Abstract
We identified a new regulator, PcaO, which is involved in regulation of the protocatechuate (PCA) branch of the β-ketoadipate pathway in Corynebacterium glutamicum. PcaO is an atypical large ATP-binding LuxR family (LAL)-type regulator and does not have a Walker A motif. A mutant of C. glutamicum in which pcaO was disrupted (RES167ΔpcaO) was unable to grow on PCA, and growth on PCA was restored by complementation with pcaO. Both an enzymatic assay of PCA 3,4-dioxygenase activity (encoded by pcaHG) and transcriptional analysis of pcaHG by reverse transcription-PCR revealed that PcaO positively regulated pcaHG. A promoter-LacZ transcriptional fusion assay suggested that PcaO interacted with the sequence upstream of pcaHG. Electrophoretic mobility shift assay (EMSA) analysis indicated that an imperfect palindromic sequence (−78AACCCCTGACCTTCGGGGTT−59) that was located upstream of the −35 region of the pcaHG promoter was essential for PcaO regulation. DNase I footprinting showed that this imperfect palindrome was protected from DNase I digestion. Site-directed mutation and EMSA tests revealed that this palindrome sequence was essential for PcaO binding to the DNA fragment. In vitro EMSA results showed that ATP weakened the binding between PcaO and its target sequence but ADP strengthened this binding, while the effect of protocatechuate on PcaO binding was dependent on the protocatechuate concentration.
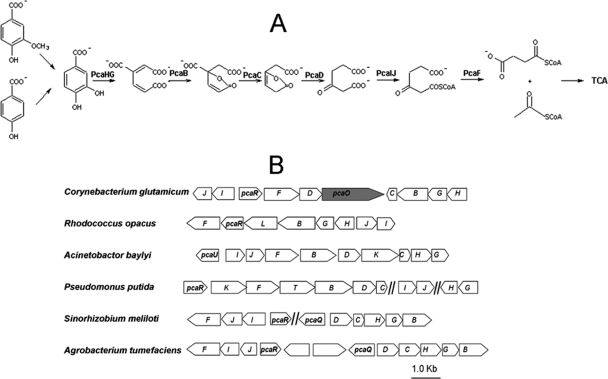
Protocatechuic acid (PCA) is an important intermediate during microbial degradation of various aromatic compounds, including natural products such as lignin monomers and chemically synthesized chemicals such as 4-chlorobenzoate. In many bacteria (5, 8, 12, 13, 16, 20, 23, 25, 30, 34), PCA is degraded via the β-ketoadipate pathway (Fig. 1A), and the ring cleavage dioxygenase (PCA 3,4-dioxygenase) is encoded by two consecutive genes, pcaHG (3). The pcaHG genes, together with other pca genes involved in PCA degradation, have been extensively investigated in Gram-negative bacteria, such as Pseudomonas putida (12, 23) and Acinetobacter baylyi (10, 28), and recently have been investigated in Gram-positive bacteria, such as Rhodococcus and Streptomyces species (8, 16) (Fig. 1B).
FIG. 1.
Schematic diagram of the protocatechuate (PCA) branch of the β-ketoadipate pathway (A) and genetic organization of the genes involved in C. glutamicum and other bacteria (B). The complete designations of all regulator genes are indicated, and the pcaO gene is indicated by shading. Other pca genes are indicated by capital letters. PcaHG, PCA 3,4-dioxygenase; PcaB, β-carboxy-cis,cis-muconate cycloimerase; PcaC, γ-carboxymuconolactone decarboxylase; PcaD, β-ketoadipate enol-lactone hydrolase; PcaIJ, β-ketoadipate succinyl-coenzyme A transferase; PcaF, β-ketoadipate coenzyme A thiolase; TCA, tricarboxylic acid cycle.
Regulation of the PCA branch of the β-ketoadipate pathway has been investigated using several Gram-negative bacteria (41), and information is accumulating. In P. putida, the IclR-type PcaR protein regulates pcaIJBDCF (11, 30); however, how pcaHG is regulated is still unknown. In A. baylyi, the IclR-type PcaU protein regulates the pcaIJFBDKCHG operon, and PCA is the effector (10, 28, 40). In Sinorhizobium meliloti, PcaR regulates pcaIJF, while the LysR-type protein PcaQ regulates pcaDCHGB (20, 21). In Agrobacterium tumefaciens, PcaQ regulates pcaDCHGB and pcaJIF with β-carboxy-cis,cis-muconate and β-ketoadipate as effectors (25, 26). Putative regulators have been identified in Gram-positive bacteria, such as Corynebacterium glutamicum and Rhodococcus opacus (2, 8, 34, 35); however, the regulation of the PCA branch of the β-ketoadipate pathway has not been well documented.
The ability of C. glutamicum to metabolize aromatic compounds has recently been characterized, and this species has been shown to be a good model for studying aromatic compound metabolism (15, 33-35). The PCA branch of the β-ketoadipate pathway in C. glutamicum is encoded by a genetic element (pca) that contains 10 pca genes (35) and one transporter gene (6) (Fig. 1B). There are two putative regulator genes in the pca genetic cluster, pcaR and pcaO. According to the CoryneRegNet 4.0 database (1), PcaR is inferred to be the regulator of the whole pathway. But Brinkrolf et al. (2) reviewed all of the possible regulators in the catabolic pathway for aromatic compounds in C. glutamicum, and they concluded that PcaR regulated only part of PCA degradation. Here, we report functional identification of a new regulator, PcaO, and describe its regulation of the PCA branch of the β-ketoadipate pathway in C. glutamicum.
MATERIALS AND METHODS
Bacterial strains, plasmids, and cultivation.
Bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli strains were grown at 37°C in Luria-Bertani (LB) broth or on LB medium plates containing 1.5% (wt/vol) agar. C. glutamicum strains were cultivated routinely in LB broth or brain heart infusion broth supplemented with 50 mM sorbitol (BHIS). To test their abilities to grow on various aromatic compounds, C. glutamicum strains were cultivated in minimal medium (MM) (pH 8.0) (18). The final concentration of aromatic compounds in MM broth was 2.0 mM. C. glutamicum strains were incubated at 30°C with rotary shaking at 150 rpm. Growth was monitored by measuring the turbidity (optical density at 600 nm [OD600]). Depending on the genetic vectors used, antibiotics were added at the following concentrations: ampicillin, 100 μg ml−1 for E. coli; chloramphenicol, 50 μg ml−1 for E. coli and 10 μg ml−1 for C. glutamicum; kanamycin, 50 μg ml−1 for E. coli and 25 μg ml−1 for C. glutamicum; and nalidixic acid, 50 μg ml−1 for C. glutamicum.
TABLE 1.
Bacterial strains, plasmids, and primers used in this study
| Strain, plasmid, or primer | Relevant characteristics or sequence | Source, reference, or comment |
|---|---|---|
| E. coli strains | ||
| JM109 | recAl supE44 endAI hsdR17 gyrA96 relA1 thi Δ(lac-proAB) F′ (traD36 proAB lacIqlacZΔM15) | Stratagene (catalog no. 200235) |
| BL21(DE3) | hsdS gal (λcIts857 ind-l Sam7 nin-5 lacUV5-T7 gene 1) | Novagen (catalog no. 69387-3) |
| C. glutamicum strains | ||
| RES167 | Restriction-deficient mutant of ATCC 13032, Δ(cglIM-cglIR-cglIIR) | University of Bielefeld |
| RES167ΔPcaO | DNA fragment encoding amino acids 106 to 421 of pcaO was deleted | This study |
| Plasmids | ||
| pK18mobsacB | Mobilizable vector for generation of C. glutamicum mutants for deletion of pcaO | 32 |
| pK18mobsacB-ΔPcaO | This study | |
| pXMJ19-lacZ | pXMJ19 carrying lacZ | This study |
| pXMJ19-PpcaHG-lacZ | pXMJ19 carrying PpcaHG and lacZ; lacIq and Ptac were deleted | This study |
| pET28a | Expression vector | Novagen |
| pET28a-PcaO | pET28a derivative for expression of pcaO | This study |
| pEASY-T1 | Cloning vector | Transgen (Beijing, People's Republic of China) |
| Primersa | ||
| 2311F | TGCGGATCCAAAGGAGGAATGTCCCCTATACTAGGTTATTG (BamHI) | Used to generate |
| 2311R | TGATCTAGATCAGACCTTCAGTAATTCTCGAAGCTC (XbaI) | pK18mobsacB-ΔPcaO and pXMJ19-PcaO |
| 2311EF | TGCGGATCCATGTCCCCTATACTAGGTTATTG (BamHI) | Used to generate pET28a- |
| 2311ER | CTATCGCCGGCGTCAGACCTTCAGTAATTC (NotI) | PcaO |
| 2315PE | TCGTTGCTTGGGTTGCGC | Used for primer extension |
| 1516F | AGCTCCGCAGTTCACTCG | of pcaHG and DNase I footprinting |
| 2314RTF | TACAGGGAAGAACGGCGAGT | Used for RT-PCR analysis |
| 2314RTR | AATCAGAGTTGTGGATGC | of ncgl2314 |
| 2315RTF | CAACCCAAGCAACGATCTCA | Used for RT-PCR analysis |
| 2315RTR | TCCTGGAAGAACAATGGATCG | of ncgl2315 |
| rpoBF | GTTGGTGTTCGTATTGAC | Used for RT-PCR analysis |
| rpoBR | TGGTCATACGCTCACGAAC | of rpoB |
| Pro15F | CACGGATCCATGTTCACAACCAGTCTCC (PstI) | Used for generation of |
| Pro15R | TATGCGGCCGCTCAGACCTTCAGTAATTCTC (NarI) | pXMJ19-PpcaHG-lacZ |
| Intr1516Fb | GCGAGACCTTTCTGCGTC | |
| Intr1516R1b | TACTTGTCAACGGCCTCAATC | |
| Intr1516R2b | CGCGATCTGACATGTTCGC | |
| Intr1516R3b | TTGCGCTGGTGGCGAATGTTC | |
| Intr1516R4b | CTTATGCACACCCCTAGTTAACC | |
| Intr1516F5b | TATGCACACCCCTAGTTAAC | |
| Intr1516R5b | GCCACCAGCGCAAACCCC | |
| FiFc | TTATGCACACCCCTAGTTAACCCCGACCTTCGGGGTTT | |
| FaFc | TTATGCACACCCCTAGTTCCAAAAGACCTTCGGGGTTT | |
| FbFc | TTATGCACACCCCTAGTTAACCCCGACCTTTTGGTGCGCTGGTGGCGAATGTT | |
| FcFc | TTATGCACACCCCTAGTTCCAAAAGACCTTCTTGGGGTGCGCTGGTGGCGAATGTT | |
| FRc | GCGCGCGAACATTCGCCA | |
| lacZF | GCCCTGCAGATGACCATGATTACGGA (PstI) | Used for PCR of lacZ gene |
| lacZR | GGGATCCCGGGGAAATACGGGCAGACA (BamHI, SmaI) |
Restriction enzyme cut sites are underlined. Ribosome-binding sites are indicated by bold type.
Primer used for amplification of divided fragments of the intergenic sequence between pcaHG and ncgl2316 for EMSA analysis (fragment F1, bases −7 to 59; fragment F2, bases −34 to 59; fragment F3, bases −59 to 59; fragment F4, bases −95 to 59; fragment F5, bases −49 to −95).
Primer used for generation of mutants with mutations in the Rep4 palindromic sequence.
Analysis of sequence data.
The genome sequence of C. glutamicum ATCC 13032 (accession no. NC_003450 and NC_006958) was retrieved from the GenBank database (http://www.ncbi.nlm.nih.gov/). Sequence comparisons and database searches were carried out using BLAST programs at the BLAST server of the NCBI website. Phylogenetic analysis of PcaO was done with the MEGA 3.1 software. SBASE was used for protein domain prediction (http://hydra.icgeb.trieste.it/servers/protein/sbase/).
