Abstract
The transcriptomes of Salmonella enterica serovar Typhimurium SL1344 lacking a functional ramA or ramR or with plasmid-mediated high-level overexpression of ramA were compared to those of the wild-type parental strain. Inactivation of ramA led to increased expression of 14 SPI-1 genes and decreased expression of three SPI-2 genes, and it altered expression of ribosomal biosynthetic genes and several amino acid biosynthetic pathways. Furthermore, disruption of ramA led to decreased survival within RAW 264.7 mouse macrophages and attenuation within the BALB/c ByJ mouse model. Highly overexpressed ramA led to increased expression of genes encoding multidrug resistance (MDR) efflux pumps, including acrAB, acrEF, and tolC. Decreased expression of 34 Salmonella pathogenicity island (SPI) 1 and 2 genes, decreased SipC production, decreased adhesion to and survival within macrophages, and decreased colonization of Caenorhabditis elegans were also seen. Disruption of ramR led to the increased expression of ramA, acrAB, and tolC, but not to the same level as when ramA was overexpressed on a plasmid. Inactivation of ramR had a more limited effect on pathogenicity gene expression. In silico analysis of a suggested RamA-binding consensus sequence identified target genes, including ramR, acrA, tolC, sipABC, and ssrA. This study demonstrates that the regulation of a mechanism of MDR and expression of virulence genes show considerable overlap, and we postulate that such a mechanism is dependent on transcriptional activator concentration and promoter sensitivity. However, we have no evidence to support the hypothesis that increased MDR via RamA regulation of AcrAB-TolC gives rise to a hypervirulent strain.
Multidrug efflux pumps, such as the resistance-nodulation-division (RND) superfamily member AcrAB-TolC in the Enterobacteriaceae, export many substrates, including antibiotics (9, 27, 29, 49, 51, 64). AcrAB-TolC and homologous efflux systems are important in the pathogenicity of Salmonella enterica serovar Typhimurium (10, 14) and other bacterial species (11, 15, 16). Recent work by our laboratory in S. Typhimurium has shown that following the disruption of acrA, acrB, or tolC, a phenotype of decreased pathogenicity was associated with global changes in expression of genes involved in virulence (such as those in SPI-1), chemotaxis, and motility (70).
Regulation of RND efflux pumps has been widely studied in Escherichia coli, with the global AraC/XylS family regulators marA, soxS, and rob having been shown to activate expression of efflux pumps (5, 8, 46, 52, 71). Mechanistically, this is achieved via the binding, and subsequent activation, of discrete but degenerate nucleotide sequences within regulon genes, known as mar-, rob-, or soxboxes (30, 35, 42, 43). E. coli marA is part of the marRAB operon, linked to another operon, marCD, of unknown function. Expression of both is controlled from the same operator site, marO (20). Regulation of E. coli marA is provided through the local repressor marR (62), which controls the transcription of marA by binding to marO, preventing autoactivation of marA, which competes with marR for the marbox of marO (43). Expression of marA can be induced by various compounds and conditions, including exposure to bile salts (54), salicylate, cyclohexane, and the antibiotics tetracycline and chloramphenicol (65). The regulon of marA in E. coli comprises up to 60 genes, including acrAB, tolC, ompF, and zwf (4, 8, 27, 52). In E. coli, the level of soxS is controlled by the activator soxR, in contrast to the repressor activity of marR upon marA. Activation of soxR by superoxide stress induces expression of soxS and the subsequent induction of the soxRS regulon (23).
Salmonellae, Klebsiella pneumoniae, and Enterobacter aerogenes contain ramA, an additional member of the AraC/XylS family of transcriptional activators, which is absent from E. coli and Shigella spp. Overexpression of ramA confers multidrug resistance (6, 19, 28, 60, 67) through induction of acrAB and tolC (6). Regulation of ramA is provided locally by ramR (STM0580), whose function can be ablated by internal point mutations and insertions within the helix-turn-helix motif (3, 57) and/or deletions within the DNA-binding region between ramR and ramA (73). All such events confer multidrug resistance (MDR), presumably through the prevention of RamR binding to an operator sequence near ramA, and therefore the subsequent relaxation of RamR repression at the ramA promoter (3, 57). This model is similar to the mechanism of repression of E. coli marRA (5, 62). Within S. Typhimurium, marA, soxS, and rob appear to play a lesser role in the regulation of MDR, and overexpression of marA and soxS is rarely observed within MDR clinical isolates (3, 29, 51). Furthermore, induction of acrAB by indole is regulated through ramA, independent of marA, soxS, or rob (48). Recently, we showed that inactivation of acrB or tolC was associated with overexpression of ramA (70). Prieto et al. (53) also showed that the presence or absence of an outer membrane protein in S. Typhimurium, AsmA, can affect marA expression. Taken together, these data suggest the existence of a feedback loop of regulation between AraC/XylS regulators such as marA and ramA and genes within their regulons.
In order to define the regulon of RamA, we investigated the effects of inactivation of ramR and the inactivation or artificial overexpression of ramA on the transcriptome of S. Typhimurium. Our data indicate that in addition to the regulation of multidrug efflux pump and accessory genes, expression of ramA influences expression of virulence genes, pathogenicity effectors, and genes within amino acid biosynthetic pathways.
MATERIALS AND METHODS
Strains and media used.
Strains used in this study are listed in Table 1. All strains were derived from S. enterica serovar Typhimurium SL1344 (72). The ramA::aph disruptant was constructed as described previously (22) and then subsequently P22-transduced into SL1344 to avoid possible λred recombinase-mediated mutations. To overexpress ramA, pTRChisA-ramA (67) was transformed into L133 (ramA::aph). Ampicillin (Sigma-Aldrich; 50 μg/ml) was used in cultures to ensure retention of the plasmid, except during growth for RNA preparations for gene expression profiling. This was to prevent altered gene expression due to ampicillin exposure. pTRChis-ramA was not lost during this short (3-h) period (data not shown). Expression of ramA was quantified in both the presence and absence of 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside), with IPTG induction giving a large increase in expression (6). Therefore, where ramA induction was required, sterile IPTG was added to the relevant growth media to a concentration of 1 mM. Luria-Bertani (LB) (Sigma-Aldrich) and MOPS (morpholinepropanesulfonic acid) minimal medium (Teknova Inc.) broth were used throughout this study.
TABLE 1.
Strains used in this study
RNA extraction and transcriptional analyses.
Overnight cultures of S. Typhimurium SL1344 and the test strain were grown in MOPS minimal medium at 37°C as previously described (7). Microarray experiments were carried out exactly as described by Webber et al., (70) using the Pan-Salmonella Generation IV array generated at the Wellcome Trust Sanger Institute (Hinxton, United Kingdom). From each strain, three biological and two technical replicate RNA preparations were made. RNA was extracted and quantified as described previously (6, 70).
Transcriptomic experiments.
RNA (25 μg) was used to generate probes labeled with either Cy-3 or Cy-5 (GE Healthcare) using Superscript III (Invitrogen). For each microarray experiment, the wild-type (SL1344) RNA was pooled after quantification to provide a common reference. Data were analyzed with Bioconductor (56) and Pathway Tools (37). Data with a B value (log odds value) of >0 and a P value of <0.05 were taken as significant.
Validation of microarray data.
Reverse transcription PCRs (RT-PCRs) were performed as previously (6, 24) with a selection of SPI genes shown to be differentially expressed in the microarray experiments: hilC, invF, sipC, sopB, ssrA, and ssaJ (Table 3). Furthermore, expression of ramA, acrB, and tolC was also determined.
TABLE 3.
