Abstract
The prion diseases seem to be caused by a conformational change of the prion protein (PrP) from the benign cellular form PrPC to the infectious scrapie form PrPSc; thus, detailed information about PrP structure may provide essential insights into the mechanism by which these diseases develop. In this study, the secondary structure of the recombinant Syrian hamster PrP of residues 29–231 [PrP(29–231)] is investigated by multidimensional heteronuclear NMR. Chemical shift index analysis and nuclear Overhauser effect data show that PrP(29–231) contains three helices and possibly one short β-strand. Most striking is the random-coil nature of chemical shifts for residues 30–124 in the full-length PrP. Although the secondary structure elements are similar to those found in mouse PrP fragment PrP(121–231), the secondary structure boundaries of PrP(29–231) are different from those in mouse PrP(121–231) but similar to those found in the structure of Syrian hamster PrP(90–231). Comparison of resonance assignments of PrP(29–231) and PrP(90–231) indicates that there may be transient interactions between the additional residues and the structured core. Backbone dynamics studies done by using the heteronuclear [1H]-15N nuclear Overhauser effect indicate that almost half of PrP(29–231), residues 29–124, is highly flexible. This plastic region could feature in the conversion of PrPC to PrPSc by template-assisted formation of β-structure.
Keywords: NMR structure, conformational change, backbone dynamics
Prion diseases are a novel class of neurodegenerative diseases, including scrapie in sheep; bovine spongiform encephalopathy in cattle; and kuru, Creutzfeldt–Jacob disease, Gerstmann–Sträussler–Scheinker syndrome, and fatal familiar insomnia in humans (1, 2). A substantial body of evidence has shown that the scrapie isoform of the prion protein (PrPSc) is the major component of prions (3, 4). The prion protein (PrP) is composed of ≈250 amino acids and is encoded by a single chromosomal gene. Cellular PrP (PrPC) is processed to remove a 22-amino acid N-terminal signal peptide and 23 C-terminal residues upon the addition of the glycosylphosphatidylinositol anchor to Ser-231. Residues 179 and 214 are linked by a disulfide bridge, and Asn-181 and Asn-197 are glycosylation sites in the native protein. The fundamental event in prion disease pathology appears to be the profound conformational change during conversion of PrPC to PrPSc (5, 6).
To understand the conformational transition that accompanies PrPSc formation, a knowledge of the three-dimensional structure of the PrP is essential. CD and Fourier transform infrared studies suggest that conversion of α-helices into β-sheets features in the formation of PrPSc (5). Recently, the solution structure of a mouse PrP fragment, MoPrP(121–231), was reported to contain three α-helices and two short antiparallel β-strands (7, 8). Although MoPrP(121–231) contains a significant portion of a minimal region of the protease-resistant core (residues 90–231) that can mediate disease, the fragment lacks the biologically important N-terminal octarepeat region ([P(Q/H)GGG(G/-)WGQ]x5) and does not contain all of the sites of pathologic point mutations (9). The recent NMR structure of the Syrian hamster (SHa) PrP(90–231) (10) corresponding to the 27–30-kDa infectious core of PrPSc (designated PrP 27–30) indicates that the longer fragment also contains three helices and a short, irregular two-strand β-sheet. All three helices are longer in PrP(90–231) than those in MoPrP(121–231). In addition, the disordered loop between the last two helices in MoPrP(121–231) shows a greater degree of order in the structure of PrP(90–231), and residues 113–125 show some interactions with the folded core of the protein. The octarepeat region is not present in either MoPrP(121–231) or PrP(90–231), but an increased number of octarepeats is known to cause inherited prion disease (11–13). Because PrP can adopt two or more conformations, it is unclear whether the residues corresponding to MoPrP(121–231) will display context-dependent structural behavior in the full protein.
We report here a structural study by NMR of a recombinant SHa PrP(29–231) that represents the entire protein, including the octarepeats. The largely conserved octarepeat region may play an important role in the conversion of PrPC to PrPSc, either by participation in the profound conformational change, or in nonspecific binding to membranes (14) in the subcellular compartment where the conversion takes place (15). An earlier study of an octarepeat peptide by CD spectroscopy suggested that the repeat region adopts a nonrandom, extended conformation with the properties of a poly-l-proline left-handed helix (14). In the present paper the secondary structure of full-length SHa PrP(29–231) has been determined, and the dynamic properties of the protein backbone have been measured. We postulate a plausible role for the octarepeat region in prion disease pathogenesis. During the preparation of this paper, a report describing a preliminary analysis of the NMR spectrum of MoPrP(23–231) appeared (16). The present paper describes an extensive analysis of the full-length SHa PrP(29–231), including complete backbone resonance assignments obtained from triple-resonance experiments and measurements of polypeptide chain dynamics.
MATERIALS AND METHODS
Protein Expression and Purification.
