, after RECQ4 depletion. Depletion of RECQ1, though not that of RECQ4, also suppresses replication fork rates in otherwise unperturbed cells. These results indicate that RECQ1 and RECQ4 are integral components of the human replication complex and play distinct roles in DNA replication initiation and replication fork progression in vivo.
The RecQ helicases are a family of DNA-unwinding enzymes essential for the maintenance of genome integrity in all kingdoms of life. Five RecQ enzymes have been found in human cells: RECQ1 (also called RECQL or RECQL1), BLM (RECQ2 or RECQL3), WRN (RECQ3 or RECQL2), RECQ4 (RECQL4), and RECQ5 (RECQL5) (3, 7). Here we refer to these helicases as RECQ1, RECQ4, and RECQ5, without the “L” that is present in the official gene names. Mutations in the BLM, WRN, and RECQ4 genes are linked to Bloom syndrome (BS), Werner syndrome (WS), and the subset of Rothmund-Thomson syndrome (RTS) patients at high risk of developing osteosarcomas, respectively (19, 31, 71). RECQ4 mutations have also been associated with RAPADILINO and Baller-Gerold syndrome (56, 61). Although these disorders are all associated with inherent genomic instability and cancer predisposition, they show distinct clinical features, suggesting that BLM, WRN, and RECQ4 are involved in different aspects of DNA metabolism. However, the molecular events underlying the pathogenesis of BS, WS, and RTS remain obscure. Mutations in the remaining two human RecQ helicase genes, RECQ1 and RECQ5, have not as yet been identified as causes of either genomic instability or heritable cancer predisposition disorders.
Several lines of evidence suggest that RecQ helicases play an important role in DNA replication control (3, 10). In particular, RecQ helicases are thought to facilitate replication by preserving the integrity of stalled replication forks and by remodeling or repairing damaged or collapsed forks to allow the resumption of replication. Consistent with these ideas, several investigators have shown that primary fibroblasts from BS, WS, and RTS patients and RecQ5-deficient mouse embryonic fibroblasts all show differential hypersensitivity to agents that perturb DNA replication (12, 14, 26, 29). Moreover, BLM and WRN are recruited to DNA replication forks after replicative stress, and DNA fiber track analyses have shown that both BLM and WRN are required for normal fork progression after DNA damage or replication arrest (11-13, 47, 54). In particular, BLM in conjunction with DNA topoisomerase III and two other accessory proteins, RMI-1 and RMI-2, has been shown to catalyze the resolution of double-Holliday-junction recombination intermediates to generate noncrossover products. This dissolution reaction could play an important role in the error-free recombinational repair of damaged or stalled forks during S phase (57, 67). WRN also appears to promote error-free repair by contributing to the resolution of gene conversion events to generate noncrossover products (46). In line with the above observations, WRN and BLM can be found associated with replication foci or other DNA damage response proteins in damaged cells. In contrast, in unperturbed cells, a majority of each protein is found in the nucleolus (WRN) or associated with PML bodies (BLM) (5, 37, 62).
RECQ4 has also been implicated in DNA replication. Recent studies have shown that hypomorphic mutants of the Drosophila melanogaster homolog of human RECQ4, DmRECQ4, have reduced DNA replication-dependent chorion gene amplification (65). These findings are thus consistent with a postulated role for Xenopus laevis RECQ4 (XRECQ4) in the initiation of DNA replication (39, 48). The N terminus of XRECQ4 bears homology to the N termini of the yeast proteins Sld2 (Saccharomyces cerevisiae [budding yeast]) and DRC1 (Schizosaccharomyces pombe [fission yeast]), which play a central role, in association with budding yeast Dpb11 and the fission yeast homolog Cut5/Rad4, in the establishment of DNA replication forks (38, 41, 63). Consistently, the N terminus of XRECQ4 has been shown to interact with the X. laevis variant of Cut5, and XRECQ4 depletion severely perturbs DNA replication initiation in X. laevis egg extracts (39, 48). The notion that the function of XRECQ4 is evolutionarily conserved in mammals is supported by the observations that the human protein can complement its Xenopus counterpart in cell-free assays for replication initiation and that depletion of human RECQ4 inhibits cellular proliferation and DNA synthesis (39, 48). Moreover, deletion of the N-terminal region of mouse RECQ4 has been shown to be an embryonic lethal mutation (27). These observations suggest that vertebrate RECQ4 might be a functional homolog of Sld2/DRC11, although its precise function during replication initiation and progression is not known. Recent results, published while this work was in progress, indicate that human RECQ4 interacts with the MCM replicative complex during replication initiation and that this interaction is regulated by CDK phosphorylation of RECQ4 (69). These findings, together with our results below, provide clues to the mechanism regulating RECQ4 interaction with the replication machinery.
RECQ1 is the most abundant of the human RecQ helicases and was the first of the human RecQ proteins to be discovered on the basis of its potent ATPase activity (50). Despite this, little is known about the cellular functions of RECQ1, and no human disease associations have been identified to date. Recent studies have shown that RECQ1 is involved in the maintenance of genome integrity and that RECQ1 depletion affects cellular proliferation (51). Moreover, biochemical studies have shown that RECQ1 and BLM display distinct substrate specificities, indicating that these helicases are likely to perform nonoverlapping functions (43). These results suggest an important—though as yet mechanistically ill-defined—role for RECQ1 in cell cycle progression and/or DNA repair (52).
In order to better delineate the role of human RecQ helicases in DNA replication, we investigated the in vivo interactions of all five human RecQ enzymes with three well-characterized human DNA replication origins in quantitative chromatin immunoprecipitation (ChIP) assays. We also determined how nascent-origin-dependent DNA synthesis, chromatin binding of replication proteins, origin firing frequency, and replication fork rates were altered by depleting specific human RecQ helicase proteins. We found that only two of the five human RecQ helicases, RECQ1 and RECQ4, specifically interact with origins in unperturbed cells. Our results provide new mechanistic insight into the distinct roles of human RECQ1 and RECQ4 in DNA replication initiation and in replication fork progression.
MATERIALS AND METHODS
Antibodies.
Polyclonal antibodies against RECQ1 (BL2074) and WRN (NB 100-471) were purchased from Bethyl Laboratories and Novus Biologicals, respectively. Polyclonal antibody raised against residues 60 to 111 of human RECQ4 was produced in Weidong Wang's laboratory as described previously (70). Polyclonal antibodies against BLM and RECQ5 were generous gifts from Ian Hickson (66) and Pavel Janscak (30), respectively. Anti-Orc2 (3B7) antibodies were from MBL. Polyclonal anti-ORC1 antisera were produced and purified by immunization of rabbits with a His-tagged ORC1 protein fragment consisting of residues 250 to 480 (59). MCM3 (N-19), MCM4 (H-300), Cdc6 (N-19), cyclin E (M-20), cyclin A1 (H-432), and PCNA (F-2) antibodies were purchased from Santa Cruz Biotechnologies. RPA (BL915) antibody was purchased from Bethyl Laboratories. Anti-α-tubulin (B-5-1-2) antibody was obtained from Sigma. Anti-p84 (3F10) and rat antichlorodeoxyuridine/bromodeoxyuridine (anti-CldU/BrdU) [BU1/75(ICR1)] antibodies were from Abcam. Mouse anti-iododeoxyuridine/BrdU (anti-IdU/BrdU) (347580) and anti-BrdU-fluorescein isothiocyanate (antiBrdU-FITC) (347583) antibodies were from BD Biosciences. Anti-rat Alexa 594-conjugated (A11007) and anti-mouse Alexa 488-conjugated (A11001) antibodies were obtained from Molecular Probes.
Cell culture, synchronization, and cell cycle analysis.
The T98G/HeLa and IMR-90 cell lines were grown in Dulbecco's modified Eagle's medium (DMEM) and minimum essential medium (MEM), respectively, supplemented with Glutamax (Life Technologies) and 10% (vol/vol) fetal bovine serum (FBS; Life Technologies). K562 cells were grown in RPMI 1640 medium supplemented with Glutamax and 10% (vol/vol) FBS. T98G cells were synchronized by serum starvation as previously reported (42, 58) or by drug arrest with 0.8 mM mimosine (Sigma), as were HeLa and K562 cells. Synchronized cells were then analyzed for their cell cycle profile (DNA content) by propidium iodide staining (Sigma) and flow cytometry on a FACSCalibur flow cytometer (Becton Dickinson). Cell cycle profile distributions were determined with Modfit LT 3.0 software. BrdU incorporation experiments were performed on transiently small interfering RNA (siRNA)-transfected cells at 72 h posttransfection. Cells were pulsed for 1 h with BrdU (Sigma) (final concentration, 10 μM), and BrdU-positive cells were detected by using a mouse anti-BrdU-FITC primary antibody followed by an anti-mouse Alexa 488-conjugated secondary antibody. Flow cytometric analysis of cells was performed on a FACSCalibur flow cytometer (Becton Dickinson) to simultaneously determine the cell cycle profile (DNA content) by incorporation of propidium iodide and the S-phase cell population by incorporation of BrdU. Cell cycle profile distributions were determined with CellQuestPro and Modfit LT 3.0 software.