DNA extraction and manipulation.
Genomic DNA of C. glutamicum was isolated by the method of Tauch et al. (37). DNA manipulation, plasmid isolation, and agarose gel electrophoresis were routinely carried out using standard methods (31). Restriction endonucleases, ligase, and DNA polymerase were used according to the manufacturer's instructions. Vectors were electroporated into cells of E. coli and C. glutamicum by using the methods of Tauch et al. (38). Restricted DNA fragments were separated by agarose gel electrophoresis and were purified by using an agarose gel DNA fragment recovery kit (Tiangen, Beijing, People's Republic of China).
Genetic disruption and complementation in C. glutamicum.
The pK18mobsacB derivatives used for gene disruption and the pXMJ19 derivatives used for complementation were constructed in this study (Table 1) and were transformed into C. glutamicum cells by electroporation (32, 38). Screening for the first and second recombination events and analyses to confirm the chromosomal deletions were performed as described by Schäfer et al. (32). Expression of a target gene in pXMJ19 derivatives in C. glutamicum was induced by addition of 0.6 mM isopropyl-β-d-thiogalactopyranoside (IPTG) to the culture broth.
Enzymatic assay.
PCA 3,4-dioxygenase activity was measured as described by Iwagami et al. (16). The absorbance at 290 nm and 25°C was recorded with a spectrophotometer, and the results were used to calculate PCA 3,4-dioxygenase activity.
RT-PCR.
Total RNA from C. glutamicum cells was isolated with a TRIzol extraction kit (Invitrogen, Carlsbad, CA). The RNA preparation first was treated with DNase I and then was used as a template for reverse transcription with Moloney murine leukemia virus (MMLV) reverse transcriptase (Promega, Madison, WI). The reverse transcription products (cDNAs) were used to amplify fragments (approximately 350 to 500 bp) of target genes. The primers used in RT-PCR are listed in Table 1. In order to exclude the possibility of DNA contamination, negative controls were run in parallel (i.e., the MMLV reverse transcriptase was omitted from the reaction mixtures). The rpoB gene (encoding the β-subunit of RNA polymerase) was used as a positive control to check the reaction system.
Construction of plasmid pXMJ19-PpcaHG-lacZ.
Plasmid pXMJ19-lacZ was constructed by replacing lacI and Ptac with a lacZ fragment at the NarI and PstI sites of plasmid pXMJ19 (17). Plasmid pXMJ19-PpcaHG-lacZ carrying the promoter region of pcaHG (PpcaHG) was constructed by ligating the amplified PpcaHG DNA fragment from the C. glutamicum genome to pXMJ19-lacZ.
Assay of β-galactosidase activity.
The activity of the pcaHG promoter in pXMJ19-PpcaHG-lacZ was monitored by assaying β-galactosidase activity. C. glutamicum was grown in LB medium and in minimal salts medium. The RES167ΔpcaO mutant was first grown in LB broth until the mid-log phase, and then cells were harvested and were transferred to mineral medium that contained 2 mM PCA for induction (2 h) of β-galactosidase activity. The β-galactosidase activity was determined using the Miller assay (22).
Heterologous expression and purification of PcaO from recombinant E. coli cells.
Plasmid pET28a-PcaO was electroporated into E. coli BL21(DE3). Synthesis of PcaO proteins was initiated by addition of 0.1 mM IPTG, and cultivation was continued for an additional 10 h at 16°C. Cells were harvested by centrifugation at 10,000 × g, washed twice with ice-cold 50 mM Tris-HCl buffer (pH 8.0), resuspended in the same buffer, and disrupted by sonication in an ice-water bath. After centrifugation at 20,000 × g for 30 min at 4°C, the supernatant was used for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis. A His-bind purification kit was used to purify the His-tagged PcaO proteins.
Primer extension.
Primer 2315PE (Table 1) was labeled with [γ-32P]ATP and T4 polynucleotide kinase. Reverse transcription was carried out with 10 pM 5′-32P-labeled 2315PE and 30 μg of total RNA from C. glutamicum cells cultivated with PCA. After annealing at 65°C for 10 min, deoxynucleoside triphosphates (dNTPs) and MMLV transcriptase were added, and the mixture was incubated at 45°C for 60 min and then at 65°C for 10 min to inactivate the transcriptase. The reaction mixture was treated with phenol, and the contents were precipitated with ethanol and resuspended in loading buffer. The dissolved DNA fragments were separated on a 6% polyacrylamide gel containing 42% urea. DNA sequencing reaction mixtures were prepared using the same 32P-labeled primers and a silver stain sequencing kit (Promega, Madison, WI).
DNase I footprinting.
The DNA segment containing the promoter of pcaHG was amplified from genomic DNA with 5′-32P-labeled primer 1516F and unlabeled primer 2315PE. The 32P-labeled DNA fragment (∼0.1 nM) was mixed with purified PcaO (10 pM) in 50 μl (total volume) of binding buffer (10 mM Tris, 50 mM KCl, 1 mM dithiothreitol; pH 7.5), and the mixture was incubated for 30 min at 25°C. Then 10 mU DNase I was added, and the mixture was incubated for 3 min at 25°C. After this, 50 μl of DNase I stop buffer was added to the reaction mixture. The reaction mixture was quickly treated with phenol, and the contents were precipitated with ethanol at −70°C overnight. The precipitates were resuspended in loading buffer. Separation of DNA fragments on a 6% polyacrylamide gel containing 42% urea and DNA sequencing were performed with a silver stain sequencing kit (Promega, Madison, WI).
Electrophoretic mobility shift assay (EMSA).