Expression of both recombinant SHa PrP(29–231) and PrP(90–231) was accomplished by using a secreted ph041 vector driven by an alkaline phosphatase promoter in a protease-deficient strain of Escherichia coli (27C7). Expression of the proteins was induced about 9 hr postinoculation by phosphate depleting of the growth medium. Uniform isotopic labeling of the protein was done by minimal modifications of the protocol described earlier (17). The bacterium was grown overnight in a shaker flask containing 100 ml of Celtone microbial growth media labeled with either 15N or 15N/13C. The culture then was used for inoculation of a 2-liter fermentation where the following components were isotopically labeled: isoleucine, ammonium sulfate, ammonium hydroxide, glucose, and 10% reconstituted Celtone powdered media for the 20% yeast extract and NZ amines. The fermentations were allowed to proceed for either 24 hr for 15N labeling or 12–14 hr for 15N/13C labeling. Wet cell paste yield was between 40–50 g, ultimately giving a minimum of 100 mg of purified protein. This yield is about half of the amount normally obtained from unlabeled fermentations.
Purification was as described except that 20 mM methionine was added to the size exclusion material before HPLC separation to reduce methionine oxidation. Purified PrP(29–231) or PrP(90–231) was lyophilized before refolding. Lyophilized PrP(29–231) or PrP(90–231) was solubilized at 1 mg/ml in 6–8 M guanidinium hydrochloride and then rapidly diluted to 100 μg/ml in 25 mM Tris⋅HCl at pH 8.0 and 5 mM EDTA concentration. The refolded protein then was dialyzed overnight against 20 mM sodium acetate at pH 5.0 and 0.005% sodium azide. Protease inhibitors are essential for maintaining full-length protein. Insoluble material was removed by filtration through a 0.2 μm filter. Samples were concentrated first by Centriprep 10 (Amicon) and then further by Centriplus 10 (Amicon). Deuterated buffer exchange was done simultaneously with the final concentrating step. Samples were analyzed by mass spectroscopy, Fourier transform infrared, and CD spectroscopy.
NMR Spectroscopy.
NMR samples were prepared in 20 mM sodium acetate-d5 buffer at a final pH of 5.2 in 90% 1H2O/10% 2H2O. All solutions contained 0.005% sodium azide to inhibit bacterial growth. All deuterated reagents were purchased from Cambridge Isotope Laboratories (Cambridge, MA). NMR data were acquired at 30°C calibrated with neat methanol. The protein concentration for the 15N-labeled sample was about 1 mM and the 15N/13C-labeled sample about 0.6 mM for PrP(29–231). The final concentration for 15N-labeled PrP(90–231) used in the heteronuclear nuclear Overhauser effect (NOE) experiments was about 1 mM. The spectrometers used in the study were Bruker DMX 750 for the 15N and 13C separated NOE spectroscopy (NOESY) experiments and Bruker DRX 600 and AMX 600 instruments for all other triple-resonance experiments. All spectrometers are equipped with 5-mm triple-axis gradient triple-resonance probes and an 8-mm actively shielded Z-gradient probe for the DRX 600 instrument. For backbone and side-chain resonance assignments, the following experiments were carried out: two-dimensional 15N and 13C heteronuclear single-quantum coherence (HSQC) (18, 19), three-dimensional HNCA (20), HNCACB (21), CBCA(CO)NH (22), 15N-total correlation spectroscopy (TOCSY)-HSQC (23), C(CO)NH-TOCSY (24), HCCH-TOCSY (25), 15N-NOESY-HSQC (23) with 50-ms and 80-ms mixing times, 13C-NOESY-HSQC (26) with an 80-ms mixing time, HNCO (20), (HCA)CO(CA)NH (27), and HBHA(CBCACO)NH (28). All experiments were modified for samples dissolved in 90% H2O with sensitivity-enhanced gradient coherence selection (29, 30) and minimal water saturation (31). The heteronuclear [1H]-15N NOE experiments (32) for both PrP(29–231) and PrP(90–231) were carried out on both Bruker DRX 600 and AMX-II 500 spectrometers. Two data sets were acquired for both PrP(29–231) and PrP(90–231). In each data set, two experiments with and without proton saturation were carried out. The final NOE values were calculated from the ratio of the saturated intensities divided by the unsaturated intensities from each data set. The mean values from the two data sets then were calculated, and the error bars were taken to be the difference between the values from the two data sets. All NMR data were processed and analyzed by using FELIX95 (Molecular Simulations, San Diego, CA) that was run on Silicon Graphics (Indigo and O2) computers. All spectra were referenced relative to 2,2-dimethyl-2-silapentane-5-sulfonate, directly for 1H and indirectly for 13C and 15N dimensions as described by Wishart et al. (33).
RESULTS
Backbone and Side-Chain Resonance Assignments of PrP(29–231).
