Abstract
Currently, one-third of the world's population is believed to be latently infected with Mycobacterium tuberculosis. The mechanisms by which M. tuberculosis establishes latent infection remain largely undefined. mprAB encodes a two-component signal transduction system required by M. tuberculosis for aspects of persistent infection. MprAB regulates a large and diverse group of genetic determinants in response to membrane stress, including the extracytoplasmic function (ECF) sigma factor sigE and the HtrA-like serine protease pepD. Recent studies have demonstrated that PepD functions as both a protease and chaperone in vitro. In addition, inactivation of pepD alters the virulence of M. tuberculosis in a mouse model system of infection. Here, we demonstrate that PepD plays an important role in the stress response network of Mycobacterium mediated through MprAB and SigE. In particular, we demonstrate that the protease activity of PepD requires the PDZ domain, in addition to the catalytic serine at position 317. pepD expression initiates from at least three promoters in M. tuberculosis, including one that is regulated by SigE and is located upstream of the mprA coding sequence. Deletion of pepD or mprAB in Mycobacterium smegmatis and M. tuberculosis alters the stress response phenotypes of these strains, including increasing sensitivity to SDS and cell wall antibiotics and upregulating the expression of stress-responsive determinants, including sigE. Taking these data together, we hypothesize that PepD utilizes its PDZ domain to recognize and process misfolded proteins at the cell membrane, leading to activation of the MprAB and SigE signaling pathways and subsequent establishment of a positive feedback loop that facilitates bacterial adaptation.
Currently, one-third of the world's population is believed to be latently infected with Mycobacterium tuberculosis, the causative agent of tuberculosis. There are an estimated 9 million new cases of infection caused by this organism each year, accounting for more than 2 million deaths annually (3). This makes M. tuberculosis one of the leading causes of death due to a single infectious agent. Humans are the only known reservoir of M. tuberculosis, and as such, part of its success as a pathogen lies in its ability to maintain a significant persistent reservoir within the population. Latently infected individuals are asymptomatic and are harder to treat, thereby complicating measures to control the spread of disease (27). In addition, the emergence of multi- and extensively drug-resistant strains has further compromised established strategies for treatment. A better understanding of the mechanisms by which M. tuberculosis establishes persistent infection is urgently needed so that better therapeutics for the latently infected population can be developed.
M. tuberculosis is a facultative intracellular pathogen that primarily infects alveolar macrophages. The organism survives within these cells by blocking key steps in the phagosome maturation process (38). Infection by M. tuberculosis triggers a cell-mediated immune response, resulting in the formation of caseous granulomas at sites of infection, an environment in which the bacillus is thought to persist for extended periods in a metabolically altered physiological state (46). It is likely that M. tuberculosis possesses determinants that mediate resistance to stressful conditions within these in vivo environments that likely include acidic pH, low oxygen, poor nutrient availability, reactive oxygen and nitrogen intermediates, and other host-generated products. Transcriptional regulators, such as two-component signal transduction systems and sigma factors, play key roles in coordinating the complex array of gene expression changes required for these adaptive responses in bacteria.
M. tuberculosis encodes 11 complete two-component systems (8). The MprAB two-component system has been shown to be important for the establishment and maintenance of persistence by M. tuberculosis in a murine model of infection (59). mprAB is located within an in vivo-expressed genomic island (52), is induced following infection of M. tuberculosis in the lungs of chronically infected mice (53), and is induced in M. tuberculosis during growth within an artificial granuloma model system (25). mprAB is also induced in vitro following exposure of M. tuberculosis to membrane-damaging stress (20, 30, 36) and nutrient starvation or depletion (6, 17). More recently, MprAB has been implicated in the stringent response of M. tuberculosis mediated through RelA (50). Microarray and quantitative reverse transcription (qRT)-PCR analyses have delineated a large MprAB regulon (19, 35, 36), including two stress-responsive sigma factors, sigB and sigE; the heat shock protein encoded by acr2; and an HtrA-like serine protease encoded by pepD, all of which are directly regulated by MprAB (19, 35, 36). Collectively, these observations suggest that MprAB functions in stress response networks necessary for M. tuberculosis adaptation and pathogenesis.
An emerging class of proteins important in stress response networks is the HtrA family of serine proteases. These proteases are evolutionarily conserved among many different species spanning both prokaryotes and eukaryotes. HtrA-like proteases contain two conserved domains: a serine protease domain and one or two C-terminal PDZ domains. These proteins often possess the activities of chaperones and/or proteases (26). In bacteria, HtrA-like family members have been shown to participate in stress response networks, including those regulated through sigma factors and two-component systems (1, 7). A subset of HtrA family members is also required for aspects of pathogenesis in both Gram-positive and Gram-negative organisms (14, 16, 45, 49, 57). M. tuberculosis possesses three HtrA-like proteases: Rv1223 (degP or htrA1), Rv0983 (pepD or htrA2), and Rv0215 (pepA or htrA3) (8, 33), all of which share regions of sequence homology with one another. Initial studies of PepD and PepA in M. tuberculosis indicate that these proteins are secreted and are present within the culture filtrate (47). More recently, PepD has been shown to possess both protease and chaperone activities (33); however, the biological activities of DegP and PepA remain unclear. ΔpepD::Hygr mutants of M. tuberculosis Erdman are altered for virulence in mice. In particular, ΔpepD::Hygr mutant-infected animals exhibit increased time to death, and infection by ΔpepD::Hygr mutants induces less tissue pathology than in animals infected with wild-type or complemented mutant derivatives (33). Taken together, these results suggest that PepD is required for aspects of M. tuberculosis adaptation within the host. Here, we build upon these initial observations of PepD and provide new evidence supporting a role for PepD in the stress response network in M. tuberculosis mediated through the MprAB and SigE signaling networks.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
All strains, plasmids, and mycobacteriophages used in the study are described in Table 1. Escherichia coli Top 10 (Invitrogen, Carlsbad, CA) or DH5α was used for all cloning procedures. E. coli HB101 was used as a transduction host for generating phasmids. E. coli BL21(DE3)/pLysS (Novagen, La Jolla, CA) was used to express and purify recombinant proteins. All E. coli strains were grown with aeration at 37°C in Luria-Bertani (LB) broth or on LB agar. When required, the medium was supplemented with 25 μg/ml chloramphenicol and/or 50 μg/ml kanamycin sulfate. Isopropyl-β-d-thiogalactopyranoside (IPTG) (Invitrogen, Carlsbad, CA) was added to E. coli BL21(DE3)/pLysS cultures at a concentration of 0.1 mM to induce protein overexpression. The Mycobacterium strains used in this study are derivatives of M. tuberculosis H37Rv (ATCC 27294) or Mycobacterium smegmatis mc2155 (ATCC 700084). The mycobacteria were grown with aeration at 37°C in Middlebrook 7H9 broth (Difco, Franklin Lakes, NJ) or on Middlebrook 7H10 agar medium (Difco, Franklin Lakes, NJ) supplemented with 0.5% glycerol, 10% albumin-dextrose-catalase (ADC) or oleic acid-albumin-dextrose-catalase (OADC) supplement, and 0.05% Tween 80. For protein expression studies, M. tuberculosis was grown in glycerol alanine salts (GAS) medium (51). Generation of mycobacteriophages and their transduction into M. tuberculosis H37Rv proceeded as described previously (5). Electrocompetent cells of M. tuberculosis or M. smegmatis were prepared as described previously (23). When required, the mycobacterium medium was supplemented with 25 μg/ml kanamycin sulfate, 50 μg/ml hygromycin B, and/or 50 μg/ml cycloheximide. Kanamycin sulfate and cycloheximide were purchased from Fisher (Waltham, MA). Hygromycin B was purchased from AG Scientific (San Diego, CA).
TABLE 1.
Strains, plasmids, and bacteriophages used in this study
| Strain, plasmid, or bacteriophage | Genotype or description | Application | Reference or source |
|---|---|---|---|
| Strains | |||
| M. tuberculosis H37Rv | Laboratory strain | ATCC 27294 | |
| M. smegmatis mc2155 | Laboratory strain | ATCC 700084 | |
| E. coli DH5α | [λ− φ80dlacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17(rK− mK−) supE44 thi-1 gyrA relA1] | Cloning | Laboratory collection |
| E. coli TOP10 | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(ara leu)7697 galU galK rpsL (StrR) endA1 nupG | Cloning | Invitrogen |
| E. coli HB101 | supE44 hsdS20(rB− mB−) recA13 ara-14 proA2 lacY1 galK2 rpsL20 xyl-5 mtl-1 leuB6 thi-1 | E. coli transduction host | Laboratory collection |
| E. coli BL21(DE3)/pLysS | F−ompT hsdSB(rB− mB−) gal dcm (DE3)/pLysE (Camr) | Protein overexpression | Novagen |
| Plasmids | |||
| pCR2.1-TOPO | 3.9-kb plasmid for cloning PCR products; Ampr Kanr | PCR cloning vector | Invitrogen |
| pET-24b | 5.3-kb plasmid allowing C-terminal protein fusion to His6; Kanr | Protein overexpression vector | Novagen |
| pMV306 | 3.9-kb plasmid for single-copy integration into mycobacterial chromosome; Kanr | Complementation vector | Laboratory collection |
| pTZ557 | pYUB854 containing pepD upstream and downstream regions; Hygr | pepD deletion | This study |
| pTZ612 | pYUB854 containing sigE upstream and downstream regions; Hygr | sigE deletion | This study |
| pTZ758 | pET-24b containing pepDΔTM coding sequence; Kanr | Overexpression | This study |
| pTZ775 | pET-24b containing pepDΔTMΔPDZ coding sequence; Kanr | Overexpression | This study |
| pTZ792 | pMV306 containing pepD and moaB2 coding sequences; Kanr | Complementation | This study |
| pTZ797 | pET-24b containing pepDΔTMS317A coding sequence; Kanr | Overexpression | This study |
| pTZ844 | pYUB854 containing mprA upstream and mprB downstream regions; Hygr | mprAB deletion | This study |
| pTZ898 | pYUB854 containing sacB coding sequence; Hygr | Mycobacterium gene deletion vector | This study |
| pTZ903 | pMV306 containing pepDS317A and moaB2 coding sequences; Kanr | Complementation | This study |
| pTZ947 | pTZ898 containing mprAB upstream and downstream regions; Hygr | mprAB deletion | This study |
| pTZ949 | pTZ898 containing pepD upstream and downstream regions; Hygr | pepD deletion | This study |
| pTZ1065 | pMV306 containing sigE coding sequence; Kanr | Complementation | This study |
| pYUB854 | 3.9-kb cosmid for generating gene deletions in Mycobacterium. Carries a Hygr cassette flanked by multiple cloning sites and a λpac site; Hygr | Mycobacterium gene deletion vector | 5 |
| pYUB870 | 8.9-kb plasmid for unmarking mutations in Mycobacterium. Carries the tnpR and sacB cassettes; Kanr | Mycobacterium gene deletion unmarking vector | 5 |
| Mycobacteriophages | |||
| phAE87 | Derivative of conditionally replicating mycobacteriophage PH101(Ts) | 5 | |
| phTZ557 | phAE87 derivative containing pTZ557 | pepD deletion | This study |
| phTZ612 | phAE87 derivative containing pTZ612 | sigE deletion | This study |
| phTZ844 | phAE87 derivative containing pTZ844 | mprAB deletion | This study |
Expression and purification of rPepD and rPepD mutants.