The 32P-labeled intergenic DNA fragment between pcaHG and ncgl2316 was obtained by PCR amplification with [γ-32P]ATP and labeled primers using T4 polynucleotide kinase. The DNA fragment (approximately 10 pM) and various amounts of PcaO (final concentrations, 0 to 0.5 μM) were mixed in 20 μl binding buffer and were incubated for 15 min at room temperature. The effectors were used at concentrations ranging from 0.5 to 2.0 mM. The binding buffer contained 10 mM Tris-HCl (pH 7.5), 50 mM KCl, 1 mM dithiothreitol, 1 μg poly(dI-dC), and 5% glycerol. The binding mixture was loaded onto a native gel (5% polyacrylamide), and 0.5× Tris-borate-EDTA buffer was used as the running buffer. After electrophoresis the gels were dried and subjected to autoradiography.
RESULTS
pcaO is required for PCA assimilation in strain RES167 and encodes a putative novel LAL-type regulator.
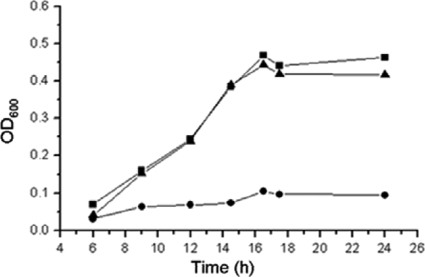
Two genes encoding putative transcriptional regulators were located in the pca gene cluster (Fig. 1B). Transcriptional analysis indicated that PcaR negatively regulated pcaIJ and pcaFDO but did not regulate pcaHGBC (2). In order to identify the function of pcaO, this gene was disrupted by deletion of the DNA fragment encoding amino acids 106 to 421, and the RES167ΔpcaO mutant was obtained (Table 1). The RES167ΔpcaO mutant was not able to grow on PCA (Fig. 2), 4-hydroxybenzoate, and vanillate, which were assimilated via the PCA branch of the β-ketoadipate pathway (34, 35). Growth of the RES167ΔpcaO mutant on benzoate and resorcinol as carbon and energy sources was not affected (data not shown). When RES167ΔpcaO was complemented with pXMJ19-PcaO, the ability to grow on protocatechuate was restored (Fig. 2). Based on these results, it was concluded that PcaO was essential for PCA degradation and the PCA branch of the β-ketoadipate pathway.
FIG. 2.
Growth of C. glutamicum strains in mineral salts medium with 2 mM protocatechuate as the sole carbon and energy source. ▪, RES167; •, RES167ΔpcaO; ▴, RES167ΔpcaO/pXMJ19-PcaO.
Nucleotide sequence analysis of pcaO predicted that this gene encodes a protein consisting of 687 amino acid residues. Based on sequence analysis, PcaO was structurally similar to MalT, a member of the large ATP-binding LuxR (LAL) subfamily of bacterial regulators that has Walker A and Walker B motifs for ATP binding, as well as a LuxR helix-turn-helix (HTH) DNA-binding domain (7, 19, 39). A putative HTH DNA-binding domain was identified at the C terminus of PcaO (amino acid residues 631 to 685), and a putative walker B motif was identified at the N terminus (amino acid residues 65 to 78). These features suggested that PcaO may be a member of the LAL subfamily. However, PcaO is short (687 amino acids), and no obvious Walker A motif was identified (data not shown) in comparisons with the previously identified LAL regulators (7, 19).
Molecular phylogenetic analysis showed that PcaO grouped with the functionally identified LAL-type regulators (data not shown). The highest level of identity found was the level of identity between PcaO and a putative regulator of benzoate degradation (BenR, NCgl2324) of C. glutamicum (28% identity) (14, 15, 35). The levels of identity between PcaO and other LAL regulators, including LipR of Streptomyces coelicolor (42), LipR of Streptomyces exfoliatus (9), PikD of Streptomycoes venezuelae (47), AcoK of Klebsiella pneumoniae (27), AlkS of Pseudomonas oleovorans (48), and MalT of E. coli (29), were less than 20%. We concluded that this PcaO was a novel and atypical LAL-type regulator.
PcaO regulatation of pcaHG transcription and determination of the promoter region.
Previous work demonstrated that pcaHG encodes PCA 3,4-dioxygenase. This enzyme is essential for vanillate, 4-hydroxybenzoate, and protocatechuate assimilation (34, 35). Previous work also demonstrated that the activity of PCA 3,4-dioxygenase was inducible in C. glutamicum RES167 upon addition of PCA (34, 35). In this study, we found that PCA 3,4-dioxygenase activity was not detectable even in the presence of PCA in culture medium of the RES167ΔpcaO mutant (data not shown). Further, an RT-PCR analysis of pcaHG showed that pcaHG was transcribed in RES167 but not in RES167ΔpcaO (data not shown). Thus, we concluded that PcaO regulated pcaHG at the transcriptional level.
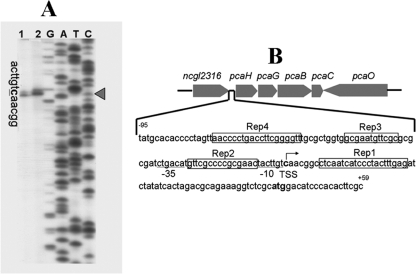
The transcriptional start site (TSS) of pcaHG was determined to be the 59th base (C) upstream of the translational start codon (ATG) (Fig. 3A). The pcaHG promoter (PpcaHG) was located between pcaHG and ncgl2316 (Fig. 3B). Thus, the sequence from base −212 to base 72 was amplified by PCR performed with primers Pro15F and Pro15R (Table 1). The PCR product was ligated to pXMJ19-lacZ, and fusion plasmid pXMJ19-PpcaHG-lacZ was obtained. The pcaHG promoter activity was evaluated by determining the β-galactosidase activity in cells of RES167 and RES167ΔpcaO. The results clearly indicated that the β-galactosidase activity relied on the presence of PcaO. In all of the experiments with RES167 β-galactosidase activity was apparent, but there was not significant β-galactosidase activity in RES167ΔpcaO (Fig. 4). Based on these results, we concluded that PcaO regulated the β-galactosidase activity in the pcaHG promoter-lacZ transcriptional fusion, and we also deduced that PcaO interacted with the promoter region of pcaHG.