15N-labeled and 13C/15N-labeled protein samples of PrP(29–231) were used for the multidimensional heteronuclear NMR studies. The protein was found to be monomeric and stable for up to 3 months at pH 5.2 and 30°C in 20 mM sodium acetate buffer with 10% D2O and 0.005% sodium azide, as monitored by CD spectroscopy and NMR. Residue types were identified by using aliphatic 13C chemical shifts from a combination of HNCA, CBCA(CO)NH, and C(CO)NH-TOCSY experiments (28). Sequential assignments of 1H, 13C, and 15N resonances of the protein were obtained by using a combination of Cα- and CO-based triple-resonance experiments. In the Cα-based approach, combined use of HNCACB and CBCA(CO)NH experiments allowed about 80% of the amide proton and nitrogen resonances of residue i to be correlated with the Cα and Cβ chemical shifts of residues i and i-1. Sequential connectivities not identified in the above combination because of low sensitivity of the HNCACB experiment were made by using HNCA and CBCA(CO)NH experiments. Ambiguities caused by Cα degeneracy were resolved by the CO-based triple-resonance experiments [HNCO and (HCA)CO(CA)NH] that take advantage of the large intrinsic dispersion of carbonyl chemical shifts (34). In these experiments, the amide proton and nitrogen resonances of residue i were correlated with the carbonyl chemical shifts of residues i and i-1. Side-chain Hα and Hβ resonances were obtained by using the HBHA(CO)NH experiment in conjunction with the 15N-TOCSY-HSQC and 15N-NOESY-HSQC experiments. Resonance assignments were verified by using sequential through-space connectivities in 15N-resolved NOESY spectra. The combined use of these experiments yielded complete backbone assignments except for the N-terminal Gly-29. Complete Cα assignments were achieved except for Leu-130, and 97% of Cβ and 98% of CO resonances were assigned, as were 93% of Hα resonances. Assignments of the aromatic ring protons have not been made at this point. The assignments for PrP(90–231) were obtained as described (10).
Resonances of corresponding residues in four of five octarepeats (PHGGGWGQ) essentially were unresolved. The first octarepeat has a slightly different sequence (PQGGGTWGQ) from the other four, and the GTW portion could be assigned sequence-specifically. Residues in the same position in the remaining four octarepeats were degenerate in all frequencies and gave one intense cross peak in Cα and Hα, Cβ, and Hβ (if any), N, NH, and even CO resonances. Comparison of the intensity of amide proton/nitrogen cross peaks in the 15N HSQC spectrum with residues on either side of the octarepeat region gave the expected 1:4 intensity ratio.
Secondary Structure of PrP(29–231).
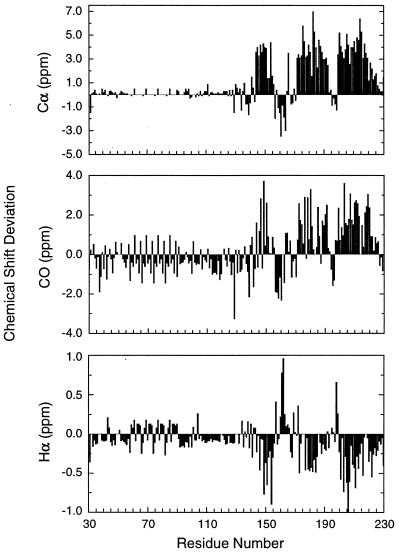
The chemical shift index (CSI) has been used to identify secondary structure elements (35–37). The deviation of Cα, Hα, Hβ, and 13CO chemical shifts from their random coil values (38, 39) suggests whether the residues are involved in α-helices or β-sheets in proteins. The presence of secondary structure elements then can be verified from short-range and medium-range NOEs that are indicative of different types of secondary structures (40). Fig. 1 shows the Cα, CO, and Hα chemical shift deviation of PrP(29–231) from random coil values. The most striking feature is that almost half of the protein is indistinguishable in chemical shift from random coil. In particular no deviations are greater than 0.5 ppm from random-coil Cα chemical shifts for residue 29–124. In contrast the C-terminal portion of PrP(29–231) displays large positive and negative deviations (typically 4 ppm) from random-coil values for Cα resonances. The same patterns are seen in the chemical shifts of Hα, CO, and Cβ resonances.
Figure 1.
Summary of Cα, Hα, and 13CO chemical shift deviations from random-coil values (38, 39) used to identify the secondary structure of PrP(29–231). The chemical shift deviations in ppm are plotted against residue numbers in PrP(29–231). Note the repeating nature of shift values in the octarepeat region.