All primers used are described in Table S1 in the supplemental material. pET-24b (Novagen, La Jolla, CA) was used to overexpress recombinant forms of PepD (rPepD) containing C-terminal His6 tags. All pepD sequences were amplified from M. tuberculosis H37Rv genomic DNA by PCR using Pfu polymerase (Invitrogen, Carlsbad, CA), cloned into pCR2.1-TOPO (Invitrogen, Carlsbad, CA), subcloned into the NdeI and XhoI sites of pET-24b, and sequenced to confirm their identities and the absence of mutations. PepDΔTMS317A carries a point mutation in the predicted catalytic serine of PepD and was generated using the QuikChange XL site-directed mutagenesis kit (Stratagene, La Jolla, CA). E. coli BL21(DE3)/pLysS strains containing overexpression constructs were grown overnight on selective LB agar medium, suspended in LB broth containing kanamycin and chloramphenicol, and grown to mid-exponential phase. Protein overexpression was induced by the addition of 0.1 mM IPTG for 3 h at 30°C. The induced cells were suspended in lysis buffer (20 mM Tris [pH 7.9], 500 mM NaCl, 5 mM imidazol, and 6 μg of DNase/ml), passed 3 times through a French press, and centrifuged at 25,000 × g for 30 min. Cellular supernatants were first passed over a nickel-nitrilotriacetic acid-agarose column (Qiagen, Valencia, CA), and then the collected fractions were pooled and placed on a Sephacryl 200 HR (Sigma, St. Louis, MO) size exclusion column with running buffer (10 mM Tris [pH 7.6] and 20 mM NaCl). Peak fractions were determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and subjected to anion-exchange chromatography (DEAE-Sephacel). Bound proteins were eluted with a gradient of 20 mM to 1 M NaCl. The purified protein fractions were pooled and stored in 2.5 M glycerol at −80°C.
Protease activity assays.
The proteolytic activities of rPepD derivatives were determined using the nonspecific protein substrate β-casein or fluorescein isothiocyanate (FITC)-labeled casein. Briefly, increasing amounts of purified rPepD-His were incubated with 1 μg of β-casein in 50 mM potassium phosphate buffer (pH 7.5) at 37°C for 16 h. The reaction mixtures were then separated on 12% SDS-PAGE gels and stained using Coomassie blue. To quantify the activity of rPepD, purified protein was incubated with 0.5% FITC-casein under similar conditions (55). The reaction mixtures were passed through a Microcon YM-100 centrifugal filter (Millipore, Billerica, MA) to remove uncleaved and large cleavage products, and the filtrate was pH buffered by the addition of 0.5 M Tris (pH 7.9). Fluorescence was measured using standard 485-nm excitation/530-nm emission filters on a Spectramax M5 plate reader (Molecular Devices, Sunnyvale, CA). For some experiments, protease inhibitors (Sigma, St. Louis, MO) were added to the reaction mixtures, including N-tosyl-l-lysine chloromethyl ketone (TLCK) (15 mg/ml in distilled H2O [dH2O]), N-tosyl-l-phenylalanine chloromethyl ketone (TPCK) (10 mg/ml in dimethyl sulfoxide [DMSO]), leupeptin (10 mg/ml in dH2O), di-isopropyl fluorophosphate (DFP) (5 mM in isopropanol), aprotinin (10 mg/ml in dH2O), EDTA (50 mM in dH2O), E64 (1 mM in dH2O), and pepstatin A (1 mM in DMSO).
Generation of unmarked M. tuberculosis and M. smegmatis deletion mutants and complemented strains.
mprAB, pepD, and sigE were deleted in M. tuberculosis H37Rv using specialized transduction as previously described (5). Regions (1.0 kb in size) upstream and downstream of genes were PCR amplified from H37Rv genomic DNA. The mprAB deletion extended from −14 bp upstream of the originally annotated mprA translational start site at +1 to +1302 downstream of the mprB translational start site, leaving intact the C-terminal 214 bp of mprB and the mprB-pepD intergenic region that included the three MprA recognition motifs (20). The pepD and sigE deletions removed the entire coding sequence for each gene. Following packaging and transduction into M. tuberculosis, hygromycin-resistant clones were screened for loss of mprAB, pepD, and sigE by PCR. The resulting ΔmprAB::Hygr, ΔpepD::Hygr, and ΔsigE::Hygr strains were electroporated with pYUB870 to unmark the mutation. Following counterselection on medium containing 10% sucrose, the resulting sucrose-resistant, kanamycin-sensitive, and hygromycin-sensitive colonies were screened for unmarking of the mprAB, pepD, and sigE mutations by PCR. Finally, to complement the pepD mutant, pepD and moaB2, along with ∼300 bp of DNA upstream of pepD, were amplified from M. tuberculosis genomic DNA. The resulting product was cloned into pCR2.1-TOPO and subcloned into pMV306, resulting in pTZ792. This plasmid was then electroporated into M. tuberculosis ΔpepD, and recombinants were selected on medium containing kanamycin. Complementation was verified by PCR. The sigE mutation was complemented by amplifying sigE, along with ∼150 bp of upstream DNA from M. tuberculosis genomic DNA. The resulting product was cloned into pCR2.1-TOPO and subcloned into pMV306, resulting in pTZ1065. This plasmid was then electroporated into M. tuberculosis ΔsigE, and recombinants were selected on medium containing kanamycin. The mprAB mutation was complemented by the introduction of pTZ215 (59), a derivative of pMV306 carrying the mprAB coding sequence and the upstream region from M. tuberculosis.
Unmarked deletion mutations of the mprAB (MSMEG5488 and MSMEG5487) and pepD (MSMEG5486) coding regions in M. smegmatis were generated using a standard two-step homologous-recombination procedure. The counterselectable marker sacB was amplified from pYUB870 (5) and cloned into the Mycobacterium suicide vector pYUB854 (5) to generate pTZ898. Regions (1.0 kb in size) upstream and downstream of the mprAB and pepD coding sequences were then amplified from M. smegmatis mc2155 genomic DNA. Both the mprAB and pepD deletions removed the complete coding sequences for these genes. The resulting recombinants were screened by PCR to confirm gene loss. The generated ΔpepD or ΔmprAB mutant was complemented with pTZ792 or pTZ215, respectively.
DNA and RNA extraction.
Mycobacterial genomic DNA was prepared as described previously (23). For isolation of RNA, M. tuberculosis was grown in 7H9 broth to early log phase (optical density at 600 nm [OD600] = 0.4). The cultures were left untreated or were exposed to 0.05% SDS for 90 min to induce expression of mprAB and/or pepD (19, 20). RNA was fixed by treating the cultures with RNAlater (Ambion, Austin, TX) for 40 min at room temperature prior to processing them. Bacteria were collected by centrifugation, washed twice with 1× phosphate-buffered saline (PBS), and then mechanically disrupted in the presence of RNA-Bee (Tel-Test, Friendswood, TX) by bead beating. RNA was then extracted with chloroform, precipitated with isopropanol, and stored at −20°C until it was used. The resulting RNA was resuspended in diethylpyrocarbonate (DEPC)-treated water, treated with Turbo DNase (Ambion, Austin, TX) to remove contaminating genomic DNA, and purified using an RNeasy Miniprep column (Qiagen, Valencia, CA).
RT-PCR, 5′ RACE, and DNA microarray studies.
For RT-PCR, cDNAs from M. tuberculosis strains were prepared by incubating 500 ng total RNA with random M. tuberculosis decamers (15) in a reverse transcription reaction mixture containing SuperScript III reverse transcriptase (Invitrogen, Carlsbad, CA). To test for the presence of DNA contamination, identical reactions lacking SuperScript III were also run. All PCRs were performed on an iCycler iQ real-time PCR detection system (Bio-Rad, Hercules, CA) with iQ SYBR green Supermix and gene-specific primers. The reaction conditions consisted of 40 rounds of denaturation at 95°C for 30 s and annealing/extension at 60°C for 15 s. Single PCR products were confirmed by performing a postamplification melt curve analysis for each reaction. The transcript levels of target genes were normalized to those of the ribosomal gene rrs or sigA. For 5′ rapid amplification of cDNA ends (RACE), the FirstChoice RLM-RACE kit (Ambion, Austin, TX) was used essentially as recommended by the manufacturer. Briefly, 3 to 5 μg of total RNA was ligated to 300 ng of 5′ RACE adaptor using 5 units of T4 RNA ligase (Ambion, Austin, TX) for 1 h at 37°C. The RNA was then reverse transcribed with M. tuberculosis-specific decamers and Maloney-murine leukemia virus (M-MLV) reverse transcriptase (Ambion, Austin, TX) for 1 h at 42°C. PCR was then performed using a primer nested within the gene of interest and a primer specific to the 5′ RACE adaptor (5′ RACE outer primer). The resulting PCR products were cloned into pCR2.1-TOPO and sequenced using the M13rev primer. Ten to 20 clones were sequenced for each primer set. For DNA microarray comparisons, RNA was isolated from three independent cultures of wild-type M. tuberculosis H37Rv or the isogenic ΔpepD mutant and used to generate labeled cDNA by reverse transcription using SuperScript III, M. tuberculosis decamers, and Cy3- or Cy5-dCTP (Amersham, Piscataway, NJ). RNA was derived from cultures grown in 7H9 medium in the absence of stress. Dye flip controls were also conducted. The labeled cDNAs were purified, mixed, and hybridized to M. tuberculosis DNA microarrays obtained through the Pathogen Functional Genomic Resource Center. The Cy3 and Cy5 intensities for each spot were quantified using ScanAlyze (Stanford), the median background values were subtracted, and the values were normalized based on the total intensity of good-quality spots over background. The data were analyzed using statistical analysis of microarrays (SAM) (54) to determine significant expression changes.
