Abstract
A major fraction of intracellular protein degradation is mediated by the proteasome. Successful degradation of these substrates requires ubiquitination and delivery to the proteasome followed by protein unfolding and disassembly of the multiubiquitin chain. Enzymes, such as Rpn11, dismantle multiubiquitin chains, and mutations can affect proteasome assembly and activity. We report that different rpn11 mutations can affect proteasome interaction with ubiquitinated proteins. Moreover, proteasomes are unstable in rpn11-1 and do not form productive interactions with multiubiquitinated proteins despite high levels in cell extracts. However, increased levels of ubiquitinated proteins were found associated with shuttle factors. In contrast to rpn11-1, proteasomes expressing a catalytically inactive mutant (rpn11AXA) were more stable and bound very high amounts of ubiquitinated substrates. Expression of the carboxyl-terminal domain of Rpn11 partially suppressed the growth and proteasome stability defects of rpn11-1. These results indicate that ubiquitinated substrates are preferentially delivered to intact proteasome.
Keywords: DNA/Repair, Proteases/Proteosomes, Proteases/Ubiqiuitin, Protein/Turnover, Protein Assembly, Rad23, Rpn11, Deubiquitination, Shuttle Factors
Introduction
A major fraction of intracellular protein degradation is mediated by the proteasome, which consists of a catalytic (20 S) complex that is bound to two regulatory (19 S) particles (1). The 20 S catalytic particle is assembled through a series of intermediates that are guided by chaperones (2–5). In contrast, insight into 19 S particle assembly is only now beginning to emerge (6–12). The 19 S regulatory particle consists of two subcomplexes, base and lid, that are linked by Rpn10 (13), a multiubiquitin chain receptor (14). Recent studies suggest that the assembly of the base subcomplex of the 19 S particle occurs in close association with the 20 S catalytic particle (8). Rpn10 is an important substrate receptor in the proteasome, and its activity is functionally linked to shuttle factors that translocate substrates to the proteasome (15, 16), such as Rad23 (17–19). The loss of both Rpn10 and Rad23 (rad23Δ rpn10Δ) severely impairs growth and proteolysis (17).
We examined the delivery of ubiquitinated substrates to the proteasome in rpn11 mutants of Saccharomyces cerevisiae. The temperature-sensitive rpn11-1 (mpr1-1) mutant displays growth, proteolytic, and mitochondrial deficiencies (20). Another mutant (rpn11AXA), which contains engineered changes in the MPR1-PAD1-N terminas (MPN) domain, does not support viability, although certain biochemical properties have been examined in an rpn11-1 mutant (21). In agreement with previous studies (21), we found that rpn11-1 harbors structurally defective proteasomes that showed reduced binding to multiubiquitinated proteins and low peptidase activity. In contrast, proteasomes containing the catalytically inactive rpn11AXA mutant protein are structurally sound and bind very high levels of multiubiquitinated proteins. Expression of the carboxyl terminus of Rpn11 (GST-CRpn11)2 partially suppressed the diverse defects of rpn11-1, consistent with other studies that showed the this domain is required for Rpn11 function (21–23). We found that the high levels of multiubiquitinated proteins in rpn11-1 were bound to shuttle factors. In contrast, multiubiquitinated proteins that accumulated in rpn11AXA were enriched in proteasomes. One interpretation of these results is that shuttle factors transfer ubiquitinated proteins to intact proteasomes, even if they are functionally defective.
EXPERIMENTAL PROCEDURES
Yeast Strains and Plasmids
RPN11, rpn11-1, and rpn11AXA were kindly provided by Drs. Deshaies and Verma (California Institute of Technology, Pasadena, CA). Plasmid DNA for generating an integrated derivative of Pre1-FLAG was provided by Dr. J. Dohmen (University of Cologne). DNA was partially digested with BsmI and transformed into the strains indicated in supplemental Table 1. Plasmid DNA for integrating Rpn1-GFP-HA into the chromosomal locus was provided Dr. C. Enenkel (Humboldt University). (Plasmids are indicated in supplemental Table 2.) A plasmid for integrating FLAG-Rpn7 was prepared by amplifying the gene with its native promoter and cloning into Yiplac22 (24). Integrative transformation was performed after the DNA was digested with MscI. To generate a fusion of the carboxyl terminus of Rpn11 with glutathione S-transferase (GST), we amplified the DNA encoding amino acids 267–306 and cloned it into pCBGST1 (GST-CRpn11). Plasmids expressing Rad23, Ddi1, and Dsk2 with an amino-terminal FLAG epitope were generated by PCR and cloned into Yeplac112. These genes were expressed from a PCUP1 promoter by the addition of 100 μm CuSO4. All the amplified DNAs were verified by sequencing both strands.
Measurement of LLVY-AMC Hydrolysis
Protein lysates (15 μg in 50 μl) were premixed with 200 ng of proteasome inhibitor epoxomicin (Boston Biochem) or an equivalent volume of dimethyl sulfoxide (DMSO) lacking the inhibitor. Proteasome assay buffer (200 μl; 25 mm HEPES, pH 7.5, 0.5 mm EDTA) contained 40 μm LLVY-AMC (Boston Biochem) with or without 0.05% SDS. Reactions were incubated at 30 °C for 1 h, and fluorescence was measured using a Tecan Infinite F200 detector. Similarly, LLVY-AMC hydrolysis by immunopurified proteasomes was performed directly on the FLAG-agarose matrix. The values represent epoxomicin-sensitive measurements that were generated from duplicate assays that were repeated three times.