CSI analysis indicates three α-helices and two short β-strands. Helix A spans residues 144–156, helix B contains residues 172–193, and helix C includes residues 200–227. The locations of the helices are confirmed by the presence of medium-range αN(i, i+3) and αN(i, i+4) NOEs and supported by continuous sequential NN NOEs in the helical regions. As shown in Fig. 2, medium-range NOEs are observed between residues 144–157, 174–192, and 200–226. A short piece of β-strand spanning residues 160–163 is identified by Cα, Hα, CO, and Cβ chemical shifts. The accompanying β-strand to pair with residues 160–163 is less clear, although residues 137–140 are suggested to contain β-structure by Cα and CO chemical shifts. There is no indication from the CSI that the β-strand identified in the solution structures of MoPrP(121–231) (7) and PrP(90–231) (10) between residues 129–131 is present in a β-conformation (Fig. 1), although interpretation of the data is complicated by missing assignments caused by exchange-broadened resonances in this region. On the other hand, there is evidence in the form of a single long-range NOE between the NH protons of residues 129 and 163 for a close contact between the 129–131 and 160–163 regions of the polypeptide; there is no such contact evident between 160–163 and 137–140, the β-region identified by the chemical shifts. For the octarepeat region, the NOEs remain ambiguous because of degeneracy in proton chemical shifts and are not shown in Fig. 2.
Figure 2.
Sequential and medium-range NOEs used to verify the secondary structure elements derived from CSI analysis. NOEs between amide protons of consecutive residues and between Hα and the amide proton of subsequent residues are represented by bars connecting the residues. For NN(i, i+1) and αN(i, i+1) NOEs, the thickness of the bar qualitatively represents the relative intensity (weak, medium, or strong) of the NOE. The NOEs for four octarepeats are not shown because they were assigned only to residue types in the repeat region because of overlap. The three unambiguously identified α-helices also are shown.
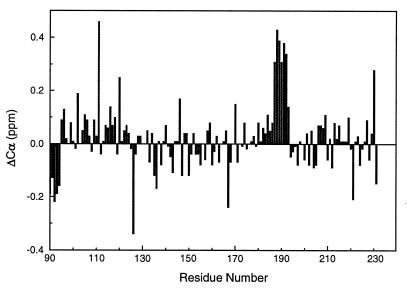
To investigate the possible influence of the additional 61 residues in PrP(29–231) upon the structure of PrP(90–231), a comparison was made of the chemical shift assignments from the two proteins. The Cα chemical shift differences are shown in Fig. 3. In addition to the expected chemical shift difference for the residues at the N terminus of PrP(90–231), residues 187–193 display consistent and significant differences (greater than 0.3 ppm) between PrP(29–231) and PrP(90–231). Amide protons and nitrogens also give the most significant changes in chemical shifts for residues 187–193, with amide nitrogen shift differences greater than 0.2 ppm in this region. Most of the Cα and amide nitrogen chemical shifts show differences less than ± 0.1 ppm between the two proteins.
Figure 3.
Difference in the Cα chemical shift between values measured for PrP(29–231) and PrP(90–231) plotted against residue number.
Backbone Dynamics of PrP(29–231) and PrP(90–231).
Heteronuclear [1H]-15N NOE experiments were carried out to assess the flexibility of PrP(29–231) and PrP(90–231). PrP(29–231) contains 13 prolines and an N-terminal amino group not available for heteronuclear NOE measurements. Amide peaks from 14 residues are severely overlapped, and the intensities could not be reliably measured. In addition, residues L130 and G131 give very weak signals and also were excluded from the NOE measurements. Of the 188 amide peaks, 172 heteronuclear NOEs are included in the analysis. For PrP(90–231), apart from prolines, residues 146, 162, 184, and 189 were excluded because of severe overlap of the peaks.
The heteronuclear NOE data are shown for PrP(29–231) and PrP(90–231) in Fig. 4 A and B, respectively. As can be seen from Fig. 4A, the 500-MHz heteronuclear NOE values for residues 31–124 of PrP(29–231) are all negative, typically −0.5. Amides in the octarepeat region give similar NOE values to those on both sides of the region. The periodicity shown for residues 59–91 arises because the NOE values were approximated by dividing (by 4) the volumes of the degenerate cross peaks corresponding to residues in analogous positions in the four octarepeats. By contrast, strong positive NOEs (typically over 0.65) are observed for residues 134–223. The C-terminal residues 228–231 give very small positive or negative NOEs. In addition, residues 190–197, corresponding to the loop between helix B and helix C have relatively lower positive NOE values. The heteronuclear NOEs obtained at 600 MHz gave comparable magnitudes, and the change in the sign of the NOEs occurs again at residue Gly-124. The linewidth of the amide peaks in the HSQC spectrum for the N-terminal region is about 6 Hz narrower than those in the core region. In addition, residues 129, 130, and 131 give very weak signals in all two-dimensional and three-dimensional experiments, presumably caused by exchange broadening of these amide peaks. The [1H]-15N NOEs for the residues in the structured core region of PrP(90–231) have similar values to those of corresponding residues of PrP(29–231) (Fig. 4). For both proteins, the dividing point for negative and positive NOEs occurs between residues 124 and 125. The heteronuclear NOEs of the loop residues (190–197) also are lowered in PrP(90–231), as in PrP(29–231).
Figure 4.