Northern blot analysis.
For Northern blot analysis, the NorthernMax-Gly Kit (Ambion, Austin, TX) was used according to the manufacturer's instructions. Briefly, ∼20 μg RNA from either untreated or SDS-treated M. tuberculosis cultures was separated by electrophoresis on a 1% agarose gel, transferred to Zeta-Probe GT Genomic Tested Blotting Membrane (Bio-Rad, Hercules, CA), and hybridized under stringent conditions. A pepD-specific probe was generated by end labeling it with [γ-32P]ATP using a PCR product generated with primers pepDtrunc and pepDcomR.
Protein extraction and Western blot analysis in M. tuberculosis.
Cultures of M. tuberculosis derivatives were grown in GAS medium to stationary phase (OD600 > 1.5), collected by centrifugation, and suspended in protein lysis buffer (100 mM Tris [pH 7.6], 100 mM NaCl, 5 mM EDTA) containing protease inhibitors. The cells were mechanically disrupted by bead beating, and the supernatants were collected. The protein lysates were passed through a 0.45-μm low-protein binding syringe filter (Corning, Lowell, MA). To collect proteins from the culture filtrate, the spent broth medium was passed through a 0.45-μm low-protein binding filter (Corning, Lowell, MA) and concentrated using an Amicon Ultracell-15 (10,000 molecular weight cutoff). Protein concentrations were determined using the BCA Protein Assay (Pierce, Rockford, IL). Approximately 10 μg of protein was added to 10 μl of 2× SDS-PAGE loading buffer containing β-mercaptoethanol (β-ME), boiled for 5 min, separated on a 12% SDS-PAGE gel, and transferred onto an Immobilon-P membrane (Millipore, Billerica, MA). The membranes were blocked in TTBS (20 mM Tris-HCl [pH 7.5], 500 mM NaCl, 0.5% Tween 20) containing 5% skim milk and 1% bovine serum albumin (BSA) for at least 1 h. The membranes were probed with rabbit polyclonal anti-PepD antibody (1:10,000 or 1:50,000; Covance, Princeton, NJ) or rabbit polyclonal anti-casein (1:10,000; Immunology Consultants Laboratory, Inc., Newberg, OR) in TTBS overnight at 4°C. After being washed, the membranes were incubated with donkey anti-rabbit IgG antibody conjugated to horseradish peroxidase (Fisher, Waltham, MA) for 30 min at room temperature. The blots were visualized using SuperSignal West Pico or Femto Chemiluminescent Substrate kits (Pierce, Rockford, IL).
Isolation and infection of macrophages.
Human monocyte-derived macrophages were isolated from buffy coats obtained from the Blood Center of Southeastern Wisconsin and were maintained as described previously (37). J774A.1 macrophages (ATCC TIB-67) were maintained in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum and 2 mM l-glutamine. All cells were incubated at 37°C in humidified air containing 5% CO2. All macrophages were seeded at a density of 1 × 105 cells/well in 12-well tissue culture plates and infected with M. tuberculosis derivatives at a multiplicity of infection (MOI) of 1 bacterium per macrophage for murine macrophages or an MOI of 0.1 for human macrophages as described previously (58). For some experiments, J774A.1 macrophages were activated by the addition of 500 U/ml murine recombinant gamma interferon (rIFN-γ) (Peprotech, Rocky Hill, NJ) and 500 ng/ml purified E. coli lipopolysaccharide (LPS) (Sigma, St. Louis, MO) for 24 h prior to infection.
In vitro survival assays.
The sensitivities of Mycobacterium derivatives to stressors were measured using broth or disc diffusion assays. For broth assays, M. tuberculosis or M. smegmatis derivatives were grown to mid-log phase (OD600 = 0.4 to 0.6) and either left untreated or exposed to 200 μg/ml d-cycloserine (Sigma, St. Louis, MO) or SDS at a final concentration of 0.4% or 0.1%, respectively, for M. tuberculosis or M. smegmatis. The cultures were incubated at 37°C with shaking, and aliquots were removed at 2 h, 4 h, 8 h, and 12 h postexposure for CFU determination. The bacteria were diluted in 1× PBS for plating onto solid agar medium. For disc diffusion assays, 25 ml 7H9 bottom agar (1.2% agar) per dish was poured into petri dishes. M. smegmatis cultures were grown to mid-log phase (OD600 = 0.6), and 100-μl aliquots were added to 3.5 ml of 0.6% 7H9 top agar that had been tempered to 55°C and poured onto bottom agar for solidification. Sterile filter discs (6 mm) were then added to the medium and impregnated with either 5 μl of a 10% SDS solution or 10 μl of 50 mg/ml cefuroxime (Sigma, St. Louis, MO). The plates were incubated at 37°C, and the zones of inhibition were measured after 3 days.
Statistical analysis.
All statistical analyses were conducted using Student's t test or a one-way analysis of variance. Values were determined to be statistically significant at P < 0.05.
Microarray data accession number.
The microarray data have been deposited in NCBI's Gene Expression Omnibus (GEO) (13) and are accessible through GEO series accession number GSE17587 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE17587).
RESULTS
Purification of M. tuberculosis PepD in E. coli.
PepD from M. tuberculosis contains several conserved domains, including a hydrophobic transmembrane domain (amino acids 102 to 124), a serine protease domain (amino acids 166 to 364), and a single PDZ interaction domain (amino acids 368 to 446) predicted to be involved in protein-protein interactions. To generate recombinant derivatives of PepD for overexpression and purification in E. coli, pepD was PCR amplified from M. tuberculosis H37Rv and cloned into pET-24b, a vector allowing incorporation of a His6 epitope tag at the C terminus of PepD (see Fig. S1A in the supplemental material). Initial attempts to overexpress and purify full-length rPepD derivatives (amino acids 1 to 464) were unsuccessful (data not shown), likely due to the presence of the hydrophobic transmembrane domain at the N terminus of the protein. To circumvent this problem, rPepD variants lacking this domain were generated. PepDΔTM lacked the first 148 amino acids, including the transmembrane domain, while PepDΔTMΔPDZ lacked both the first 148 amino acids and the last 98 amino acids, including the PDZ domain. A derivative of PepDΔTM was also constructed in which the predicted catalytic serine residue, S317, was mutated to an alanine residue (PepDΔTMS317A). Removal of the transmembrane domain resulted in protein derivatives that were partially soluble and could be purified to near homogeneity by a three-step strategy, including nickel-nitrilotriacetic acid-agarose column chromatography, size exclusion chromatography, and anion-exchange chromatography (see Fig. S1B in the supplemental material; data not shown).
Protease activities of rPepD derivatives.
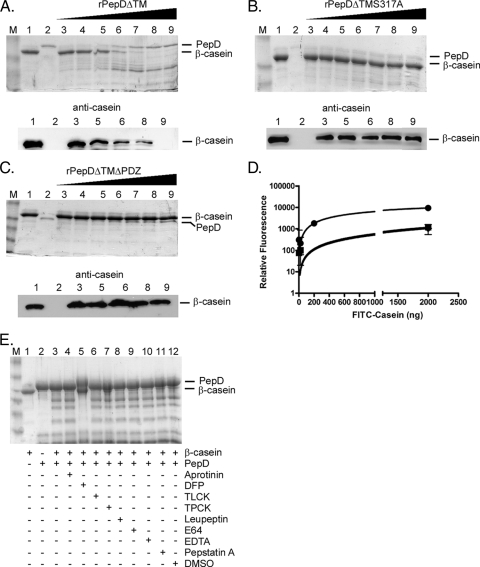
A previous study indicated that M. tuberculosis PepD possessed protease activity (33). To corroborate these data and to determine whether the PDZ domain was required, protease activity assays of rPepD derivatives were conducted using β-casein as an artificial protein substrate (33, 56). Increasing amounts of rPepD were incubated with fixed amounts of β-casein at 37°C for 16 h, and the reaction products were separated by SDS-PAGE, followed by Coomassie blue staining or by Western blotting using a polyclonal antibody generated against casein. Degradation of β-casein was observed in reactions following incubation with increasing amounts of PepDΔTM (Fig. 1A). This degradation was due specifically to the catalytic activity of PepDΔTM, as β-casein cleavage was not observed in analogous reaction mixtures containing PepDΔTMS317A, a derivative lacking the catalytic serine residue (Fig. 1B). In addition, the observed protease activity required the PDZ domain, as incubation with PepDΔTMΔPDZ failed to cleave β-casein to detectable levels (Fig. 1C). To compare the extents of cleavage mediated by rPepD variants, the protease reactions were repeated using a quantitative fluorescence-based assay with FITC-labeled casein. Although PepDΔTM cleaved FITC-casein in a concentration-dependent manner, PepDΔTMS317A and PepDΔTMΔPDZ possessed ∼10% of the activity that was observed with the parental protein (Fig. 1D). Finally, to confirm that PepD functioned as a serine protease, β-casein cleavage studies were repeated in the presence of various protease inhibitors. The serine protease inhibitor DFP inhibited the activity of PepDΔTM, as observed on Coomassie-stained gels, while other inhibitor classes failed to prevent β-casein cleavage (Fig. 1E). Further work demonstrated that PepDΔTM functions optimally as a protease at 37°C, at a pH between 7.0 and 8.5, and in a reaction buffer containing 50 mM phosphate (see Fig. S2 in the supplemental material). Thus, PepD is a serine protease that requires both its catalytic serine at position 317 and its PDZ domain for efficient proteolytic activity.