Pathogenicity genes and known regulatorsa
| Gene/operon | Fold change in expression relative to SL1344b |
||
|---|---|---|---|
| L133 | L786 | L1007 | |
| Regulators of SPI-1 | |||
| fis | 2.27 | - | - |
| iagA | - | 0.52 | - |
| invF | 5.34 | 0.35 | - |
| sprA | 3.73 | 0.34 | - |
| SPI-1 | |||
| orgAa | 1.64 | 0.59 | - |
| prgH | 4.61 | 0.50 | - |
| prgI | 3.24 | 0.34 | - |
| prgJ | 4.86 | 0.38 | - |
| prgK | - | 0.48 | - |
| sicP | 2.42 | - | - |
| sipA | 4.70 | 0.23 | - |
| sipB | 3.80 | 0.22 | - |
| sipC | 3.00 | 0.18 | - |
| sipD | 3.80 | 0.32 | - |
| sipF | 2.77 | 0.36 | - |
| spaI | - | 0.32 | - |
| spaK | 6.81 | 0.27 | - |
| spaN | - | 0.30 | - |
| spaO | 3.24 | - | - |
| spaT | - | 0.30 | - |
| stpA | - | 0.45 | - |
| Regulators of SPI-2 | |||
| slyA | - | 3.90 | - |
| ssrA | - | 0.45 | - |
| SPI-2 | |||
| SL1323 | - | 0.57 | - |
| ssaD | - | 0.45 | - |
| ssaH | - | 0.49 | - |
| ssaI | 0.39 | - | - |
| ssaJ | 0.55 | 0.19 | - |
| ssaL | - | 0.42 | - |
| ssaN | - | 0.35 | - |
| ssaO | - | 0.34 | - |
| ssaP | - | 0.56 | - |
| ssaV | - | 0.43 | - |
| sscA | - | 0.25 | - |
| sseA | - | 0.25 | - |
| sseC | - | 0.26 | - |
| sseD | - | 0.27 | - |
| sseE | - | 0.25 | 0.51 |
| STM1410 | 0.45 | 0.29 | - |
| Other pathogenicity genes | |||
| mig-14 | - | 0.53 | - |
| pagC | - | 0.14 | - |
| sigD | 3.17 | 0.18 | - |
| sigE | - | 0.19 | - |
| sopD2 | - | 0.45 | - |
| sopE | 5.36 | 0.16 | - |
| sopE2 | 4.76 | 0.16 | - |
| STM0306 | 0.45 | 0.41 | - |
| STM2780 | 0.51 | 0.19 | - |
| STM4258 | - | 0.32 | - |
| STM4260 | 3.48 | 0.39 | - |
| STM4261 | - | 0.39 | - |
| virK | - | 0.36 | - |
Genes with significantly (B > 0) changed expression in L133 (ramA::aph), L786 (L133 pTRChisA-ramA), and L1007 (ramR::aph) compared with SL1344.
-, no significant change in expression.
In silico analysis of putative rambox.
Nikaido et al. (48) described a putative degenerate rambox sequence, 5′ ATGGCACG[A/T]AA[A/C][A/G]CCAA[A/C][C/T][A/T] 3′. To establish whether this rambox was common to genes found to be differentially regulated by modification of ramAR, an in silico pattern search was performed against the S. Typhimurium LT2 genome (45) within xBase (17) and using the “fuzznuc” pattern search algorithm within EMBOSS (59). Given the degree of degeneracy known to be present in mar- and soxboxes (32, 44), various allowances of up to 6 mismatches were permitted and no restrictions were placed upon the search location. Data were cross-compared with transcriptomic data using the Microsoft Excel VLOOKUP function.
Protein purification and Western blotting of SipC.
Protein purification and Western blotting were performed exactly as described previously (47, 70).
Adhesion and intracellular survival assays.
Adhesion to and intracellular survival within eukaryotic macrophages (RAW 264.7) by S. Typhimurium SL1344 and strains derived therefrom were assessed as described previously (14). A two-tailed Student's t test was used to assess significance, using a P value of <0.05 as a cutoff.
Mouse model experiments.
Experimental infections using the murine model of infection to compare SL1344 (wild type) and L133 (ramA::aph) were conducted essentially as previously described (39). Briefly, female 6- to 8-week-old BALB/c ByJ or C57/BL6 mice (five mice per group) were inoculated by the intragastric (i.g.) route with approximately 5 × 107 CFU of SL1344 or L133 (ramA::aph) in PBS (pH 7.4). Alternatively, mice were inoculated by the intravenous route with approximately 2.4 × 103 CFU of strain SL1344 or L133. Five days (i.g. inoculation) or four days (intravenous inoculation) after inoculation, mice were sacrificed and organs recovered by dissection. The number of CFU per organ was determined by plating serial 10-fold dilutions of homogenized tissue on LB agar plates or LB plus kanamycin (50 μg/ml). The statistical significance of differences in colonization of each organ was calculated using the Mann-Whitney test, using GraphPad Prism software. A P value of <0.05 was considered significant.
Caenorhabditis elegans survival assays.
C. elegans strain Bristol N2 was kindly donated by Robin May (University of Birmingham) and was cultured using standard methods as described previously (13, 61). Survival assays were performed essentially as previously (1, 2), with 20 larval stage 4 (L-4) C. elegans worms being picked and transferred onto each assay plate and with a minimum of 120 worms being used on three independent occasions. Plates were incubated at 25°C and scored daily for survival. No effects on worm survival were seen with IPTG, kanamycin, and ampicillin at the concentrations used in the experiments. Worms were transferred onto a fresh nematode growth medium (NGM) (33) plate containing the same bacterial test or control strain from the same original culture during the fertile period and reincubated at 25°C. A Kaplan-Meier estimate was used to determine the probability of C. elegans survival to the next day. Survival curves were generated by plotting probability of survival (y) against time (x). These curves were then compared using the log rank test to establish whether the difference between two curves was statistically significant, using a chi-squared test and a derived P value of <0.05 as the significance cutoff.
Microarray data accession numbers.
Microarray datasets have been deposited in ArrayExpress (http://www.ebi.ac.uk/microarray-as/ae/) with the following experiment identifiers: for ramA::aph, E-MEXP-2193; for ramR::aph, E-MEXP-2194; for pTRChisA-ramA, E-MEXP-2195.
RESULTS
Inactivation of ramA and ramR and overexpression of ramA result in some common gene expression changes.
Comparison of the transcriptome of L133 (ramA::aph) with that of SL1344 revealed that disruption of ramA resulted in significantly (log odds value > 0 and P < 0.05) altered expression of 223 genes (6% of the SL1344 genome). Of these 223, 119 genes were increased in expression and 104 were decreased in expression (Fig. 1; also, see Table S1 in the supplemental material). Comparison of the transcriptome of L1007 (ramR::aph) with that of SL1344 revealed that disruption of ramR resulted in significantly altered expression of 187 genes (5% of the SL1344 genome). Of these 187, 173 genes were increased in expression and 14 were decreased in expression (Fig. 1; also, see Table S2 in the supplemental material). Comparison of the transcriptome of L786 (L133 pTRChisA-ramA) with that of SL1344 revealed that overexpression of ramA resulted in significantly altered expression of 310 genes (8% of the SL1344 genome). Of these 310, 158 genes were increased in expression and 152 were decreased in expression (Fig. 1; also, see Table S3 in the supplemental material).
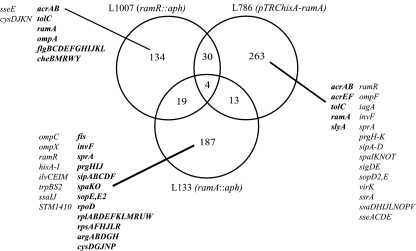
FIG. 1.
Venn diagram of all significant (B > 0) gene expression changes within the comparison of L354 (S. Typhimurium SL1344) against L133 (S. Typhimurium ramA::aph), L1007 (S. Typhimurium ramR::aph), and L786 (S. Typhimurium pTRChisA-ramA). Selected genes from each strain are displayed. Boldface shows increased gene expression; lightface shows decreased gene expression.