Growth Assays and Sensitivity to Translation Inhibitors
Yeast cultures were grown in selective medium and normalized to an optical density at A600 of ∼1. Ten-fold serial dilutions were prepared and spotted on YPD agar plates and then incubated at either 23 °C or 37 °C. Aliquots were also spotted onto medium containing 0.8 mm paromomycin or 0.1 mm hygromycin-B (25).
UV Survival Assays
Yeast strains were grown in synthetic medium (∼107 cells/ml), washed with sterile water, and resuspended at A600 of 1. Ten-fold serial dilutions were spread on YPD plates and exposed to 254-nm UV light at 1 J/m2/s. The plates were exposed for 0, 30, 60, and 90 s and incubated in the dark at 30 °C. The number of colonies was counted after 4 days, and the average value of duplicate platings from two experiments was plotted.
Pulse-Chase Measurement of Protein Stability
Protein stability measurements were performed as described previously (26, 27) by metabolically labeling yeast cells with 0.5 mCi of EXPRE35S35S (PerkinElmer Life Sciences). Following incubation for 5 min at 30 °C, the cells were suspended in chase medium containing cycloheximide and unlabeled methionine and cysteine. Aliquots were withdrawn at 0, 10, 30, and 60 min. Equal amounts of [35S]cpm were incubated with antibodies against β-galactosidase and protein A-Sepharose. The gels were treated with salicylic acid, dried, and exposed to x-ray film.
Native PAGE
Measurement of peptidase activity of proteasomes was examined in a native polyacrylamide gel, as described (28). Protein lysates (50 μg) were separated in steps of 3, 4, and 5% polyacrylamide and resolved at 4 °C (125 V × 3 h). The gel was overlaid with buffer containing LLVY-AMC, and the fluorescence signal was captured with a Kodak GelLogic imager. Peptidase activity of the 20 S core was stimulated by 0.05% SDS.
Immunoprecipitation/Immunoblotting
Yeast cells were suspended in buffer A (50 mm HEPES, pH 7.5, 150 mm NaCl, 5 mm EDTA and 1% Triton X-100) containing Complete protease inhibitor (Roche) and lysed by glass bead disruption (Thermo-Savant Fast Prep FP120). Protein extracts were normalized using the Bradford reagent (Bio-Rad) and incubated with FLAG-M2 affinity agarose (Sigma). The bound proteins were released in SDS gel loading buffer, separated in 10 or 12% SDS-Tricine PAGE, and characterized by immunoblotting.
Formaldehyde Cross-linking
Cells were grown in YPD medium at 30 °C. Cell pellets were suspended in 1% formaldehyde for 15 min on ice and then washed four times with 1 ml of phosphate-buffered saline.
Antibodies
Polyclonal anti-Rpn12 and anti-Rpn10 antibodies were provided by Dr. D. Skowyra (St. Louis University). Anti-Pab1 was a gift from Dr. S. Peltz (University of Medicine and Dentistry of New Jersey (UMDNJ)). Antibodies against ubiquitin and FLAG were from Sigma. Monoclonal antibodies against β-galactosidase were from Promega (Madison, WI). Antibodies against GST-tagged Rpt1, Rad23, Rpn11, and Dsk2 were made at Pocono Rabbit Farm and Laboratory, Inc. (Canadensis, PA).
Reagents
Epoxomicin and Suc-LLVY-AMC were purchased from Boston Biochem. Enhanced chemiluminescent (ECL) reagents were from PerkinElmer Life Sciences, and the signals were detected using a Kodak GelLogic 1500 imaging system.
RESULTS
Proteasome Assembly Influences Interaction with Multiubiquitinated Proteins
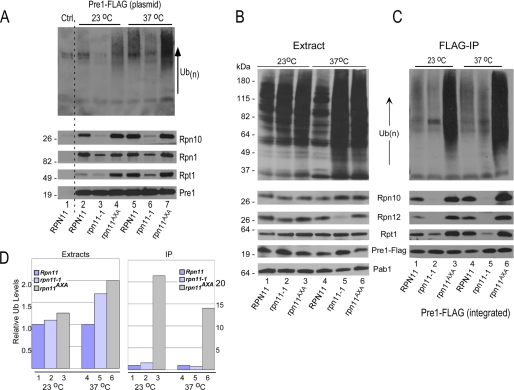
A high copy (2 μm) plasmid expressing Pre1-FLAG, an epitope-tagged subunit in the 20 S catalytic particle, was transformed into yeast strains RPN11, rpn11-1, and rpn11AXA. These strains also contained an integrated, epitope-tagged derivative of Rpn1 (Rpn1-GFP-HA). Yeast strains were grown at the permissive temperature (23 °C), and a part of the culture was transferred to the non-permissive temperature (37 °C) for 4 h. Extracts were prepared, and proteasomes were purified on FLAG-agarose. The precipitated proteins were resolved by SDS-PAGE and examined by immunoblotting (Fig. 1A). Incubation of the filter with anti-ubiquitin antibodies showed that proteasomes purified from rpn11AXA contained high levels of multiubiquitinated proteins at 23 °C (Fig. 1A, lane 4). A further increase in the levels of multiubiquitinated proteins was observed at 37 °C (lane 7), consistent with the growth and proteolytic defects of this mutant. In contrast, the amount of multiubiquitinated proteins that was co-purified with proteasomes from rpn11-1 was noticeably reduced (lanes 3 and 6). The immunoblots were probed sequentially with antibodies against other proteasome subunits, including Rpn10, Rpn1 (anti-HA), and Rpt1. Intriguingly, although similar levels of Pre1-FLAG were recovered from each strain (lower panel, Pre1), the amount of 19 S subunits co-purified from rpn11-1 was significantly lower than from RPN11 and rpn11AXA. Nonspecific precipitation was not detected in a strain lacking Pre1-FLAG (Ctrl, lane 1).