FIG. 1.
rPepDΔTM exhibits proteolytic activity against the artificial substrate β-casein. Protease assays were carried out with the artificial substrate β-casein and rPepDΔTM (A), rPepDΔTMS317A (B), or rPepDΔTMΔPDZ (C) for 16 h at 37°C. The reaction mixtures contained 1 μg β-casein alone (lanes 1), 500 ng rPepD variant alone (lanes 2), or 1 μg β-casein incubated with increasing concentrations (67 to 500 ng) of rPepD variants (lanes 3 to 9). The reaction products were resolved by SDS-PAGE and visualized by either Coomassie staining or Western blotting using a polyclonal antibody against casein. (D) The cleavage of β-casein by rPepD variants was quantified using 41.5 μg FITC-labeled casein as a substrate and measuring the relative fluorescence of reaction products (loss of FITC quenching) as a function of the rPepD concentration. The rPepDΔTMS317A and rPepDΔTMΔPDZ variants exhibited approximately 10% of the activity of rPepDΔTM. •, rPepDΔTM; ▪, rPepDΔTMS317A; ⧫, rPepDΔTMΔPDZ. The error bars indicate standard errors. (E) The specificity of rPepDΔTM-mediated β-casein cleavage was determined by the inclusion of various protease inhibitors in the reaction mixture. The reaction mixtures contained 1 μg β-casein alone (lane 1), 500 ng rPepDΔTM alone (lane 2), or 1 μg β-casein and 500 ng rPepDΔTM (lane 3) incubated with 1.5 mM aprotinin (lane 4), 5 mM DFP (lane 5), 27 mM TLCK (lane 6), 28 mM TPCK (lane 7), 23 mM leupeptin (lane 8), 1 mM E64 (lane 9), 50 mM EDTA (lane 10), 1 mM pepstatin A (lane 11), or DMSO (lane 12). Inhibition was observed in the reaction mixture containing DFP. Lanes M, molecular weight marker.
Transcriptional organization of pepD.
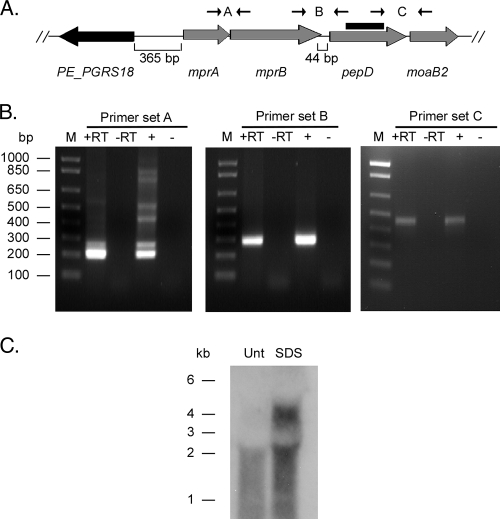
To determine the transcriptional organization of pepD, RNA was prepared from M. tuberculosis H37Rv and used in RT-PCRs with primer sets spanning the intergenic regions between the genes (Fig. 2A). PCR products of the correct size were observed with cDNA using the mprA-mprB intergenic primer set (212 bp expected), the mprB-pepD intergenic primer set (278 bp expected), and the pepD-moaB2 intergenic primer set (464 bp expected), indicating that mprA and mprB are coexpressed and that both pepD and moaB2 are coexpressed on a transcript that includes mprAB (Fig. 2B). Products were not observed in control reactions lacking either reverse transcriptase or nucleic acid template. As further confirmation, Northern blot analysis was performed using a pepD-specific probe. A single transcript of approximately 2 kb was observed with RNA isolated from M. tuberculosis cultures grown in 7H9 medium (Fig. 2C). The size of this transcript is consistent with that expected for a message carrying the open reading frames for pepD and moaB2 (1,939 bp predicted). In contrast, two transcripts (∼2 kb and ∼4 kb) were observed when RNA was extracted from M. tuberculosis cultures previously exposed to 0.05% SDS (Fig. 2C). The size of the larger transcript was consistent with that expected from the message carrying the mprA-mprB-pepD-moaB2 coding sequences (4,189 bp predicted). When taken together, these results suggest that pepD and the downstream gene moaB2 are expressed from at least two promoters, one that is located immediately upstream of pepD and one that is SDS responsive and positioned near or upstream of mprA.
FIG. 2.
Transcriptional organization of the pepD locus in M. tuberculosis. (A) Chromosomal organization of pepD and surrounding genes in M. tuberculosis H37Rv. The primers used to determine the operon structure and the probe generated for Northern blot hybridization are shown. (B) Agarose gels depicting RT-PCRs with primer sets spanning intergenic regions. Lanes: M, marker lane; +RT, RNA treated with reverse transcriptase; −RT, RNA treated without reverse transcriptase; +, genomic DNA; −, no template. (C) Northern blot of RNA samples obtained from either untreated (Unt) or SDS-treated M. tuberculosis H37Rv cultures.
Identification and characterization of mprA and pepD promoter elements.
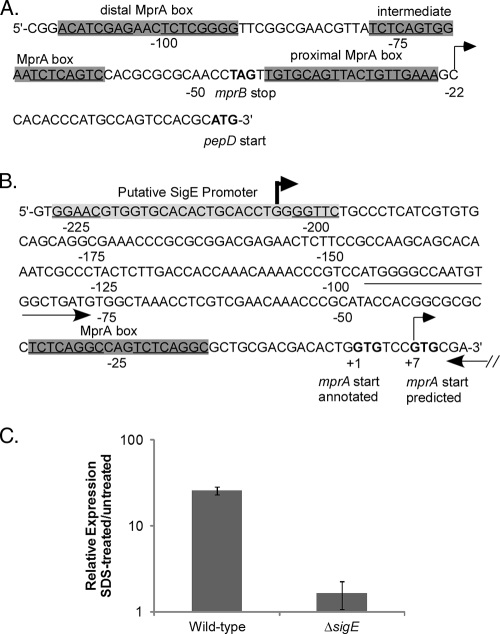
To delineate the transcriptional start site(s) for mprAB-pepD-moaB2, 5′ RACE was carried out on RNA isolated from M. tuberculosis H37Rv. Two gene-specific reverse primers separated by ∼50 bp were generated to the 5′ end of either the pepD or the mprA coding sequence and used in combination with a forward primer generated to the 5′ RNA adaptor. A predominate transcript initiating at −22 (cytosine) relative to the pepD translational start site was identified in reaction mixtures that contained either pepD-specific primer (Fig. 3A and data not shown). Transcripts beginning with this nucleotide were also identified when RNA from M. tuberculosis cultures induced in the presence of 0.05% SDS was used. A predominant transcript initiating at +7 (guanine) relative to the previously annotated translation start site for mprA was identified with each mprA-specific primer and RNA samples from untreated cultures of M. tuberculosis (Fig. 3B and data not shown). This proximal mprA transcriptional start site is consistent with that recently reported by Pang et al. (36), suggesting that transcription and translation of mprA likely initiate from the same nucleotide, a site downstream of the one originally annotated in the H37Rv genome sequence. A different predominant transcriptional start site for mprA was identified further upstream at −204 (guanine) when RNA from M. tuberculosis cultures exposed to 0.05% SDS was utilized in combination with mprA-specific primers (Fig. 3B and data not shown). These results agree with findings by Manganelli et al. (30) that describe a putative SigE binding site in the same region upstream of mprA (Fig. 3B). To verify that transcripts initiating from this distal mprA promoter element were SDS responsive and regulated by SigE, qRT-PCR analyses were carried out with RNA from wild-type H37Rv or an isogenic ΔsigE mutant and a primer set that amplified a region immediately downstream of the distal mprA promoter. Strains were either left untreated or exposed to 0.05% SDS for 90 min. Consistent with results from Northern blot and 5′ RACE analyses, the relative expression levels of the amplified region were significantly higher in wild-type M. tuberculosis following SDS treatment than in untreated cultures (Fig. 3C). In contrast, the expression levels of the amplified region were not elevated in the M. tuberculosis ΔsigE mutant strain following SDS treatment, and overall expression levels in this strain were significantly lower than that observed with the wild-type M. tuberculosis (Fig. 3C). These results suggest that under SDS stress, pepD and moaB (in addition to mprAB) are expressed from a distal SigE-regulated promoter element located at −204 relative to the originally annotated mprA translational start site. pepD and moaB are also expressed from two promoters, one at +7 relative to the originally annotated mprA translational start site and the other at −22 relative to the pepD translational start site, during growth under physiological conditions.
FIG. 3.
Characterization of the pepD promoter region. (A and B) Genomic depiction of pepD (A) and mprA (B) upstream regions, including MprA binding sites (gray boxes), determined transcriptional start sites (arrows), and annotated or predicted translational start and stop sites (boldface letters). Note that one of the transcriptional start sites for mprA is downstream of the translational start site previously annotated. The location of the primer set used to measure expression from the distal mprA promoter is also shown by arrows. (C) The wild type and the ΔsigE M. tuberculosis mutant were grown in the absence of SDS or were exposed to 0.05% SDS for 90 min, and the relative amounts of expression from the distal mprA promoter were determined. Expression levels were normalized to those of the 16S ribosomal gene, rrs, and are expressed as relative expression from cultures exposed to SDS versus untreated cultures. The results represent the means ± standard errors of the mean (SEM) from three independent experiments performed in triplicate.
Mycobacterial ΔmprAB and ΔpepD strains exhibit increased sensitivity to cell wall-perturbing stress.