Summary of heteronuclear [1H]-15N NOE data used to assess the backbone flexibility of the PrP(29–231) and PrP(90–231). (A) The mean values of heteronuclear NOE from two sets of data are plotted against residue number in the full-length SHa PrP(29–231). The error bars represent the difference between the two data sets. The NOEs in four of the octarepeats (residues 59–91) are approximate values estimated by dividing the degenerate peak volumes by four. (B) Heteronuclear NOE values for PrP(90–231). All spectroscopic parameters and processing procedures are the same as those of PrP(29–231).
DISCUSSION
For the identification of secondary structure elements, the Hα-based and 13C-based CSI analyses were used. By using the majority rule (two of three or three of four) (37) and considering that 13Cα chemical shift has the highest predicting power, we unambiguously have identified three α-helices, with helix A spanning residues 144–156, helix B including residues 172–193, and helix C containing residues 200–227. By using a consensus from Cα, CO, and Hα CSI a 92% confidence in prediction has been reported (37). Furthermore, the helical residues as indicated by CSI analysis are almost identical to those derived from medium range [αN(i, i+3) and αN(i, i+4)] NOEs (Fig. 2). The lengths of the three helices derived for SHa PrP(29–231) are similar to those reported for PrP(90–231) (10) but are different from those of MoPrP(121–231) (7, 8). In MoPrP(121–231), the three helices A, B, and C span residues 144–154, 179–193, and 200–217, respectively (7). The differences in the lengths of the three helices presumably are caused by the absence of critical residues in MoPrP(121–231) (10). In addition, the differences in buffer conditions and the more acidic pH for the study of MoPrP(121–231) also may have contributed to the variations in secondary structure.
The chemical shift and NOE-derived helices of PrP(29–231) are very similar to those in the recently determined NMR structure of PrP(90–231) (10). A short antiparallel β-sheet has been reported for MoPrP(121–231) (7) and an irregular β-sheet for SHa PrP(90–231) (10). All of the chemical shift indices suggest a β-strand between residues 160–163 of PrP(29–231). However, the location of the other strand is not clear from the chemical shift data, with the segment between residues 137–140 being the only region where both the Cα and CO chemical shifts suggest a possibility of a β-strand. In the reported NMR structures of MoPrP(121–231) and PrP(90–231), the β-sheet consists of residues 161–164 and 128–131; the presence of this latter strand in PrP(29–231) cannot be inferred from the CSIs. Whether this discrepancy reflects poor predictive power of the CSI index method in this case, where the β-strands are short and some assignments are missing, or whether it shows the presence of small populations of alternative conformations for the prion proteins in the presence of the 29–129 region must await further structural work on PrP(29–231). Residues 137–140 might simply be in an extended conformation that fulfills the geometric characteristics of a β-strand. It has been speculated that the short β-sheet in the MoPrP(121–231) serves as a nucleation site for the conversion of PrPC to PrPSc (7). It is attractive to consider the possibility that a β-strand consisting of residues 137–140 in PrP(29–231) could form part of a β-sheet during the conversion of PrPC to PrPSc. This is consistent with computational studies of PrPSc, in which one of the four plausible β-strands spans residues 138–144 (41).
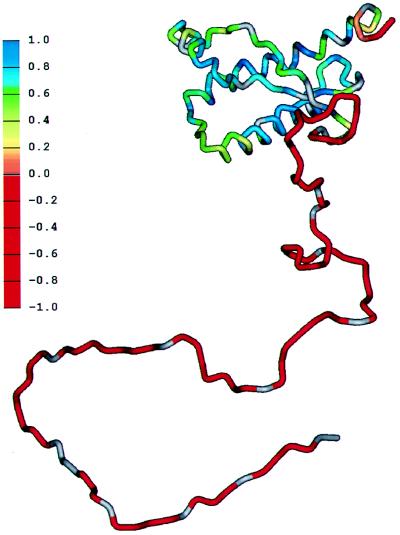
The heteronuclear [1H]-15N NOE is an excellent measure of flexibility in proteins (42). Most of the residues between 130–227 of PrP(29–231) give large positive NOEs, indicating these residues are within the structured core. Residues 190–197, corresponding to the loop between helix B and helix C, have smaller, but still positive, NOE values, indicating these residues are relatively more flexible than those in regular secondary structure. Five residues at the C terminus of both PrP(29–231) and PrP(90–231) are flexible, a property common to the termini of many proteins. The region from 90–124 is clearly flexible in both PrP(29–231) and PrP(90–231), with large negative NOEs and local correlation times less than 1 ns, indicating that these residues do not form part of any stable secondary or tertiary fold and are highly flexible (43). The flexibility of the various parts of the polypeptide chain in PrP(29–231) is shown schematically in Fig. 5. The presence of a folded C-terminal domain of residues 124–231, with disorder in the remainder of the chain, also has been noted in MoPrP(23–231) (16).
Figure 5.
Schematic diagram showing the flexibility of the polypeptide chain for PrP(29–231). The structure of the portion of the protein representing residues 90–231 was taken from the coordinates of PrP(90–231) (10). The remainder of the sequence was hand-built for illustration purposes only. The color scale shows the heteronuclear [1H]-15N NOE data from Fig. 4A, from red for the lowest (most negative) values, where the polypeptide is most flexible, to blue for the highest (most positive) values in the most structured and rigid regions of the protein.
