Abstract
Amplification of FGFR1 occurs in approximately 10% of breast cancers and is associated with poor prognosis. However, it is uncertain whether over-expression of FGFR1 is causally linked to the poor prognosis of amplified cancers. Here, we show that FGFR1 over-expression is robustly associated with FGFR1 amplification in two independent series of breast cancers. Breast cancer cell lines with FGFR1 over-expression, and amplification, show enhanced ligand dependent signalling, with increased activation of the MAPK and PI3K-AKT signalling pathways in response to FGF2, but also show basal ligand independent signalling, and are dependent on FGFR signalling for anchorage independent growth. FGFR1 amplified cell lines demonstrate resistance to 4-OH-Tamoxifen, that is reversed by siRNA silencing of FGFR1, suggesting that FGFR1 over-expression also promotes endocrine therapy resistance. FGFR1 signalling suppresses progesterone receptor (PR) expression in vitro and likewise amplified cancers are frequently PR negative, identifying a potential biomarker for FGFR1 activity. Furthermore, we show that amplified cancers have a high proliferative rate assessed by Ki67 staining, and that FGFR1 amplification is found in 16-27% of luminal B type breast cancers. Our data suggests that amplification and over-expression of FGFR1 may be a major contributor to poor prognosis in luminal type breast cancers, driving anchorage independent proliferation and endocrine therapy resistance.
Keywords: Breast Cancer, FGFR1, Endocrine therapy
Introduction
Despite substantial improvements in the treatment of breast cancer, resistance to therapy is a major clinical problem. Understanding the mechanisms of resistance, and the identification of novel therapeutic targets is of vital importance if the prognosis of breast cancer is to be further improved. Molecular subtyping of breast cancer has identified distinct subtypes of breast cancer. Cancers that express the oestrogen receptor (ER) divide into two broad categories of luminal A and B types(1), largely depending on whether the tumour has low or high proliferation respectively(2, 3). Although, in general, ER positive tumours are considered to have a good prognosis, highly proliferative, luminal B type tumours have a poor prognosis in patients treated with adjuvant endocrine therapy(4). Resistance to endocrine therapy, whether acquired or intrinsic, is a major factor implicated in the relapse of these breast cancers, and understanding the factors that result in endocrine therapy resistance is important if outcome is to be improved.
A number of recurrent high-level amplifications have been identified in breast cancer(5), and for some of these amplifications the driver gene has been identified, for example HER2 at chromosomal region 17q21 and CCND1 and 11q13. Along with these genes, FGFR1 was one of the first to be shown to be amplified in breast cancer(6), amplified in approximately 10% cancers(5). Amplification of FGFR1 is associated with early relapse, and poor survival, specifically in ER positive breast cancer(7). Amplification of FGFR1 is uncommon in HER2 amplified tumours, suggesting that amplification of FGFR1 and HER2 may be mutually exclusive ways of activating similar downstream pathways(7).
However, amplifications at chromosomal region 8p11-12, the genomic locus of FGFR1, are complex with at least two separate regions, or cores, of amplification(8). Some studies have found that FGFR1 expression correlates with FGFR1 amplification(8-12), but others have not(13-16). Although we have previously demonstrated that the FGFR1 amplified cell line MDA-MB-134 is dependent on FGFR1 for proliferation(17), it is not universally accepted that FGFR1 is a driver of cancers harbouring 8p11-12 amplification.
Here, we have comprehensively evaluated FGFR1 amplified breast cancer, demonstrating that FGFR1 expression is highly correlated with FGFR1 copy number. We show that MDA-MB-134 cells have acquired a KRAS mutation that compromises them as a model cell line, and identified a number of breast cancer cell lines with FGFR1 over-expression and amplification. We show that over-expression of FGFR1 results in both enhanced ligand dependent, and independent, signalling with important consequences for anchorage independent growth and response to endocrine therapy. Finally we provide evidence that FGFR1 amplification is a frequent in proliferative, luminal B subtype ER positive cancers, suggesting that FGFR1 over-expression may be a major factor contributing to the poor prognosis of these tumours.
Materials and Methods
Cell lines, materials and antibodies
Cell lines were obtained from ATCC or Asterand, and maintained in phenol red free DMEM or RPMI with 10% FBS (PAA gold) and 2mM L-glutamine (Sigma-Aldrich, Dorset, UK). S68 was a kind gift of Veronique Catros, Rennes, France. MDA-MB-134 was originally obtained directly from MD Anderson by Mike O' Hare, Ludwig Institute, London. PD173074, 4-OH-Tamoxifen (4-OHT) and ICI-182780 were from Sigma and U0126 Calbiochem (Merck KGaA, Darmstadt, Germany). siRNA were from Dharmacon (Lafayette, CO): FGFR1 siGenome SMARTpool (siFGFR1,M-003131-03), siGenome Non-Targeting siRNA Pool#1 (siCON,D-001206-13), and PLK1 siGenome SMARTpool (siPLK1,M-003290-01). Antibodies used were phosphorylated FRS2-Tyr196 (3864), phosphorylated AKT1-Ser473 (4058), phosphorylated ERK1/2-Thr202/Tyr204 (4370), phosphorylated RSK-Thr359/Ser363 (9344), phosphorylated PLCgamma1-Tyr783 (2821), ERK1/2 (9102), CCND1 (2978), PR (3172) (all Cell Signaling Technology, Danvers, MA), and FGFR1 (sc-121), ER (sc-543), β-ACTIN (sc-1616) (all Santa-Cruz Biotechnology, Santa Cruz, CA).
Tumour samples and Microarray-Based Comparative Genomic Hybridisation,
The Guy's series of 87 oestrogen receptor positive primary breast cancers all treated with adjuvant tamoxifen has been described previously(3). Analysis of FGFR1 CISH and Ki67 was on 3μM thick formalin fixed paraffin embedded (FFPE) tissue sections. The tissue microarray (TMA) series of 245 invasive breast cancers has been described previously(18). RNA was extracted from tumour sections with RNeasy FFPE RNA Isolation Kit (Qiagen, Crawley, UK). RNA was extracted from cell lines using Trizol (Invitrogen, Paisley, UK). DNA was extracted using DNeasy blood and tissue kit (Invitrogen). The 32K BAC re-array collection (CHORI) tiling path aCGH platform was constructed at the Breakthrough Breast Cancer Research Centre, and arrays hybridised and analysed as previously described(19, 20).