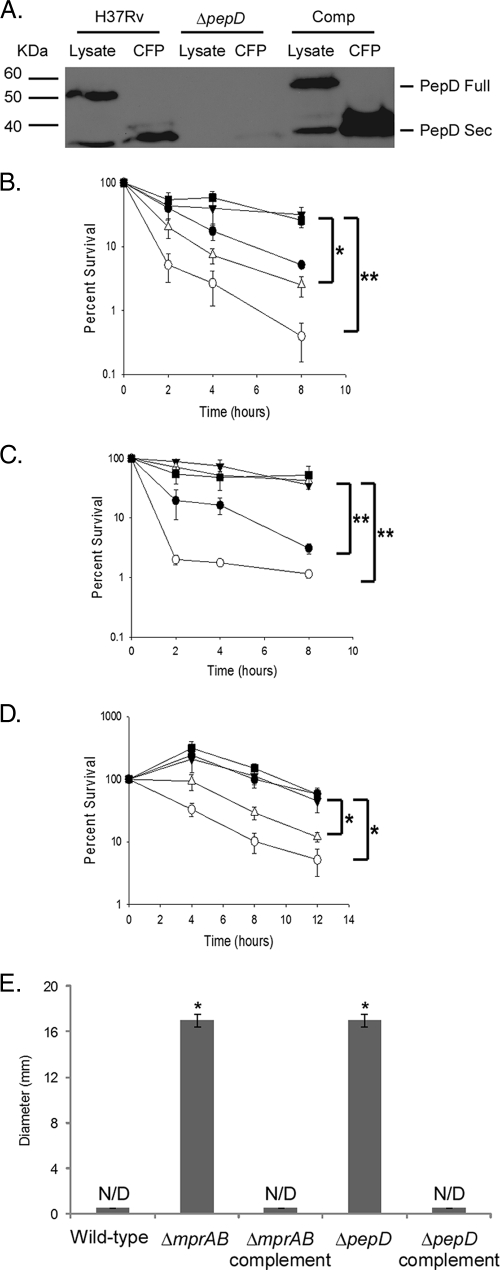
To determine whether PepD functions in a stress response pathway with MprAB, unmarked mprAB and pepD deletion derivatives were generated in M. smegmatis and M. tuberculosis (see Fig. S3 in the supplemental material), and the mutants were assayed for survival following exposure to cell wall-perturbing stress. M. smegmatis is a fast-growing nonpathogenic mycobacterial species, and it possesses a locus (MSMEG5488-MSMEG5484) with high amino acid homology to MprA, MprB, PepD, and MoaB2 from M. tuberculosis and with a similar genetic arrangement. It is important to note that the region deleted in the M. tuberculosis ΔmprAB mutant included the proximal promoter element identified at the beginning of mprA. To confirm the loss of PepD in the generated M. tuberculosis ΔpepD mutant, Western blots were conducted using a polyclonal antibody specific for PepD. Loss of the predicted ∼50-kDa full-length form of PepD was observed in cytoplasmic fractions from the M. tuberculosis ΔpepD mutant (Fig. 4A). Interestingly, loss of a less abundant, lower-molecular-mass (∼35-kDa) protein was also observed in the culture filtrate proteins from this strain (Fig. 4A), consistent with previous reports indicating that PepD may be secreted in M. tuberculosis (47). While deletion of mprAB or pepD did not affect the growth of M. smegmatis or M. tuberculosis in 7H9 medium in vitro (see Fig. S3 in the supplemental material), M. smegmatis ΔmprAB (Fig. 4B) and M. tuberculosis ΔmprAB (Fig. 4C) mutants exhibited significantly increased sensitivity to SDS relative to their wild-type counterparts. Resistance to SDS was partially restored in both mutant derivatives following expression of M. tuberculosis mprAB in trans (Fig. 4B and C). The inability to fully complement the M. smegmatis ΔmprAB mutant may be a consequence of differences in expression of M. tuberculosis mprAB in M. smegmatis or differences in recognition of M. smegmatis MprA-regulated genes by M. tuberculosis MprA. The inability to fully complement the M. tuberculosis ΔmprAB mutant is likely to be due to removal of the proximal mprA promoter element.
FIG. 4.
Characterization of mycobacterial ΔmprAB and ΔpepD mutants. (A) Western blot analysis of cell lysates and culture filtrate protein (CFP) from wild-type, ΔpepD, and complemented M. tuberculosis strains probed with the α-PepD antibody. (B and C) Sensitivities of wild-type, mutant, and complemented M. smegmatis derivatives (B) or M. tuberculosis derivatives (C) to SDS as determined by broth sensitivity assays. The results are expressed as percent survival between untreated cultures and cultures treated with 0.1% or 0.4% SDS, respectively. (D) Survival of M. smegmatis derivatives after exposure to 200 μg/ml d-cycloserine expressed as percent survival between untreated and treated cultures. Survival for broth sensitivity assays was determined by plating aliquots onto 7H10 agar medium and assaying them for CFU. (E) Sensitivities of M. smegmatis derivatives to cefuroxime as determined by disc diffusion assay. The results are expressed as zones of inhibition (N/D, not detected). ▪, wild-type; ○, ΔmprAB; ▵, ΔpepD; •, ΔmprAB complement; ▴, ΔpepD complement. The results represent the means ± SEM from three independent experiments performed in triplicate. *, P < 0.05; **, P < 0.01.
Sensitivity to SDS was also examined in ΔpepD M. smegmatis or M. tuberculosis mutants. M. smegmatis ΔpepD mutants exhibited increased sensitivity to SDS relative to the wild-type parent (Fig. 4B). This phenotype was fully complemented using pepD from M. tuberculosis (Fig. 4B). In contrast, no difference in SDS sensitivity was observed in the M. tuberculosis ΔpepD mutant relative to the wild-type parent (Fig. 4C). A similar lack of phenotype was also observed with the M. tuberculosis ΔpepD mutant following infection of primary human monocyte-derived macrophages or the J774A.1 murine macrophage-like cell line (see Fig. S4 in the supplemental material). To investigate whether the observed sensitivity of ΔmprAB and ΔpepD mutants of M. smegmatis was specific to cell wall-perturbing agents like SDS, the sensitivity assays were repeated with different cell wall inhibitors. Cycloserine prevents the synthesis of peptidoglycan by inhibiting the action of alanine racemase and d-alanine-d-alanine ligase (2). Cefuroxime is a cephalosporin that acts on penicillin binding proteins. M. smegmatis ΔmprAB and ΔpepD mutants also exhibited increased sensitivity to these cell wall agents compared to the wild-type parent (Fig. 4D and E). Furthermore, both the complemented ΔmprAB and ΔpepD M. smegmatis mutants were resistant to the activities of these antibiotics (Fig. 4D and E). Taken together, these results indicate that deletion of pepD or mprAB in M. smegmatis increases sensitivity to various cell wall perturbants. However, deletion of mprAB, but not pepD, increases the sensitivity of M. tuberculosis to SDS-mediated stress.
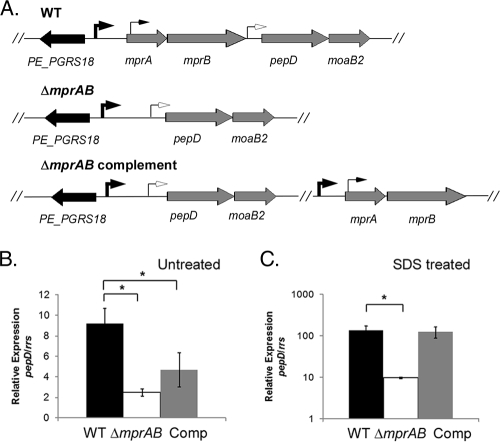
The ΔmprAB allele in M. tuberculosis is polar on pepD expression.
To confirm that the failure to fully complement the M. tuberculosis ΔmprAB mutant was due to removal of the mprA-proximal promoter element and partial polarity on pepD (Fig. 5A), pepD expression levels were quantified by RT-PCR in wild-type, ΔmprAB mutant, or complemented mutant derivatives grown in the absence or presence of 0.05% SDS. In the absence of SDS, expression of pepD was significantly reduced in the ΔmprAB mutant relative to the wild-type strain (Fig. 5B). pepD expression levels could be only partially restored in the ΔmprAB mutant following expression of mprAB from an exogenous site in trans (Fig. 5B). In contrast, the expression levels of pepD were also lower in the ΔmprAB mutant following exposure to SDS; however, expression of this gene was restored to wild-type levels following the introduction of mprAB in trans (Fig. 5C). Taken together, these results support the contention that expression of mprAB-pepD-moaB2 proceeds from promoters in the mprB-pepD intergenic region and at the start of mprA in the absence of SDS stress and from an SDS-responsive promoter element further upstream of mprA during growth in the presence of SDS stress.
FIG. 5.
Polarity resulting from the ΔmprAB mutation. (A) Genomic organization of the mprA-mprB-pepD-moaB2 locus in M. tuberculosis H37Rv (wild type [WT], the ΔmprAB mutant, and complemented strains. The arrows depict the identified transcriptional start sites in the region. (B and C) Wild-type M. tuberculosis, the ΔmprAB mutant, and the ΔmprAB complemented strain were grown without SDS (B) or were exposed to 0.05% SDS for 90 min (C). The relative amounts of pepD in the strains were compared. The expression levels were normalized to that of the 16S ribosomal gene, rrs. The results represent the means ± SEM from three independent experiments performed in triplicate. *, P < 0.05.
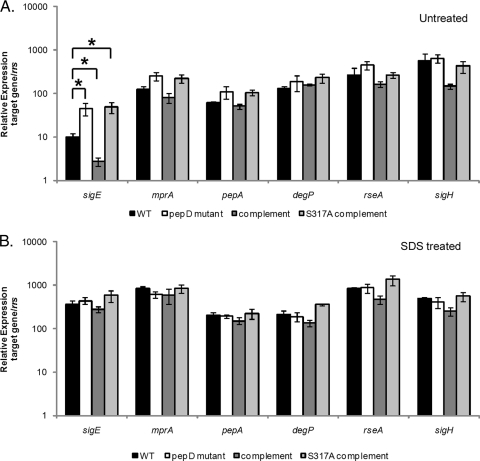
Deletion of pepD in M. tuberculosis leads to an upregulation in sigE expression.