FIGURE 1.
Proteasome assembly affects interaction with multiubiquitinated proteins. A, RPN11, rpn11-1, and rpn11AXA expressing 20 S subunit Pre1-FLAG from a plasmid were grown for 4 h at either 23 °C or 37 °C. An equal amount of protein extract was incubated with FLAG-agarose to purify proteasomes, and the bound proteins were resolved in SDS-10% polyacrylamide gels. Immunoblots were incubated sequentially with antibodies against ubiquitin (Ub) (upper panel), Rpn10, Rpn1, Rpt1, and Pre1-FLAG. B, a gene encoding Pre1-FLAG was integrated into RPN11, rpn11-1, and rpn11AXA and was expressed at physiological levels. Equal amounts of protein extracts were separated by SDS-PAGE and examined with antibodies. A control lane (Ctrl) represents the RPN11 wild-type strain lacking Pre1-FLAG. With the exception of Rpn12, equivalent levels of the other proteasome subunits were detected. C, equal amounts of protein extract were incubated with FLAG-agarose, and the bound proteins were resolved by SDS-PAGE. Immunoblots were treated with the antibodies against the indicated proteins. IP, immunoprecipitation. D, the amount of ubiquitin detected in panels B and C was quantified by densitometry. The levels of ubiquitin in extracts prepared from rpn11-1 and rpn11AXA were compared with RPN11 (where the value is set to 1). The levels of ubiquitin in proteasomes purified from rpn11-1 and rpn11AXA are also compared with RPN11 (where the amount is arbitrarily set to 1).
Our initial studies were performed using yeast strains that overexpressed Pre1-FLAG (Fig. 1A). To avoid the possibility that these findings were affected by the high levels of Pre1-FLAG, we generated integrated derivatives of Pre1-FLAG in RPN11, rpn11-1, and rpn11AXA (Fig. 1, B and C). Protein extracts were prepared from cells grown at 23 and 37 °C, and proteasomes were purified on FLAG-agarose. Total protein extracts were examined by immunoblotting (Fig. 1B), and anti-ubiquitin antibodies showed high levels of multiubiquitinated proteins in both rpn11-1 and rpn11AXA at 23 °C (lanes 2 and 3) and much higher levels at 37 °C (lanes 5 and 6). The immunoblots were probed with antibodies against proteasome subunits (indicated on the right). A control reaction (anti-Pab1) revealed equal protein loading. Despite moderate variability in Pre1-FLAG levels, the overall abundance of other proteasome subunits was similar, with the exception of Rpn12, whose levels were significantly reduced in rpn11-1 at 37 °C (lane 5).
Protein extracts were applied to FLAG-agarose to purify proteasomes (Fig. 1C). Very low amounts of multiubiquitinated species were isolated in association with proteasomes purified from rpn11-1 (lanes 2 and 5), despite the high levels that were detected in total extracts (Fig. 1B). This is evident when Fig. 1B, lanes 4 and 5, are compared with Fig. 1C, lanes 4 and 5. Rpn10 and Rpn12 were not co-purified with proteasomes from rpn11-1 at either 23 °C (lane 2) or 37 °C (lane 5). Similarly, the ATPase subunit Rpt1 was inefficiently purified with proteasomes at 23 °C (lane 2) and 37 °C (lane 5). These results reveal a faulty interaction between the 20 S particle and components of the 19 S particle in rpn11-1. The high levels of multiubiquitinated proteins that were purified with proteasomes in rpn11AXA (lanes 3 and 6) could reflect higher levels in total extract (Fig. 1B, lanes 3 and 6), as well as more stable proteasomes (Fig. 1C, lanes 3 and 6). Equivalent amount of Pre1-FLAG was isolated from each strain, indicating that the results are not caused by variable abundance of the 20 S particle.
The levels of high molecular weight multiubiquitinated proteins shown in Fig. 1, B and C, were quantified (Fig. 1D). The densitometry results do not fully reflect the magnitude of difference in total multiubiquitin levels in rpn11-1 and rpn11AXA due to saturation of the signal in Fig. 1B. Nonetheless, the quantified data clearly show that the amount of ubiquitinated proteins that were co-purified with proteasomes from rpn11AXA was ∼20-fold higher than in rpn11-1.
Proteasomes Purified with a Lid Subunit Confirmed an Assembly Defect in rpn11-1
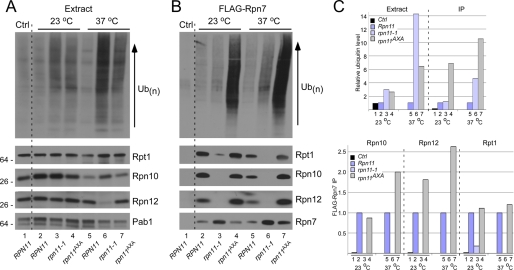
The results in Fig. 1 suggested that the association between 19 S and the 20 S particles is significantly reduced in rpn11-1. To corroborate these findings, we integrated a lid subunit, FLAG-Rpn7, in RPNP11, rpn11-1, and rpn11AXA. Protein extracts were prepared from cultures grown at 23 and 37 °C (for 4 h) and examined by immunoblotting. A control strain lacking FLAG-Rpn7, and the same strain containing integrated FLAG-Rpn7, showed low levels of multiubiquitinated proteins in total extracts (Fig. 2A, lanes 1 and 2). The levels of Rpt1, Rpn10, and Rpn12 were also similar in these two strains, demonstrating that the expression of FLAG-Rpn7 had no adverse effects on proteasome abundance. High levels of multiubiquitinated proteins were detected in total extracts prepared from rpn11-1 and rpn11AXA at both 23 °C (lanes 3 and 4) and 37 °C (lanes 6 and 7). As noted earlier, the abundance of Rpn12 was significantly reduced in rpn11-1 at 37 °C (Fig. 2A, lane 6). Pab1 levels provide a loading control (bottom panel).