FIG. 3.
Determination of the transcription start site (TSS) of pcaHG (A) and diagram of intergenic sequences between pcaHG and ncgl2316 (B). In panel A, the transcription start base C is indicated by a triangle. Primer extension was performed using DNase I-treated RNA (10 μg) isolated from cells cultivated in mineral salts medium containing protocatechuate. The labeled primer (2315PE) was complementary to a sequence 96 bp downstream of the translation start codon of pcaH. The sequencing products of PpcaHG obtained with the labeled primer (lanes G, A, C, and T) were simultaneously electrophoresed with the reverse transcribed products. In panel B, the palindromic sequences (Rep1, Rep2, Rep3, and Rep4), putative ribosome binding site (RBS), and putative −10 and −35 regions are indicated.
FIG. 4.
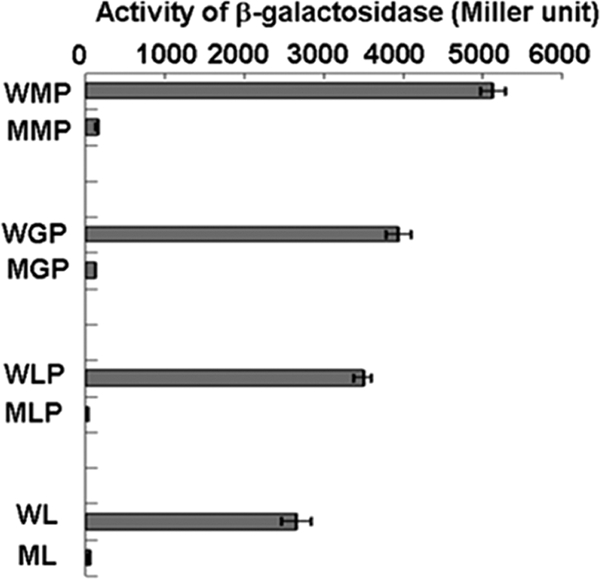
Determination of pcaHGBC promoter activity in C. glutamicum RES167 and RES167ΔpcaO, as reflected by β-galactosidase activity. WMP, wild-type strain in mineral medium with 2.0 mM protocatechuate; MMP, mutant strain in mineral medium with 2 mM protocatechuate; WGP, wild-type strain in mineral medium with protocatechuate and 1 mM glucose; MPG, mutant strain in mineral medium with 2 mM protocatechuate and 1 mM glucose; WLP, wild-type strain in LB broth with 2.0 mM protocatechuate; MLP, mutant strain in LB broth with 2.0 mM protocatechuate; WL, wild-type strain in LB broth; ML, mutant strain in LB broth.
Determination of the PcaO binding region and sequence.
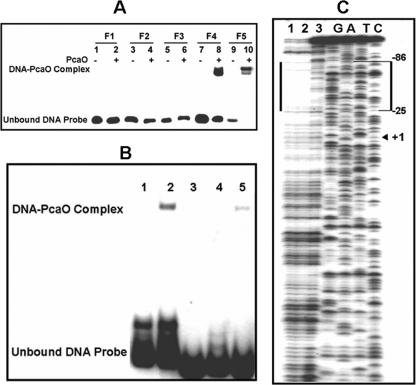
Analysis of the intergenic region (ITS) between pcaHG and ncgl2316 (bases −212 to 72, a total of 284 bases) revealed 4 palindromic sequences in the region between bases −95 and 59 (154 bases) (Fig. 3B). The importance of palindromic sequences for regulator binding was established previously for various regulators (36, 44, 46). Thus, we deduced that the 4 palindromic sequences might be involved in PcaO binding to a DNA fragment. Four DNA fragments (fragments F1 to F4) that carried one or more of these palindromes were tested to determine their binding to PcaO. The results indicated that only fragment F4 (bases −95 to 59, including all four palindromes) bound to PcaO, and binding between PcaO and fragments F1, F2, and F3, which carried the Rep1, Rep1 and Rep2, and Rep1, Rep2, and Rep3 palindromes, respectively, was not observed (Fig. 5A). This indicated that the Rep1, Rep2, and Rep3 palindromes were not involved in binding to PcaO. Further, DNA fragment F5 (bases −95 to −49), which contained only the fourth palindromic sequence (Rep4), bound to PcaO (Fig. 5A). DNase I footprinting suggested that the region from base −86 to base −25 with respect to the transcription start site of pcaH was protected (Fig. 5B). By combining the EMSA and DNase I footprinting results, the PcaO binding area was identified as the region from base −86 to base −49 that carried the fourth palindromic sequence (Rep4).
FIG. 5.
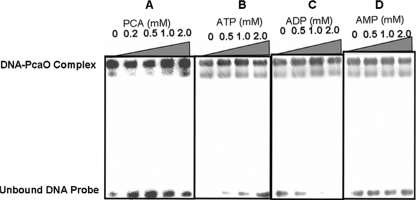
Determination of PcaO DNA-binding region and sequences by gel retardation (A), DNase I footprinting (C), and site-directed mutation of the palindromic sequence (B). (A) 32P-end-labeled promoter fragments (approximately 0.02 pM) were incubated with PcaO (5 pM) at 25°C for 20 min. The positions of fragments F1, F2, F3, F4, and F5 are bases −7 to 59, −34 to 59, −59 to 59, −95 to 59, and −49 to −93, respectively (Fig. 3B). (B) Lane 1, 32P-labeled pcaHG promoter sequence (bases −96 to −33); lane 2, same as lane 1 but with 5 pM PcaO added; lanes 3 to 5, 32P-labeled promoter sequences with different mutations in the palindromic sequence with 5 pM PcaO added. For lane 3, AACCCC(N)8GGGGTT was replaced by CCAAAA(N)8GGGGTT; for lane 4, AACCCC(N)8GGGGTT was replaced by AACCCC(N)8TTTTGG; and for lane 5, AACCCC(N)8GGGGTT was replaced by CCAAAA(N)8TTTTGG. The concentration of the 32P-labeled DNA sequences used in the experiment was 0.02 pM. (C) The 289-bp promoter sequence of the nontemplate strand was labeled using 32P. Lanes G, A, T, C contained sequencing ladders. Lanes 1 and 2 show the DNase I protection patterns in the presence of 10 pM PcaO, and lane 3 shows the DNase I protection pattern in the absence of PcaO. The concentration of the 32P-labeled DNA sequence used in the experiment was 0.5 pM.