To determine whether the failure to observe SDS sensitivity in the M. tuberculosis ΔpepD mutant was a result of a compensatory upregulation in the expression of sigE, mprA, or one of the other HtrA-like serine protease genes (degP and pepA), qRT-PCR analyses were carried out in the wild type, the ΔpepD mutant, or the complemented ΔpepD mutant strain of M. tuberculosis grown in the absence or presence of 0.05% SDS. sigE was found to be significantly upregulated in the ΔpepD mutant compared with wild-type H37Rv in the absence of stress (Fig. 6A). This phenotype was complemented to below wild-type levels following the introduction of pepD-moaB2 in trans and was lost upon mutation of the catalytic serine (S317) in pepD (Fig. 6A). Deletion of pepD also resulted in a modest but reproducible increase in mprA and pepA expression levels, although this difference did not reach statistical significance (Fig. 6A). In contrast, differences in degP expression were not observed between the ΔpepD mutant and other strains examined under these conditions. To gain further insights into the mechanism(s) of sigE upregulation, expression analyses were expanded to include other known regulators of sigE, including RseA and SigH. RseA is the cognate anti-sigma factor for SigE (12), and SigH is a sigma factor that regulates sigE in response to heat shock and diamide exposure (29). Neither gene displayed any difference in expression in the ΔpepD mutant relative to the wild-type strain under these conditions (Fig. 6A). Since rseA is located approximately 158 bases downstream from sigE and could potentially be regulated along with sigE, we also investigated whether these genes were cotranscribed. No PCR products were amplified using cDNA from these strains and primers nested within sigE and rseA (data not shown), supporting the contention that sigE and rseA are not in an operon together. To determine whether similar differences in expression were observed following exposure to cell wall-perturbing stress, profiling was repeated on strains exposed to 0.05% SDS. While the relative sigE, mprA, pepA, and rseA expression levels were significantly elevated in all M. tuberculosis derivatives compared with the untreated controls (Fig. 6B), the overall expression levels observed between strains were similar. In contrast, the expression levels of degP and sigH were not responsive to SDS exposure in any of the strains examined, and all strains exhibited similar expression levels under these conditions (Fig. 6B). Thus, loss of pepD in M. tuberculosis results in the upregulation of sigE under physiological growth conditions.
FIG. 6.
Impact of pepD deletion on M. tuberculosis gene expression in the absence or presence of SDS stress. (A and B) Wild-type M. tuberculosis, the ΔpepD mutant, the ΔpepD complemented strain, and the ΔpepD S317A complemented strain were grown in the absence of stress (A) or were exposed to 0.05% SDS for 90 min (B). The relative amounts of sigE, mprA, degP, pepA, rseA, or sigH expression in the strains were then compared. The expression level for each gene under each condition was normalized to that of the 16S ribosomal gene, rrs. The results represent the means ± SEM from three independent experiments performed in triplicate. *, P < 0.05.
Identification of genes differentially regulated in the M. tuberculosis ΔpepD mutant strain.
To determine whether loss of PepD results in a general stress response under physiological conditions, global gene expression profiling studies were carried out on the wild-type parent and the ΔpepD mutant of M. tuberculosis. Overall, 16 genes were differentially regulated between these strains following growth in 7H9 medium (Table 2). Of these, 13 genes were upregulated in the ΔpepD mutant, including sigE and the heat shock protein hspX (Table 2). In addition, three SigE-regulated genes were also identified, including Rv0789c, Rv0997, and Rv1057. In contrast, three genes were significantly downregulated in the ΔpepD mutant compared with the wild-type strain, including pepD and moaB2 (Table 2). qRT-PCR analyses of these genes confirmed the differential expression observed (Table 2). Thus, these data suggest that loss of PepD triggers a stress response in M. tuberculosis under physiological conditions that results in the upregulation of a number of gene products, including sigE, and a subset that are regulated by SigE.
TABLE 2.
Genes determined to be differentially regulated between wild-type and ΔpepD strains by microarray analysis
| Locus | Gene | Microarray ratioa | RT-PCR ratiob | Gene productc |
|---|---|---|---|---|
| Rv0079 | 3.0 | 1.5 | Hypothetical protein | |
| Rv0789c | 13.0 | 2.0 | Hypothetical protein | |
| Rv0983 | pepD | 0.1 | N/A | Serine protease |
| Rv0984 | moaB2 | 0.06 | 0.6 | Pterin carbinolamine dehydratase |
| Rv0997 | 8.6 | 2.1 | Hypothetical protein | |
| Rv1057 | 4.6 | 4.0 | Hypothetical protein | |
| Rv1076 | lipU | 0.3 | 0.6 | Possible lipase |
| Rv1221 | sigE | 3.0 | 4.9 | Sigma factor E |
| Rv2031c | hspX | 2.8 | 1.6 | Heat shock protein |
| Rv2784c | lppU | 3.2 | 1.6 | Probably lipoprotein |
| Rv2959c | 4.6 | 1.8 | Possible methyltransferase | |
| Rv2987c | leuD | 4.0 | 2.7 | Isopropylmalate isomerase |
| Rv2988c | leuC | 3.7 | 3.1 | Isopropylmalate isomerase |
| Rv3095 | 3.5 | 6.0 | Hypothetical transcriptional regulatory protein | |
| Rv3135 | PPE50 | 3.0 | 2.3 | PPE family protein |
| Rv3841 | bfrB | 3.0 | 1.3 | Possible bacterioferritin |
Microarray ratios were determined using SAM comparing ΔpepD and wild-type (wt) RNA (ΔpepD/wt).
RT-PCR ratios for the genes were determined using the ΔCT method to compare RNA levels between strains (ΔpepD/wt). The values were first normalized to the housekeeping sigma factor, sigA.
Gene products are annotated based on the Pasteur Institutes' Tuberculist website (http://genolist.pasteur.fr/Tuberculist).
DISCUSSION
M. tuberculosis is capable of adapting to and surviving within challenging environments inside the host during infection. While the mechanism(s) by which the organism achieves this is not well understood, several two-component systems and stress-responsive sigma factors are believed to participate in the process. One such system, the MprAB two-component system, regulates a number of stress-responsive genes, including the extracytoplasmic function (ECF) sigma factor sigE and the HtrA-family member pepD. Evidence presented in this study suggests overlap between the MprAB system and the SigE stress regulon that is at least partially mediated through the proteolytic activity of PepD.
PepD possesses an architecture that is conserved with other HtrA family members, including a transmembrane domain, a serine protease domain, and a single PDZ domain (26). PepD exhibits both protease and chaperone activities (33). In vitro, PepD functions optimally as a protease at 37°C, at near-neutral pH, and in a buffer containing 50 mM phosphate. PepD protease activity is also inhibited by the general protease inhibitor DFP, providing further evidence that this protein is a serine protease. In vivo, PepD is present in the cytoplasm but is also secreted from the bacterium in a mature form with a reduced molecular mass. Recent studies carried out by Mohamedmohaideen et al. in which PepD was crystallized indicate that the single PDZ domain predicted to be involved in protein-protein interactions binds to an autoproteolytic peptide and that this binding could be important for the regulation of PepD activity (33). Furthermore, this group found that PepD removes its PDZ domain in an autoproteolytic event. Consistent with these observations, we observed that removal of the PDZ domain from PepD diminishes its protease activity by >90%, suggesting that this may be a mechanism employed by M. tuberculosis to regulate protease activity following substrate recognition or once PepD is secreted from the bacterium. Additional studies are currently under way to better define the regions of PepD removed in the mature secreted form.
In many Mycobacterium species, including Mycobacterium leprae, Mycobacterium marinum, Mycobacterium bovis, and Mycobacterium avium, mprAB is located immediately upstream of pepD and a gene encoding a hypothetical protein with homology to pterin-4-alpha-carbinolamine dehydratases. Cotranscription and promoter-mapping studies of M. tuberculosis have indicated that pepD is transcribed from three distinct promoters, one that is located in the intergenic region between mprB and pepD, one that overlaps with the translational start site for mprA, and one upstream of mprA that resides in a predicted SigE-regulated promoter region (30). Thus, pepD is regulated transcriptionally with mprAB and is also positively regulated by MprA posttranscriptionally through interaction at three sites located between the end of mprB and the beginning of pepD (20, 36). In an effort to examine the role of PepD in the context of the MprAB two-component system, mprAB and pepD deletion derivatives were generated in both M. smegmatis and M. tuberculosis and resistance to membrane-perturbing stresses was examined. The M. tuberculosis ΔmprAB mutant exhibited increased sensitivity to SDS compared with wild-type or complemented mutant strains, conflicting with a previous report in which an M. tuberculosis ΔmprAB mutant was found to be more resistant to SDS stress (36). While the reasons for this discrepancy are unclear, the M. tuberculosis ΔmprAB mutants carried different deletion alleles and the deletion mutations were generated using different techniques. However, a similar defect in resistance to SDS was also observed with the M. smegmatis ΔmprAB mutant, as well as increased sensitivity to cell wall inhibitors. The increased sensitivity to SDS observed in ΔmprAB mutants of Mycobacterium is consistent with previous reports demonstrating that MprAB positively regulates sigE expression (19, 36). Furthermore, mutants of M. tuberculosis unable to make SigE are hypersensitive to SDS stress (28, 30).
In addition to mprAB, deletion of pepD in M. smegmatis and M. tuberculosis also alters the response of these strains to membrane-perturbing stress. The ΔpepD mutant of M. smegmatis is more sensitive to killing by SDS, cycloserine, and cefuroxime. Interestingly, deletion of pepD does not alter the ability of M. tuberculosis to resist SDS-mediated killing. Loss of pepD also does not alter the growth and/or survival characteristics of M. tuberculosis within peripheral human blood monocyte-derived macrophages or within resting or activated J774A.1 macrophages in vitro. Rather, deletion of pepD in M. tuberculosis upregulates the expression of sigE and other stress-responsive determinants, which may compensate, at least in part, for the loss of this protein. Regardless, additional studies are needed to better delineate the complex interactions between the MprAB and SigE stress response pathways and to better define the role of PepD in these systems.