Quantitative RT-PCR
cDNA was synthesised from RNA using Superscript III and random hexamers (Invitrogen). Quantitative PCR was performed using Taqman chemistry (Applied Biosystems, Foster City, CA) on the ABI Prism 7900T system (Applied biosystems) using standard curve method. Expression of FGFR1 (Hs00241111_m1), PR (Hs01556701_m1), or CCND1 (Hs00277039_m1) was expressed relative to the mean of three endogenous controls S18 (4310893E), MRPL19 (Hs00608519_m1) and β-ACTIN (4310881E).
Definition of FGFR1 over-expression
Gene expression analysis using Affymetrix U133A Genechips (Affymetrix, Santa Clara, CA) was performed and normalised as previously described(3). The median-weighted mean of five FGFR1 probes (207822_at, 210973_s_at, 211535_s_at, 222164_at, 226705_at) was used to assess FGFR1 expression.
To define FGFR1 over-expression, for both quantitative PCR or Affymetrix data, the standard deviation of the data was estimated from the median absolute deviation. A sample was considered FGFR1 over-expressed when level exceeded 3 standard deviations from the median, identifying samples with outlier over-expression.
Chromogenic in situ hybridisation (CISH) and Immunohistochemistry
FGFR1 CISH was performed on the TMA series with an in-house biotin-labelled probe(21), and on the Guy's series(3) with FGFR1 ZytoDot-SPEC Probe (Zytovision GMbH, Bremerhaven, Germany) and the SPoT-Light CISH Polymer Detection Kit (Invitrogen). FGFR1 signals per cell were counted in 100 tumour nuclei, considered amplified if >50% of the neoplastic cells harboured either >5 copies of the gene or large gene clusters(22). All amplified tumours were also amplified according to criteria outlined in the ASCO/CAP guidelines for HER2 gene amplification. The Ki67 staining was performed using the MIB-1 clone (Dako, code M7240). Antigen retrieval was with 2 minutes pressure cooking in 0.01M citrate pH6. Bound antibody was detected using the Vector ABC kit (Vector laboratories, PK-7200), with DAB as chromogen (K3468, Dako). Both FGFR1 CISH and Ki67 IHC were assessed blinded to FGFR1 expression.
Cell line drug sensitivity and siRNA transfection
All experiments were performed in 10% serum unless stated otherwise. Cell lines were transfected with siRNA (50nM final concentration) in 96 well plates with RNAiMax (Invitrogen). Survival was assessed with Cell Titre-Glo cell viability assay (Promega, Madison, WI). For sensitivity to PD173074, cell lines were plated in 96 well plates, the following day media supplemented with PD173074 at increasing concentrations, and survival assessed after 96 hours exposure. For assessment of endocrine therapy sensitivity cells were maintained in Phenol Red free media, supplemented with 10% charcoal/DCC stripped serum (HyClone, South Logan, UT), 2mM L-glutmaine, and 1nM oestradiol (Sigma), plated in 96 well plates and treated for 6 days with a range of doses of 4-OHT.
FGFR1 stable cell lines
cDNA coding for full length FGFR1-IIIc was cloned into the p-LEX-MCS vector (Open Biosystems, Huntsville, AL). The vector was packaged into Lentivirus in 293-T cells, and T47D cells were infected with p-LEX-MCS (T47D-EV) or p-LEX-FGFR1 (T47D-FGFR1). At 96 hour post infection 1μg/ml puromycin was added, and a polyclonal stable pool established under continuous selection.
Anchorage independent growth
CAL120 cells were seeded in 4% agarose (Sigma) in 6 well plates (5000cells per well), on a base layer of 5% agarose, in 1X RPMI, 10% FBS, and glutamine with or without 1μM PD173074. The top layer was left to set, following which a covering of media with or without 1μM PD173074 was added, and replaced every 3-4 days. After two weeks growth colonies of cells were visualised by light microscopy, followed by staining with crystal violet and counting of colony number.
Western blotting and FACS
Indicated cell lines where grown on 10cm plates, treated as indicated, and lysed in NP40 lysis buffer. Western blots were carried out with precast TA or Bis-Tris gels (Invitrogen) as previously described (23). FACS analysis was performed as described previously (23).
ER-directed transcription
SUM44 were transfected with EREIItkLuc and pCH110 using GeneJuice (EMD Biosciences, Inc., Madison, WI USA), and ERE-luciferase and β-galactosidase assayed after 48 hrs FGF2 treatment, or not, as previously described(24).
Statistical analysis
All statistical analysis was two sided and performed with GraphPab Prism version 5.0.
Results
FGFR1 is robustly over-expressed in FGFR1 amplified tumours
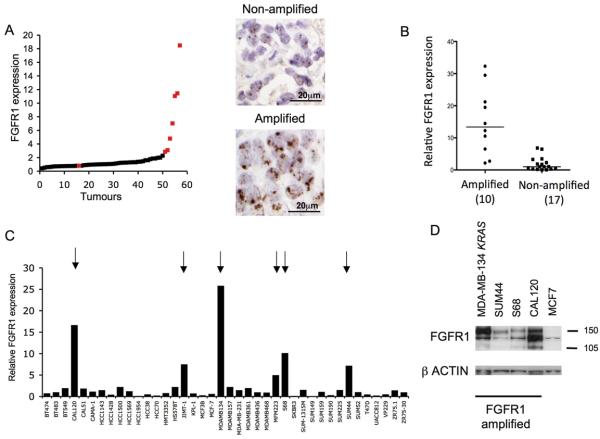
We examined the relationship between FGFR1 amplification and FGFR1 mRNA expression in two independent series of breast cancers. The first series consisted of 87 oestrogen (ER) positive tumours (Guy's series)(3), with FGFR1 copy number assessed in 58 cases by chromogenic in situ hybridisation (CISH). FGFR1 mRNA expression, assessed by gene expression profiling, was substantially higher in amplified tumours compared to non-amplified tumours (5.91 vs 1.0 respectively, p<0.0001 Mann Whitney U Test) (Figure 1A). The second series consisted of invasive breast cancers in a tissue microarray (TMA)(18). FGFR1 amplification was present in 11.8% tumours (11/93 analysable cores). RNA was extracted from corresponding formalin fixed paraffin embedded tissue sections and FGFR1 expression assessed by quantitative PCR, in 10 FGFR1 amplified tumour sections and 17 grade and ER matched controls. FGFR1 was substantially higher in amplified tumours compared to non-amplified controls (Median 13.4 vs 1.0 respectively, p<0.0001 Mann Whitney U Test) (Figure 1B). In both series over-expression of FGFR1 mRNA was only found in cancers with FGFR1 amplification. In the first series 88% (7/8), and the second series 80% (8/10), amplified tumours displayed FGFR1 expression higher than any non-amplified control, validating a tight relationship between amplification and over-expression.