To further characterize the Rep4 palindromic sequence and to determine if it is essential for PcaO binding, site mutations in this sequence were made. When AACCCC was replaced by CCAAAA or when GGGGTT was replaced by TTTTGG, there was no binding to PcaO. We also observed that when AACCCC and GGGGTT were simultaneously replaced by CCAAAA and TTTTGG (thus creating a new palindromic sequence), the binding affinity was partially restored (Fig. 5C).
PCA and ATP/ADP concentrations affect PcaO binding.
Since PCA induced PcaHG activity (34) and PcaO regulated the key step in aromatic cleavage during PCA degradation at the pcaHG transcriptional level (this study), PCA was tested as a potential effector to determine its influence on the binding of PcaO to its target sequence (−46AACCCCTGACCTTCGGGGTT −59). As shown in Fig. 6A, the binding affinity was different at different PCA concentrations. As a positive regulator, PcaO bound to its target sequence in the absence of PCA. PCA enhanced PcaO binding to its target sequence at high PCA concentrations (1.0 and 2.0 mM) but reduced the binding at lower concentrations (0.2 and 0.5 mM).
FIG. 6.
Effects of PCA (A), ATP (B), ADP (C), and AMP (D) on the binding affinity of PcaO for its target sequence. PCA was not included in the experiments whose results are shown in panels B to D. In all experiments, the PcaO concentration was 0.5 μM and the concentration of the 32P-labeled pcaHG promoter sequence was 1 nM.
Due to identification of a putative ATP-binding domain (Walker B) (45) in the PcaO molecule, we tested ATP and its derivatives (ADP and AMP) as potential effectors. The results showed that the binding affinity of PcaO for its target sequence decreased as the ATP concentration increased from 0.5 to 2.0 mM (Fig. 6B). In contrast, ADP (0.5 to 2 mM) enhanced the binding affinity of PcaO for its target sequence (Fig. 6C). AMP had no observable effect on the binding affinity of PcaO for its target sequence (Fig. 6D).
DISCUSSION
In this study, we identified a novel transcriptional regulator, PcaO, which is involved in the regulation of the PCA branch of the β-ketoadipate pathway in C. glutamicum. According to amino acid sequence analysis, PcaO is phylogenetically related to members of the previously proposed LAL subfamily of bacterial regulators (7). PcaO has unique features. It is a positive regulatory protein that responds to PCA, ATP, and ADP, and it is an atypical LAL subfamily member that does not have the Walker A domain. To our knowledge, this is the first study that identified such a regulator involved in the regulation of the PCA branch of the β-ketoadipate pathway. PcaO also is the first LAL-type positive-regulation protein identified in C. glutamicum.
PcaO is the first LAL-type regulator involved in catabolism of aromatic compounds, although the involvement of LAL-type regulators in regulation of different metabolic activities has been intensively characterized. The MalT protein in E. coli regulates catabolism of maltose (24, 29), the AlkS protein in P. oleovorans regulates degradation of medium-chain-length alkanes (43, 48), the LipR proteins in S. exfoliatus and S. coelicolor regulate transcription of lipase (9, 42), the AcoK protein in K. pneumoniae regulates acetoin degradation (27), and the PikD protein in S. venezuelae regulates pikromycin synthesis (47). It has been proposed that LAL-type regulators are involved in regulation of diverse types of metabolism in both Gram-negative and Gram-positive bacteria, but their regulatory mechanisms might be different. In C. glutamicum, ATP reduced the ability of PcaO to bind to its target sequence, but ATP activated mal genes in E. coli (24, 29).
The β-ketoadipate pathway occurs in a wide range of organisms, and the pca genes involved in this pathway are tightly regulated in many bacteria (4, 10, 11, 20, 21, 25, 26, 28, 30, 40, 41). This study clearly demonstrated that in C. glutamicum PcaO positively regulated the PCA branch of the β-ketoadipate pathway and that PcaO responded to the effectors of PCA, ATP, and ADP. Interestingly, PCA reduced the binding between PcaO and its target sequence at low concentrations (0.2 and 0.5 mM), while it increased the binding at higher concentrations (1.0 and 2.0 mM). A tentative hypothesis to explain this observation is that in C. glutamicum cells the transcription of genes involved in PCA assimilation gradually slows down as the PCA concentration in the environment decreases. Another new observation is that ADP increased, but ATP reduced, the binding of PcaO to its targeted DNA sequence. Under physiological conditions and in the presence of PCA, it is probable that the ratio of ATP to ADP in cells controls the transcription of pcaHG in C. glutamicum. Previous work demonstrated that in C. glutamicum PcaR repressed all of the pca genes except pcaHGBC (2). We demonstrated that PcaO regulates pcaHG. Because some of the pca genes, such as pcaIJ and pcaFD, are also involved in the resorcinol and catechol degradation pathway (15, 35), the PcaR-PcaO regulatory system in C. glutamicum allows precise control of degradation of aromatic compounds.
Acknowledgments
This work was supported by grants 20877095, 30730002, and 30725001 from the National Natural Science Foundation of China.
Footnotes
Published ahead of print on 14 January 2010.