FIGURE 2.
Proteasomes purified with a lid subunit show assembly defects in rpn11-1. A, a FLAG epitope-tagged derivative of Rpn7 was inserted into the chromosomal locus in RPN11, rpn11-1, and rpn11AXA and expressed at physiological levels. Total protein extracts were prepared from strains grown at 23 and 37 °C and examined by immunoblotting. Lane 1 contains extract prepared from a wild-type strain lacking a FLAG-tagged protein (Ctrl). The level of Rpn12 is lower in rpn11-1 at 37 °C (lane 6). The reaction against Pab1 represents a protein loading control. Ub, ubiquitin. B, FLAG-Rpn7 was immunoprecipitated on FLAG-agarose, and the bound proteins were examined by immunoblotting with antibodies against the proteins indicted on the right. The control lane (Ctrl, lane 1) shows no reaction against any of the proteasome subunits. The expression of FLAG-Rpn7 is shown in the bottom panel. Despite the recovery of high levels of FLAG-Rpn7 from rpn11-1 at 37 °C, 19 S subunits were not co-purified (lane 6). C, the levels of ubiquitin cross-reacting material in panels A and B were quantified by densitometry (upper panel). Ubiquitin levels in lane 1 (Ctrl) were set to an arbitrary value of 1. The lower panel shows the quantified levels of proteasome subunits Rpn10, Rpn12, and Rpt1 from panel B. For each protein, the signal in the wild-type strain is set to 1. IP, immunoprecipitation.
FLAG-Rpn7 was immunoprecipitated to investigate proteasome composition (Fig. 2B). Large amounts of multiubiquitinated proteins were co-purified with FLAG-Rpn7 from rpn11AXA at both 23 °C (lane 4) and 37 °C (lane 7). However, the amount of Rpt1, Rpn10, and Rpn12 that was co-purified with FLAG-Rpn7 from RPN11 and rpn11AXA was similar despite the strikingly different amounts of associated multiubiquitinated proteins (compare lanes 2 and 4). Although high amounts of FLAG-Rpn7 were immunoprecipitated from rpn11-1 (Fig. 2B, lane 3 and 6, bottom panel), very low levels of Rpt1, Rpn10, Rpn12, and multiubiquitinated proteins were recovered.
The levels of ubiquitinated species were quantified by densitometry (Fig. 2C). Wild-type yeast, either lacking or expressing integrated FLAG-Rpn7, contained equivalent amounts of ubiquitin cross-reacting material at 23 °C (Extract, lanes 1 and 2) and 37 °C (lane 5). However, high levels were measured in rpn11-1 and rpn11AXA. The amount of ubiquitinated proteins that was immunoprecipitated with FLAG-Rpn7 from rpn11AXA was significantly increased at both 23 °C (IP, lane 4) and 37 °C (lane 7). The co-purification of Rpn10, Rpn12, and Rpt1 with FLAG-Rpn7 was also quantified (Fig. 2C, lower panel).
Proteasome Stability Is Reduced in rpn11-1
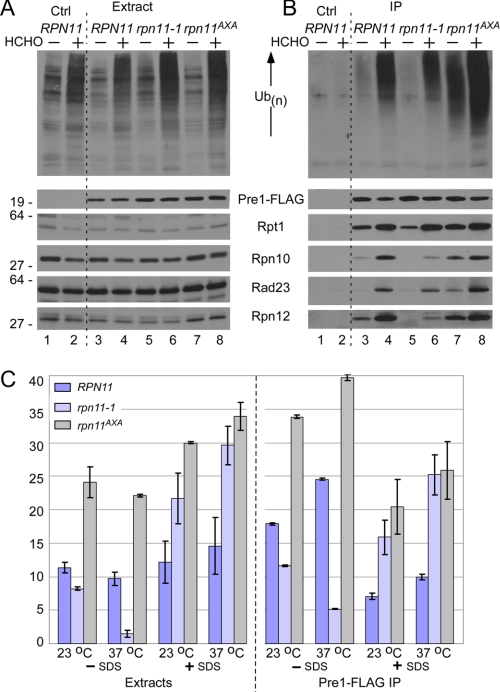
The failure to purify 19 S subunits from rpn11-1, using Pre1-FLAG (Fig. 1C) or FLAG-Rpn7 (Fig. 2B), suggested that proteasomes might be unstable in this mutant. Because proteasomes are essential for viability, the apparent dissociation is likely to occur during purification. A similar instability of 19 S particle was previously detected in rpn10Δ (13). To test this idea, yeast cells were grown at 23 °C and treated with formaldehyde to cross-link proteasome subunits and associated factors (Fig. 3). Total protein extracts were prepared from RPN11, rpn11-1, and rpn11AXA containing integrated Pre1-FLAG, as well as a control strain lacking Pre1-FLAG (Ctrl, lanes 1 and 2). Extracts were prepared from untreated and formaldehyde-treated cells and examined by immunoblotting. The levels of proteasome subunits (Rpt1, Rpn12, and Rpn10), and a shuttle factor (Rad23) were unchanged upon formaldehyde treatment (Fig. 3A). Although Pre1-FLAG was expressed at equivalent levels (Fig. 3A, lanes 3–8), multiubiquitinated proteins were detected at significantly higher levels in the formaldehyde-treated cell extracts (upper panel). The levels detected in rpn11-1 and rpn11AXA were higher than in RPN11 (lanes 6 and 8).