Chromatin immunoprecipitation assays.
Cells were fixed by adding formaldehyde (Fluka) directly to the cell culture medium to a 1% (vol/vol) final concentration. Cross-linking was allowed to proceed for 7 min at 37°C and then stopped by the addition of glycine (Sigma) at a final concentration of 125 mM. Cells were washed and digested with 125 U micrococcal nuclease S7 (Roche)/1 × 107 to 2 × 107 cells at 37°C for 15 min. Reactions were stopped by adding EDTA (20 mM final concentration). Cells were washed and resuspended in HNNG buffer (20 mM HEPES, pH 7.5, 250 mM NaCl, 0.5% NP-40, 10% glycerol, 1 mM phenylmethylsulfonyl fluoride [PMSF]) supplemented with protease inhibitors, and then chromatin was further sheared by sonication to generate DNA fragments of <0.5 kb. The resulting sonicated lysates were used for immunoprecipitation with anti-RECQ1, -BLM, -WRN, -RECQ4, -RECQ5, -ORC2, -ORC1, -MCM3, and -CDC6 antibodies. A rabbit IgG was used as a negative control. Immunocomplexes were collected with protein A beads, washed sequentially with HNNG buffer and HLNG buffer (15 mM HEPES, pH 7.5, 250 mM LiCl, 0.5% NP-40, 10% glycerol, 1 mM PMSF), resuspended in Tris-EDTA (TE) buffer, and treated with 100 μg/ml RNase A (Roche) for 30 min at 37°C. Samples were incubated for 1 h at 56°C with 0.5 mg/ml proteinase K (Sigma), followed by 15 h at 65°C to revert formaldehyde cross-links. DNA was extracted with phenol-chloroform-isoamyl alcohol (25:24:1) (Invitrogen), ethanol precipitated, and resuspended in 10 mM Tris-HCl, pH 7.5.
Real-time and competitive PCR.
The abundance of specific immunoprecipitated DNA sequences, prepared as described above, was determined by quantitative real-time PCR and/or competitive PCR. Sequence-specific primers for real-time PCR analysis were designed to amplify and detect origins as well as nonorigin control regions near the human lamin B2, granulocyte-macrophage colony-stimulating factor 1/2 (GM-CSF1/2), and beta-globin replication origins (Table 1). Real-time PCR experiments were carried out in triplicate, using IQ SYBR green master mix (Bio-Rad) on a CFX96 real-time PCR system (Bio-Rad). Enrichments were calculated using the ΔΔCT method. Competitive PCR experiments were performed using previously described primers and competitors specific for the lamin B2 replication origin and nonorigin control sequences (17).
TABLE 1.
Oligonucleotides used for real-time PCR analysis of lamin B2, GM-CSF, and β-globin gene origins
| Primer | Sequence (5′→3′) | Position in human genome (NCBI sequence accession no.) | Annealing temp (°C) |
|---|---|---|---|
| B48 RT For | CTCCACCCCCAAGGAAAAAG | 2368081-2368062 (NT_011255.14) | 60 |
| B48 RT Rev | GGCAGGGTCCCATGCA | 2368005-2368020 (NT_011255.14) | |
| B13 RT For | CCCCAGGGAGTAGGTTGTGA | 2363991-2363972 (NT_011255.14) | 60 |
| B13 RT Rev | TGTTATTTGAGAAAAGCCCAAAGAC | 2363892-2363916 (NT_011255.14) | |
| 17 RT For | AGCCAAGGTGCCTCTTTCAG | 33833183-33833202 (NT_034772.5) | 60 |
| 17 RT Rev | GTCCGGGAGGAGCAGACA | 33833240-33833223 (NT_034772.5) | |
| 21 RT For | TGTACAAATGAAGTGCACAAATCTTAAG | 33834440-33834467 (NT_034772.5) | 60 |
| 21 RT Rev | CCTGGGTGCCAGTTATTTATATAGG | 33834522-33834498 (NT_034772.5) | |
| 23 RT For | GTCCACCTCACTAATGCAGACAAT | 33836142-33836165 (NT_034772.5) | 60 |
| 23 RT Rev | AGAGAAGCAGGAAGGGCTTAGAGA | 33836213-33836193 (NT_034772.5) | |
| BG-9 For | TGTGTTCACGACTGACATCACCG | 52790-52812 (U01317.1) | 62 |
| BG-9 Rev | GCTGGGCTTCTGTTGCAGTAGGG | 52879-52857 (U01317.1) | |
| BG-3 For | CTGCCGTTACTGCCCTGTGGG | 62215-62235 (U01317.1) | 62 |
| BG-3 Rev | ACCAACCTGCCCAGGGCCTC | 62284-62265 (U01317.1) |
RNAi and colony forming assays.
Cells were transiently transfected for 72 h with a SMART pool siRNA against RECQ1 (NM_032941) or RECQ4 (NM_004260) (Dharmacon) at a final concentration of 100 nM by use of HiPerFect reagent (Qiagen) following the manufacturer's instructions. RNA interference (RNAi) control experiments were performed using an siRNA against luciferase (Dharmacon). RECQ1 depletion was also obtained by transfection with the pcDNASup expression vector, encoding a short hairpin RNA (shRNA) against RECQ1 mRNA (target sequence, 5′-GAGCUUAUGUUACCAGUUA-3′), using the calcium phosphate method. The RECQ1-downregulated clones were selected using G418 (0.5 mg/liter) after 48 h of transient transfection, for 2 weeks of selection. Colony forming assays were conducted as previously described (21), using six-well plates and T98G glioblastoma cells that had been transfected with RNAi pools targeting RECQ1, RECQ4, or luciferase as a negative control. Transfected cells were seeded at different dilutions (200, 400, and 800 cells/well). Colonies formed after growth for at least 7 days were counted by a VersaDoc 4000 imaging system (Bio-Rad). The colony-forming capacity was calculated as the average ratio of the number of formed colonies to the number of cells seeded, expressed as a percentage.
Nascent-DNA quantitation.
Nascent DNA was obtained from RECQ1, RECQ4, or luciferase RNAi-transfected T98G cells by neutral sucrose gradient centrifugation as previously described (25). Quantification of the abundances of the different origin and control sequences in gradient fractions was performed by quantitative real-time PCR analysis as described above.
maRTA.
Origin firing frequency and replication fork progression rates were determined by using a recently described microfluidic-assisted replication track analysis protocol (maRTA) (55). In brief, RECQ1-, RECQ4-, or mock-depleted T98G glioblastoma cells were labeled for 40 min each with 100 μM IdU followed by 100 μM CldU and then collected by trypsinization and used to prepare agarose plugs as previously described (55). High-molecular-weight DNA was isolated from cells embedded in agarose by brief heating to 75°C to melt the agarose, followed by agarose digestion. The resulting high-molecular-weight DNA was then loaded by capillary tension into microchannels to uniformly stretch and capture DNA on glass coverslips for immunostaining and fluorescence microscopy (55). Origin firing efficiency was determined by counting the fraction of origin firing events among all active replication events (i.e., ongoing forks and converging forks). Replication elongation efficiency was determined by measuring the mean length of first-label replication tracks in double-labeled tracks in order to unambiguously analyze active/ongoing fork rates. Track lengths were measured in digital images of tracks by using the AxioVision software package (Carl Zeiss). Three replicate samples of RECQ1-, RECQ4-, or mock-depleted cells were analyzed for each determination, where 250 to 450 replication tracks were measured in each sample.
Biochemical fractionation and immunoprecipitation.
Biochemical fractionation was performed following a previously described procedure (40). Immunoprecipitation experiments were performed as described above for ChIP experiments. After being washed with HLNG buffer, the sample was boiled for 30 min in SDS-PAGE sample buffer to reverse formaldehyde cross-links. Protein samples were loaded and run in 8% or 10% SDS-PAGE gels prior to transfer to polyvinylidene difluoride (PVDF) or nitrocellulose membranes (Amersham) for Western analysis and detection by ECL (Amersham).
RESULTS
Association of RecQ helicases with human DNA replication origins.
We investigated the in vivo association of different RecQ helicases with three well-characterized human DNA replication origins by ChIP and real-time PCR analysis. One origin is located on chromosome 19q, in the 3′ end of the lamin B2 gene and upstream of the promoter of the TIMM13 gene (2, 25) (Fig. 1A). The other two origins (GM-CSF Ori1 and Ori2) are located 5 kb apart and approximately 5 kb downstream of the 3′ end of the GM-CSF gene, on chromosome 5q (59) (Fig. 1B). ChIP experiments were initially done using asynchronous T98G human glioblastoma cells and then repeated with a normal human lung fibroblast cell line (IMR-90) to demonstrate that our results were not cell type specific (Fig. 1C and D). Cross-linked chromatin was immunoprecipitated with specific anti-RecQ antibodies (Fig. 1C). After reversal of cross-links, the associated DNA was used for quantitative real-time PCR to determine the amount of origin-containing DNA bound by the immunoprecipitated protein (region B48 of the lamin B2 origin and regions 17 and 23 of GM-CSF Ori1 and Ori2, respectively) compared with adjacent regions that do not contain origins (regions B13, for the lamin B2 origin, and 21, for the GM-CSF origins) (Fig. 1A and B). Origin-specific DNA was enriched three- to sixfold for all three origins in untreated T98G cells relative to control sequences after immunoprecipitation with anti-RECQ1 and anti-RECQ4 antibodies (Fig. 1C and D). Of note, we observed no enrichment for origin-containing regions above control levels when immunoprecipitations were performed with anti-BLM, anti-WRN, and anti-RECQ5 antibodies (Fig. 1C and D).