REFERENCES
- 1.Baumbach, J. 2007. CoryneRegNet 4.0—a reference database for corynebacterial gene regulatory networks. BMC Bioinformatics 8:429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brinkrolf, K., I. Brune, and A. Tauch. 2006. Transcriptional regulation of catabolic pathways for aromatic compounds in Corynebacterium glutamicum. Genet. Mol. Res. 5:773-789. [PubMed] [Google Scholar]
- 3.Brown, C. K., M. W. Vetting, C. A. Earhart, and D. H. Ohlendorf. 2004. Biophysical analyses of designed and selected mutants of PCA 3,4-dioxygenase. Annu. Rev. Microbiol. 58:555-585. [DOI] [PubMed] [Google Scholar]
- 4.Brzostowicz, P. C., A. B. Reams, T. J. Clark, and E. L. Neidle. 2003. Transcriptional cross-regulation of the catechol and PCA branches of the β-ketoadipate pathway contributes to carbon source-dependent expression of the Acinetobacter sp. strain ADP1 pobA gene. J. Bacteriol. 69:1598-1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buchan, A., E. L. Neidle, and M. A. Moran. 2004. Diverse organization of genes of the β-ketoadipate pathway in members of the marine Roseobacter lineage. Appl. Environ. Microbiol. 70:1658-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chaudhry, M. T., Y. Huang, X.-H. Shen, A. Poetsch, C.-Y. Jiang, and S.-J. Liu. 2007. Genome-wide investigation of aromatic acid transporters in Corynebacterium glutamicum. Microbiology 153:857-865. [DOI] [PubMed] [Google Scholar]
- 7.De Schrijver, A., and R. De Mot. 1999. A subfamily of MalT-related ATP-dependent regulators in the LuxR family. Microbiology 145:1287-1288. [DOI] [PubMed] [Google Scholar]
- 8.Eulberg, D., S. Lakner, L. A. Golovleva, and M. Schlömann. 1998. Characterization of a PCA catabolic gene cluster from Rhodococcus opacus 1CP: evidence for a merged enzyme with 4-carboxymuconolactone decarboxylating and 3-oxoadipate enol-lactone-hydrolyzing activity. J. Bacteriol. 180:1072-1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Evangelista-Martínez, Z., G. González-Cerón, and L. Servín-González. 2006. A conserved inverted repeat, the LipR box, mediates transcriptional activation of the Streptomyces exfoliatus lipase gene by LipR, a member of the STAND class of P-loop nucleoside triphosphatases. J. Bacteriol. 188:7082-7089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gerischer, U., A. Segura, and L. N. Ornston. 1998. PcaU, a transcriptional activator of genes for PCA utilization in Acinetobacter. J. Bacteriol. 180:1512-1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo, Z., and J. E. Houghton. 1999. PcaR-mediated activation and repression of pca genes from Pseudomonas putida are propagated by its binding to both the −35 and the −10 promoter elements. Mol. Microbiol. 32:253-263. [DOI] [PubMed] [Google Scholar]
- 12.Harwood, C. S., N. N. Nichols, M. K. Kim, J. L. Ditty, and R. E. Parales. 1994. Identification of the pcaRKF gene cluster from Pseudomonas putida: involvement in chemotaxis, biodegradation, and transport of 4-hydroxybenzoate. J. Bacteriol. 176:6479-6488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harwood, C. S., and R. E. Parales. 1996. The β-ketoadipate pathway and the biology of self-identity. Annu. Rev. Microbiol. 50:553-590. [DOI] [PubMed] [Google Scholar]
- 14.Haussmann, U., S. W. Qi, D. Wolters, M. Rögner, S. J. Liu, and A. Poetsch. 2009. Physiological adaptation of Corynebacterium glutamicum to benzoate as alternative carbon source—a membrane proteome-centric view. Proteomics 9:3635-3651. [DOI] [PubMed] [Google Scholar]
- 15.Huang, Y., K.-X. Zhao, X.-H. Shen, M. T. Chaudhry, C.-Y. Jiang, and S.-J. Liu. 2006. Genetic characterization of resorcinol catabolic pathway in Corynebacterium glutamicum. Appl. Environ. Microbiol. 72:7238-7245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iwagami, S. G., K. Yang, and J. Davies. 2000. Characterization of the protocatechuic acid catabolic gene cluster from Streptomyces sp. strain 2065. Appl. Environ. Microbiol. 66:1499-1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jakoby, M., C. E. Ngouoto-Nkili, and A. Burkovski. 1999. Construction and application of new Corynebacterium glutamicum vectors. Biotechnol. Tech. 13:437-441. [Google Scholar]
- 18.Konopka, A. 1993. Isolation and characterization of a subsurface bacterium that degrades anilines and methylalanines. FEMS. Microbiol. Lett. 111:93-99. [Google Scholar]
- 19.Leipe, D. D., E. V. Koonin, and L. Aravind. 2004. STAND, a class of P-loop NTPases including animal and plant regulators of programmed cell death: multiple, complex domain architectures, unusual phyletic patterns, and evolution by horizontal gene transfer. J. Mol. Biol. 343:1-28. [DOI] [PubMed] [Google Scholar]
- 20.MacLean, A. M., G. MacPherson, P. Aneja, and T. M. Finan. 2006. Characterization of the β-ketoadipate pathway in Sinorhizobium meliloti. Appl. Environ. Microbiol. 72:5403-5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.MacLean, A. M., M. I. Anstey, and T. M. Finan. 2008. Binding site determinants for the LysR-type transcriptional regulator PcaQ in the legume endosymbiont Sinorhizobium meliloti. J. Bacteriol. 190:1237-1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller, L. H. 1972. Experiments in molecular genetics, p. 352-355. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 23.Ornston, L. N., and R. Y. Stanier. 1966. The conversion of catechol and PCA to β-ketoadipate by Pseudomonas putida. J. Biol. Chem. 241:3776-3786. [PubMed] [Google Scholar]
- 24.Panagiotidis, C. H., W. Boos, and H. A. Shuman. 1998. The ATP-binding cassette subunit of the maltose transporter MalK antagonizes MalT, the activator of the Escherichia coli mal regulon. Mol. Microbiol. 30:535-546. [DOI] [PubMed] [Google Scholar]