FIGURE 3.
Proteasome stability is reduced in rpn11-1. A, actively growing RPN11, rpn11-1, and rpn11AXA yeast strains expressing Pre1-FLAG at physiological levels were suspended in medium containing 1% formaldehyde (HCHO) on ice. Protein extracts were prepared from untreated and treated cells and resolved by SDS-PAGE. An immunoblot was incubated with antibodies against the proteins indicated on the right. A strain lacking Pre1-FLAG is shown in lanes 1 and 2. A control lane (Ctrl) represents the RPN11 wild-type strain lacking Pre1-FLAG. B, protein extracts were incubated with FLAG-agarose to immunoprecipitate (IP) Pre1-FLAG and proteasome-associated Rpt1, Rpn10, Rpn12, and the shuttle factor Rad23. The upper panel shows the levels of multiubiquitinated proteins that were recovered in each strain. No proteasome subunits were isolated from a strain lacking Pre1-FLAG (lanes 1 and 2). Ub, ubiquitin. C, chymotryptic peptidase activity of proteasomes isolated from RPN11, rpn11-1, and rpn11AXA was measured using the fluorogenic substrate LLVY-AMC. Protein extracts were prepared from cells grown at either 23 °C or 37 °C and measured in the presence or absence of 0.05% SDS. Epoxomicin-sensitive values were determined in duplicate. The right panel represents activity that was co-precipitated with Pre1-FLAG. Error bars represent S.D.
Pre1-FLAG was immunoprecipitated from untreated and formaldehyde-treated cells, and the co-purified proteins were examined by immunoblotting (Fig. 3B). Nonspecific precipitation of proteasome subunits or multiubiquitinated proteins was not observed in the control strain (Ctrl, lanes 1 and 2), demonstrating the effectiveness of the procedure and the specificity of the antibody reaction. Similar amounts of Pre1-FLAG were recovered from all strains, in both treated and untreated preparations. High levels of multiubiquitinated proteins were co-purified with proteasomes from wild-type cells following formaldehyde treatment (compare lanes 3 and 4). High levels of multiubiquitinated proteins were purified with proteasomes from rpn11AXA, both in the presence and in the absence of cross-linking (lanes 8 and 7). The co-purification of Rpn10, Rpn12, and Rad23 with proteasomes from rpn11AXA was similarly elevated. Significantly, chemical cross-linking allowed us to detect proteasome subunits and ubiquitinated proteins in proteasomes isolated from rpn11-1 (compare lanes 5 and 6). This finding is consistent with the idea that proteasomes are unstable in rpn11-1.
The base subcomplex of the 19 S particle can regulate protein degradation by controlling the translocation of substrates into the 20 S particle (29–32). However, the peptidase activity of the free 20 S particle can also be stimulated by low concentration of detergent. We measured peptidase activity to determine whether the altered stability of proteasomes in rpn11-1 affected its catalytic function. Protein extracts were prepared from RPN11, rpn11-1, and rpn11AXA and incubated with LLVY-AMC to measure chymotryptic activity (Fig. 3C). Although proteasomes are equally abundant in RPN11 and rpn11AXA (Fig. 3A), peptidase activity was noticeably higher in rpn11AXA at both 23 °C and 37 °C. The chymotryptic activity in rpn11-1 was similar to the wild type at 23 °C but was significantly reduced at 37 °C. We speculate that high temperature might further destabilize proteasomes in rpn11-1, leading to reduced hydrolysis of LLVY-AMC. We measured proteasome activity in rpn11-1 extracts after the addition of SDS and detected ∼4- and ∼15-fold higher levels at 23 and 37 °C. We propose that proteasome dissociation in rpn11-1 might result in the accumulation of 20 S particles. SDS-stimulated activity was also higher in rpn11AXA than in RPN11, demonstrating that both mutants retained robust hydrolytic activity.
Protein extracts were incubated with FLAG-agarose, and peptidase activity that was purified with Pre1-FLAG was measured. In the absence of SDS, proteasomes purified from rpn11-1 showed slightly lower peptidase activity than in wild-type proteasomes (at 23 °C). However, peptidase activity was markedly reduced at 37 °C, consistent with the increased instability of proteasomes in rpn11-1 at higher temperature. Proteasomes purified from rpn11AXA showed high chymotryptic activity at both 23 °C and 37 °C. At 37 °C, peptidase activity of rpn11AXA proteasomes was almost 8-fold higher than rpn11-1 proteasomes. The addition of SDS resulted in higher activity in both rpn11-1 and rpn11AXA proteasomes when compared with RPN11. However, an overall decrease in LLVY-AMC hydrolysis (when compared with extract) could be due to an inhibitory effect of SDS on immobilized proteasomes (see Fig. 7C). Nonetheless, the pattern of peptidase activities in the three strains, in the presence or absence of SDS, is similar in extracts and purified proteasomes.