Figure 1. FGFR1 amplified breast tumours and cancer cell lines over-express FGFR1.
A. 58 ER positive breast cancers distributed in order of FGFR1 mRNA level, expressed relative to the median expression level; Red - tumours with FGFR1 amplification assessed by CISH; Black - tumours without FGFR1 amplification. Right panel example photomicrographs from a tumour without and with FGFR1 amplification.
B. FGFR1 amplification status assessed in a second series of 93 invasive breast cancers. FGFR1 gene expression was assessed by quantitative RT-PCR from FGFR1 amplified tumours, and grade and ER matched controls, and expressed relative to the median expression level of controls. FGFR1 amplified tumours had substantially higher median FGFR1 expression than non-amplified controls (13.4 vs 1, p=0.0002 Mann Whitney U test).
C. FGFR1 expression assessed by quantitative RT-PCR in a panel of 40 breast cancer cell lines. Six cell lines over-express FGFR1 (indicated by arrows), all off which have high level FGFR1 amplification (Supplementary Figure 3). FGFR1 expression displayed relative to median expression.
D. Western blot confirming over-expression of FGFR1 protein in cell lines with FGFR1 amplification compared to non-amplified control cell line MCF7.
Identification of FGFR1 amplified cell line models
We had previously shown that the MDA-MB-134 cell line, obtained directly from originating lab in MD Anderson, harbours FGFR1 amplification and over-expression, and was sensitive to FGFR1 inhibitors and silencing of FGFR1 by RNA interference (siRNA)(17). Multiple different siRNA targeting FGFR1 reduced the survival of MDA-MB-134, demonstrating that the effect was on-target (Supplementary Figure 1). However, over time we were unable to propagate the original cell line, and we obtained a new sample from the American Type Culture Collection (ATCC). To our surprise, the re-supplied MDA-MB-134 was no longer sensitive to FGFR1 siRNA (Figure 2A), despite efficient knockdown of FGFR1 (Supplementary Figure 1). We confirmed by both array CGH and gene expression profiling that the re-supplied line was indeed MDA-MB-134 (Supplementary Figure 3). The re-supplied line over-expressed FGFR1 (Figure 1D), but was also found to be heterozygous for a G12R KRAS mutation (Supplementary Figure 1). This mutation has always been present in MDA-MB-134, but originally only at low frequency(25). Blockade of MEK signalling with U0126 in the KRAS mutant MDA-MB-134 restored dependence on FGFR signalling (Supplementary Figure 2), suggesting that the KRAS mutation explained the resistance to FGFR targeting.
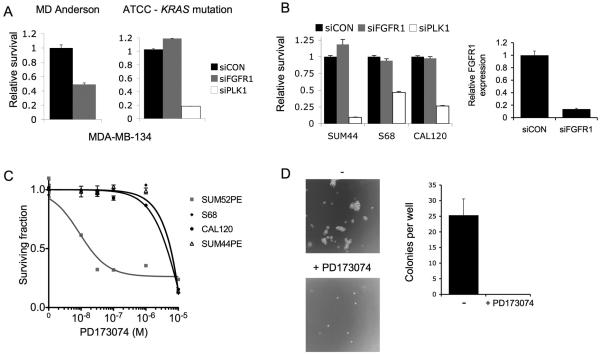
Figure 2. Assessment of the FGFR dependence of FGFR1 amplified cell lines.
A. Sensitivity of MDA-MB-134 cell lines to FGFR1 siRNA (siFGFR1). Cells were transfected with siFGFR1, or siCON non-targeting control, and survival assessed six days later with Cell Titre-Glo® cell viability assay (Promega). MDA-MB-134 obtained directly from MD Anderson were sensitive to FGFR1 knockdown (p<0.001, Student's T Test), but not MDA-MB-134 obtained from ATCC. Error bars SEM of three repeat experiments.
B. Left: Transfection of FGFR1 amplified cell lines with siCON or siFGFR1, and siPLK1 as a positive toxicity control, with survival assessed at 5-7 days transfection. Right: FGFR1 expression by quantitative RT-PCR in SUM44 cells transfected with siFGFR1, or siCON, 72 hours prior to RNA extraction.
C. FGFR1 amplified cell lines were grown for 96 hrs in media supplemented with a range of concentrations of PD173074 pan FGFR tyrosine kinase inhibitor, and survival expressed relative to that of untreated cells. The SUM52PE breast cancer cell line that harbours FGFR2 amplification, and is highly sensitive to FGFR inhibitors, was used a positive control (26).
D. CAL120 cells were grown in soft agar with or without continuous exposure to 1μM PD173074. Example micrographs at 4X power from wells with and without PD173074. Bar chart: mean colonies per well from 3 repeats (without 25.3 colonies vs with PD173074 0 colonies, p=0.008, Student's T test).
With MDA-MB-134 partially compromised as a model of FGFR1 amplification, we set out to identify new cell line models. We screened a panel of 40 breast cancer cell lines by quantitative RT-PCR for FGFR1, and identified six breast cancer cell lines that over-expressed FGFR1 (CAL120, JIMT-1, MDA-MB-134 KRAS, MFM223, S68, and SUM44, Figure 1C). All cell lines that over-expressed FGFR1 harboured high level FGFR1 amplification as assessed by arrayCGH (Supplementary Figure 3). Two cell lines were not investigated further: JIMT-1 due to co-amplification of HER2, and MFM223 due to co-amplification of FGFR2 (Turner et al submitted), either of which might complicate assessment. All amplified cell lines over-expressed FGFR1 protein by western blot (Figure 1D).
FGFR1 amplification is required for anchorage independent growth
We examined the effect of targeting FGFR1 in the newly identified cell line models of FGFR1 amplification. In routine 2D culture, silencing of FGFR1 by siRNA did not effect survival of SUM44, CAL120, or S68 (Figure 2B) despite achieving substantial knockdown of FGFR1 (Figure 2B). The cell lines were also not sensitive to the potent pan-FGFR inhibitor PD173074 in 2D culture, unlike the FGFR2 amplified positive control cell line SUM52PE(26) (Figure 2C). Therefore, we investigated the requirement of FGFR signalling in 3D culture by growing cells in soft agar. This demonstrated that FGFR activity was required for anchorage independent growth of CAL120, and colony formation was completely abolished by PD173074 (Figure 2D). We were unable to assess S68 and SUM44 in this assay, as they did not grow in soft agar (data not shown).