FIG. 1.
RECQ1 and RECQ4 helicases are recruited to human origins of DNA replication. (A and B) Genomic regions containing the lamin B2 origin (A) and two GM-CSF replication origins (B) are shown together with the locations of sets of primers (converging arrow pairs) used for quantitative real-time PCR analysis. (C) Quantification of cross-linked lamin B2 origin DNA immunoprecipitated by ChIP from T98G and IMR-90 cells, using the antibodies indicated at the bottom. The inset shows the results of Western analysis after immunoprecipitation of the cross-linked material with specific antibodies against the five human RecQ proteins. ORC2-specific immunoprecipitation served as a positive control, and rabbit IgG immunoprecipitation served as a negative control. Whole T98G cell lysates (WCL) were used to confirm the endogenous expression of the five RecQ helicases. (D) Quantification of cross-linked GM-CSF1 and GM-CSF2 origin DNAs immunoprecipitated by ChIP from T98G cells, using the antibodies indicated at the bottom. Fold enrichments of origin sequences were determined versus nonorigin control sequences, and the dashed line indicates the threshold enrichment level obtained by using a negative-control antibody (normal rabbit IgG). Results are reported as means ± standard errors of the means (SEM) (indicated by error bars) for at least three independent experiments.
We used the ORC2 replication protein as a positive control in all experiments, since ORC2 has been shown to bind to the lamin B2 origin (1). Results of real-time PCR analysis were also confirmed using different anti-RecQ antibodies and by competitive PCR experiments in which the amount of origin sequence was estimated by comparing the intensities of corresponding amplified bands to those of bands generated from known amounts of competitor present in the same reaction tubes (data not shown). The same experiments repeated in the presence of the replication inhibitor hydroxyurea (HU) showed that all five human RecQ helicases, including RECQ1 and RECQ4, interacted with the lamin B2 origin upon replication stress, providing further support for the proposed roles of these helicases in promoting fork recovery or repair (Fig. 2). However, our results indicate that of the five human RecQ helicases, only RECQ1 and RECQ4 specifically bind replication origin regions in unperturbed cells, and that origin binding is not origin or cell type specific, suggesting that these two RecQ helicases are required during normal replication.
FIG. 2.
Comparison of association of RECQ1 and RECQ4 helicases with the lamin B2 origin in untreated versus HU-treated cells. T98G cells were treated with 2 mM HU for 24 h. The bar graph shows the quantification of cross-linked lamin B2 origin DNA immunoprecipitated from T98G glioblastoma cells, using antibodies specific to the proteins shown across the bottom. Fold enrichments of origin sequences were determined versus nonorigin control sequences, and the dashed line indicates the threshold enrichment level obtained by using a negative-control antibody (normal rabbit IgG). Results are reported as means ± SEM for at least three independent experiments.
Cell cycle-regulated association of RECQ1 and RECQ4 helicases with replication origins.
Our finding that RECQ1 and RECQ4 associate with DNA replication origins raises the possibility that these two helicases may regulate replication origin function. In order to address this possibility, we used ChIP analyses to determine whether RECQ1 or RECQ4 was recruited to origins in a cell cycle phase-dependent fashion. T98G cells were synchronized by serum starvation in G0, released from the resting state by serum addition, and harvested at different time points (Fig. 3). Synchronization and cell cycle progression were monitored by fluorescence-activated cell sorter (FACS) analysis as well as by cyclin E and cyclin A1 expression (Fig. 3A and B). In agreement with previous findings, cyclin E and cyclin A1 protein levels started to rise in middle G1 (12 h) and early S (22 h), respectively, while both cyclins were present at the G1/S boundary (≥20 h) (36). RECQ1 was present throughout synchronization and release, with a small but reproducible increase at the G1/S border (20 h), a pattern similar to, though not as pronounced as, that observed for cyclin A1. In contrast, RECQ4 was almost undetectable in G0 or resting cells but was readily detectable in late G1 and early S (≥18 h) (Fig. 3A).
FIG. 3.
Cell cycle-dependent association of RECQ1 and RECQ4 with the human lamin B2 replication origin. (A) Western blot analysis of whole-cell extracts of asynchronous T98G cells (AS) or cells from different times after synchronization and release. (B) Cell cycle phase timing was determined by flow cytometry profiling of synchronized T98G cells that had been cultured without serum for 72 h and then sampled over a 28-h time course after the addition of serum. PI, propidium iodide. (C) Quantification of cross-linked lamin B2 origin DNA immunoprecipitated by ChIP from synchronized cells. The key on top indicates the antibodies used for ChIP analyses. Fold enrichments of lamin B2 origin region (B48) DNA over control B13 region DNA are reported for each antibody, where the dashed line indicates the threshold enrichment obtained using a negative-control normal rabbit IgG antibody. Histogram bars report the mean ± SEM for at least three independent experiments for each antibody and cell cycle fraction. (D) Cell cycle-dependent subcellular distribution of RECQ1 and RECQ4. Whole-cell lysates were fractionated to generate cytosolic (S2), soluble nuclear (S3), and chromatin-enriched (P3) fractions, in which protein levels were assessed by Western blotting. Antibodies against p84 and α-tubulin were used as controls for nuclear and cytoplasmic localization, respectively.
ChIP experiments showed that neither RECQ1 nor RECQ4 was associated with replication origins in G0 or early G1. A threefold enrichment of RECQ4 binding to the lamin B2 origin region could be detected at the G1/S boundary and increased to approximately sixfold above background with the onset of S phase (Fig. 3C). Conversely, RECQ1 was found enriched at origin sequences only in early S, after RECQ4 had been recruited. For comparison, ORC2, a well-characterized component of the prereplication complex (pre-RC), was found on origin regions beginning in early G1 and remained bound through early S (1). We repeated these ChIP experiments using T98G cells blocked at late G1 by mimosine treatment (60). These experiments confirmed that ORC2 and RECQ4 could be found on origins already in late G1, when RECQ1 binding was not detected (data not shown). These results indicate that RECQ4 is loaded onto origins beginning in late G1 and is followed by RECQ1 loading when cells enter S phase and the early lamin B2 origin is licensed for firing.
A detailed analysis of the cell cycle-dependent association of RECQ4, RECQ1, and other known replication factors with the lamin B2 origin is shown in Fig. 4. We found, consistent with published results, that CDC6, ORC1, ORC2, and MCM4 assembled on the origin in G1 to complete the prereplication complex (1, 24, 49). RECQ4 loading was detected in late G1 as part of the prereplication complex and was most abundant at the G1/S border as CDC6 was lost. In early S phase, ORC1 was also lost from origins, as expected (1), whereas RECQ1 and additional RECQ4 could now be detected on the lamin B2 origin. RECQ4, RECQ1, and MCM4 binding to the origin region of lamin B2 was no longer detectable after mid-S phase (Fig. 4). In agreement with our findings, a recent study showed that a Flag-tagged version of RECQ4 specifically interacted with the lamin B2 origin during the G1 and S phases of the cell cycle in human 293T cells (69). In this analysis, the lamin B2 origin appeared to be enriched fivefold at G1 compared with S phase. These results are difficult to assess in the absence of detailed ChIP analysis time course data. The apparent enrichment in origin binding at G1 versus S may reflect the fact that the sample used for S-phase experiments corresponded to mid- or late S, as suggested from the flow cytometry data, where we showed that RECQ4 was almost undetectable on the lamin B2 origin. Collectively, our results and those reported by Xu et al. (69) indicate that RECQ4 is part of the prereplication complex and thus might play a role in the transition from the prereplication to the preinitiation complex. RECQ1, in contrast, may be assembled on origins only at the start of bidirectional DNA synthesis. The subsequent loss of both RECQ1 and RECQ4 from origin regions could indicate either disassembly or the tracking of one or both RecQ helicases with the newly formed replisome after origin firing.
FIG. 4.
G1- and S-phase-specific loading of RECQ4 and RECQ1 on the lamin B2 origin. Paired columns show flow cytometry profiles of synchronized T98G cells at different times after release from serum starvation, together with bar graphs of cross-linked lamin B2 origin DNA immunoprecipitated by ChIP with antibodies to each of the proteins shown across the bottom of the bar graph at that time point. Histograms report the mean ± SEM for at least three independent experiments at each time point, where fold enrichment of lamin B2 origin region (B48) DNA was determined as described in the legend to Fig. 3.