Comparison of the transcriptomes of L133 (ramA::aph) and L1007 (ramR::aph) revealed 19 common genes, 16 of which were oppositely expressed (Fig. 1). Comparison of the transcriptomes of L133 and L786 (pTRChis-ramA) revealed 13 common genes; all are pathogenicity genes. Twelve were oppositely expressed, suggesting that they are directly affected by ramA expression (Fig. 1). Overexpression of ramA via inactivation of ramR (L1007), or artificially on pTRC (L786), showed that 30 genes were common to both. Of these, 23 were similarly increased or decreased in expression (Fig. 1). Four genes were significantly differentially expressed by all three strains; ilvE, ilvM, napF, and pckA (Fig. 1). Of these, two (napF and pckA) were increased in expression in all three strains, compared with SL1344. Two genes (ilvE and ilvM) were increased in expression when ramR was inactivated but decreased in expression when ramA was inactivated or overexpressed.
Overexpression of ramA and disruption of ramRA affect expression of genes encoding efflux pumps and porins.
When ramR was inactivated, expression of ramA was increased 25-fold (Table 2). High-level overexpression in L786 (pTRChisA-ramA) resulted in even greater expression (144.3-fold [Table 2]). These data were confirmed by RT-PCR (Fig. 2D) (6). After ramR disruption, expression of acrA, acrB, and tolC was increased 2.1- to 3.4-fold; even greater expression of these genes (5.2- to 8.7-fold [Table 2]) was seen when ramA was highly overexpressed. Increased expression of acrB and tolC was confirmed by RT-PCR (14.9-fold and 2.6-fold, respectively) (Fig. 2D) (6). Increased expression of genes encoding an additional RND efflux pump, AcrEF (2.5- and 3.0-fold, respectively [Table 2]) was also seen for L786 but not L133. Expression of ramR, the local repressor of ramA, was decreased 0.4-fold when ramA was highly overexpressed (Table 2), and expression of the porin gene ompF was decreased 0.2-fold (Table 2). No differential expression of the porin regulators ompR and envZ was observed.
TABLE 2.
Genes encoding cell envelope,a ribosomal, and amino acid biosynthetic proteins
| Gene or operon | Fold change in expression relative to SL1344b |
||
|---|---|---|---|
| L133 | L786 | L1007 | |
| Multidrug transporters/regulators | |||
| acrA | - | 8.66 | 3.36 |
| acrB | - | 8.19 | 2.13 |
| acrE | - | 2.47 | - |
| acrF | - | 3.00 | - |
| tolC | - | 5.16 | 3.35 |
| ompA | - | - | 1.43 |
| ompC | 0.46 | - | - |
| ompF | - | 0.21 | - |
| ompX | 0.61 | - | - |
| ramA | - | 144.32 | 25.04 |
| ramR | 0.21 | 0.39 | - |
| rpoD | 1.36 | - | - |
| Ribosomal biosynthesis | |||
| rplA/B | 1.98/1.65 | - | - |
| rplD-F | 1.44-1.84 | - | - |
| rplK-M | 1.74-2.04 | - | - |
| rplR | 2.33 | - | - |
| rplU | 1.80 | - | - |
| rplW | 1.82 | - | - |
| rpsA | 2.15 | - | - |
| rpsF | 1.97 | - | - |
| rpsH | 2.05 | - | - |
| rpsJ | 1.95 | - | - |
| rpsL | 2.08 | - | - |
| rpsR | 2.01 | - | - |
| Amino acid biosynthesis | |||
| argA/B | 4.29/7.32 | - | - |
| argD | 12.09 | - | - |
| argG/H | 8.88/5.94 | - | - |
| argR | 4.16 | - | - |
| aroA | 0.47 | - | - |
| aroH | 0.16 | - | - |
| artI/J | 2.42/12.99 | - | - |
| artP | 3.25 | - | - |
| cysC | - | - | 0.42 |
| cysD | 11.32 | - | 0.25 |
| cysG | 3.75 | - | - |
| cysH | - | - | 0.53 |
| cysJ | 2.93 | - | 0.37 |
| cysK | 0.43 | - | 0.47 |
| cysN | 2.81 | - | 0.38 |
| cysP | 12.60 | - | - |
| hisA-D | 0.17-0.23 | - | - |
| hisF-I | 0.20-0.23 | - | - |
| hisQ | 2.72 | - | - |
| ilvC | 0.42 | - | - |
| ilvE | 0.64 | - | - |
| ilvI | 0.53 | - | - |
| ilvM | 0.54 | - | - |
| trpB | 0.11 | - | - |
| trpS2 | 0.04 | - | - |
Genes relevant to antibiotic transport and regulation thereof with significantly (B > 0) different expression compared with SL1344.
-, no significant change in expression.
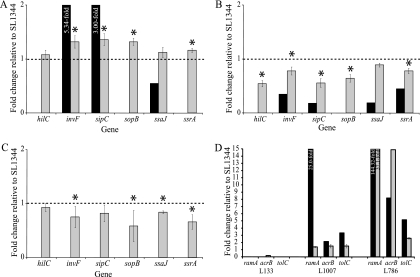
FIG. 2.
RT-PCR for six SPI genes for ramA::aph (A), pTRChisA-ramA (B), and ramR::aph (C) (asterisks show significant [P < 0.05] differences) and RT-PCR for acrB, tolC, and ramA (D) (all RT-PCR data shown are significant; P < 0.05). Black bars, microarray data; gray bars, RT-PCR data; dashed line, fold change of 1 (i.e., no change); for all array changes, a B value of >0 is considered significant.
RT-PCR revealed that upon inactivation of ramA, expression of acrB was decreased 0.2-fold compared to that in SL1344 (Fig. 2D). Furthermore, after ramA inactivation, expression of genes encoding the porins OmpC and OmpX was decreased (0.5- and 0.6-fold, respectively [Table 2]).
Inactivation of ramA increases expression of ribosomal biosynthetic genes and alters expression of genes encoding some amino acid biosynthetic pathways.
Expression of 17 genes within the rpl and rps operons was statistically significantly increased after ramA inactivation (1.4- to 2.3-fold [Table 2]). Expression of 15 genes in the arg, art, and cys operons, responsible for arginine and cysteine biosynthesis, was also increased (4.2- to 12.1-fold [Table 2]). However, expression of 14 genes within the his, ilv, and trp operons, responsible for histidine, isoleucine, and tryptophan biosynthesis, respectively, was decreased (0.04- to 0.6-fold [Table 2]). Expression of genes encoding components of the glycolytic and gluconeogenic pathways, including adh, zwf, and pckA, was also affected by the disruption of ramA, with increased expression (≤4.1-fold) and decreased expression (≤0.2-fold) being observed (see Table S1 in the supplemental material). After disruption of ramR, expression of genes encoding proteins involved in cysteine biosynthesis, cysCDHJKNU, was statistically significantly decreased (0.3- to 0.6-fold [Table 2]).
Disruption or high-level overexpression of ramA differentially affects expression of pathogenicity genes and proteins.
The microarray experiments with L133 (ramA::aph) and L786 (L133 pTRChisA-ramA) revealed significantly altered expression of Salmonella pathogenicity island genes. Inactivation of ramA increased expression of 14 genes within SPI-1, including effectors such as sipABCDF and regulators such as sprA and invF (1.6- to 6.8-fold [Table 3]). Increased expression of invF, sipC, sopB and ssrA was confirmed by RT-PCR (Fig. 2A). No changes in expression of other known regulators of SPI-1, such as phoPQ, pmrA, and hilC, were seen; no differential expression of the latter was confirmed by RT-PCR (Fig. 2A). Expression of some genes within SPI-2, -4, -5, -6, and -7a was also altered in L133, with expression of genes from SPI-2, -6, and -7a (e.g., ssaI and STM1410) being decreased and that of genes within SPI-4 and -5 (e.g., sigD and sopE) being increased (Table 3). However, in contrast, when ramA was highly overexpressed (L786), decreased expression of 47 pathogenicity genes was observed, including many of the same SPI-1 genes for which increased expression upon ramA inactivation was seen; this was also confirmed by RT-PCR (Fig. 2B). Furthermore, RT-PCR validation also showed decreased expression of the regulatory gene hilC and the gene encoding the effector protein SopB (Fig. 2B). The RamA-dependent genes with decreased expression included those in SPI-1, -2, -4, -5, -6, and -7a as well as other genes whose products are also associated with the ability of salmonellae to be pathogenic (0.1- to 0.6-fold [Table 3]). Of interest, high-level overexpression of ramA in L786 was associated with increased expression of slyA, a known activator of SPI-2 (3.9-fold [Table 3]), despite the generally decreased expression of SPI-2 genes after ramA overexpression. Disruption of ramR, giving modest overexpression of ramA, altered expression of only one SPI-2 gene, sseE, which was decreased 0.51-fold (Table 3). However, RT-PCR validation revealed similarly (to high-level ramA overexpression) decreased expression of SPI genes (invF, sopB, ssrA, and ssaJ) (Fig. 2C).