The octarepeat region (five repeats in most species) is a functionally important section of the PrP (9). Addition of two, four, five, six, seven, eight, nine, and 18 octarepeats in humans results in an inherited prion disease (11–13). One additional octarepeat has been reported in some cattle but it does not seem to alter the susceptibility of those animals to bovine spongiform encephalopathy (44, 45). Deletion of one repeat in humans creates a polymorphism that apparently does not cause disease (9). From a structural viewpoint, it is clear that the presence of the 61 additional residues in PrP(29–231), relative to PrP(90–231), does not have a significant effect on the secondary structure elements. In particular, residues 90–124 remain highly flexible in the full-length protein. This is consistent with recent studies of the antigenic structures of PrPC and PrPSc, which document that the structure of the region between 90–124 varies between the cellular and scrapie isoforms whereas the C-terminal region is unaltered (46).
The only major difference between PrP(29–231) and PrP(90–231) is an apparent consistent difference in 13Cα chemical shifts for residues 187–193 (Fig. 3), suggesting that stabilization of helical structure has occurred in helix B in PrP(29–231). This effect may well be caused by transient tertiary interactions between the N-terminal residues containing the octarepeats and helix B in the longer fragment.
Although the function of the octarepeat region remains uncertain, several possibilities can be re-evaluated in light of our structural observations. First, the octarepeats may participate in binding to the membrane or another protein. In either case, the flexibility of this region, as documented by heteronuclear NOE data, would facilitate binding to a partner macromolecule. Alternatively, the octarepeats may be in an apoprotein conformation that will adopt a more organized structure in the presence of a cofactor. Candidates for this cofactor would include Cu(II) (J. Stöckel, F.E.C., and S.B.P., unpublished observations). Finally, in spite of the flexibility of this region, chemical shift data indicate that the 29–125 region stabilizes the conformational ensemble of the helix between residues 187 and 193. This region is important in PrPC/PrPSc recognition but also must serve a normal physiologic function. Although the function of PrP remains unknown, the large unstructured N terminus of PrP(29–231) might provide a clue to its function. Although it is unusual for almost half of any protein to be unstructured, the phenomenon is not unprecedented. Whether the discovery of the unstructured N terminus of full-length PrP will provide insight into the function of PrPC remains to be established, but this highly flexible region probably provides the plasticity required for the conformational transition of PrPC into PrPSc. Studies with recombinant mAbs support this postulate in that epitopes located at the N terminus, but not the C terminus, of PrPSc are buried whereas epitopes throughout native PrPC are accessible (46).
We conclude from an analysis of four relevant NMR studies of PrPC molecules (refs. 7, 10, and 16, and this work) that 29–124 forms a flexible region, and that 125–231 forms a disulfide-bridged three-helix bundle. A small amount of context-dependent β-structure may exist in PrPC, but its relevance is uncertain. The flexible regions are clearly biologically important, if only in the process of conversion from PrPC to PrPSc. It is not clear whether they participate in the normal cellular function of the PrP in the same unstructured state. Comparison of biologically inactive (121–231) and active (90–231) fragments of PrPC highlights the role of the 165–175 loop in PrPSc replication, which may be mediated by another factor, termed protein X (47). The surface of PrPC contains distinct PrPSc and protein X recognition domains that are constructed from spatially proximal, but sequentially distant regions of the polypeptide chain. PrPSc recognizes a surface that includes residues from the regions 90–144 and 180–205, whereas protein X recognizes an antipodal surface that includes residues from the regions 160–180 and 205–231 (48). Thus, the biological and structural data presently available on the PrPs indicate that conversion of PrPC to PrPSc is a complex process that involves context-dependent conformational changes in the N-terminal region of the protein (46). The discovery that this region is largely unstructured in solution is important for our understanding of the conversion process, because the energy barrier to the formation of the β-sheet containing PrPSc will be much lower from an initial “random coil” than from a structure that already contains stable secondary structure.
Acknowledgments
We thank Dr. John Chung for assistance with NMR experiments and Mike Pique for preparing Fig. 5. This work was supported by grants from the National Institutes of Health.