In agreement with the above results, biochemical fractionation experiments showed that RECQ4 was enriched in the chromatin fraction during late G1 and early S phases (Fig. 3D). The continued presence of RECQ4 bound to chromatin in late S is consistent with the idea that RECQ4 might leave origin regions to travel with the replisome after replication initiation. Although most of the RECQ1 protein was bound to chromatin throughout all phases of the cell cycle, ChIP experiments showed that RECQ1 interacted with replication origins only at the onset of S phase, when origins are licensed for firing. Interestingly, the partitioning of RECQ1 and RECQ4 between cytosolic and nucleoplasmic fractions changed over the cell cycle. This suggests that RECQ1 and RECQ4 function might be regulated by a combination of cell cycle-dependent synthesis and subcellular localization (Fig. 3A and D).
Analysis of the replication timing-regulated association of RECQ1 and RECQ4 with replication origins.
The lamin B2 and GM-CSF origins are early firing replication origins (2, 59). To test if the temporal loading of RECQ1 and RECQ4 to replication origins might change as a function of replication timing, we compared the loading of these two helicases on the human beta-globin replication origin in HeLa versus K562 cell lines in ChIP experiments (Fig. 5A). The beta-globin origin is located on chromosome 11, in the region encompassing the human beta-globin gene, and it is known to replicate in early S phase in hematopoietic cells (K562 cells) and later in S in nonhematopoietic cells (HeLa cells) (8, 20, 32). The K562 and HeLa cell lines were synchronized by mimosine treatment in late G1 and then released from the drug and harvested at different time points during S phase. Cellular synchronization and progression were monitored by FACS analysis (Fig. 5B and C).
FIG. 5.
Association of RECQ1 and RECQ4 with beta-globin replication origin as a function of origin timing. (A) Genomic region containing the beta-globin replication origin, together with the locations of sets of primers (converging arrow pairs) used for quantitative real-time PCR analysis. (B) Flow cytometry profiling of synchronized K562 cells that had been cultured with mimosine for 24 h and then sampled 3 (early S) and 9 (late S) h after the removal of the drug is shown at the top. The histograms below quantify cross-linked lamin B2 and beta-globin origin DNAs immunoprecipitated by ChIP from early- and late-S-phase-synchronized K562 cells. (C) Flow cytometry profiling of synchronized HeLa cells that had been cultured with mimosine for 24 h and then sampled 3 (early S) and 9 (late S) h after the removal of the drug is shown at the top. The histograms below quantify cross-linked lamin B2 and beta-globin origin DNAs immunoprecipitated by ChIP from early- and late-S-phase-synchronized HeLa cells. Histograms report the means ± SEM for at least three independent experiments, where fold enrichment of lamin B2 and beta-globin origin region DNAs was determined as described in the legends to Fig. 1 and 2.
Control experiments confirmed that RECQ1 and RECQ4 were enriched at the lamin B2 origin sequence in early S and were lost from the lamin B2 origin in late S in both cell lines. These results support our previous conclusion that the interaction of RECQ1 and RECQ4 with the early-firing lamin B2 origin is not cell type specific (Fig. 5B and C). Our results using the beta-globin origin in K562 cells showed that the timing of the interaction of RECQ1 and RECQ4 was identical to what we observed for the lamin B2 origin (Fig. 5B). Conversely, in HeLa cells, where beta-globin origin firing occurs later during S, RECQ1, RECQ4, and ORC2 were already detected on the origin in early S and remained bound in late S (Fig. 5C). The presence of origin-bound RECQ1, RECQ4, and ORC2 already in early S phase indicates that the temporal loading of these factors is likely to be independent of origin replication timing. These results suggest a model in which replication factors assemble on most or all origins during G1/early S, when cyclin-dependent kinase levels are permissive, but that further signals are necessary to start replicating at a specific point in S phase. Further analyses will be required to test this model, as to our knowledge there have been no reports describing the assembly of the replicative complex on the beta-globin replication origin in hematopoietic versus nonhematopoietic cells.
Analysis of proliferation and cell cycle progression of RECQ1- and RECQ4-depleted cells.
Consistent with the idea that RECQ1 and RECQ4 may play a role in replication initiation and with previous reports, we observed a significant reduction in cell proliferation following the siRNA-mediated depletion of RECQ1 or RECQ4 from T98G cells (23, 48, 51). In particular, we compared the colony-forming properties of T98G glioblastoma cells transfected with RECQ1 or RECQ4 siRNA versus those of a T98G cell line transfected with a control siRNA against luciferase (mock-depleted control cells) (Fig. 6A). RECQ1- and RECQ4-depleted T98G cells showed fivefold and sixfold reductions in proliferation, respectively, as measured by colony formation, versus mock-depleted control cells (Fig. 6A). Western blot verification of the extent of depletion indicated marked and near-complete depletion of RECQ1 and RECQ4, respectively (Fig. 6B). The same experiment repeated with the human GM00637 SV40 fibroblast cell line indicated that RECQ1 and RECQ4 depletion was growth suppressive. Thus, the effect of RECQ1 or RECQ4 depletion is not cell type specific (data not shown). To exclude any off-target effect of the siRNA used for RECQ1 silencing, the results were also confirmed using T98G and GM00637 SV40 cell lines transfected with a specific short hairpin RNA (shRNA) against RECQ1 (data not shown).
FIG. 6.
Downregulation of RECQ1 and RECQ4 inhibits cellular proliferation and DNA synthesis. (A) Colony-forming efficiency of T98G cells after siRNA-mediated transfection. Plate photos show representative colony formation after plating of cells transfected with siRNA pools directed against the gene named across the top. The cell number plated is indicated by the code along the left, as follows: I, 200 cells/well; II, 400 cells/well; and III, 800 cells/well. Colonies were stained and counted after 7 days of growth to determine colony-forming efficiencies (means ± SEM) for three independent experiments. (B) Western blot analysis of RNAi-mediated depletion of RECQ1 or RECQ4 from T98G cells transfected with an siRNA pool against RECQ1, RECQ4, or luciferase (Luc) at 72 h posttransfection. α-Tubulin was used as a blot control. (C) Flow cytometry profiles of DNA content (x axis; propidium iodide [PI] staining) versus BrdU incorporation (y axis; anti-BrdU immunostaining) 72 h after siRNA transfection. Boxes are labeled to indicate cell cycle phases, where the mean percent S-phase cells is shown in parentheses across the top of each histogram. The bar graph at bottom reports the percentages of G0/G1, S-phase/BrdU-positive, and G2/M cells in cultures that had been transfected with a RECQ1, RECQ4, or luciferase (control) siRNA pool (indicated in the key). Results shown are the means ± SEM for three independent experiments.
The results above suggest a role for RECQ1 and RECQ4 in regulating cell proliferation by controlling DNA replication. Consistent with this idea, FACS analysis of RECQ1- or RECQ4-depleted cells that had been labeled with BrdU demonstrated >50% reductions in both BrdU labeling and the S-phase fraction, together with an increased G1 fraction, versus those in controls (Fig. 6C). These results are consistent with previous reports of an increased G1 fraction and decreased BrdU incorporation in RECQ4-depleted cells (48). Similar results were again obtained using RECQ1- or RECQ4-depleted GM00637 fibroblasts (data not shown). These results demonstrate that RECQ1 or RECQ4 depletion suppresses cell proliferation and may do so by interfering with DNA replication. In order to provide mechanistic insight into how RECQ1 or RECQ4 depletion reduced BrdU incorporation and cell proliferation, we quantified nascent-DNA production from origins and measured origin firing frequency and replication fork rates in RECQ1- and RECQ4-depleted cells.
Nascent-DNA analysis and chromatin loading of replication factors.
The nascent-DNA assay used quantitative real-time PCR to measure newly synthesized DNA corresponding to the lamin B2 origin and the GM-CSF Ori1 and Ori2 regions in neutral sucrose gradient centrifugation fractions from RECQ1- and RECQ4-depleted cells (25). We found that origin-sequence DNA was reduced by approximately 50% in RECQ1-depleted cells and by >90% in RECQ4-depleted cells 72 h after siRNA transfection (Fig. 7). In order to provide additional mechanistic insight into the loss of nascent DNA, we also examined the order of chromatin loading of replication factors in RECQ1- or RECQ4-depleted T98G cells at the onset of S phase. Cells were synchronized by serum starvation in G0 prior to release by the addition of serum and concurrent transfection with RECQ1 or RECQ4 siRNA (Fig. 8A). Cells were harvested 24 h after serum stimulation and transfection in order to isolate RECQ-depleted cell populations highly enriched in S phase. Using this protocol, we could achieve >90% RECQ4 depletion and marked, though less efficient, depletion of RECQ1, consistent with our previous observation that RECQ1 is already expressed in G0 (Fig. 3A and D). Our results demonstrate that RECQ1 and RECQ4 depletion does not affect chromatin recruitment of MCM3, ORC1, or ORC2. This indicates that RECQ1 and RECQ4 are loaded on origins after assembly of the prereplication complex (Fig. 8B). In contrast, the recruitment of PCNA to chromatin was decreased in RECQ1-depleted—and more markedly in RECQ4-depleted—cells, while RPA loading was suppressed only in RECQ4-depleted cells (Fig. 8B). These results are again consistent with RECQ4 being loaded onto origins before origin firing, followed by RECQ1 loading with or immediately after RPA recruitment.