FGFR1 amplification drives both ligand dependent and independent signalling
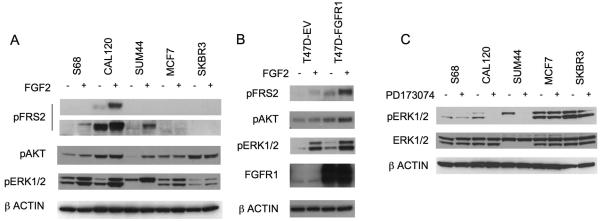
Fibroblast growth factors (FGFs) are present in very low concentrations in normal serum; FGFs bind heparin proteoglycans (HSPGs) avidly and are sequestered in the extracellular matrix and cell surface at the site of production(27). We examined down-stream signalling in response to the addition of relatively low dose FGF2 (1ng/ml) to routine culture media (Figure 3A). Signalling through the FGFR family is reliant on the adapter protein Fibroblast Receptor Substrate 2 alpha (FRS2) to activate MAPK and PI3K-AKT signalling. Phosphorylation of FRS2 in response to FGF2 was only seen in amplified cell lines (Figure 3A). Amplified cell lines displayed substantially enhanced ERK1/2 phosphorylation in comparison with control cell lines, with a minor increase in ERK1/2 phosphorylation in MCF7 and no change in SKBR3. Ribosomal S6 kinase (RSK), a down stream effector of MAPK signalling(28-30), was also phosphorylated in response to FGF2 only in amplified cell lines (Supplementary Figure 4). Phosphorylation of ERK1/2 and AKT was almost exclusively FGF2 dependent in SUM44 (Figure 3A).
Figure 3. FGFR1 amplification drives both ligand dependent and independent signalling.
A. Indicated cell lines growing in 10% serum were treated for 15 minutes prior to lysis with 1ng/ml FGF2 (+), or not (−). Lysates were subject to SDS-PAGE and western blotting with antibodies against phosphorylated-FRS2-Tyr196, phosphorylated-AKT1-Ser473, phosphorylated-ERK1/2-Thr202/Tyr204, and β-ACTIN. Two different exposures of FRS2-Tyr196 are shown.
B. Stable polyclonal pool of T47D cells were established with empty vector (T47D-EV) or FGFR1 expression vector (T47D-FGFR1). Western blots of T47D-EV or T47D-FGFR1 cells treated for 15 minutes prior to lysis with 1ng/ml FGF2, or no treatment (−), and blotted with indicated antibodies.
C. Indicated cell lines were serum starved for 24 hours, and lysates were made after 1hr exposure to 1μM PD173074 (+), or no exposure (−), as indicated. Lysates were subjected to western blotting and blotted with indicated antibodies.
To confirm that over-expression of FGFR1 can enhance downstream signalling in response to ligand, we generated T47D cells that stably over-expressed FGFR1. T47D were infected with a lentiviral FGFR1 expression vector, or control empty vector, and a stable polyclonal pool established. T47D-FGFR1 cells demonstrated both increased FGF2 dependent signalling, with an induction of AKT phosphorylation not seen in control cells, but also an increase in basal-unstimulated signalling (Figure 3B) suggesting that over-expression of FGFR1, to high levels, could induce ligand independent signalling.
We examined whether FGFR1 cancer cell lines demonstrated basal signalling in serum starved conditions. After serum starvation, phosphorylation of ERK1/2 was blocked by PD173074 in both CAL120 and SUM44PE (Figure 3C). To differentiate ligand independent from autocrine mediated signalling, we examined for FGF ligand expression in cell lines. CAL120 expressed both FGF2 mRNA and conditioned media with FGF2 ligand (Supplementary Figure 4). In contrast T47D, SUM44, MDA-MB-134, and other FGFR1 amplified cell lines did not appreciably express any of the FGFR1 ligands (data not shown), indicating that in these cell lines FGFR1 over-expression resulted in low-level basal ligand independent signalling.
FGFR1 amplification drives endocrine therapy resistance
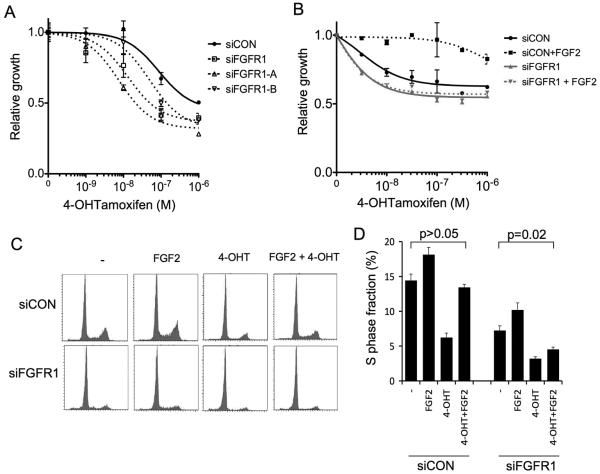
We have previously shown that amplification of FGFR1 is associated with poor prognosis specifically in patients with ER positive breast cancer(7). One potential explanation for this observation could be resistance to endocrine therapies. To investigate this, we examined the effect of silencing of FGFR1 on endocrine therapy sensitivity in the FGFR1 amplified ER positive cell lines. We initially studied the FGFR sensitive MDA-MB-134 sub-line, which was partially resistant to 4-HydroxyTamoxifen (4-OHT) (Figure 4A). Silencing of FGFR1 in this cell line with FGFR1 siRNA SMARTpool, or two individual siRNA, increased 4-OHT sensitivity compared to cells transfected with siCON non-targeting control (Figure 4A). At both 10−7M and 10−6M 4-OHT, siCON transfected cells were less sensitive than siFGFR1, and both individual siRNA, transfected cells (p<0.05 Student's T Test).
Figure 4. FGFR1 drives endocrine therapy resistance in amplified lines.
A. FGFR sensitive MDA-MB-134 cells, obtained from MD Anderson, were transfected with siCON, siFGFR1, or two individual siRNA targeting FGFR1 (siFGFR1-A and siFGFR1-B). Starting at 48hours post transfection, cells were treated with range of concentrations of 4-hyrdoxytamoxifen (4-OHT), and survival assessed at 6 days exposure. Error bars represent SEM of 3 repeat experiments.
B. SUM44 cells were transfected with siCON, or siFGFR1, and 48 hours after transfection treated with range of concentrations of 4-OHT in the presence of 10ng/ml FGF2, or with no FGF2. Survival was assessed after 6 days exposure.
C. Propidium Iodide FACS profiles in SUM44 cells transfected six days earlier with siCON, or siFGFR1, and treated for 72 hours with 10ng/ml FGF2, 10nM 4OH-tamoxifen, the combination, or no treatment (−).