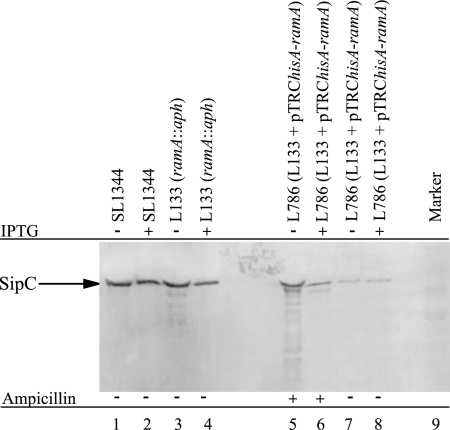
To validate the decreased gene expression in L786, expression of SipC, a SPI-1 protein, was determined by Western blotting. No change in SipC production was observed for SL1344 or L133 (ramA::aph) (Fig. 3, lanes 1 and 3). As a control, IPTG was also added to both strains; no effect was seen upon SipC production (Fig. 3, lanes 2 and 4). L786 decreased production of SipC, in both the absence (0.76-fold) and presence (0.30-fold) of IPTG, compared to SL1344 (Fig. 3, lanes 5 and 6). Furthermore, in the absence of ampicillin, SipC production was decreased further; 0.24-fold (no IPTG) and 0.20-fold (with IPTG) (Fig. 3, lanes 7 and 8) compared to SL1344.
FIG. 3.
Western blotting for SipC after growth of strains SL1344, L133, and L786 in minimal medium, in the presence and absence of IPTG (to induce overexpression of ramA) and ampicillin (to ensure maintenance of the pTRC plasmid).
The ability of S. Typhimurium to adhere to and survive within RAW macrophages and to colonize mice and C. elegans is RamA dependent.
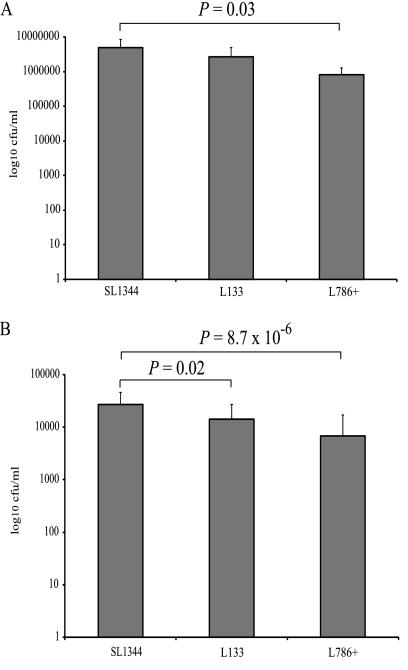
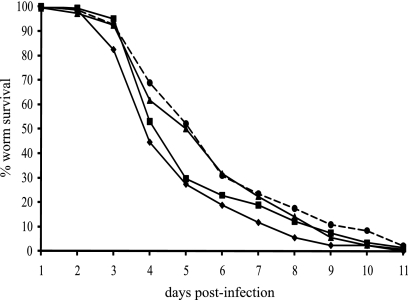
As the expression of SPI-1 and SPI-2 genes was differentially altered by the overexpression and inactivation of ramA, and as production of SipC was decreased by overexpression of ramA, the ability of ramA mutants to adhere to and survive within RAW 264.7 macrophages and to colonize mice and C. elegans was investigated. Compared with SL1344, inactivation of ramA in L133 had no significant effect on adherence (Fig. 4A). The ability of L133 to survive within RAW 264.7 macrophages was significantly impaired, however (0.53-fold decrease [Fig. 4B]). Furthermore, L133 was recovered in significantly lower numbers (>100-fold difference in geometric mean) from the livers and spleens of BALB/c mice than the parental strain, following oral inoculation (Fig. 5A). The ability of this strain to colonize the livers and spleens of BALB/c mice following intravenous inoculation was not significantly different (Fig. 5B). These data were confirmed for the C57/BL6 mice (data not shown). When ramA was highly overexpressed in L786, the ability to adhere and survive within RAW 264.7 macrophages was significantly attenuated (Fig. 4). Unfortunately, in the mouse experiments, pTRChisA-ramA could not be maintained, so the C. elegans model was used to further support the transcriptomic and cell culture data. In this model, L786 was significantly (χ2 = 7.39, P = 0.0066) attenuated in its ability to kill the worm compared to SL1344 (Fig. 6). L133 had no significant difference in pathogenicity compared to SL1344 (χ2 = 1.83, P = 0.18) (Fig. 6). Interestingly, even though limited SPI gene expression changes were observed for L1007, this strain was also significantly (χ2 = 10.24, P = 0.0014) attenuated compared to SL1344 (Fig. 6).
FIG. 4.
Adhesion to (A) and survival in (B) RAW 264.7 mouse macrophages of SL1344, L133, and L786 (with IPTG).
FIG. 5.
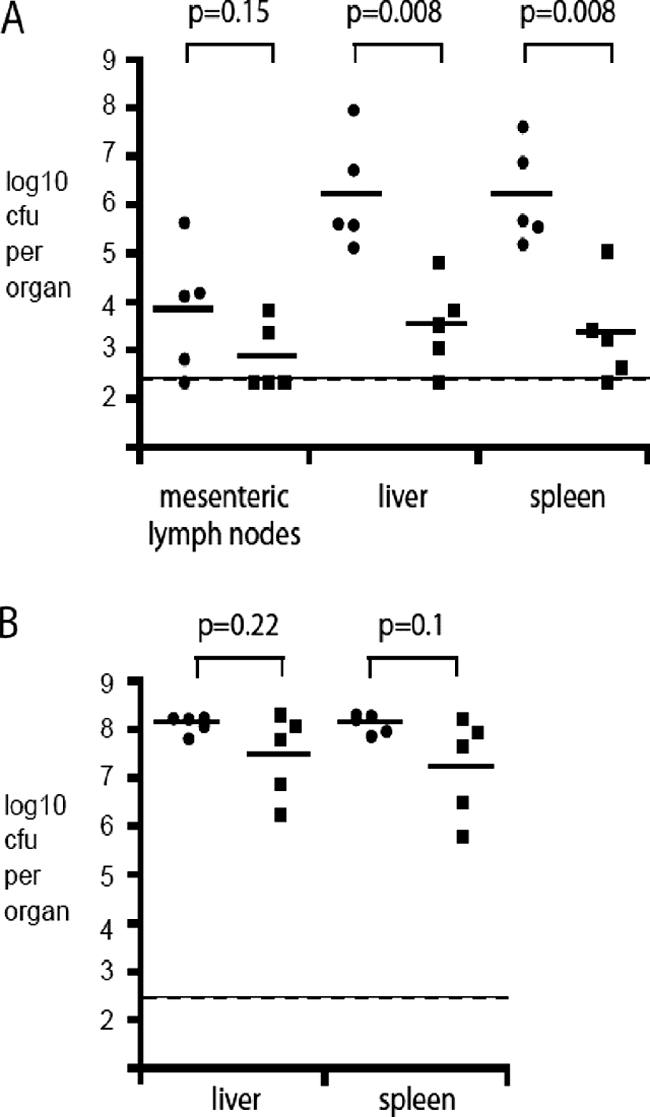
Colonization of BALB/c mice by S. Typhimurium SL1344 and L133 (ramA::aph). Mice were inoculated orally with 4 × 107 CFU (A) or intravenously with 2.4 × 103 CFU (B). The CFU per organ of SL1344 (circles) and L133 (squares) and the geometric mean (horizontal bar) are indicated. P values were calculated using the Mann-Whitney test.