FIG. 7.
RECQ1 and RECQ4 depletion reduces newly synthesized nascent DNA from early-firing human replication origins. Fold enrichment of nascent DNA from lamin B2 (A) and two GM-CSF (B) origin regions versus that from adjacent nonorigin regions in neutral sucrose gradient fractions was determined for T98G cells depleted of RECQ1 (middle row) or RECQ4 (bottom row) by RNAi transfection and compared to that for controls (luciferase RNAi-transfected cells). Graphs report the means ± SEM for at least three independent experiments.
FIG. 8.
RECQ1 and RECQ4 depletion impairs replication factor loading onto chromatin at the start of S phase. (A) Experimental outline for synchronization and release of human T98G cells to generate highly enriched S-phase cell populations. Asynchronous cells were grown for 72 h without serum and then refed, RNAi transfected, and harvested after 24 h for blot analysis. (B) Western blot analysis of whole and fractionated cell extracts from synchronized T98G cells. Left and right panels show the subcellular distribution of RECQ1 (left column, top), RECQ4 (right column, top), and four additional replication proteins, together with an α-tubulin control (both columns), in whole-cell lysates (WCL) and in cytosolic (S2), soluble nuclear (S3), and chromatin-enriched (P3) fractions prepared from RECQ1-, RECQ4-, or control (luciferase)-depleted cells.
Replication origin use and fork progression rate.
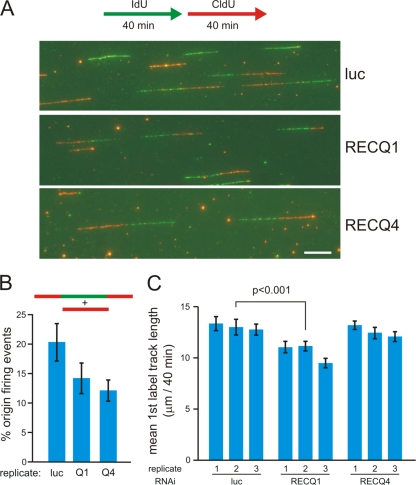
The sequential loading of RECQ4 and RECQ1 on origins at different times after pre-RC formation suggests that these two proteins could play distinct roles during or after replication initiation. In order to determine whether this might be the case, we performed replication track analyses of DNA from T98G cells transfected with RECQ1 or RECQ4 siRNA. In these experiments, cells were labeled sequentially for 40 min each with IdU and with CldU, after which genomic high-molecular-weight DNA was isolated and stretched with the aid of microfluidic channels for replication track analysis (Fig. 9) (54, 55).
FIG. 9.
Depletion of human RECQ1 or RECQ4 affects DNA replication dynamics. (A) An outline of the experimental protocol is shown at the top, in which asynchronous cells were labeled consecutively with IdU (green) and then with CldU (red) for 40 min each prior to isolating and stretching DNA for immunostaining as described in Materials and Methods. Shown below this are representative images of replication tracks in control (luciferase)-, RECQ1-, and RECQ4-depleted cells. All track photos are shown at identical magnifications, where the white scale bar (lower right) is 10 μm long, representing approximately 39 kb of DNA. (B) RECQ1 or RECQ4 depletion reduces the probability of origin firing in stretched DNA samples. Origin firing events among all tracks labeled during the protocol for panel A were identified as CldU-only (red only) or CldU-IdU-CldU (red-green-red) triple-segment tracks (see diagram at top). The mean percentages of new origin firing events defined by these two track types among all labeled tracks are shown for three independent experiments in which 200 to 450 tracks/experiment were typed for control-, RECQ1-, or RECQ4-depleted cells. Error bars show standard deviations. The P values, calculated using Student's t test, are 0.074 between luciferase and RECQ1 and 0.021 between luciferase and RECQ4. (C) RECQ1 depletion, but not RECQ4 depletion, slows ongoing replication forks. The bar graph summarizes mean lengths of first-label IdU (green) segments labeled for 40 min in two-segment (green-red) tracks to ensure that fork rate measurements were made from active replication forks (see the diagram in panel A). Track lengths were measured in μm, using AxioVision software (Carl Zeiss), for three independent experiments in which 150 to 370 first-segment track lengths were measured for each sample. Error bars show 95% confidence intervals for sample means. The statistical significance of differences in mean track lengths was determined by a two-sample Kolmogorov-Smirnov test. A representative P value is shown for a control (luciferase)- versus RECQ1-depleted sample pair.
To evaluate origin firing efficiency, we estimated the fraction of origin firing events among all replication events during the 80-min labeling period. Origin firing events corresponded to tracks containing CldU only or that had a central IdU segment flanked by two CldU segments. Other ongoing replication events were elongating forks (IdU-CldU tracks) and converging forks (IdU-CldU-IdU tracks). This analysis revealed that RECQ1 and RECQ4 depletion reduced the probability of origin firing during the labeling period, from 20% (in luciferase controls) to 14 and 12%, respectively (Fig. 9B). These results are consistent with our nascent-DNA assay results and indicate that RECQ1 and, to a greater extent, RECQ4 are important for efficient replication initiation.
We also determined whether replication elongation was affected in RECQ1- or RECQ4-depleted cells. We measured the lengths of IdU segments in ongoing forks represented by two-segment (IdU-CldU) tracks. Interestingly, this analysis showed that in RECQ1- but not RECQ4-depleted cells, first-label segments were significantly (P < 0.001) shorter than in control cells (Fig. 9C). Lengths of second-label CldU segments in the same tracks were also shorter, as were those of CldU-only tracks in RECQ1-depleted cells (data not shown). These data indicate that RECQ1 may play an additional role in maintaining replication fork progression after initiation.
DISCUSSION
DNA replication is a tightly regulated process essential for the faithful transmission of genetic information in all living organisms (6, 16, 22). Given the centrality of replication, it is not surprising that cells have evolved complex mechanisms to regulate DNA replication origin firing and fork progression, together with a variety of pathways to prevent replication defects, repair damaged replication forks, and enable fork reactivation. The different cellular and DNA metabolic defects associated with the loss of RecQ helicase function in many organisms suggest that the five human RecQ helicases play important and potentially distinct roles in DNA replication. However, the nature and mechanistic details of these postulated roles remain controversial.
Our work extends existing knowledge on the roles of specific human RecQ helicase proteins in DNA replication. We found that only two of the five human RecQ helicases, RECQ1 and RECQ4, bind specifically in vivo to three well-defined human replication origins in unperturbed cells. RECQ4 is bound to origins in late G1, when the pre-RC is assembled (Fig. 10). Additional RECQ4 is found at origins at the G1/S border, when CDC6 leaves origins. This suggests that RECQ4 may be an integral component of the human preinitiation complex. RECQ1, despite its presence throughout the cell cycle, is first reliably detected at origins at the onset of S phase, when ORC1 is lost and origins are licensed for firing. The amounts of origin-bound RECQ1 and RECQ4 are maximal in early S phase, and by mid-S phase, both are lost from the early-firing human lamin B2 and GM-CSF replication origins. An intriguing mechanism for the loss of RECQ1 and RECQ4 from origins is that one or both proteins become associated with active replisomes. Interestingly, we found that RECQ1 and RECQ4 persist on origins during late S, including the late-firing beta-globin origin in HeLa cells. This suggests that RECQ1 and RECQ4 are likely to play the same roles at both early- and late-firing origins. Of note, we did not detect the three other human RecQ helicases, BLM, WRN, and RECQ5, at replication origins. However, all three could be found at origins after treating cells with the replication inhibitor HU. This is consistent with the idea that BLM, WRN, and RECQ5 function in S phase in response to replication stress to promote fork recovery or repair (3).
FIG. 10.
Model of cell cycle-dependent loading of RECQ1 and RECQ4 proteins onto DNA replication origins. RECQ4 is recruited to origins in late G1 as part of pre-RC assembly. At the G1/S transition, CDC6 release signals preinitiation complex (pre-IC) formation. RECQ1, as well as additional RECQ4, is recruited in early S phase, after the release of ORC1. Both RECQ1 and RECQ4 are no longer detected on the lamin B2 origin by mid-S phase, when either or both may be associated with active replisomes. This cell cycle phase-dependent loading and the subsequent loss of RECQ1 and RECQ4 for origins of replication suggest specific roles for each protein in replication initiation and, potentially, other specific aspects of DNA replication, such as fork progression (see text for additional discussion).
Our results indicate that RECQ1 and RECQ4 are integral components of the replication complex and play an important role in DNA replication initiation. Consistent with this idea, nascent-DNA experiments showed that RECQ1-depleted, and even more strikingly, RECQ4-depleted cells have reduced amounts of nascent, newly synthesized DNA containing the lamin B2 and GM-CSF Ori1 and Ori2 origin sequences. RECQ1- and RECQ4-depleted cells also show reduced proliferation and an elevated G1 fraction. Similar analyses, in which levels of other known replication initiation proteins, such as ORC2, were reduced, also showed a decrease in proliferation and an elevated G1 fraction (35). These responses may protect cells from premature S-phase entry without the proper number of activated replication origins.