D. Quantification of S phase fraction from three independent experiments. Fraction in S phase; siCON transfected no treatment 14.4% vs FGF2/4OH-T treated 13.4% (p=0.3, Student's T test); siFGFR1 transfected 7.2% vs 4.5% respectively (p=0.02).
SUM44 is an ER positive cell line that is sensitive to 4-OHT, although addition of FGF2 to media abolished sensitivity to 4-OHT (Figure 4B). Silencing of FGFR1 in SUM44 only modestly increased sensitivity to 4-OHT but blocked the ability of FGF2 to cause resistance (Figure 4B), suggesting that the effect of FGF2 was mediated by FGFR1. This observation concurs with the strong ligand dependence of downstream signalling in SUM44 (Figure 3A). FGF2 also induced resistance to the oestrogen receptor antagonist ICI-182780 (data not shown). Similarly, T47D-FGFR1 cells showed greater resistance to 4-OHT and oestrogen deprivation in response to FGF2, in comparison with T47D-EV cells (Supplementary Figure 5).
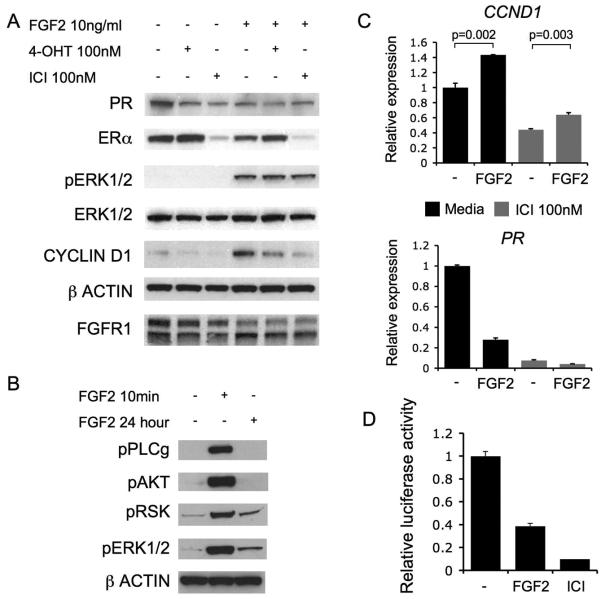
We examined the mechanism of resistance. In SUM44, 4-OHT reduced S phase fraction, but dual treatment with FGF2 and 4-OHT increased S phase fraction to levels comparable with untreated cells (Figure 4C and D). In contrast in FGFR1 siRNA transfected cells, 4-OHT reduced S phase fraction, but FGF2 was unable to restore levels comparable with untreated cells (relative increase FGF2+tamoxifen vs tamoxifen: siCON 120% vs siFGFR1 43% p=0.047, Student's T test). We examined signalling in SUM44 cells treated with 4-OHT and ICI-182780, with or without FGF2 (Figure 5). Treatment with FGF2 alone decreased progesterone receptor (PR) expression, to a comparable level to 4-OHT. In contrast, cyclin D1 (CCND1) was substantially elevated in the presence of FGF2 (Figure 5A), and remained elevated after treatment with 4-OHT. Treatment with ICI-182780 lead to ER degradation and partial loss of CCND1 expression in FGF2 treated cells, although CCND1 remained elevated compared to ICI-182780 treated cells without FGF2 (Figure 5A/B).
Figure 5. Signalling in SUM44 cells is response to endocrine therapies.
A. Western blots of PR, ER, phosphorylated-ERK1/2, ERK1/2, CCND1, β-ACTIN, and FGFR1. SUM44 cell lysates treated for 24 hours prior to lysis with 100nM 4-OHT, 100nM ICI-182780, or no treatment (−), with or without 10ng/ml FGF2. Phosphorylated-AKT1 was not detected.
B. Western blots of phosphorylated-PLCγ1 (Tyr783), phosphorylated-AKT, phosphorylated-p90RSK (Thr359/Ser363), phosphorylated-ERK1/2, and β-ACTIN on SUM44 cell lysates treated for either 10 minutes or 24 hours with 10ng/ml FGF2 prior to lysis.
C. Quantitative RT-PCR analysis of cyclin D1 (CCND1 - Top) and (PR - Bottom) expression in SUM44 cells treated with or without 10ng/ml FGF2 for 24 hours prior to RNA isolation, without (Black bars) or in the presence of 100nM ICI-182780 (Grey bars).
D. SUM44 cells were co-transfected with EREIItkLuc (ERE-luciferase reporter construct) and pCH110 (β-galactosidase reporter construct) and treated for 48 hours with 10ng/ml FGF2, or no treatment, with 100nM ICI-182780 as positive control. Luciferase activity was expressed relative to β-galactosidase activity. Error bars SEM of 3 repeats, p values Student's T-test.
Quantitative PCR assessment of PGR mRNA (PR) confirmed that PR expression was suppressed by FGF2 (Figure 5C). We, therefore, examined the effect of FGF2 on an ERE (estrogen response element) luciferase reporter construct. FGF2 inhibited ER-directed transcription (Figure 5D), confirming that the decrease in PR expression reflected a suppression of ER-dependent transcriptional activity. Both ERK1/2 and RSK phosphorylation was persistently phosphorylated by FGF2 stimulation after 24hr, whereas AKT and phospholipase C phosphorylation was undetectable (Figure 5B). This data suggests that FGFR1 signalling induced endocrine resistance through persistent MAPK activation, which promoted CCND1 expression both in an ER dependent and independent manner to cause resistance to endocrine therapy.
Clinical features of FGFR1 amplified tumours
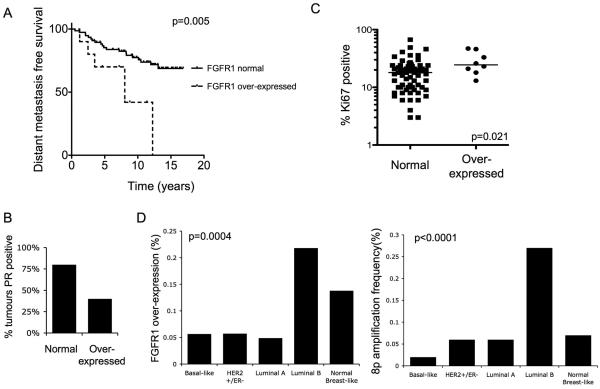
In the Guy's series of 87 ER positive tumours all treated with tamoxifen as sole adjuvant therapy(3), distant metastasis free survival was significantly worse for FGFR1 over-expressing tumours compared to tumours lacking over-expression (Figure 6A). In this series, FGFR1 over-expressing tumours were frequently ER positive but PR negative (PR negative 20% non-amplified vs 60% amplified, p=0.032 Fisher's exact test) (Figure 6B). This concurs with the in vitro observations with SUM44 (Figure 5).