ABBREVIATIONS
- PrP
prion protein
- PrPSc
scrapie PrP isoform
- PrPC
cellular PrP isoform
- Mo
mouse
- SHa
Syrian hamster
- HSQC
heteronuclear single-quantum coherence
- NOE
nuclear Overhauser effect
- NOESY
NOE spectroscopy
- TOCSY
total correlation spectroscopy
- CSI
chemical shift index
References
- 1.Gajdusek D C. Science. 1977;197:943–960. doi: 10.1126/science.142303. [DOI] [PubMed] [Google Scholar]
- 2.Prusiner S B. Science. 1997;278:245–251. doi: 10.1126/science.278.5336.245. [DOI] [PubMed] [Google Scholar]
- 3.Prusiner S B. Science. 1982;216:136–144. doi: 10.1126/science.6801762. [DOI] [PubMed] [Google Scholar]
- 4.Prusiner S B. Science. 1991;252:1515–1522. doi: 10.1126/science.1675487. [DOI] [PubMed] [Google Scholar]
- 5.Pan K-M, Baldwin M, Nguyen J, Gasset M, Serban A, Groth D, Mehlhorn I, Huang Z, Fletterick R J, Cohen F E, Prusiner S B. Proc Natl Acad Sci USA. 1993;90:10962–10966. doi: 10.1073/pnas.90.23.10962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Horwich A L, Weissman J S. Cell. 1997;89:499–510. doi: 10.1016/s0092-8674(00)80232-9. [DOI] [PubMed] [Google Scholar]
- 7.Riek R, Hornemann S, Wider G, Billeter M, Glockshuber R, Wüthrich K. Nature (London) 1996;382:180–182. doi: 10.1038/382180a0. [DOI] [PubMed] [Google Scholar]
- 8.Billeter M, Riek R, Wider G, Hornemann S, Glockshuber R, Wüthrich K. Proc Natl Acad Sci USA. 1997;94:7281–7285. doi: 10.1073/pnas.94.14.7281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bamborough P, Wille H, Telling G C, Yehiely F, Prusiner S B, Cohen F E. Cold Spring Harbor Symp Quant Biol. 1996;61:495–509. [PubMed] [Google Scholar]
- 10.James T L, Liu H, Ulyanov N B, Farr-Jones S, Zhang H, Donne D G, Kaneko K, Groth D, Mehlhorn I, Prusiner S B, Cohen F E. Proc Natl Acad Sci USA. 1997;94:10086–10091. doi: 10.1073/pnas.94.19.10086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goldfarb L G, Brown P, McCombie W R, Goldgaber D, Swergold G D, Wils P R, Cervenakova L, Baron H, Gibbs C J, Jr, Gajdusek D C. Proc Natl Acad Sci USA. 1991;88:10926–10930. doi: 10.1073/pnas.88.23.10926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Poulter M, Baker H F, Frith C D, Leach M, Lofthouse R, Ridley R M, Shah T, Owen F, Collinge J, Brown J, Hardy J, Mullan M J, Harding A E, Bennett C, Doshi R, Crow T J. Brain. 1992;115:675–685. doi: 10.1093/brain/115.3.675. [DOI] [PubMed] [Google Scholar]
- 13.Capellari S, Vital D, Parchi P, Petersen R B, Ferrer X, Jarnier D, Pegoraro E, Gambetti P, Julien J. Neurology. 1997;49:133–141. doi: 10.1212/wnl.49.1.133. [DOI] [PubMed] [Google Scholar]
- 14.Smith C J, Drake A F, Banfield B A, Bloomberg G B, Palmer M S, Clarke A R, Collinge J. FEBS Lett. 1997;405:378–384. doi: 10.1016/s0014-5793(97)00220-2. [DOI] [PubMed] [Google Scholar]
- 15.Kaneko K, Vey M, Scott M, Pilkuhn S, Cohen F E, Prusiner S B. Proc Natl Acad Sci USA. 1997;94:2333–2338. doi: 10.1073/pnas.94.6.2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Riek R, Hornemann S, Wider G, Glockshuber R, Wüthrich K. FEBS Lett. 1997;413:282–288. doi: 10.1016/s0014-5793(97)00920-4. [DOI] [PubMed] [Google Scholar]
- 17.Mehlhorn I, Groth D, Stockel J, Moffat B, Reilly D, Yansura D, Willett W S, Baldwin M, Fletterick R, Cohen F E, Vandlen R, Henner D, Prusiner S B. Biochemistry. 1996;35:5528–5537. doi: 10.1021/bi952965e. [DOI] [PubMed] [Google Scholar]
- 18.Bodenhausen G, Ruben D J. Chem Phys Lett. 1980;69:185–189. [Google Scholar]
- 19.Vuister G W, Bax A. J Magn Reson. 1992;98:428–435. [Google Scholar]
- 20.Grzesiek S, Bax A. J Magn Reson. 1992;96:432–440. [Google Scholar]
- 21.Wittekind M, Mueller L. J Magn Reson. 1993;101:201–205. [Google Scholar]