In E. coli, adaptation to envelope stress is mediated through the combined activities of three signal transduction systems: the CpxRA and BaeRS two-component signal transduction systems and the RpoE sigma factor (42). The CpxRA response regulator and sensor kinase and the associated periplasmically localized inhibitory molecule, CpxP, are activated in response to a variety of stimuli, including alkaline pH (9); overexpression of envelope proteins, including NlpE and P pilus subunits (24, 48); and alterations in membrane composition (31). The Cpx regulon is comprised of over 50 genes from different transcriptional units (40), most of which are thought to assist in envelope protein folding and/or degradation. Of note, the HtrA-like serine protease degP is one of the genes most highly upregulated by CpxR following membrane stress (39). Similarly, the BaeRS response regulator and sensor kinase mediate resistance in E. coli to various toxic compounds, including antibiotics and other membrane-perturbing agents, through induction of at least two independent efflux pumps (4, 21, 34, 41). Finally, rpoE encodes an essential alternative sigma factor (11) that is responsive to cell envelope stress and is thought to play a critical role in maintaining cell envelope integrity (18). RpoE is activated by β-barrel proteins that have become unfolded in the periplasmic space prior to insertion into the outer membrane. Conditions that activate RpoE include exposure to heat, ethanol, or other membrane-damaging conditions (32, 44). RpoE regulates a number of outer membrane proteins and genes involved in outer membrane biogenesis (43). RpoE also regulates degP, the peptidylprolyl isomerases fpkA and surA, and the periplasmic chaperone skp (10, 22). Thus, DegP is jointly regulated by both the CpxRA and RpoE pathways in E. coli.
The functional and regulatory similarities between the MprAB and SigE signal transduction systems in M. tuberculosis and those of the CpxAR and RpoE systems from E. coli are striking for a number of reasons. First, all of these systems mediate resistance to membrane-perturbing stress. Second, both MprAB and CpxAR directly activate HtrA family proteases that are coregulated by SigE and RpoE, respectively. Finally, regulons controlled by all four transcription factors include genes that have been shown or are predicted to be involved in aspects of periplasmic or cellular envelope integrity. Interestingly, the MprAB and SigE networks differ in one important aspect from that seen with the Cpx and RpoE systems. In M. tuberculosis, MprA directly regulates expression of sigE and vice versa. In E. coli, the Cpx and RpoE networks are integrated further downstream, at degP.
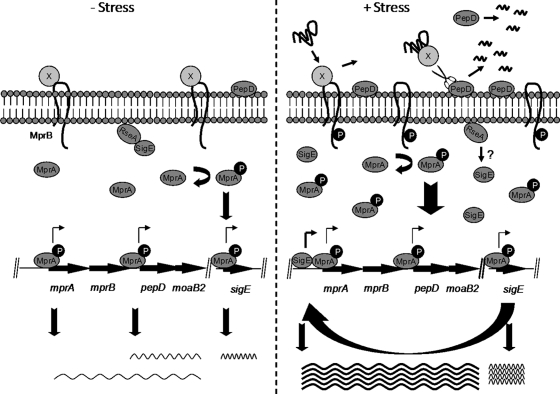
Based on this information, we propose a model in which the MprAB and SigE systems sense and process stress in M. tuberculosis resulting from the accumulation of as yet unidentified unfolded or misfolded protein substrates (Fig. 7). Similar to the Cpx system, we propose that MprAB processes stress in the extracytoplasmic space. We predict that MprB is kept inactive under nonstress conditions by a protein repressor. Mutagenesis studies are currently under way to address this possibility. Following exposure to membrane-perturbing stress, this repressor disengages from MprB, binds to misfolded protein substrates, and presents them to PepD for subsequent processing. This derepression event leads to the activation of the MprAB system and subsequent upregulation of genes comprising the MprAB regulon, including mprAB, pepD, and sigE. As the newly synthesized SigE is abundant relative to its cognate antisigma factor RseA, it would be available to mediate downstream signaling events through the SigE regulon. SigE now functions in a positive feedback loop to drive the expression of the mprA-mprB-pepD-moaB2 operon, as well as other genes in its regulon. Conversely, loss of PepD results in the accumulation of misfolded protein substrates in the extracytoplasmic space, leading to activation of both MprAB and SigE. Continued work on PepD is expected to provide important insights into how the MprAB two-component system and the SigE regulon function in the physiology and pathogenesis of this medically significant organism.
FIG. 7.
Model of the activation of the MprAB system. Under nonstress conditions (left), an unidentified protein repressor (protein X) binds to MprB, keeping it in an inactive conformation and resulting in basal-level expression of mprAB transcription. In the presence of membrane stress (right), misfolded proteins accumulate in the extracytoplasmic space, resulting in the release of protein X from MprB and activation of the MprAB signaling pathway, including upregulation of sigE and subsequent induction of mprAB-pepD-moaB. PepD is secreted into the extracytoplasmic space between the cell membrane and the cell wall, where it functions to degrade and/or refold damaged substrate targets. Following proteolysis, the PDZ domain of PepD is removed in an autocatalytic event, resulting in release of PepD into the culture supernatant. Wavy lines indicate transcripts, with darker lines indicating higher expression lavels.
Supplementary Material
Acknowledgments
We are grateful to members of the Zahrt laboratory for useful discussions regarding these studies. We thank the Pathogen Functional Genomics Resource Center and TB Vaccine Testing and Research Materials Contract for providing materials necessary for this research.
This work was supported by RO1 AI51669 from the National Institutes of Health and Infectious Disease and an Investigator in Pathogenesis award from the Burroughs Wellcome Fund.
Footnotes
Published ahead of print on 8 January 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Alba, B. M., H. J. Zhong, J. C. Pelayo, and C. A. Gross. 2001. degS (hhoB) is an essential Escherichia coli gene whose indispensable function is to provide sigma (E) activity. Mol. Microbiol. 40:1323-1333. [DOI] [PubMed] [Google Scholar]
- 2.Anonymous. 2008. Cycloserine. Tuberculosis (Edinb.) 88:100-101. [DOI] [PubMed] [Google Scholar]
- 3.Anonymous. 2008. Global tuberculosis control. Surveillance, planning, financing. World Health Organization, Geneva, Switzerland.
- 4.Baranova, N., and H. Nikaido. 2002. The baeSR two-component regulatory system activates transcription of the yegMNOB (mdtABCD) transporter gene cluster in Escherichia coli and increases its resistance to novobiocin and deoxycholate. J. Bacteriol. 184:4168-4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bardarov, S., S. Bardarov, Jr., M. S. Pavelka, Jr., V. Sambandamurthy, M. Larsen, J. Tufariello, J. Chan, G. Hatfull, and W. R. Jacobs, Jr. 2002. Specialized transduction: an efficient method for generating marked and unmarked targeted gene disruptions in Mycobacterium tuberculosis, M. bovis BCG and M. smegmatis. Microbiology 148:3007-3017. [DOI] [PubMed] [Google Scholar]
- 6.Betts, J. C., P. T. Lukey, L. C. Robb, R. A. McAdam, and K. Duncan. 2002. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol. Microbiol. 43:717-731. [DOI] [PubMed] [Google Scholar]
- 7.Buelow, D. R., and T. L. Raivio. 2005. Cpx signal transduction is influenced by a conserved N-terminal domain in the novel inhibitor CpxP and the periplasmic protease DegP. J. Bacteriol. 187:6622-6630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, A. Krogh, J. McLean, S. Moule, L. Murphy, K. Oliver, J. Osborne, M. A. Quail, M. A. Rajandream, J. Rogers, S. Rutter, K. Seeger, J. Skelton, R. Squares, S. Squares, J. E. Sulston, K. Taylor, S. Whitehead, and B. G. Barrell. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 9.Danese, P. N., and T. J. Silhavy. 1998. CpxP, a stress-combative member of the Cpx regulon. J. Bacteriol. 180:831-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dartigalongue, C., D. Missiakas, and S. Raina. 2001. Characterization of the Escherichia coli sigma E regulon. J. Biol. Chem. 276:20866-20875. [DOI] [PubMed] [Google Scholar]
- 11.De Las Penas, A., L. Connolly, and C. A. Gross. 1997. SigmaE is an essential sigma factor in Escherichia coli. J. Bacteriol. 179:6862-6864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dona, V., S. Rodrigue, E. Dainese, G. Palu, L. Gaudreau, R. Manganelli, and R. Provvedi. 2008. Evidence of complex transcriptional, translational, and posttranslational regulation of the extracytoplasmic function sigma factor sigmaE in Mycobacterium tuberculosis. J. Bacteriol. 190:5963-5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Edgar, R., M. Domrachev, and A. E. Lash. 2002. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30:207-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Farn, J., and M. Roberts. 2004. Effect of inactivation of the HtrA-like serine protease DegQ on the virulence of Salmonella enterica serovar Typhimurium in mice. Infect. Immun. 72:7357-7359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fisher, M. A., B. B. Plikaytis, and T. M. Shinnick. 2002. Microarray analysis of the Mycobacterium tuberculosis transcriptional response to the acidic conditions found in phagosomes. J. Bacteriol. 184:4025-4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flannagan, R. S., D. Aubert, C. Kooi, P. A. Sokol, and M. A. Valvano. 2007. Burkholderia cenocepacia requires a periplasmic HtrA protease for growth under thermal and osmotic stress and for survival in vivo. Infect. Immun. 75:1679-1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hampshire, T., S. Soneji, J. Bacon, B. W. James, J. Hinds, K. Laing, R. A. Stabler, P. D. Marsh, and P. D. Butcher. 2004. Stationary phase gene expression of Mycobacterium tuberculosis following a progressive nutrient depletion: a model for persistent organisms? Tuberculosis (Edinb.). 84:228-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hayden, J. D., and S. E. Ades. 2008. The extracytoplasmic stress factor, sigmaE, is required to maintain cell envelope integrity in Escherichia coli. PLoS One 3:e1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He, H., R. Hovey, J. Kane, V. Singh, and T. C. Zahrt. 2006. MprAB is a stress-responsive two-component system that directly regulates expression of sigma factors SigB and SigE in Mycobacterium tuberculosis. J. Bacteriol. 188:2134-2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He, H., and T. C. Zahrt. 2005. Identification and characterization of a regulatory sequence recognized by Mycobacterium tuberculosis persistence regulator MprA. J. Bacteriol. 187:202-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hirakawa, H., K. Nishino, T. Hirata, and A. Yamaguchi. 2003. Comprehensive studies of drug resistance mediated by overexpression of response regulators of two-component signal transduction systems in Escherichia coli. J. Bacteriol. 185:1851-1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hiratsu, K., M. Amemura, H. Nashimoto, H. Shinagawa, and K. Makino. 1995. The rpoE gene of Escherichia coli, which encodes sigma E, is essential for bacterial growth at high temperature. J. Bacteriol. 177:2918-2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jacobs, W. R., Jr., G. V. Kalpana, J. D. Cirillo, L. Pascopella, S. B. Snapper, R. A. Udani, W. Jones, R. G. Barletta, and B. R. Bloom. 1991. Genetic systems for mycobacteria. Methods Enzymol. 204:537-555. [DOI] [PubMed] [Google Scholar]