Figure 6. FGFR1 over-expression is common in high risk ER positive breast cancer.
A. Kaplan-Meier curves of distant metastasis free survival from Guys series of ER positive tumours with FGFR1 over-expression (n=10) versus normal FGFR1 expression (76). FGFR1 over-expressing tumours have substantially worse survival (HR 7.4, 95% CI 1.8 to 30.5, p=0.0053, log rank test).
B. Proportion of tumours with progesterone receptor expression in the same cohort (all tumours were ER positive, p=0.03 Fisher's exact test).
C. Ki67 was assessed by IHC in the same cohort. FGFR1 over-expressing tumours have higher Ki67 (P=0.021, Mann Whitney U Test). Of tumours with high proliferation (≥14% Ki67 as a surrogate for luminal B subtype(2)) 16% have FGFR1 over-expression, compared to 3.5% of low proliferation cancers.
D. Left: Incidence of FGFR1 over-expression in breast cancers according to intrinsic subtype (23% Luminal B over-express FGFR1). Right: Incidence of 8p11-12 amplification, defined by co-overexpression of neighbouring genes (Supplementary methods), according to intrinsic subtype (27% Luminal B have 8p11-12 amplification). Analysis of data on 295 invasive breast cancers from van de Vijver et al 2002(31). Statistical comparison across groups was with the Chi squared test.
We examined whether FGFR1 amplification / over-expression was associated with any one specific breast cancer subtype(1). We next interrogated a published gene expression array data set of 295 breast cancers from van de Vijver et al(31). Over-expression of FGFR1 was strongly associated with luminal B type breast cancers (Figure 6D), as was the incidence of 8p11-12 amplification (the genomic locus of FGFR1, Figure 6D). Similarly, in the data set of Chin et al (12), FGFR1 over-expression and high level amplification of FGFR1 was found specifically in ER positive tumours (Supplementary Figure 6). To confirm these findings we assessed Ki67 expression, as a surrogate of proliferative rate, in the Guy's series of ER positive breast cancers. FGFR1 over-expressing cancers had a significantly higher proliferative rate (Figure 6C) with 87.5% (8/9) tumours having high Ki67 (≥14% as defined by Cheang et al 2009(2)). These data suggests that FGFR1 amplification is particularly important in the high proliferative, poor prognosis, luminal B subtype of ER positive breast cancers.
Discussion
In this study we have shown that amplification of FGFR1 promotes anchorage independent proliferation and resistance to endocrine therapies, and this may be reflected in the poor prognosis of amplified cancers. Over-expression of FGFR1 in amplified cell lines results in aberrant ligand dependent signalling, with persistent activation of MAPK signalling and engagement of PI3K-AKT signalling in response to ligand, which is not seen in un-amplified cell lines. In addition, we show that higher levels FGFR1 expression result in basal ligand independent signalling further enhancing down-stream signalling. The amplified cell lines MDA-MB-134 and CAL120 are sensitive to targeting of FGFR1, demonstrating that FGFR1 is a potential therapeutic target in amplified cancers.
Our study adds to the increasing evidence linking aberrant FGF signalling to breast cancer(32). A single nucleotide polymorphism in FGFR2 is associated with an increased risk of ER positive breast cancer(33, 34). Mouse models have demonstrated that expression and constitutive activation of FGFR1 in the mouse mammary epithelium induced proliferation and invasive lesions(35). Activation of the same construct in vitro in murine HC11 cells drove proliferation, survival, and invasion(36), confirming the potential oncogenic nature of FGFR1 signalling. Interestingly, expression of activated FGFR1 in murine prostate induced carcinoma, but a similar FGFR2 construct did not(37), suggesting that FGFR1 may have enhanced oncogenic potential in comparison with FGFR2.
Our data suggest that FGF ligand, potentially in an epithelial-stromal interaction, is important in the promotion of breast cancer progression by FGFR1. FGF2 is expressed at substantially higher levels in breast cancers(38), and in serum and nipple aspirate fluid(39) of patients with breast cancer compared to women without cancer(39, 40). In addition, breast cancer cells express HSPGs on their cell surface that bind and promote the FGFR1-FGF2 interaction(41). A consequence of the low FGF ligand concentration of serum is that FGFR1 amplified cancer cell lines have effectively been derived in conditions of either very low ligand (42). It is plausible, that the lack of dependence on FGFR1 most amplified cell lines display in 2D culture could simply reflect the conditions under which these cell lines were derived.
It is important to emphasise that genes other than FGFR1 in the 8p11-12 amplicon are also likely to contribute to oncogenesis(43-45), potentially acting in collaboration with FGFR1(45). In addition, FGFR1 is commonly co-amplified with CCND1(5), which may cooperate in oncogenesis(45, 46). We have shown a tight relationship between FGFR1 mRNA expression and FGFR1 amplification. In particular, tumours lacking FGFR1 amplification did not over-express FGFR1 to a level comparable with amplified tumours, in contrast to previous data(9). A small fraction of FGFR1 amplified cancers do not appear to over-express FGFR1 mRNA. Although this may constitute a false positive CISH result, or a false negative qPCR result, we consider it more likely that these cancers are driven by other genes within the 8p11-12 amplicon(8).
Our data suggests that FGFR1 amplification drives resistance to endocrine therapy in vitro, and that this observation is reflected in the poor prognosis of FGFR1 over-expression tumours treated with adjuvant tamoxifen (Figure 5). We have also previously studied FGFR1 amplification in a large series of 800 breast cancers(7). In this data set, the ER positive tumours treated tamoxifen as sole adjuvant therapy had an equally poor prognosis (FGFR1 amplified overall survival HR 5.58, 95% CI 1.47 - 21.1). Although the decision to give tamoxifen was unrandomised in both the data sets (Figure 6A), the data does not support there being substantial benefit with adjuvant tamoxifen. The observation that ER positive FGFR1 amplified tumours are more likely to be PR negative than controls (Figure 6) provides further support for the role of FGFR1 in the growth of these tumours; loss of PR expression is thought to reflect activation of growth factor signalling(47) and may provide a biomarker of FGFR1 activity in amplified cancers.
Taken together, we provide strong circumstantial evidence that FGFR1 amplification is one of the major drivers of highly proliferative, poor prognosis, luminal B subtype ER positive breast cancers. This provides a strong rational for the investigation of drugs that target FGFR1 in breast cancer, in particular in combination with endocrine therapy. A number of studies of FGFR tyrosine kinase inhibitors have commenced, or are planned, in breast cancer and the results of these studies are awaited with interest.