FIG. 6.
Survival of Caenorhabditis elegans after infection with SL1344 (diamonds), L133 (squares), L1007 (circles), or L786 (triangles).
The SPI genes pckA and ilvM contain a rambox.
In silico analyses using the degenerative rambox sequence proposed by Nikaido et al. (48) revealed several targets for RamA. These included acrA and tolC, whose expression has previously been shown to be increased by ramA overexpression (48) (Table 4). The SPI-1 effector genes sipA, sipB, and sipC and the SPI-1 regulator invF all contained a putative rambox, as did the SPI-2 gene ssaJ and the putative virulence effect gene srfB (Table 4). However, the expression of these RamA targets was variable, suggesting that RamA can activate or repress target gene expression. Only two genes containing a putative rambox, pckA and ilvM, were commonly expressed in all three strains (Table 4). The expression of pckA, which encodes the gluconeogenic pathway enzyme phosphoenolpyruvate carboxykinase, was increased in all three strains (1.6- to 2.3-fold), whereas the expression of ilvM, which encodes acetolactate synthase, was decreased in L133 (ramA::aph) and L786 (L133 pTRChisA-ramA) 0.5- and 0.4-fold, respectively, but increased in L1007 (ramR::aph) (1.9-fold [Table 4]).
TABLE 4.
Selected genes commonly differentially regulated in two or more arrays which contain a putative rambox
| Gene | Product | Fold changea |
||
|---|---|---|---|---|
| L133 (ramA::aph) | L1007 (ramR::aph) | L786 (L133 pTRChisA-ramA) | ||
| acrA | Multidrug resistance accessory protein | - | 3.36 | 8.66 |
| ilvM | Acetohydroxy acid synthase II, small subunit | 0.54 | 1.87 | 0.37 |
| invF | Possible AraC-family regulatory protein | 5.34 | - | 0.35 |
| pckA | Phosphoenolpyruvate carboxykinase | 2.31 | 1.60 | 2.27 |
| ramR | Putative tetR-family transcriptional regulator | 0.21 | - | 0.39 |
| sipA | Pathogenicity island 1 effector protein | 4.70 | - | 0.23 |
| sipB | Pathogenicity island 1 effector protein | 3.80 | - | 0.22 |
| sipC | Pathogenicity island 1 effector protein | 3.00 | - | 0.18 |
| srfB | Putative virulence effector protein | - | 1.93 | 0.62 |
| ssaJ | Putative pathogenicity island lipoprotein | 0.55 | - | 0.19 |
| tolC | Outer membrane protein | - | 3.35 | 5.16 |
Relative to S. Typhimurium SL1344. -, no significant change.
DISCUSSION
There is mounting evidence that RamA is a transcriptional activator that regulates genes conferring MDR in various species, including the salmonellae (6, 19, 28, 38, 48, 58, 67, 73). Therefore, we sought to resolve the regulon of ramA by comparing the transcriptomes of the wild-type SL1344 parental strain with those of three isogenic mutants in which ramA or ramR (the local repressor of ramA) was inactivated or in which ramA was highly overexpressed. Given that when genes which confer MDR, such as the those encoding the RND efflux pump AcrAB-TolC, are deleted, the virulence of the bacterium is attenuated (3, 14), it has been hypothesized that if overexpression of the same genes occurs, there may be a concurrent increase in virulence. Support for this hypothesis is derived from work with Neisseria gonorrhoeae (69, 68) showing that mutants overexpressing the Mtr efflux pump genes were more virulent than their wild-type parental strain.
As hypothesized, and confirming prior studies (3, 57), the microarray experiments revealed that inactivation of ramR resulted in overexpression of ramA. This is likely responsible for the increased expression of acrAB and tolC and decreased expression of porin genes, such as ompA. As with overproduction of E. coli marA (21, 31), this gives rise to MDR due to reduced intracellular concentrations of antibiotic brought about by the decreased uptake of multiple agents and increased drug efflux. The magnitude of increased expression of acrAB and tolC was greater when ramA was highly overexpressed through an IPTG-inducible plasmid, which in addition increased expression of genes encoding an additional efflux pump, AcrEF, and the porin gene ompF. These data suggest an intracellular concentration-dependent response of the bacterium to increasing overexpression of ramA/RamA. We postulate that such a mechanism is dependent on transcriptional activator concentration and promoter sensitivity, similar to that established previously for E. coli marA, soxS, and rob (41). Our data further suggest that bacterial cells carefully “orchestrate” the level of RamA, with too much or none being deleterious to the bacterium. This is reminiscent of levels of the PhoP/Q system, where expression of these genes and those within its regulon are produced, and at the correct level, only under the appropriate conditions (50). Conversely, inactivation of ramA affected the expression of very few drug efflux pump genes, and ompC, rather than ompF, was decreased. These data lend further weight to the hypothesis that RamA regulates expression of MDR genes encoding porins and efflux pumps.
Nikaido et al. (48) showed the presence of a putative rambox in acrAB and tolC. In this study, we found that ramR also contains a rambox, and our data support previous hypotheses that ramR acts as a local repressor of ramA (3, 57). We also hypothesize that the absence of functional RamA reduces the expression of ramR in a mechanism aimed to restore wild-type expression of ramA, whereas overexpression of ramA successfully competes with RamR and suppresses further expression of ramR. The ramR-independent mechanism by which ramA expression is returned to basal levels is unclear. However, our data support the hypothesis that ramRA works through a mechanism similar to that of marRA within E. coli, with RamA being able to compete with RamR in binding to a hypothetical operator sequence ramO, leading to autoinduction of self-expression. Further work is required in order to elucidate the exact composition of any “ramO” sequence within ramR, as a significant amount of degeneracy (6 base mismatches) was used to generate the sequence of the rambox used in this study. Putative ramboxes were also identified within the pathogenicity effector genes sipABC and ssaJ and the AraC/XylS family SPI regulator invF. This coincided with the decreased expression of SPI-1 and SPI-2 genes upon high-level ramA overexpression, while ramA disruption induced SPI-1 expression. Previous work with AraC/XylS DNA regulators has suggested that these are transcriptional activators (55, 66), whereas in our study RamA appeared to repress the expression of invF, ssrA and sprA, all additional AraC/XylS family regulators. The association of an AraC/XylS-family DNA regulator with control of virulence genes has been reported previously for InvF and ToxT (34, 36), but purely as an activator of further genes. This suggests that the general downregulation of many SPI-1 and SPI-2 genes may not be due to direct interaction with RamA but may be due to the downregulation of invF and ssrA. The concentration-dependent regulation of the ramA regulon also appeared to extend to genes involved in mediating the pathogenicity of S. Typhimurium, as inactivation of ramR, and therefore lower-level overexpression of ramA, did not significantly affect the expression of SPI genes.