Replication track analyses confirmed these results by showing that RECQ1 and, especially, RECQ4 are required for efficient replication initiation. In addition, RECQ1-depleted cells display shorter replication tracks than do control cells, indicating that RECQ1, though not RECQ4, might play an additional role in replication fork progression in unperturbed cells. Similar DNA fiber analyses previously identified a role for BLM in efficient restart of replication forks and in the suppression of new origin firing after replication stress (13, 44). WRN, in contrast, is not required for efficient restart but is required to ensure normal fork progression after recovery from HU-mediated arrest (54). Thus, RECQ1, BLM, and WRN may play distinct functions in modulating fork activity and fork rate during S phase.
Our work identifies new parallels and provides new mechanistic insight into the comparative roles of human RECQ4 and X. laevis RECQ4 proteins in replication. X. laevis RECQ4 is required to establish active replication forks and appears to act by facilitating the loading of replication factors at origins (39, 48). The recent demonstration that RECQ4, as well as RECQ1, possesses intrinsic ATP-dependent helicase activity (68) suggests that both proteins might facilitate replication initiation by promoting origin unwinding with the MCM helicase complex. Consistent with this, a recent study reported that RECQ4 physically interacts with the MCM2-7 replicative complex. This interaction is mediated by MCM10, which may regulate the helicase activity of RECQ4 to prevent unlicensed replication initiation (69). Interestingly, the same authors showed that the MCM10-RECQ4 interaction is not shared by other human RecQ helicases, including RECQ1, thus suggesting that RECQ1 might interact with a different component of the replication complex. These results and our data indicate that human RECQ4 and RECQ1 recruitment following assembly of the pre-RC might facilitate replication initiation in the following two ways: by origin unwinding and by the recruitment of proteins required to assemble the replisome.
Consistent with this idea, Xu and collaborators reported that RECQ4 downregulation does not affect the chromatin binding of MCM and CDC6, which supports the idea that RECQ4 is loaded on the origin downstream of these pre-RC complex factors, but does affect chromatin recruitment of the GINS complex (69). This finding was also confirmed by a recent study showing that the assembly of the Cdc45-MCM2-7-GINS complex requires RECQ4 and MCM10 (28). Moreover, we found that RECQ4 and RECQ1 are required for efficient PCNA loading that precedes, and is required for, polymerase loading onto the replication fork. RECQ4 also facilitates the loading of the single-stranded-DNA-binding protein RPA, as has already been shown for X. laevis (48). Since RPA is likely to act in concert with helicases to stabilize unwound replication origins, a lack of RECQ4 helicase activity could hamper origin unwinding with the generation of single-stranded DNA and the loading of RPA and PCNA onto nascent replication forks.
Previous studies have demonstrated that recombinant human RECQ4 can replace X. laevis RECQ4 in DNA replication in egg extracts (48) and that deletion of the N terminus of mouse RECQ4 leads to early embryonic lethality (27). Of note, the N terminus of X. laevis RECQ4 shares homology with the N termini of the yeast proteins Sld2 and DRC1. These proteins are essential for DNA replication in yeast but do not appear to have readily identifiable, conserved vertebrate homologs (39). Our results support the idea that there may be functional conservation between human and Xenopus RECQ4s and these essential yeast proteins for DNA replication initiation. Moreover, our finding that RECQ4 has a role in replication initiation, though not in elongation, provides new insight into the function of this helicase during the replication process. Of note, this replication function of RECQ4 is likely to be preserved in RTS patients, since most of the mutations associated with RTS are located in the central helicase domain of RECQ4 and leave the N-terminal domain of RECQ4 intact.
Cellular functions of RECQ1, in contrast to those of RECQ4, are not as well defined. We and others have shown that acute depletion of RECQ1 affects cellular proliferation (23, 51, 52). Moreover, RECQ1 depletion renders cells sensitive to DNA damage and leads to spontaneous γ-H2AX focus formation as well as to elevated levels of sister chromatid exchanges. These results indicate that RECQ1 plays an important role in the maintenance of genome stability (51). Surprisingly, RECQ1-deficient mice do not show any apparent phenotypic difference compared to wild-type mice, although embryonic fibroblasts derived from the RECQ1-deficient mice show hypersensitivity to ionizing radiation and spontaneous chromosomal breakage (53). This discrepancy between cellular and organismal phenotypes could be explained by functional compensation for the loss of RECQ1 during development, as has been reported for proteins such as the tumor suppressor pRb (45). Alternatively, loss of RECQ1 alone may not reveal a phenotype in the absence of additional defects or exogenous DNA damage. This has been the case for Wrn-deficient mice, which develop accelerated tumorigenesis only on a p53-negative background (34) and have an organismal phenotype resembling Werner syndrome, with features of premature aging, only when combined with later-generation telomerase deficiency (9, 18). Another explanation for the lack of an organismal phenotype in RECQ1-deficient mice is that they may not be true null mutants: this mouse model has a deletion of a portion of the gene that includes the helicase domain IV and part of helicase domain V and thus could still express a portion of RECQ1 required for replication. A similar story may be the case for RECQ4 mutant mice carrying deletions or nonsense mutations of the RecQ4 helicase domain that are still viable. Only deletion of the murine RECQ4 N-terminal domain leads to embryonic lethality (15, 27).
One potential role for RECQ1 in genomic stability assurance was suggested by the recent identification of RECQ1 as part of a Piwi (P-element-induced wimpy testis)-associated RNA (piRNA) complex implicated in transcriptional gene silencing (33). The size, single-stranded nature, and strand specificity of short noncoding piRNAs suggest that piRNA generation could occur during DNA replication and thus be influenced by RECQ1 or other replication proteins (4). Moreover, a recent study demonstrated that RECQ1 is an integral component of the prereplication complex required for Kaposi's sarcoma-associated herpesvirus replication (64). Our results are consistent with these findings and provide new evidence that RECQ1 plays an important role in replication initiation and replication fork progression. An intriguing possibility that cannot be excluded at this stage is that RECQ1 might be required for the firing of a subset of origins that lack RECQ4. This function could be particularly relevant for the firing of dormant origins after DNA replication stress that either slows or stalls normal replication forks. RECQ1, together with RECQ4, may also play a role in early S phase to stabilize or repair replication forks. This role would be consistent with the DNA damage sensitivity of both RECQ1-deficient HeLa cells and RECQ4-deficient primary fibroblasts (29, 53).
In summary, our work identifies important and distinct roles for two human RecQ helicase proteins, RECQ1 and RECQ4, in DNA replication. RECQ4 appears to function early, in replication initiation, when prereplication complex assembly takes place and active replisomes are assembled. RECQ1 is also required for efficient replication initiation and may play an additional role during replication elongation. The mechanisms by which RECQ1 and RECQ4 participate in replication initiation and determining whether helicase activities are required in this process demand further investigation. Our studies also begin to suggest how the loss of specific human RecQ helicases may promote genomic instability and promote tumorigenesis as part of characteristic, heritable human disease phenotypes.
Acknowledgments
We are grateful to I. Hickson and P. Janscak for providing the anti-BLM and anti-RECQ5 antibodies, respectively. We thank A. Carrer for help with the nascent-DNA analysis and G. Triolo for technical assistance.
This work was supported by a grant from the Associazione Italiana per la Ricerca sul Cancro (AIRC) to A.V., by NCI grant PO1 CA77852 to R.J.M., by grants from the AIRC and the Instituto Toscano dei Tumori to A.F., and by the Intramural Research Program of the National Institute on Aging (grant AG000661-06), National Institutes of Health, to W.W.
Footnotes
Published ahead of print on 11 January 2010.