Supplementary Material
Acknowledgements
This work was supported by grants from Cancer Research UK and Breakthrough Breast Cancer. Dr Nicholas Turner is a CRUK clinician scientist. We acknowledge NHS funding to the NIHR Biomedical Research Centre. We thank Lesley Ann-Martin for assistance with ERE-luciferase reporter assays, and Jonathon Welti and Andrew Reynolds for assistance with FGF2 ELISA.
References
- 1.Perou CM, Sorlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–52. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 2.Cheang MC, Chia SK, Voduc D, et al. Ki67 index, HER2 status, and prognosis of patients with luminal B breast cancer. J Natl Cancer Inst. 2009;101:736–50. doi: 10.1093/jnci/djp082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Loi S, Haibe-Kains B, Desmedt C, et al. Definition of clinically distinct molecular subtypes in estrogen receptor-positive breast carcinomas through genomic grade. J Clin Oncol. 2007;25:1239–46. doi: 10.1200/JCO.2006.07.1522. [DOI] [PubMed] [Google Scholar]
- 4.Paik S, Shak S, Tang G, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351:2817–26. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 5.Courjal F, Cuny M, Simony-Lafontaine J, et al. Mapping of DNA amplifications at 15 chromosomal localizations in 1875 breast tumors: definition of phenotypic groups. Cancer research. 1997;57:4360–7. [PubMed] [Google Scholar]
- 6.Theillet C, Adelaide J, Louason G, et al. FGFRI and PLAT genes and DNA amplification at 8p12 in breast and ovarian cancers. Genes Chromosomes Cancer. 1993;7:219–26. doi: 10.1002/gcc.2870070407. [DOI] [PubMed] [Google Scholar]
- 7.Elbauomy Elsheikh S, Green AR, Lambros MB, et al. FGFR1 amplification in breast carcinomas: a chromogenic in situ hybridisation analysis. Breast Cancer Res. 2007;9:R23. doi: 10.1186/bcr1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gelsi-Boyer V, Orsetti B, Cervera N, et al. Comprehensive profiling of 8p11-12 amplification in breast cancer. Mol Cancer Res. 2005;3:655–67. doi: 10.1158/1541-7786.MCR-05-0128. [DOI] [PubMed] [Google Scholar]
- 9.Jacquemier J, Adelaide J, Parc P, et al. Expression of the FGFR1 gene in human breast-carcinoma cells. Int J Cancer. 1994;59:373–8. doi: 10.1002/ijc.2910590314. [DOI] [PubMed] [Google Scholar]
- 10.Ugolini F, Adelaide J, Charafe-Jauffret E, et al. Differential expression assay of chromosome arm 8p genes identifies Frizzled-related (FRP1/FRZB) and Fibroblast Growth Factor Receptor 1 (FGFR1) as candidate breast cancer genes. Oncogene. 1999;18:1903–10. doi: 10.1038/sj.onc.1202739. [DOI] [PubMed] [Google Scholar]
- 11.Andre F, Job B, Dessen P, et al. Molecular characterization of breast cancer with high-resolution oligonucleotide comparative genomic hybridization array. Clin Cancer Res. 2009;15:441–51. doi: 10.1158/1078-0432.CCR-08-1791. [DOI] [PubMed] [Google Scholar]
- 12.Chin K, DeVries S, Fridlyand J, et al. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell. 2006;10:529–41. doi: 10.1016/j.ccr.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 13.Ray ME, Yang ZQ, Albertson D, et al. Genomic and expression analysis of the 8p11-12 amplicon in human breast cancer cell lines. Cancer research. 2004;64:40–7. doi: 10.1158/0008-5472.can-03-1022. [DOI] [PubMed] [Google Scholar]
- 14.Garcia MJ, Pole JC, Chin SF, et al. A 1 Mb minimal amplicon at 8p11-12 in breast cancer identifies new candidate oncogenes. Oncogene. 2005;24:5235–45. doi: 10.1038/sj.onc.1208741. [DOI] [PubMed] [Google Scholar]
- 15.Bernard-Pierrot I, Gruel N, Stransky N, et al. Characterization of the recurrent 8p11-12 amplicon identifies PPAPDC1B, a phosphatase protein, as a new therapeutic target in breast cancer. Cancer research. 2008;68:7165–75. doi: 10.1158/0008-5472.CAN-08-1360. [DOI] [PubMed] [Google Scholar]
- 16.Adelaide J, Finetti P, Bekhouche I, et al. Integrated profiling of basal and luminal breast cancers. Cancer research. 2007;67:11565–75. doi: 10.1158/0008-5472.CAN-07-2536. [DOI] [PubMed] [Google Scholar]
- 17.Reis-Filho JS, Simpson PT, Turner NC, et al. FGFR1 emerges as a potential therapeutic target for lobular breast carcinomas. Clin Cancer Res. 2006;12:6652–62. doi: 10.1158/1078-0432.CCR-06-1164. [DOI] [PubMed] [Google Scholar]
- 18.Reis-Filho JS, Steele D, Di Palma S, et al. Distribution and significance of nerve growth factor receptor (NGFR/p75NTR) in normal, benign and malignant breast tissue. Mod Pathol. 2006;19:307–19. doi: 10.1038/modpathol.3800542. [DOI] [PubMed] [Google Scholar]
- 19.Mackay A, Tamber N, Fenwick K, et al. A high-resolution integrated analysis of genetic and expression profiles of breast cancer cell lines. Breast Cancer Res Treat. 2009 doi: 10.1007/s10549-008-0296-7. [DOI] [PubMed] [Google Scholar]
- 20.Natrajan R, Weigelt B, Mackay A, et al. An integrative genomic and transcriptomic analysis reveals molecular pathways and networks regulated by copy number aberrations in basal-like, HER2 and luminal cancers. Breast Cancer Res Treat. 2009 doi: 10.1007/s10549-009-0501-3. epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 21.Marchio C, Iravani M, Natrajan R, et al. Mixed micropapillary-ductal carcinomas of the breast: a genomic and immunohistochemical analysis of morphologically distinct components. J Pathol. 2009;218:301–15. doi: 10.1002/path.2572. [DOI] [PubMed] [Google Scholar]
- 22.Isola J, Tanner M, Forsyth A, Cooke TG, Watters AD, Bartlett JM. Interlaboratory comparison of HER-2 oncogene amplification as detected by chromogenic and fluorescence in situ hybridization. Clin Cancer Res. 2004;10:4793–8. doi: 10.1158/1078-0432.CCR-0428-03. [DOI] [PubMed] [Google Scholar]