- 24.Jones, C. H., P. N. Danese, J. S. Pinkner, T. J. Silhavy, and S. J. Hultgren. 1997. The chaperone-assisted membrane release and folding pathway is sensed by two signal transduction systems. EMBO J. 16:6394-6406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karakousis, P. C., T. Yoshimatsu, G. Lamichhane, S. C. Woolwine, E. L. Nuermberger, J. Grosset, and W. R. Bishai. 2004. Dormancy phenotype displayed by extracellular Mycobacterium tuberculosis within artificial granulomas in mice. J. Exp. Med. 200:647-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim, D. Y., and K. K. Kim. 2005. Structure and function of HtrA family proteins, the key players in protein quality control. J. Biochem. Mol. Biol. 38:266-274. [DOI] [PubMed] [Google Scholar]
- 27.Levin, B. R., and D. E. Rozen. 2006. Non-inherited antibiotic resistance. Nat. Rev. Microbiol. 4:556-562. [DOI] [PubMed] [Google Scholar]
- 28.Manganelli, R., E. Dubnau, S. Tyagi, F. R. Kramer, and I. Smith. 1999. Differential expression of 10 sigma factor genes in Mycobacterium tuberculosis. Mol. Microbiol. 31:715-724. [DOI] [PubMed] [Google Scholar]
- 29.Manganelli, R., M. I. Voskuil, G. K. Schoolnik, E. Dubnau, M. Gomez, and I. Smith. 2002. Role of the extracytoplasmic-function sigma factor sigma(H) in Mycobacterium tuberculosis global gene expression. Mol. Microbiol. 45:365-374. [DOI] [PubMed] [Google Scholar]
- 30.Manganelli, R., M. I. Voskuil, G. K. Schoolnik, and I. Smith. 2001. The Mycobacterium tuberculosis ECF sigma factor sigmaE: role in global gene expression and survival in macrophages. Mol. Microbiol. 41:423-437. [DOI] [PubMed] [Google Scholar]
- 31.Mileykovskaya, E., and W. Dowhan. 1997. The Cpx two-component signal transduction pathway is activated in Escherichia coli mutant strains lacking phosphatidylethanolamine. J. Bacteriol. 179:1029-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Missiakas, D., J. M. Betton, and S. Raina. 1996. New components of protein folding in extracytoplasmic compartments of Escherichia coli SurA, FkpA and Skp/OmpH. Mol. Microbiol. 21:871-884. [DOI] [PubMed] [Google Scholar]
- 33.Mohamedmohaideen, N. N., S. K. Palaninathan, P. M. Morin, B. J. Williams, M. Braunstein, S. E. Tichy, J. Locker, D. H. Russell, W. R. Jacobs, Jr., and J. C. Sacchettini. 2008. Structure and function of the virulence-associated high-temperature requirement A of Mycobacterium tuberculosis. Biochemistry 47:6092-6102. [DOI] [PubMed] [Google Scholar]
- 34.Nagakubo, S., K. Nishino, T. Hirata, and A. Yamaguchi. 2002. The putative response regulator BaeR stimulates multidrug resistance of Escherichia coli via a novel multidrug exporter system, MdtABC. J. Bacteriol. 184:4161-4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pang, X., and S. T. Howard. 2007. Regulation of the alpha-crystallin gene acr2 by the MprAB two-component system of Mycobacterium tuberculosis. J. Bacteriol. 189:6213-6221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pang, X., P. Vu, T. F. Byrd, S. Ghanny, P. Soteropoulos, G. V. Mukamolova, S. Wu, B. Samten, and S. T. Howard. 2007. Evidence for complex interactions of stress-associated regulons in an mprAB deletion mutant of Mycobacterium tuberculosis. Microbiology 153:1229-1242. [DOI] [PubMed] [Google Scholar]
- 37.Pechous, R. D., T. R. McCarthy, N. P. Mohapatra, S. Soni, R. M. Penoske, N. H. Salzman, D. W. Frank, J. S. Gunn, and T. C. Zahrt. 2008. A Francisella tularensis Schu S4 purine auxotroph is highly attenuated in mice but offers limited protection against homologous intranasal challenge. PLoS One 3:e2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Philips, J. A. 2008. Mycobacterial manipulation of vacuolar sorting. Cell Microbiol. 10:2408-2415. [DOI] [PubMed] [Google Scholar]
- 39.Pogliano, J., A. S. Lynch, D. Belin, E. C. Lin, and J. Beckwith. 1997. Regulation of Escherichia coli cell envelope proteins involved in protein folding and degradation by the Cpx two-component system. Genes Dev. 11:1169-1182. [DOI] [PubMed] [Google Scholar]
- 40.Price, N. L., and T. L. Raivio. 2008. Characterization of the Cpx regulon in Escherichia coli strain MC4100. J. Bacteriol. [DOI] [PMC free article] [PubMed]
- 41.Raffa, R. G., and T. L. Raivio. 2002. A third envelope stress signal transduction pathway in Escherichia coli. Mol. Microbiol. 45:1599-1611. [DOI] [PubMed] [Google Scholar]
- 42.Raivio, T. L. 2005. Envelope stress responses and Gram-negative bacterial pathogenesis. Mol. Microbiol. 56:1119-1128. [DOI] [PubMed] [Google Scholar]
- 43.Rezuchova, B., H. Miticka, D. Homerova, M. Roberts, and J. Kormanec. 2003. New members of the Escherichia coli sigmaE regulon identified by a two-plasmid system. FEMS Microbiol. Lett. 225:1-7. [DOI] [PubMed] [Google Scholar]
- 44.Rouviere, P. E., A. De Las Penas, J. Mecsas, C. Z. Lu, K. E. Rudd, and C. A. Gross. 1995. rpoE, the gene encoding the second heat-shock sigma factor, sigma E, in Escherichia coli. EMBO J. 14:1032-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rowley, G., A. Stevenson, J. Kormanec, and M. Roberts. 2005. Effect of inactivation of degS on Salmonella enterica serovar typhimurium in vitro and in vivo. Infect. Immun. 73:459-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schluger, N. W., and W. N. Rom. 1998. The host immune response to tuberculosis. Am. J. Respir. Crit. Care Med. 157:679-691. [DOI] [PubMed] [Google Scholar]
- 47.Skeiky, Y. A., M. J. Lodes, J. A. Guderian, R. Mohamath, T. Bement, M. R. Alderson, and S. G. Reed. 1999. Cloning, expression, and immunological evaluation of two putative secreted serine protease antigens of Mycobacterium tuberculosis. Infect. Immun. 67:3998-4007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Snyder, W. B., L. J. Davis, P. N. Danese, C. L. Cosma, and T. J. Silhavy. 1995. Overproduction of NlpE, a new outer membrane lipoprotein, suppresses the toxicity of periplasmic LacZ by activation of the Cpx signal transduction pathway. J. Bacteriol. 177:4216-4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stack, H. M., R. D. Sleator, M. Bowers, C. Hill, and C. G. Gahan. 2005. Role for HtrA in stress induction and virulence potential in Listeria monocytogenes. Appl. Environ. Microbiol. 71:4241-4247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sureka, K., B. Ghosh, A. Dasgupta, J. Basu, M. Kundu, and I. Bose. 2008. Positive feedback and noise activate the stringent response regulator rel in mycobacteria. PLoS One 3:e1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Takayama, K., H. K. Schnoes, E. L. Armstrong, and R. W. Boyle. 1975. Site of inhibitory action of isoniazid in the synthesis of mycolic acids in Mycobacterium tuberculosis. J. Lipid Res. 16:308-317. [PubMed] [Google Scholar]
- 52.Talaat, A. M., R. Lyons, S. T. Howard, and S. A. Johnston. 2004. The temporal expression profile of Mycobacterium tuberculosis infection in mice. Proc. Natl. Acad. Sci. U. S. A. 101:4602-4607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Talaat, A. M., S. K. Ward, C. W. Wu, E. Rondon, C. Tavano, J. P. Bannantine, R. Lyons, and S. A. Johnston. 2007. Mycobacterial bacilli are metabolically active during chronic tuberculosis in murine lungs: insights from genome-wide transcriptional profiling. J. Bacteriol. 189:4265-4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tusher, V. G., R. Tibshirani, and G. Chu. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. U. S. A. 98:5116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Twining, S. S. 1984. Fluorescein isothiocyanate-labeled casein assay for proteolytic enzymes. Anal. Biochem. 143:30-34. [DOI] [PubMed] [Google Scholar]
- 56.Waller, P. R., and R. T. Sauer. 1996. Characterization of degQ and degS, Escherichia coli genes encoding homologs of the DegP protease. J. Bacteriol. 178:1146-1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wilson, R. L., L. L. Brown, D. Kirkwood-Watts, T. K. Warren, S. A. Lund, D. S. King, K. F. Jones, and D. E. Hruby. 2006. Listeria monocytogenes 10403S HtrA is necessary for resistance to cellular stress and virulence. Infect. Immun. 74:765-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zahrt, T. C. 2003. Molecular mechanisms regulating persistent Mycobacterium tuberculosis infection. Microbes Infect. 5:159-167. [DOI] [PubMed] [Google Scholar]
- 59.Zahrt, T. C., and V. Deretic. 2001. Mycobacterium tuberculosis signal transduction system required for persistent infections. Proc. Natl. Acad. Sci. U. S. A. 98:12706-12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.