REFERENCES
- 1.Abdurashidova, G., M. B. Danailov, A. Ochem, G. Triolo, V. Djeliova, S. Radulescu, A. Vindigni, S. Riva, and A. Falaschi. 2003. Localization of proteins bound to a replication origin of human DNA along the cell cycle. EMBO J. 22:4294-4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abdurashidova, G., M. Deganuto, R. Klima, S. Riva, G. Biamonti, M. Giacca, and A. Falaschi. 2000. Start sites of bidirectional DNA synthesis at the human lamin B2 origin. Science 287:2023-2026. [DOI] [PubMed] [Google Scholar]
- 3.Bachrati, C. Z., and I. D. Hickson. 2008. RecQ helicases: guardian angels of the DNA replication fork. Chromosoma 117:219-233. [DOI] [PubMed] [Google Scholar]
- 4.Bateman, J. R., and C. T. Wu. 2007. DNA replication and models for the origin of piRNAs. Bioessays 29:382-385. [DOI] [PubMed] [Google Scholar]
- 5.Bischof, O., S. H. Kim, J. Irving, S. Beresten, N. A. Ellis, and J. Campisi. 2001. Regulation and localization of the Bloom syndrome protein in response to DNA damage. J. Cell Biol. 153:367-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blow, J. J., and X. Q. Ge. 2009. A model for DNA replication showing how dormant origins safeguard against replication fork failure. EMBO Rep. 10:406-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bohr, V. A. 2008. Rising from the RecQ-age: the role of human RecQ helicases in genome maintenance. Trends Biochem. Sci. 33:609-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buzina, A., M. I. Aladjem, J. L. Kolman, G. M. Wahl, and J. Ellis. 2005. Initiation of DNA replication at the human beta-globin 3′ enhancer. Nucleic Acids Res. 33:4412-4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang, S., A. S. Multani, N. G. Cabrera, M. L. Naylor, P. Laud, D. Lombard, S. Pathak, L. Guarente, and R. A. DePinho. 2004. Essential role of limiting telomeres in the pathogenesis of Werner syndrome. Nat. Genet. 36:877-882. [DOI] [PubMed] [Google Scholar]
- 10.Chu, W. K., and I. D. Hickson. 2009. RecQ helicases: multifunctional genome caretakers. Nat. Rev. Cancer 9:644-654. [DOI] [PubMed] [Google Scholar]
- 11.Constantinou, A., M. Tarsounas, J. K. Karow, R. M. Brosh, V. A. Bohr, I. D. Hickson, and S. C. West. 2000. Werner's syndrome protein (WRN) migrates Holliday junctions and co-localizes with RPA upon replication arrest. EMBO Rep. 1:80-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davalos, A. R., P. Kaminker, R. K. Hansen, and J. Campisi. 2004. ATR and ATM-dependent movement of BLM helicase during replication stress ensures optimal ATM activation and 53BP1 focus formation. Cell Cycle 3:1579-1586. [DOI] [PubMed] [Google Scholar]
- 13.Davies, S. L., P. S. North, and I. D. Hickson. 2007. Role for BLM in replication-fork restart and suppression of origin firing after replicative stress. Nat. Struct. Mol. Biol. 14:677-679. [DOI] [PubMed] [Google Scholar]
- 14.Dhillon, K. K., J. Sidorova, Y. Saintigny, M. Poot, K. Gollahon, P. S. Rabinovitch, and R. J. Monnat, Jr. 2007. Functional role of the Werner syndrome RecQ helicase in human fibroblasts. Aging Cell 6:53-61. [DOI] [PubMed] [Google Scholar]
- 15.Dietschy, T., I. Shevelev, and I. Stagljar. 2007. The molecular role of the Rothmund-Thomson-, RAPADILINO- and Baller-Gerold-gene product, RECQL4: recent progress. Cell. Mol. Life Sci. 64:796-802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Diffley, J. F. 2004. Regulation of early events in chromosome replication. Curr. Biol. 14:R778-R786. [DOI] [PubMed] [Google Scholar]
- 17.Diviacco, S., P. Norio, L. Zentilin, S. Menzo, M. Clementi, G. Biamonti, S. Riva, A. Falaschi, and M. Giacca. 1992. A novel procedure for quantitative polymerase chain reaction by coamplification of competitive templates. Gene 122:313-320. [DOI] [PubMed] [Google Scholar]
- 18.Du, X., J. Shen, N. Kugan, E. E. Furth, D. B. Lombard, C. Cheung, S. Pak, G. Luo, R. J. Pignolo, R. A. DePinho, L. Guarente, and F. B. Johnson. 2004. Telomere shortening exposes functions for the mouse Werner and Bloom syndrome genes. Mol. Cell. Biol. 24:8437-8446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ellis, N. A., J. Groden, T. Z. Ye, J. Straughen, D. J. Lennon, S. Ciocci, M. Proytcheva, and J. German. 1995. The Bloom's syndrome gene product is homologous to RecQ helicases. Cell 83:655-666. [DOI] [PubMed] [Google Scholar]
- 20.Epner, E., W. C. Forrester, and M. Groudine. 1988. Asynchronous DNA replication within the human beta-globin gene locus. Proc. Natl. Acad. Sci. USA 85:8081-8085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Franken, N. A., H. M. Rodermond, J. Stap, J. Haveman, and C. van Bree. 2006. Clonogenic assay of cells in vitro. Nat. Protoc. 1:2315-2319. [DOI] [PubMed] [Google Scholar]
- 22.Friedel, A. M., B. L. Pike, and S. M. Gasser. 2009. ATR/Mec1: coordinating fork stability and repair. Curr. Opin. Cell Biol. 21:237-244. [DOI] [PubMed] [Google Scholar]
- 23.Futami, K., E. Kumagai, H. Makino, H. Goto, M. Takagi, A. Shimamoto, and Y. Furuichi. 2008. Induction of mitotic cell death in cancer cells by small interference RNA suppressing the expression of RecQL1 helicase. Cancer Sci. 99:71-80. [DOI] [PubMed] [Google Scholar]
- 24.Ghosh, M., M. Kemp, G. Liu, M. Ritzi, A. Schepers, and M. Leffak. 2006. Differential binding of replication proteins across the human c-myc replicator. Mol. Cell. Biol. 26:5270-5283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Giacca, M., L. Zentilin, P. Norio, S. Diviacco, D. Dimitrova, G. Contreas, G. Biamonti, G. Perini, F. Weighardt, S. Riva, et al. 1994. Fine mapping of a replication origin of human DNA. Proc. Natl. Acad. Sci. USA 91:7119-7123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu, Y., X. Lu, G. Zhou, E. L. Barnes, and G. Luo. 2009. Recql5 plays an important role in DNA replication and cell survival after camptothecin treatment. Mol. Biol. Cell 20:114-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ichikawa, K., T. Noda, and Y. Furuichi. 2002. Preparation of gene targeted knockout mice for human premature aging diseases, Werner syndrome, and Rothmund-Thomson syndrome caused by the mutation of DNA helicases. Nippon Yakurigaku Zasshi 119:219-226. [DOI] [PubMed] [Google Scholar]
- 28.Im, J. S., S. H. Ki, A. Farina, D. S. Jung, J. Hurwitz, and J. K. Lee. 2009. Assembly of the Cdc45-Mcm2-7-GINS complex in human cells requires the Ctf4/And-1, RecQL4, and Mcm10 proteins. Proc. Natl. Acad. Sci. USA 106:15628-15632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jin, W., H. Liu, Y. Zhang, S. K. Otta, S. E. Plon, and L. L. Wang. 2008. Sensitivity of RECQL4-deficient fibroblasts from Rothmund-Thomson syndrome patients to genotoxic agents. Hum. Genet. 123:643-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kanagaraj, R., N. Saydam, P. L. Garcia, L. Zheng, and P. Janscak. 2006. Human RECQ5beta helicase promotes strand exchange on synthetic DNA structures resembling a stalled replication fork. Nucleic Acids Res. 34:5217-5231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kitao, S., N. M. Lindor, M. Shiratori, Y. Furuichi, and A. Shimamoto. 1999. Rothmund-Thomson syndrome responsible gene, RECQL4: genomic structure and products. Genomics 61:268-276. [DOI] [PubMed] [Google Scholar]
- 32.Kitsberg, D., S. Selig, I. Keshet, and H. Cedar. 1993. Replication structure of the human beta-globin gene domain. Nature 366:588-590. [DOI] [PubMed] [Google Scholar]
- 33.Lau, N. C., A. G. Seto, J. Kim, S. Kuramochi-Miyagawa, T. Nakano, D. P. Bartel, and R. E. Kingston. 2006. Characterization of the piRNA complex from rat testes. Science 313:363-367. [DOI] [PubMed] [Google Scholar]
- 34.Lebel, M., R. D. Cardiff, and P. Leder. 2001. Tumorigenic effect of nonfunctional p53 or p21 in mice mutant in the Werner syndrome helicase. Cancer Res. 61:1816-1819. [PubMed] [Google Scholar]
- 35.Machida, Y. J., J. K. Teer, and A. Dutta. 2005. Acute reduction of an origin recognition complex (ORC) subunit in human cells reveals a requirement of ORC for Cdk2 activation. J. Biol. Chem. 280:27624-27630. [DOI] [PubMed] [Google Scholar]
- 36.Mailand, N., and J. F. Diffley. 2005. CDKs promote DNA replication origin licensing in human cells by protecting Cdc6 from APC/C-dependent proteolysis. Cell 122:915-926. [DOI] [PubMed] [Google Scholar]
- 37.Marciniak, R. A., D. B. Lombard, F. B. Johnson, and L. Guarente. 1998. Nucleolar localization of the Werner syndrome protein in human cells. Proc. Natl. Acad. Sci. USA 95:6887-6892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Masumoto, H., S. Muramatsu, Y. Kamimura, and H. Araki. 2002. S-Cdk-dependent phosphorylation of Sld2 essential for chromosomal DNA replication in budding yeast. Nature 415:651-655. [DOI] [PubMed] [Google Scholar]