- 23.Turner NC, Lord CJ, Iorns E, et al. A synthetic lethal siRNA screen identifying genes mediating sensitivity to a PARP inhibitor. Embo J. 2008;27:1368–77. doi: 10.1038/emboj.2008.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martin LA, Farmer I, Johnston SR, Ali S, Marshall C, Dowsett M. Enhanced estrogen receptor (ER) alpha, ERBB2, and MAPK signal transduction pathways operate during the adaptation of MCF-7 cells to long term estrogen deprivation. J Biol Chem. 2003;278:30458–68. doi: 10.1074/jbc.M305226200. [DOI] [PubMed] [Google Scholar]
- 25.Prosperi MT, Dupre G, Lidereau R, Goubin G. Point mutation at codon 12 of the Kiras gene in a primary breast carcinoma and the MDA-MB-134 human mammary carcinoma cell line. Cancer Lett. 1990;51:169–74. doi: 10.1016/0304-3835(90)90053-z. [DOI] [PubMed] [Google Scholar]
- 26.Tannheimer SL, Rehemtulla A, Ethier SP. Characterization of fibroblast growth factor receptor 2 overexpression in the human breast cancer cell line SUM-52PE. Breast Cancer Res. 2000;2:311–20. doi: 10.1186/bcr73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ori A, Wilkinson MC, Fernig DG. The heparanome and regulation of cell function: structures, functions and challenges. Front Biosci. 2008;13:4309–38. doi: 10.2741/3007. [DOI] [PubMed] [Google Scholar]
- 28.Anjum R, Blenis J. The RSK family of kinases: emerging roles in cellular signalling. Nat Rev Mol Cell Biol. 2008;9:747–58. doi: 10.1038/nrm2509. [DOI] [PubMed] [Google Scholar]
- 29.Xian W, Pappas L, Pandya D, et al. Fibroblast growth factor receptor 1-transformed mammary epithelial cells are dependent on RSK activity for growth and survival. Cancer research. 2009;69:2244–51. doi: 10.1158/0008-5472.CAN-08-3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kang S, Dong S, Gu TL, et al. FGFR3 activates RSK2 to mediate hematopoietic transformation through tyrosine phosphorylation of RSK2 and activation of the MEK/ERK pathway. Cancer Cell. 2007;12:201–14. doi: 10.1016/j.ccr.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van de Vijver MJ, He YD, van't Veer LJ, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 32.Grose R, Dickson C. Fibroblast growth factor signaling in tumorigenesis. Cytokine & growth factor reviews. 2005;16:179–86. doi: 10.1016/j.cytogfr.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 33.Easton DF, Pooley KA, Dunning AM, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–93. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garcia-Closas M, Hall P, Nevanlinna H, et al. Heterogeneity of breast cancer associations with five susceptibility Loci by clinical and pathological characteristics. PLoS Genet. 2008;4:e1000054. doi: 10.1371/journal.pgen.1000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Welm BE, Freeman KW, Chen M, Contreras A, Spencer DM, Rosen JM. Inducible dimerization of FGFR1: development of a mouse model to analyze progressive transformation of the mammary gland. J Cell Biol. 2002;157:703–14. doi: 10.1083/jcb.200107119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xian W, Schwertfeger KL, Vargo-Gogola T, Rosen JM. Pleiotropic effects of FGFR1 on cell proliferation, survival, and migration in a 3D mammary epithelial cell model. J Cell Biol. 2005;171:663–73. doi: 10.1083/jcb.200505098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Freeman KW, Welm BE, Gangula RD, et al. Inducible prostate intraepithelial neoplasia with reversible hyperplasia in conditional FGFR1-expressing mice. Cancer research. 2003;63:8256–63. [PubMed] [Google Scholar]
- 38.Relf M, LeJeune S, Scott PA, et al. Expression of the angiogenic factors vascular endothelial cell growth factor, acidic and basic fibroblast growth factor, tumor growth factor beta-1, platelet-derived endothelial cell growth factor, placenta growth factor, and pleiotrophin in human primary breast cancer and its relation to angiogenesis. Cancer research. 1997;57:963–9. [PubMed] [Google Scholar]
- 39.Hsiung R, Zhu W, Klein G, et al. High basic fibroblast growth factor levels in nipple aspirate fluid are correlated with breast cancer. Cancer J. 2002;8:303–10. doi: 10.1097/00130404-200207000-00006. [DOI] [PubMed] [Google Scholar]
- 40.Takei Y, Kurobe M, Uchida A, Hayashi K. Serum concentrations of basic fibroblast growth factor in breast cancer. Clin Chem. 1994;40:1980–1. [PubMed] [Google Scholar]
- 41.Mundhenke C, Meyer K, Drew S, Friedl A. Heparan sulfate proteoglycans as regulators of fibroblast growth factor-2 receptor binding in breast carcinomas. The American journal of pathology. 2002;160:185–94. doi: 10.1016/S0002-9440(10)64362-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ethier SP, Mahacek ML, Gullick WJ, Frank TS, Weber BL. Differential isolation of normal luminal mammary epithelial cells and breast cancer cells from primary and metastatic sites using selective media. Cancer Res. 1993 Feb 1;53(3):627–635. [PubMed] [Google Scholar]
- 43.Yang ZQ, Streicher KL, Ray ME, Abrams J, Ethier SP. Multiple interacting oncogenes on the 8p11-p12 amplicon in human breast cancer. Cancer research. 2006;66:11632–43. doi: 10.1158/0008-5472.CAN-06-2946. [DOI] [PubMed] [Google Scholar]
- 44.Streicher KL, Yang ZQ, Draghici S, Ethier SP. Transforming function of the LSM1 oncogene in human breast cancers with the 8p11-12 amplicon. Oncogene. 2007;26:2104–14. doi: 10.1038/sj.onc.1210002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kwek SS, Roy R, Zhou H, et al. Co-amplified genes at 8p12 and 11q13 in breast tumors cooperate with two major pathways in oncogenesis. Oncogene. 2009;28:1892–903. doi: 10.1038/onc.2009.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koziczak M, Holbro T, Hynes NE. Blocking of FGFR signaling inhibits breast cancer cell proliferation through downregulation of D-type cyclins. Oncogene. 2004;23:3501–8. doi: 10.1038/sj.onc.1207331. [DOI] [PubMed] [Google Scholar]
- 47.Cui X, Schiff R, Arpino G, Osborne CK, Lee AV. Biology of progesterone receptor loss in breast cancer and its implications for endocrine therapy. J Clin Oncol. 2005;23:7721–35. doi: 10.1200/JCO.2005.09.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.