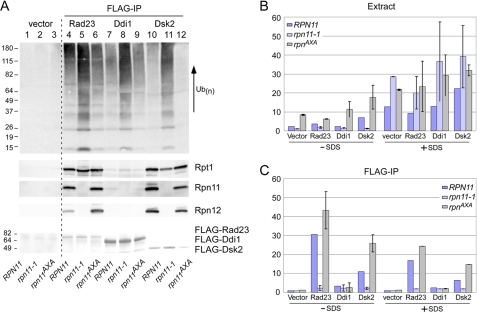
FIGURE 7.
High levels of multiubiquitinated proteins are bound to shuttle factors in rpn11-1. A, FLAG-tagged Rad23, Ddi1, and Dsk2 were expressed in RPN11, rpn11-1, and rpn11AXA. A wild-type strain lacking a FLAG-tagged protein was also examined (vector, lanes 1–3). Protein extracts were incubated with FLAG-agarose, and the levels of multiubiquitinated proteins and proteasome subunits (Rpt1, Rpn11, and Rpn12) that were purified with the shuttle factors were determined. The lower panel shows the levels of the shuttle factors by Ponceau S staining. The levels of multiubiquitinated proteins and proteasome subunits Rpt1, Rpn11, and Rpn12 are shown. IP, immunoprecipitation. B, chymotryptic activity was measured in extracts, in the presence and absence of SDS. In these assays, the extracts were prepared from yeast cells grown at 30 °C to permit semipermissive growth for rpn11-1 and rpn11AXA. C, the same extracts were incubated with FLAG-agarose, and peptidase activity co-precipitated with the shuttle factors was determined. (The assays were performed in duplicate.) Error bars in panels B and C represent S.D.
Autonomous Expression of Carboxyl-terminal Residues Can Partially Suppress the Defects of rpn11-1
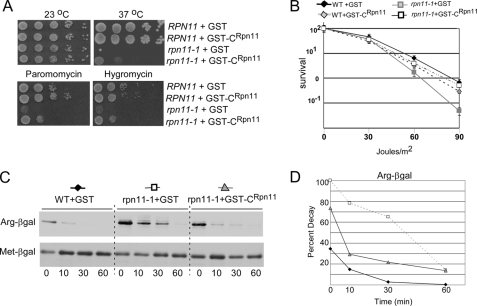
The defects of rpn11-1 are caused by the loss of 31 residues from the carboxyl terminus. We investigated whether the expression of this domain in trans could alleviate the growth and proteolytic defects of rpn11-1. A recent study showed that the C-terminal one-third of Rpn11 could overcome the mitochondrial and DNA repair defects of rpn11-1 (22) but not its proteolytic deficiency. We expressed the carboxyl-terminal 39 residues of rpn11-1 (GST-CRpn11) in RPN11 and rpn11-1 (Fig. 4). GST-CRpn11 did not cause any adverse effects in the wild-type strain. Ten-fold dilutions were spotted onto agar-containing medium and incubated at either 23 °C or 37 °C. The temperature-sensitive growth defect of rpn11-1 was partially suppressed by GST-CRpn11 but not by GST (Fig. 4A). GST-CRpn11 also enabled growth at the semipermissive temperature (30 °C) on medium containing the translational inhibitors paromomycin and hygromycin-B. Both drugs generate high levels of damaged proteins and provide a rapid way to examine proteolytic proficiency (25). The partial suppression of the growth defect of rpn11-1 by GST-CRpn11 was accompanied by a moderate recovery of proteasome function (see Fig. 5). The UV sensitivity of rpn11-1 was restored to wild-type levels by GST-CRpn11 (Fig. 4B), consistent with a recent report that showed that the carboxyl-terminal 100 amino acid residues suppressed the methyl methane sulfonate sensitivity of rpn11-1 (23). To measure proteolytic activity, we transformed RPN11 and rpn11-1 with plasmids encoding the proteolytic substrate Arg-β-galactosidase or a control protein Met-β-galactosidase (26). We also expressed GST in RPN11 and either GST or GST-CRpn11 in rpn11-1. GST had no effect on the stability of Arg-β-galactosidase in the wild-type strain (RPN11). Arg-β-galactosidase was stabilized in rpn11-1 containing GST (Fig. 4C, middle panel), but degradation was partially restored by expressing GST-CRpn11. The half-life of Arg-β-galactosidase was determined to be <5 min in the wild-type strain and ∼30 min in rpn11-1. Expression of GST-CRpn11 reduced the stability of Arg-β-galactosidase to ∼10 min.
FIGURE 4.
Autonomous expression of carboxyl-terminal residues of Rpn11 can partially suppress the defects of rpn11-1. A, the carboxyl terminus of Rpn11 was expressed as a fusion with glutathione S-transferase (GST-CRpn11) in RPN11 and rpn11-1. Ten-fold dilutions of yeast cells expressing either GST or GST-CRpn11 were spotted on agar medium, and suppression of rpn11-1 phenotypes was determined. B, the UV sensitivity of RPN11 and rpn11-1, containing either GST or GST-CRpn11, was measured. Exponential phase yeast cells were diluted and plated on agar medium and exposed to 254-nm UV light at a fluence of 1 J/m2/s. Survival was determined following 4 days of incubation at 23 °C in the dark. The figure is representative of two independent experiments. Error bars represent S.D. WT, wild type. C, RPN11 expressing GST and rpn11-1 expressing either GST or GST-CRpn11 were transformed with plasmids expressing the engineered substrate Arg-β-galactosidase (Arg-βgal) or the stable protein Met-β-galactosidase (Met-βgal). In vivo stability of these proteins was measured in a [35S]methionine pulse-chase experiment. Radiolabeled proteins were immunoprecipitated and resolved by SDS-PAGE, and the autoradiographic data were quantified (panel D).