Given that disruption of ramA led to differential pathogenicity gene regulation, we assessed the effect(s) upon pathogenicity within various models. Overexpression of ramA decreased expression of SPI-1 and SPI-2 genes, decreased production of SipC, decreased adhesion and survival within RAW 264.7 macrophages, and attenuated pathogenicity within the C. elegans worm model. These findings suggest that RamA overexpression not only affects expression of efflux pumps but also affects the pathogenicity of S. Typhimurium, via altered SPI gene expression. Expression of slyA, a known regulator of SPI-2 (40), was increased upon overexpression of ramA and in opposition to the expression of the pathogenicity island genes it regulates (26). These observations again suggest that some of the transcriptional changes that appear to be RamA dependent may be related to the levels of other regulators. Furthermore, these data suggest that the intracellular concentration of RamA defines not only the scope of its regulon but also the level of differential expression of targets within it. Disruption of ramR, leading to modest overexpression of ramA, also decreased expression of pathogenicity genes, though for far fewer genes than seen with plasmid-driven ramA overexpression. Given the attenuation of L1007 within the C. elegans model, this again supports our hypothesis that ramA/RamA expression influences not only efflux pump gene expression but also pathogenicity.
Less clear is the effect upon pathogenicity after ramA disruption. As increased expression of a large number of SPI-1 genes was observed, it was anticipated that increased adhesion to the mouse macrophage would be observed. However, no effect upon adhesion to macrophages or attenuation within the C. elegans model was observed. Interestingly, attenuation in the ability to survive both within the macrophage and within the mouse model was seen. This attenuation may suggest that the decreased expression of only three SPI-2 genes plays a greater role in defining pathogenicity in these two models than the increased expression of many more genes in SPI-1. This is supported by the lack of attenuation within the C. elegans of the ramA::aph strain. Prior work with C. elegans (2) has shown that salmonella does not cause an intracellular infection, and our data and those of others (63) suggest that adhesion, rather than intracellular survival, is measured within this model. This also correlates with recent work in salmonella that demonstrated that SPI-1 genes are less critical for pathogenicity within the mouse systemic infection model (18) than SPI-2 genes.
Inactivation of ramA revealed large changes in expression of ribosomal, glycolytic/gluconeogenic, and amino acid biosynthetic pathways, whereas overexpression of ramA had little effect on these genes. This could be due to the increased expression of SPI-1 genes prompting cellular metabolism to switch to an intracellular-adapted form. Bowden et al. (12) showed that glycolysis and glucose transport are important in the infection of mouse macrophages (12). Furthermore, transcriptomic analyses by Eriksson et al. (25) of S. Typhimurium grown intracellularly within macrophages revealed that the expression of 20 to 30% of genes in ribosomal and amino acid biosynthetic pathways was affected by intracellular growth, with an approximately equal split between increased and decreased expression. Within the subset of amino acid biosynthetic genes whose expression was altered after intracellular macrophage growth (25), cysteine biosynthesis was generally increased. Within this study, expression of cysteine biosynthetic genes was generally increased after inactivation of ramA, but decreased after inactivation of ramR. This lends further support to the hypothesis that RamA represses expression of genes contributing to pathogenicity and intracellular survival genes. Further interrogation of data obtained by Eriksson et al. (25) revealed that expression of ramA was increased 4, 8, and 12 h after macrophage infection between 7.6- and 24.3-fold, again suggesting a role for RamA in the control of salmonella pathogenicity. However, expression of ramR was relatively constant compared to the control, increasing to a maximum of only 1.5-fold.
In this study, we show that production of AcrAB-TolC and proteins that confer the ability for Salmonella enterica serovar Typhimurium to be pathogenic is RamA dependent. This demonstrates that the regulation of a mechanism of MDR and expression of virulence genes show considerable overlap. However, we have no evidence to support the hypothesis that increased drug resistance via AcrAB-TolC and RamA production gives rise to a hypervirulent strain.
Supplementary Material
Acknowledgments
We thank Maria Fookes and Gemma Landgridge (The Wellcome Trust Sanger Institute) for their help with the microarrays and in silico work; Eirwen Morgan and Mark Stevens (Institute for Animal Health) for the Western blotting; and Sarah Coleman (University of Birmingham) for the mouse macrophage assays.
This work was supported by the Bristol Myers Squibb Foundation Non-Restrictive Grant in Infectious Diseases and by MRC grants GO501415 and GO801977 to L.J.V.P. and by the Wellcome Trust for J.W., R.K., and A.I.
Footnotes
Published ahead of print on 14 January 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Aballay, A., and F. M. Ausubel. 2001. Programmed cell death mediated by ced-3 and ced-4 protects Caenorhabditis elegans from Salmonella typhimurium-mediated killing. Proc. Natl. Acad. Sci. U. S. A. 98:2735-2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aballay, A., P. Yorgey, and F. M. Ausubel. 2000. Salmonella typhimurium proliferates and establishes a persistent infection in the intestine of Caenorhabditis elegans. Curr. Biol. 10:1539-1542. [DOI] [PubMed] [Google Scholar]
- 3.Abouzeed, Y. M., S. Baucheron, and A. Cloeckaert. 2008. ramR mutations involved in efflux-mediated multidrug resistance in Salmonella enterica serovar Typhimurium. Antimicrob. Agents Chemother. 52:2428-2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aono, R., N. Tsukagoshi, and M. Yamamoto. 1998. Involvement of outer membrane protein TolC, a possible member of the mar-sox regulon, in maintenance and improvement of organic solvent tolerance of Escherichia coli K-12. J. Bacteriol. 180:938-944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ariza, R. R., Z. Li, N. Ringstad, and B. Demple. 1995. Activation of multiple antibiotic resistance and binding of stress-inducible promoters by Escherichia coli Rob protein. J. Bacteriol. 177:1655-1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bailey, A. M., I. T. Paulsen, and L. J. Piddock. 2008. RamA confers multidrug resistance in Salmonella enterica via increased expression of acrB, which is inhibited by chlorpromazine. Antimicrob. Agents Chemother. 52:3604-3611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bailey, A. M., M. A. Webber, and L. J. Piddock. 2006. Medium plays a role in determining expression of acrB, marA, and soxS in Escherichia coli. Antimicrob. Agents Chemother. 50:1071-1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barbosa, T. M., and S. B. Levy. 2000. Differential expression of over 60 chromosomal genes in Escherichia coli by constitutive expression of MarA. J. Bacteriol. 182:3467-3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baucheron, S., H. Imberechts, E. Chaslus-Dancla, and A. Cloeckaert. 2002. The AcrB multidrug transporter plays a major role in high-level fluoroquinolone resistance in Salmonella enterica serovar typhimurium phage type DT204. Microb. Drug Resist. 8:281-289. [DOI] [PubMed] [Google Scholar]
- 10.Baucheron, S., C. Mouline, K. Praud, E. Chaslus-Dancla, and A. Cloeckaert. 2005. TolC but not AcrB is essential for multidrug-resistant Salmonella enterica serotype Typhimurium colonization of chicks. J. Antimicrob. Chemother. 55:707-712. [DOI] [PubMed] [Google Scholar]