- 39.Matsuno, K., M. Kumano, Y. Kubota, Y. Hashimoto, and H. Takisawa. 2006. The N-terminal noncatalytic region of Xenopus RecQ4 is required for chromatin binding of DNA polymerase alpha in the initiation of DNA replication. Mol. Cell. Biol. 26:4843-4852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mendez, J., and B. Stillman. 2000. Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol. Cell. Biol. 20:8602-8612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Noguchi, E., P. Shanahan, C. Noguchi, and P. Russell. 2002. CDK phosphorylation of Drc1 regulates DNA replication in fission yeast. Curr. Biol. 12:599-605. [DOI] [PubMed] [Google Scholar]
- 42.Paolinelli, R., R. Mendoza-Maldonado, A. Cereseto, and M. Giacca. 2009. Acetylation by GCN5 regulates CDC6 phosphorylation in the S phase of the cell cycle. Nat. Struct. Mol. Biol. 16:412-420. [DOI] [PubMed] [Google Scholar]
- 43.Popuri, V., C. Z. Bachrati, L. Muzzolini, G. Mosedale, S. Costantini, E. Giacomini, I. D. Hickson, and A. Vindigni. 2008. The human RecQ helicases, BLM and RECQ1, display distinct DNA substrate specificities. J. Biol. Chem. 283:17766-17776. [DOI] [PubMed] [Google Scholar]
- 44.Rao, V. A., C. Conti, J. Guirouilh-Barbat, A. Nakamura, Z. H. Miao, S. L. Davies, B. Sacca, I. D. Hickson, A. Bensimon, and Y. Pommier. 2007. Endogenous gamma-H2AX-ATM-Chk2 checkpoint activation in Bloom's syndrome helicase deficient cells is related to DNA replication arrested forks. Mol. Cancer Res. 5:713-724. [DOI] [PubMed] [Google Scholar]
- 45.Sage, J., A. L. Miller, P. A. Perez-Mancera, J. M. Wysocki, and T. Jacks. 2003. Acute mutation of retinoblastoma gene function is sufficient for cell cycle re-entry. Nature 424:223-228. [DOI] [PubMed] [Google Scholar]
- 46.Saintigny, Y., K. Makienko, C. Swanson, M. J. Emond, and R. J. Monnat, Jr. 2002. Homologous recombination resolution defect in Werner syndrome. Mol. Cell. Biol. 22:6971-6978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sakamoto, S., K. Nishikawa, S. J. Heo, M. Goto, Y. Furuichi, and A. Shimamoto. 2001. Werner helicase relocates into nuclear foci in response to DNA damaging agents and co-localizes with RPA and Rad51. Genes Cells 6:421-430. [DOI] [PubMed] [Google Scholar]
- 48.Sangrithi, M. N., J. A. Bernal, M. Madine, A. Philpott, J. Lee, W. G. Dunphy, and A. R. Venkitaraman. 2005. Initiation of DNA replication requires the RECQL4 protein mutated in Rothmund-Thomson syndrome. Cell 121:887-898. [DOI] [PubMed] [Google Scholar]
- 49.Schaarschmidt, D., E. M. Ladenburger, C. Keller, and R. Knippers. 2002. Human Mcm proteins at a replication origin during the G1 to S phase transition. Nucleic Acids Res. 30:4176-4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Seki, M., J. Yanagisawa, T. Kohda, T. Sonoyama, M. Ui, and T. Enomoto. 1994. Purification of two DNA-dependent adenosine triphosphatases having DNA helicase activity from HeLa cells and comparison of the properties of the two enzymes. J. Biochem. 115:523-531. [DOI] [PubMed] [Google Scholar]
- 51.Sharma, S., and R. M. Brosh, Jr. 2007. Human RECQ1 is a DNA damage responsive protein required for genotoxic stress resistance and suppression of sister chromatid exchanges. PLoS One 2:e1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sharma, S., and R. M. Brosh, Jr. 2008. Unique and important consequences of RECQ1 deficiency in mammalian cells. Cell Cycle 7:989-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sharma, S., D. J. Stumpo, A. S. Balajee, C. B. Bock, P. M. Lansdorp, R. M. Brosh, Jr., and P. J. Blackshear. 2007. RECQL, a member of the RecQ family of DNA helicases, suppresses chromosomal instability. Mol. Cell. Biol. 27:1784-1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sidorova, J. M., N. Li, A. Folch, and R. J. Monnat, Jr. 2008. The RecQ helicase WRN is required for normal replication fork progression after DNA damage or replication fork arrest. Cell Cycle 7:796-807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sidorova, J. M., N. Li, D. C. Schwartz, A. Folch, and R. J. Monnat, Jr. 2009. Microfluidic-assisted analysis of replicating DNA molecules. Nat. Protoc. 4:849-861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Siitonen, H. A., O. Kopra, H. Kaariainen, H. Haravuori, R. M. Winter, A. M. Saamanen, L. Peltonen, and M. Kestila. 2003. Molecular defect of RAPADILINO syndrome expands the phenotype spectrum of RECQL diseases. Hum. Mol. Genet. 12:2837-2844. [DOI] [PubMed] [Google Scholar]
- 57.Singh, T. R., A. M. Ali, V. Busygina, S. Raynard, Q. Fan, C. H. Du, P. R. Andreassen, P. Sung, and A. R. Meetei. 2008. BLAP18/RMI2, a novel OB-fold-containing protein, is an essential component of the Bloom helicase-double Holliday junction dissolvasome. Genes Dev. 22:2856-2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Takahashi, Y., J. B. Rayman, and B. D. Dynlacht. 2000. Analysis of promoter binding by the E2F and pRB families in vivo: distinct E2F proteins mediate activation and repression. Genes Dev. 14:804-816. [PMC free article] [PubMed] [Google Scholar]
- 59.Todorovic, V., S. Giadrossi, C. Pelizon, R. Mendoza-Maldonado, H. Masai, and M. Giacca. 2005. Human origins of DNA replication selected from a library of nascent DNA. Mol. Cell 19:567-575. [DOI] [PubMed] [Google Scholar]
- 60.Tsang, W. Y., L. Wang, Z. Chen, I. Sanchez, and B. D. Dynlacht. 2007. SCAPER, a novel cyclin A-interacting protein that regulates cell cycle progression. J. Cell Biol. 178:621-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Van Maldergem, L., H. A. Siitonen, N. Jalkh, E. Chouery, M. De Roy, V. Delague, M. Muenke, E. W. Jabs, J. Cai, L. L. Wang, S. E. Plon, C. Fourneau, M. Kestila, Y. Gillerot, A. Megarbane, and A. Verloes. 2006. Revisiting the craniosynostosis-radial ray hypoplasia association: Baller-Gerold syndrome caused by mutations in the RECQL4 gene. J. Med. Genet. 43:148-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.von Kobbe, C., and V. A. Bohr. 2002. A nucleolar targeting sequence in the Werner syndrome protein resides within residues 949-1092. J. Cell Sci. 115:3901-3907. [DOI] [PubMed] [Google Scholar]
- 63.Wang, H., and S. J. Elledge. 1999. DRC1, DNA replication and checkpoint protein 1, functions with DPB11 to control DNA replication and the S-phase checkpoint in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 96:3824-3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang, Y., H. Li, Q. Tang, G. G. Maul, and Y. Yuan. 2008. Kaposi's sarcoma-associated herpesvirus ori-Lyt-dependent DNA replication: involvement of host cellular factors. J. Virol. 82:2867-2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wu, J., C. Capp, L. Feng, and T. S. Hsieh. 2008. Drosophila homologue of the Rothmund-Thomson syndrome gene: essential function in DNA replication during development. Dev. Biol. 323:130-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wu, L., S. L. Davies, N. C. Levitt, and I. D. Hickson. 2001. Potential role for the BLM helicase in recombinational repair via a conserved interaction with RAD51. J. Biol. Chem. 276:19375-19381. [DOI] [PubMed] [Google Scholar]
- 67.Xu, D., R. Guo, A. Sobeck, C. Z. Bachrati, J. Yang, T. Enomoto, G. W. Brown, M. E. Hoatlin, I. D. Hickson, and W. Wang. 2008. RMI, a new OB-fold complex essential for Bloom syndrome protein to maintain genome stability. Genes Dev. 22:2843-2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xu, X., and Y. Liu. 2009. Dual DNA unwinding activities of the Rothmund-Thomson syndrome protein, RECQ4. EMBO J. 28:568-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xu, X., P. J. Rochette, E. A. Feyissa, T. V. Su, and Y. Liu. 2009. MCM10 mediates RECQ4 association with MCM2-7 helicase complex during DNA replication. EMBO J. 28:3005-3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yin, J., Y. T. Kwon, A. Varshavsky, and W. Wang. 2004. RECQL4, mutated in the Rothmund-Thomson and RAPADILINO syndromes, interacts with ubiquitin ligases UBR1 and UBR2 of the N-end rule pathway. Hum. Mol. Genet. 13:2421-2430. [DOI] [PubMed] [Google Scholar]
- 71.Yu, C. E., J. Oshima, Y. H. Fu, E. M. Wijsman, F. Hisama, R. Alisch, S. Matthews, J. Nakura, T. Miki, S. Ouais, G. M. Martin, J. Mulligan, and G. D. Schellenberg. 1996. Positional cloning of the Werner's syndrome gene. Science 272:258-262. [DOI] [PubMed] [Google Scholar]