FIGURE 5.
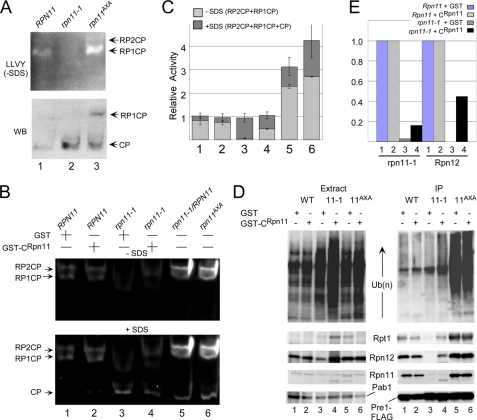
The carboxyl terminus of Rpn11 increases the levels of intact proteasomes. A, we used a fluorogenic assay to measures the cleavage of LLVY-AMC in wild type, rpn11-1, and rpn11AXA. Proteasomes appear unstable or poorly assembled in rpn11-1, as judged by the high levels of free catalytic 20 S particles and low levels of intact proteasomes (upper panel, lane 2). Proteins resolved in the native gel were transferred to nitrocellulose and then probed with antibodies against the 20 S subunit Pre1-FLAG (lower panel, WB). B, to determine the mechanism of suppression of rpn11-1 by GST-CRpn11, peptidase activity was measured in RPN11 and rpn11-1 extracts containing either GST or GST-CRpn11. The level of intact proteasomes was partially restored in rpn11-1 expressing GST-CRpn11 but not GST (lane 4). Expression of the full-length FLAG-Rpn11 protein in rpn11-1 resulted in complete recovery of proteasome assembly and activity (lane 5, FLAG-Rpn11). Total chymotryptic activity was also measured in rpn11AXA, and high peptidase activity and increased levels of intact proteasomes were observed. The positions of 20 S particle bound to one or two regulatory particles (RP1CP and RP2CP) are indicated. Activation of the free 20 S particle by SDS is shown in the lower panel (CP). C, the fluorescent signals in panel B were quantified. The lightly shaded bar represents the activity of proteasomes in the absence of SDS, whereas the addition of the dark shaded region represents the total chymotryptic activity that was detected upon the addition of SDS. Error bars represent S.D. D, the suppression of rpn11-1 defects by GST-CRpn11 is associated with the partial restoration of intact proteasomes. Pre1-FLAG was immunoprecipitated, and the expression of 19 S proteasome subunits in total extract is shown in the left panel. Proteasomes were purified on FLAG-agarose, and the 19 S subunits Rpt1, Rpn12, and Rpn11 were detected in proteasomes isolated from rpn11-1 (right panel, compare lanes 3 and 4). WT, wild type. E, the effect of GST-CRpn11 on proteasome assembly in rpn11-1 was quantified. The co-precipitation of native rpn11-1 and Rpn12 proteins with Pre1-FLAG was compared in wild-type and rpn11-1 cells expressing either GST or GST-CRpn11 (shown in panel D, lanes 1–4).
The Carboxyl Terminus of Rpn11 Promotes Proteasome Stability
The suppression of rpn11-1 defects by GST-CRpn11 could be due to increased stability of proteasomes in this mutant. We therefore examined the integrity of proteasomes in RPN11, rpn11-1, and rpn11AXA using a well described in-gel activity assay. Protein extracts were resolved in a native gel in the presence of ATP and incubated in buffer containing LLVY-AMC (Fig. 5). The hydrolysis of this substrate releases a localized fluorescent signal that can provide qualitative data on the levels of intact proteasomes, as well as assembly intermediates. Proteasomes isolated from rpn11-1 were primarily dissociated or present in altered structural form (Fig. 5A, lane 2) (21, 23). An immunoblot probing for Pre1-FLAG showed higher levels of the free 20 S catalytic particle (CP) in rpn11-1 (lower panel). The peptidase activity in rpn11AXA was detected primarily in the slow migrating form, representing a 20 S complex bound to one (RP1CP) or two regulatory particles (RP2CP), verifying the increased abundance of the intact proteasomes in this mutant.
To determine whether the partial suppression of the growth and proteolytic defects of rpn11-1 by GST-CRpn11 was due to improved proteasome assembly, we examined its integrity using an in-gel assay. Wild-type (RPN11) proteasomes were detected predominantly as RP1CP and RP2CP (Fig. 5B) in the presence of either GST or GST-CRpn11 (lanes 1 and 2). The expression of GST-CRpn11 in rpn11-1 noticeably improved the level of intact proteasomes (lane 4), although it was not restored to wild-type levels. Proteasomes in rpn11AXA were highly active (lane 6), and the expression of GST-CRpn11 had no effect (data not shown). Interestingly, overexpressing full-length FLAG-Rpn11 in rpn11-1 yielded peptidase activity that was higher than in RPN11 (compare lanes 1 and 5). The signals detected in Fig. 5B were quantified using data from two independent studies (Fig. 5C). The lightly shaded part of the column represents activity detected in the absence of SDS, whereas the SDS-induced activity is represented by the dark shaded part of the column. The very low level of proteasome activity in rpn11-1, in the absence of SDS, is clearly evident.