- 11.Bina, X. R., C. L. Lavine, M. A. Miller, and J. E. Bina. 2008. The AcrAB RND efflux system from the live vaccine strain of Francisella tularensis is a multiple drug efflux system that is required for virulence in mice. FEMS Microbiol. Lett. 279:226-233. [DOI] [PubMed] [Google Scholar]
- 12.Bowden, S. D., G. Rowley, J. C. Hinton, and A. Thompson. 2009. Glucose and glycolysis are required for the successful infection of macrophages and mice by Salmonella enterica serovar Typhimurium. Infect. Immun. 77:3117-3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brenner, S. 1974. The genetics of Caenorhabditis elegans. Genetics 77:71-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Buckley, A. M., M. A. Webber, S. Cooles, L. P. Randall, R. M. La Ragione, M. J. Woodward, and L. J. Piddock. 2006. The AcrAB-TolC efflux system of Salmonella enterica serovar Typhimurium plays a role in pathogenesis. Cell Microbiol. 8:847-856. [DOI] [PubMed] [Google Scholar]
- 15.Bunikis, I., K. Denker, Y. Ostberg, C. Andersen, R. Benz, and S. Bergstrom. 2008. An RND-type efflux system in Borrelia burgdorferi is involved in virulence and resistance to antimicrobial compounds. PLoS Pathog. 4:e1000009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burse, A., H. Weingart, and M. S. Ullrich. 2004. The phytoalexin-inducible multidrug efflux pump AcrAB contributes to virulence in the fire blight pathogen, Erwinia amylovora. Mol. Plant Microbe. Interact. 17:43-54. [DOI] [PubMed] [Google Scholar]
- 17.Chaudhuri, R. R., N. J. Loman, L. A. S. Snyder, C. M. Bailey, D. J. Stekel, and M. J. Pallen. 2008. xBASE2: a comprehensive resource for comparative bacterial genomics. Nucleic Acids Res. 36:D543-546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chaudhuri, R. R., S. E. Peters, S. J. Pleasance, H. Northen, C. Willers, G. K. Paterson, D. B. Cone, A. G. Allen, P. J. Owen, G. Shalom, D. J. Stekel, I. G. Charles, and D. J. Maskell. 2009. Comprehensive identification of Salmonella enterica serovar Typhimurium genes required for infection of BALB/c mice. PLoS Pathog. 5:e1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chollet, R., J. Chevalier, C. Bollet, J. M. Pages, and A. Davin-Regli. 2004. RamA is an alternate activator of the multidrug resistance cascade in Enterobacter aerogenes. Antimicrob. Agents Chemother. 48:2518-2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cohen, S. P., H. Hachler, and S. B. Levy. 1993. Genetic and functional analysis of the multiple antibiotic resistance (mar) locus in Escherichia coli. J. Bacteriol. 175:1484-1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cohen, S. P., L. M. McMurry, and S. B. Levy. 1988. marA locus causes decreased expression of OmpF porin in multiple-antibiotic-resistant (Mar) mutants of Escherichia coli. J. Bacteriol. 170:5416-5422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Demple, B., and C. F. Amábile-Cuevas. 1991. Redox redux: The control of oxidative stress responses. Cell 67:837-839. [DOI] [PubMed] [Google Scholar]
- 24.Eaves, D. J., V. Ricci, and L. J. V. Piddock. 2004. Expression of acrB, acrF, acrD, marA, and soxS in Salmonella enterica serovar Typhimurium: role in multiple antibiotic resistance. Antimicrob. Agents Chemother. 48:1145-1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eriksson, S., S. Lucchini, A. Thompson, M. Rhen, and J. C. Hinton. 2003. Unravelling the biology of macrophage infection by gene expression profiling of intracellular Salmonella enterica. Mol. Microbiol. 47:103-118. [DOI] [PubMed] [Google Scholar]
- 26.Fass, E., and E. A. Groisman. 2009. Control of Salmonella pathogenicity island-2 gene expression. Curr. Opin. Microbiol. 12:199-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fralick, J. A. 1996. Evidence that TolC is required for functioning of the Mar/AcrAB efflux pump of Escherichia coli. J. Bacteriol. 178:5803-5805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.George, A. M., R. M. Hall, and H. W. Stokes. 1995. Multidrug resistance in Klebsiella pneumoniae: a novel gene, ramA, confers a multidrug resistance phenotype in Escherichia coli. Microbiology 141(Pt. 8):1909-1920. [DOI] [PubMed] [Google Scholar]
- 29.Giraud, E., A. Cloeckaert, D. Kerboeuf, and E. Chaslus-Dancla. 2000. Evidence for active efflux as the primary mechanism of resistance to ciprofloxacin in Salmonella enterica serovar Typhimurium. Antimicrob. Agents Chemother. 44:1223-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Griffith, K. L., and R. E. Wolf, Jr. 2001. Systematic mutagenesis of the DNA binding sites for SoxS in the Escherichia coli zwf and fpr promoters: identifying nucleotides required for DNA binding and transcription activation. Mol. Microbiol. 40:1141-1154. [DOI] [PubMed] [Google Scholar]
- 31.Hachler, H., S. P. Cohen, and S. B. Levy. 1991. marA, a regulated locus which controls expression of chromosomal multiple antibiotic resistance in Escherichia coli. J. Bacteriol. 173:5532-5538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hernandez-Urzua, E., D. S. Zamorano-Sanchez, J. Ponce-Coria, E. Morett, S. Grogan, R. K. Poole, and J. Membrillo-Hernandez. 2007. Multiple regulators of the flavohaemoglobin (hmp) gene of Salmonella enterica serovar Typhimurium include RamA, a transcriptional regulator conferring the multidrug resistance phenotype. Arch. Microbiol. 187:67-77. [DOI] [PubMed] [Google Scholar]
- 33.Hope, I. A. (ed.). 1999. C. elegans: a practical approach. Oxford University Press, Oxford, United Kingdom.
- 34.Hulbert, R. R., and R. K. Taylor. 2002. Mechanism of ToxT-dependent transcriptional activation at the Vibrio cholerae tcpA promoter. J. Bacteriol. 184:5533-5544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jair, K. W., X. Yu, K. Skarstad, B. Thony, N. Fujita, A. Ishihama, and R. E. Wolf, Jr. 1996. Transcriptional activation of promoters of the superoxide and multiple antibiotic resistance regulons by Rob, a binding protein of the Escherichia coli origin of chromosomal replication. J. Bacteriol. 178:2507-2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaniga, K., J. C. Bossio, and J. E. Galan. 1994. The Salmonella typhimurium invasion genes invF and invG encode homologues of the AraC and PulD family of proteins. Mol. Microbiol. 13:555-568. [DOI] [PubMed] [Google Scholar]
- 37.Karp, P. D., S. Paley, and P. Romero. 2002. The Pathway Tools software. Bioinformatics 18:S225-232. [DOI] [PubMed] [Google Scholar]
- 38.Keeney, D., A. Ruzin, and P. A. Bradford. 2007. RamA, a transcriptional regulator, and AcrAB, an RND-type efflux pump, are associated with decreased susceptibility to tigecycline in Enterobacter cloacae. Microb. Drug Resist. 13:1-6. [DOI] [PubMed] [Google Scholar]
- 39.Kingsley, R. A., A. D. Humphries, E. H. Weening, M. R. de Zoete, S. Winter, A. Papaconstantinopoulou, G. Dougan, and A. J. Baumler. 2003. Molecular and phenotypic analysis of the CS54 island of Salmonella enterica serotype Typhimurium: identification of intestinal colonization and persistence determinants. Infect. Immun. 71:629-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Linehan, S. A., A. Rytkonen, X.-J. Yu, M. Liu, and D. W. Holden. 2005. SlyA regulates function of Salmonella pathogenicity island 2 (SPI-2) and expression of SPI-2-associated genes. Infect. Immun. 73:4354-4362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martin, R. G., E. S. Bartlett, J. L. Rosner, and M. E. Wall. 2008. Activation of the Escherichia coli marA/soxS/rob regulon in response to transcriptional activator concentration. J. Mol. Biol. 380:278-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martin, R. G., W. K. Gillette, and J. L. Rosner. 2000. Promoter discrimination by the related transcriptional activators MarA and SoxS: differential regulation by differential binding. Mol. Microbiol. 35:623-634. [DOI] [PubMed] [Google Scholar]