- 25.Parke, D. 1993. Positive regulation of phenolic catabolism in Agrobacterium tumefaciens by the pcaQ gene in response to beta-carboxy-cis,cis-muconate. J. Bacteriol. 175:3529-3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parke, D. 1995. Supraoperonic clustering of pca genes for catabolism of the phenolic compound PCA in Agrobacterium tumefaciens. J. Bacteriol. 177:3808-3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peng, H. L., Y.-H. Yang, W.-L Deng, and H.-Y Chang. 1997. Identification and characterization of acoK, a regulatory gene of the Klebsiella pneumoniae acoABCD operon. J. Bacteriol. 179:1497-1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Popp, R., T. Kohl, P. Patz, G. Trautwein, and U. Gerischer. 2002. Differential DNA binding of transcriptional regulator PcaU from Acinetobacter sp. strain ADP1. J. Bacteriol. 184:1988-1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Richet, E., and O. Raibaud. 1989. MalT, the regulatory protein of the Escherichia coli maltose system, is an ATP-dependent transcriptional activator. EMBO J. 8:981-987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Romero-Steiner, S., R. E. Parales, C. S. Harwood, and J. E. Houghton. 1994. Characterization of the pcaR regulatory gene from Pseudomonas putida, which is required for the complete degradation of p-hydroxybenzoate. J. Bacteriol. 176:5771-5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 32.Schäfer, A., A. Tauch, W. Jager, J. Kalinowski, G. Thierbatch, and A. Pühler. 1994. Small mobilizable multi-purpose cloning vector derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69-73. [DOI] [PubMed] [Google Scholar]
- 33.Shen, X.-H., C.-Y. Jiang, Y. Huang, Z.-P. Liu, and S.-J. Liu. 2005. Functional identification of novel genes involved in the glutathione-independent gentisate pathway in Corynebacterium glutamicum. Appl. Environ. Microbiol. 71:3442-3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shen, X.-H., and S.-J. Liu. 2005. Key enzymes of the PCA branch of the β-ketoadipate pathway for aromatic degradation in Corynebacterium glutamicum. Sci. China 48:241-249. [DOI] [PubMed] [Google Scholar]
- 35.Shen, X.-H., Y. Huang, and S.-J. Liu. 2005. Genomic analysis and identification of catabolic pathways for aromatic compounds in Corynebacterium glutamicum. Microbes Environ. 20:160-167. [Google Scholar]
- 36.Siehler, S. Y., S. Dal, R. Fischer, P. Patz, and U. Gerischer. 2007. Multiple-level regulation of genes for PCA degradation in Acinetobacter baylyi includes cross-regulation. Appl. Environ. Microbiol. 73:232-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tauch, A., F. Kassing, J. Kalinowski, and A. Pühler. 1995. The Corynebacterium xerosis composite transposon Tn5432 consists of two identical insertion sequences, designated IS1249, flanking the erythromycin resistance gene emrCX. Plasmid 34:119-131. [DOI] [PubMed] [Google Scholar]
- 38.Tauch, A., O. Kirchner, B. Loffler, S. Gotker, A. Pühler, and J. Kalinowski. 2002. Efficient transformation of Corynebacterium glutamicum with a mini-replicon derived from the Corynebacterium glutamicum plasmid pGA1. Curr. Microbiol. 45:362-367. [DOI] [PubMed] [Google Scholar]
- 39.Toyoda, K., H. Teramoto, M. Inui, and H. Yukawa. 2009. Involvement of the LuxR-type transcriptional regulator RamA in regulation of expression of the gapA gene, encoding glyceraldehyde-3-phosphate dehydrogenase of Corynebacterium glutamicum. J. Bacteriol. 191:968-977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trautwein, G., and U. Gerischer. 2001. Effects exerted by transcriptional regulator PcaU from Acinetobacter sp. strain ADP1. J. Bacteriol. 183:873-881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tropel, D., and J. R. van der Meer. 2004. Bacterial transcriptional regulators for degradation pathways of aromatic compounds. Microbiol. Mol. Biol. Rev. 68:474-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Valdez, F., G. González-Cerón, H. M. Kieser, and L. Servín-González. 1999. The Streptomyces coelicolor A3(2) lipAR operon encodes an extracellular lipase and a new type of transcriptional regulator. Microbiology 145:2365-2374. [DOI] [PubMed] [Google Scholar]
- 43.Van Beilen, J. B., S. Panke, S. Lucchini, A. G. Franchini, M. Röthlisberger, and B. Witholt. 2001. Analysis of Pseudomonas putida alkane-degradation gene clusters and flanking insertion sequences: evolution and regulation of the alk genes. Microbiology 147:1621-1630. [DOI] [PubMed] [Google Scholar]
- 44.Vannini, A., C. Volpari, C. Gargioli, E. Muraglia, R. Cortese, R. De Francesco, P. Neddermann, and S. D. Marco. 2002. The crystal structure of the quorum sensing protein TraR bound to its autoinducer and target DNA. EMBO J. 21:4393-4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Walker, J. E., M. Saraste, M. J. Runswick, and N. J. Gay. 1982. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1:945-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weingart, C. L., C. E. White, S. Liu, Y. Chai, H. Cho, C. S. Tsai, Y. Wei, N. R. Delay, M. R. Gronquist, A. Eberhard, and S. C. Winans. 2005. Direct binding of the quorum sensing regulator CepR of Burkholderia cenocepacia to two target promoters in vitro. Mol. Microbiol. 57:452-467. [DOI] [PubMed] [Google Scholar]
- 47.Wilson, D. J., Y. Xue, K. A. Reynolds, and D. H. Sherman. 2001. Characterization and analysis of the PikD regulatory factor in the pikromycin biosynthetic pathway of Streptomyces venezuelae. J. Bacteriol. 183:3468-3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yuste, L., I. Canosa, and F. Rojo. 1998. Carbon-source-dependent expression of the PalkB promoter from the Pseudomonas oleovorans alkane degradation pathway. J. Bacteriol. 180:5218-5226. [DOI] [PMC free article] [PubMed] [Google Scholar]