- 22.Grzesiek S, Bax A. J Am Chem Soc. 1992;114:6291–6293. [Google Scholar]
- 23.Marion D, Driscoll P C, Kay L E, Wingfield P T, Bax A, Gronenborn A M, Clore G M. Biochemistry. 1989;28:6150–6156. doi: 10.1021/bi00441a004. [DOI] [PubMed] [Google Scholar]
- 24.Logan T M, Olejniczak E T, Xu R X, Fesik S W. J Biomol NMR. 1993;3:225–231. doi: 10.1007/BF00178264. [DOI] [PubMed] [Google Scholar]
- 25.Bax A, Clore G M, Gronenborn A M. J Magn Reson. 1990;88:425–431. [Google Scholar]
- 26.Muhandiram D R, Farrow N A, Xu G-Y, Smallcombe S H, Kay L E. J Magn Reson Series B. 1993;102:317–321. [Google Scholar]
- 27.Löhr F, Rüterjans H. J Biomol NMR. 1995;6:189–197. doi: 10.1007/BF00211783. [DOI] [PubMed] [Google Scholar]
- 28.Grzesiek S, Bax A. J Biomol NMR. 1993;3:185–204. doi: 10.1007/BF00178261. [DOI] [PubMed] [Google Scholar]
- 29.Palmer A G, III, Cavanagh J, Wright P E, Rance M. J Magn Reson. 1991;93:151–170. [Google Scholar]
- 30.Kay L E, Keifer P, Saarinen T. J Am Chem Soc. 1992;114:10663–10665. [Google Scholar]
- 31.Grzesiek S, Bax A. J Am Chem Soc. 1993;115:12593–12594. [Google Scholar]
- 32.Farrow N A, Muhandiram R, Singer A U, Pascal S M, Kay C M, Gish G, Shoelson S E, Pawson T, Forman-Kay J D, Kay L E. Biochemistry. 1994;33:5984–6003. doi: 10.1021/bi00185a040. [DOI] [PubMed] [Google Scholar]
- 33.Wishart D S, Bigam C G, Yao J, Abildgaard F, Dyson H J, Oldfield E, Markley J L, Sykes B D. J Biomol NMR. 1995;6:135–140. doi: 10.1007/BF00211777. [DOI] [PubMed] [Google Scholar]
- 34.Yao, J., Dyson, H. J. & Wright, P. E. (1997) FEBS Lett. in press. [DOI] [PubMed]
- 35.Spera S, Bax A. J Am Chem Soc. 1991;113:5490–5492. [Google Scholar]
- 36.Wishart D S, Sykes B D, Richards F M. Biochemistry. 1992;31:1647–1651. doi: 10.1021/bi00121a010. [DOI] [PubMed] [Google Scholar]
- 37.Wishart D S, Sykes B D. J Biomol NMR. 1994;4:171–180. doi: 10.1007/BF00175245. [DOI] [PubMed] [Google Scholar]
- 38.Wishart D S, Bigam C G, Holm A, Hodges R S, Sykes B D. J Biomol NMR. 1995;5:67–81. doi: 10.1007/BF00227471. [DOI] [PubMed] [Google Scholar]
- 39.Merutka G, Dyson H J, Wright P E. J Biomol NMR. 1995;5:14–24. doi: 10.1007/BF00227466. [DOI] [PubMed] [Google Scholar]
- 40.Wüthrich K. NMR of Proteins and Nucleic Acids. New York: Wiley; 1986. [Google Scholar]
- 41.Huang Z, Prusiner S B, Cohen F E. Fold Design. 1995;1:13–19. [PubMed] [Google Scholar]
- 42.Kay L E, Torchia D A, Bax A. Biochemistry. 1989;28:8972–8979. doi: 10.1021/bi00449a003. [DOI] [PubMed] [Google Scholar]
- 43.Fushman D, Cahill S, Cowburn D. J Mol Biol. 1997;266:173–194. doi: 10.1006/jmbi.1996.0771. [DOI] [PubMed] [Google Scholar]
- 44.Goldmann W, Hunter N, Martin T, Dawson M, Hope J. J Gen Virol. 1991;72:201–204. doi: 10.1099/0022-1317-72-1-201. [DOI] [PubMed] [Google Scholar]
- 45.Prusiner S B, Fuzi M, Scott M, Serban D, Serban H, Taraboulos A, Gabriel J M, Wells G A, Wilesmith J W, Bradley R, DeArmond S J, Kristensson K. J Infect Dis. 1993;167:602–613. doi: 10.1093/infdis/167.3.602. [DOI] [PubMed] [Google Scholar]
- 46.Peretz, D., Williamson, R. A., Matsunaga, Y., Serban, H., Pinilla, C., Bastidas, R. B., Rozenshteyn, R., James, T. L., Houghten, R. A., Cohen, F. E., Prusiner, S. B. & Burton, D. R. (1997) J. Mol. Biol., in press. [DOI] [PubMed]
- 47.Telling G C, Scott M, Mastrianni J, Gabizon R, Torchia M, Cohen F E, DeArmond S J, Prusiner S B. Cell. 1995;83:79–90. doi: 10.1016/0092-8674(95)90236-8. [DOI] [PubMed] [Google Scholar]
- 48.Kaneko K, Zulianello L, Scott M, Cooper C M, Wallace A C, James T L, Cohen F E, Prusiner S B. Proc Natl Acad Sci USA. 1997;94:10069–10074. doi: 10.1073/pnas.94.19.10069. [DOI] [PMC free article] [PubMed] [Google Scholar]