- 43.Martin, R. G., K. W. Jair, R. E. Wolf, Jr., and J. L. Rosner. 1996. Autoactivation of the marRAB multiple antibiotic resistance operon by the MarA transcriptional activator in Escherichia coli. J. Bacteriol. 178:2216-2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martin, R. G., and J. L. Rosner. 2002. Genomics of the marA/soxS/rob regulon of Escherichia coli: identification of directly activated promoters by application of molecular genetics and informatics to microarray data. Mol. Microbiol. 44:1611-1624. [DOI] [PubMed] [Google Scholar]
- 45.McClelland, M., K. E. Sanderson, J. Spieth, S. W. Clifton, P. Latreille, L. Courtney, S. Porwollik, J. Ali, M. Dante, F. Du, S. Hou, D. Layman, S. Leonard, C. Nguyen, K. Scott, A. Holmes, N. Grewal, E. Mulvaney, E. Ryan, H. Sun, L. Florea, W. Miller, T. Stoneking, M. Nhan, R. Waterston, and R. K. Wilson. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852-856. [DOI] [PubMed] [Google Scholar]
- 46.McMurry, L. M., M. Oethinger, and S. B. Levy. 1998. Overexpression of marA, soxS, or acrAB produces resistance to triclosan in laboratory and clinical strains of Escherichia coli. FEMS Microbiol. Lett. 166:305-309. [DOI] [PubMed] [Google Scholar]
- 47.Morgan, E., A. J. Bowen, S. C. Carnell, T. S. Wallis, and M. P. Stevens. 2007. SiiE Is Secreted by the Salmonella enterica serovar Typhimurium pathogenicity island 4-encoded secretion system and contributes to intestinal colonization in cattle. Infect. Immun. 75:1524-1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nikaido, E., A. Yamaguchi, and K. Nishino. 2008. AcrAB multidrug efflux pump regulation in Salmonella enterica serovar Typhimurium by RamA in response to environmental signals. J. Biol. Chem. 283:24245-24253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nikaido, H., M. Basina, V. Nguyen, and E. Y. Rosenberg. 1998. Multidrug efflux pump AcrAB of Salmonella typhimurium excretes only those beta-lactam antibiotics containing lipophilic side chains. J. Bacteriol. 180:4686-4692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Perez, J. C., D. Shin, I. Zwir, T. Latifi, T. J. Hadley, and E. A. Groisman. 2009. Evolution of a bacterial regulon controlling virulence and Mg2+ homeostasis. PLoS Genet 5:e1000428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Piddock, L. J., D. G. White, K. Gensberg, L. Pumbwe, and D. J. Griggs. 2000. Evidence for an efflux pump mediating multiple antibiotic resistance in Salmonella enterica serovar Typhimurium. Antimicrob. Agents Chemother. 44:3118-3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pomposiello, P. J., M. H. J. Bennik, and B. Demple. 2001. Genome-Wide Transcriptional Profiling of the Escherichia coli Responses to Superoxide Stress and Sodium Salicylate. J. Bacteriol. 183:3890-3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Prieto, A. I., S. B. Hernandez, I. Cota, M. G. Pucciarelli, Y. Orlov, F. Ramos-Morales, F. Garcia-del Portillo, and J. Casadesus. 2009. Roles of the outer membrane protein AsmA of Salmonella enterica in control of marRAB expression and invasion of epithelial cells. J. Bacteriol. 191:3615-3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Prouty, A. M., I. E. Brodsky, S. Falkow, and J. S. Gunn. 2004. Bile-salt-mediated induction of antimicrobial and bile resistance in Salmonella typhimurium. Microbiology 150:775-783. [DOI] [PubMed] [Google Scholar]
- 55.Ramos, J. L., F. Rojo, L. Zhou, and K. N. Timmis. 1990. A family of positive regulators related to the Pseudomonas putida TOL plasmid XylS and the Escherichia coli AraC activators. Nucleic Acids Res. 18:2149-2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reimers, M., and V. J. Carey. 2006. Bioconductor: an open source framework for bioinformatics and computational biology. Methods Enzymol. 411:119-134. [DOI] [PubMed] [Google Scholar]
- 57.Ricci, V., and L. J. V. Piddock. 2009. Ciprofloxacin selects for multidrug resistance in Salmonella enterica serovar Typhimurium mediated by at least two different pathways. J. Antimicrob. Chemother. 63:909-916. [DOI] [PubMed] [Google Scholar]
- 58.Ricci, V., P. Tzakas, A. Buckley, N. C. Coldham, and L. J. V. Piddock. 2006. Ciprofloxacin-resistant Salmonella enterica serovar Typhimurium strains are difficult to select in the absence of AcrB and TolC. Antimicrob. Agents Chemother. 50:38-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rice, P., I. Longden, and A. Bleasby. 2000. EMBOSS: The European Molecular Biology open software suite. Trends Genet. 16:276-277. [DOI] [PubMed] [Google Scholar]
- 60.Schneiders, T., S. G. B. Amyes, and S. B. Levy. 2003. Role of AcrR and RamA in fluoroquinolone resistance in clinical Klebsiella pneumoniae isolates from Singapore. Antimicrob. Agents Chemother. 47:2831-2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stiernagle, T. 1999. Maintenance of C. elegans, p. 53-55. In I. A. Hope (ed.), C. elegans: a practical approach. Oxford University Press, Oxford, United Kingdom.
- 62.Sulavik, M. C., L. F. Gambino, and P. F. Miller. 1995. The MarR repressor of the multiple antibiotic resistance (mar) operon in Escherichia coli: prototypic member of a family of bacterial regulatory proteins involved in sensing phenolic compounds. Mol. Med. 1:436-446. [PMC free article] [PubMed] [Google Scholar]
- 63.Tenor, J. L., B. A. McCormick, F. M. Ausubel, and A. Aballay. 2004. Caenorhabditis elegans-based screen identifies Salmonella virulence factors required for conserved host-pathogen interactions. Curr. Biol. 14:1018-1024. [DOI] [PubMed] [Google Scholar]
- 64.Thanassi, D. G., L. W. Cheng, and H. Nikaido. 1997. Active efflux of bile salts by Escherichia coli. J. Bacteriol. 179:2512-2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tibbetts, R. J., T. L. Lin, and C. C. Wu. 2003. Phenotypic evidence for inducible multiple antimicrobial resistance in Salmonella choleraesuis. FEMS Microbiol. Lett. 218:333-338. [DOI] [PubMed] [Google Scholar]
- 66.Tobin, J. F., and R. F. Schleif. 1987. Positive regulation of the Escherichia coli l-rhamnose operon is mediated by the products of tandemly repeated regulatory genes. J. Mol. Biol. 196:789-799. [DOI] [PubMed] [Google Scholar]
- 67.van der Straaten, T., R. Janssen, D. J. Mevius, and J. T. van Dissel. 2004. Salmonella gene rma (ramA) and multiple-drug-resistant Salmonella enterica serovar typhimurium. Antimicrob. Agents Chemother. 48:2292-2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Warner, D., J. Folster, W. Shafer, and A. Jerse. 2007. Regulation of the MtrC-MtrD-MtrE efflux pump system modulates the in vivo fitness of Neisseria gonorrhoeae. J. Infect. Dis. 196:1804-1812. [DOI] [PubMed] [Google Scholar]
- 69.Warner, D. M., W. M. Shafer, and A. E. Jerse. 2008. Clinically relevant mutations that cause derepression of the Neisseria gonorrhoeae MtrC-MtrD-MtrE efflux pump system confer different levels of antimicrobial resistance and in vivo fitness. Mol. Microbiol. 70:462-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Webber, M. A., A. M. Bailey, J. M. A. Blair, E. Morgan, M. P. Stevens, J. C. D. Hinton, A. Ivens, J. Wain, and L. J. V. Piddock. 2009. The global consequence of disruption of the AcrAB-TolC efflux pump in Salmonella enterica includes reduced expression of SPI-1 and other attributes required to infect the host. J. Bacteriol. 191:4276-4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.White, D. G., J. D. Goldman, B. Demple, and S. B. Levy. 1997. Role of the acrAB locus in organic solvent tolerance mediated by expression of marA, soxS, or robA in Escherichia coli. J. Bacteriol. 179:6122-6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wray, C., and W. J. Sojka. 1978. Experimental Salmonella typhimurium infection in calves. Res. Vet. Sci. 25:139-143. [PubMed] [Google Scholar]
- 73.Zheng, J., S. Cui, and J. Meng. 2009. Effect of transcriptional activators RamA and SoxS on expression of multidrug efflux pumps AcrAB and AcrEF in fluoroquinolone-resistant Salmonella Typhimurium. J. Antimicrob. Chemother. 63:95-102. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.