The effect of the GST-CRpn11 on proteasome stability was also investigated by immunoprecipitating Pre1-FLAG (Fig. 5D). The overall levels of multiubiquitinated proteins remained elevated in rpn11-1 expressing either GST or GST-CRpn11 (see lanes 3 and 4). However, in the presence of GST-CRpn11, low levels of multiubiquitinated proteins were detected in proteasomes purified from rpn11-1 (IP, right panel, compare lanes 3 and 4). GST-CRpn11 did not have a discernible effect in rpn11AXA (compare lanes 5 and 6). The filter was also incubated with antibodies against Rpt1, Rpn12, and Rpn11, and low levels were detected in proteasomes purified from rpn11-1 expressing GST-CRpn11 but not GST (Fig. 5D, IP, compare lanes 3 and 4). Intriguingly, the truncated rpn11-1 protein was detected in proteasomes purified from rpn11-1 (lane 4). These data were quantified by densitometry (Fig. 5E), and the improved stability of proteasomes was demonstrated by the recovery of Rpn12 with Pre1-FLAG from rpn11-1 only in cells expressing GST-CRpn11.
Substrate Shuttle Factors Show Differential Binding to Proteasomes in rpn11-1 and rpn11AXA
Although high levels of multiubiquitinated proteins were present in rpn11-1 and rpn11AXA, the distinct stabilities of proteasomes in these mutants resulted in dissimilar interaction with ubiquitinated proteins. Specifically, proteasomes purified from rpn11AXA contained very high levels of multiubiquitinated substrates (Fig. 1), whereas reduced levels were recovered in rpn11-1. Because shuttle factors play a key role in the delivery of proteolytic substrates to the proteasome (33), we examined their interaction with substrates and the proteasome in rpn11-1 and rpn11AXA. The best described shuttle factors resemble Rad23, which contains an amino-terminal ubiquitin-like (UbL) domain that interacts with the proteasome (34) and ubiquitin-associated (UBA) domains that bind multiubiquitin chains (35, 36). These features enable UbL-UBA shuttle factors to deliver substrates through transient interaction with Rpn1 in the base of the proteasome (38) followed by release and recycling.
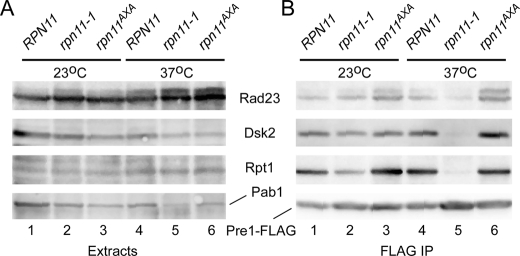
Dsk2 plays an overlapping role with Rad23 (37). Both proteins were expressed at physiological levels, and their interaction with the proteasome was examined in RPN11, rpn11-1, and rpn11AXA containing integrated Pre1-FLAG. Protein extracts were examined by immunoblotting with antibodies against Rpt1, Rad23, Dsk2, and Pab1 (Fig. 6A). The expression of Pre1-FLAG was similar in RPN11, rpn11-1, and rpn11AXA strains at both 23 °C and 37 °C (data not shown). Equivalent expression of Rpt1 and the shuttle factors Rad23 and Dsk2 were detected at 23 and 37 °C in total protein extracts (Fig. 6A). Moreover, the amount of Rad23 and Dsk2 that was co-purified with proteasomes at 23 °C was similar in RPN11, rpn11-1, and rpn11AXA (Fig. 6B). However, the level of both shuttle factors in proteasomes was strongly decreased in rpn11-1 at 37 °C (compare lanes 2 and 5), whereas their association with proteasomes from rpn11AXA at 37 °C was unaffected. Although equivalent amounts of Pre1-FLAG were immunoprecipitated from each strain, significantly lower amounts of Rpt1 were co-precipitated with the proteasome from rpn11-1.
FIGURE 6.
Substrate shuttle factors show differential binding to proteasomes in rpn11-1 and rpn11AXA. A, the interaction between proteasomes and shuttle factors (Rad23 and Dsk2) was determined by immunoprecipitating Pre1-FLAG. Yeast cells were grown at 23 and 37 °C, and total protein extracts were examined. B, proteasomes were immunoprecipitated (IP) on FLAG-agarose, and the levels of Rad23, Dsk2, and Rpt1 were determined by immunoblotting.
The UbL-UBA shuttle factors bind the Rpn1 subunit in the base complex (38), which also contains the hexameric ring of ATPases that binds the 20 S particle. However, we found that lower amounts of Rpn1 (Fig. 1A) and Rpt1 (Fig. 6B) were recovered with Pre1-FLAG in rpn11-1, which could underlie the reduced proteasome interaction with shuttle factors. Rpt1 was efficiently recovered with proteasomes in rpn11AXA, consistent with the high levels of ubiquitinated proteins and shuttle factors that were observed (Fig. 6B, and see Fig. 7).
High Levels of Multiubiquitinated Proteins Remain Bound to Shuttle Factors in rpn11-1
Studies described here reveal a failure of multiubiquitinated proteins to be efficiently delivered to or retained on proteasomes in rpn11-1. The accumulation of high levels of ubiquitinated proteins in total cell extracts in rpn11-1 was unexpected because multiubiquitin chains can be rapidly dismantled. However, an interaction with UBA domains in shuttle factors, such as Rad23, can reduce the disassembly of multiubiquitin chains (39). Therefore, we investigated whether multiubiquitinated proteins accumulated on shuttle factors in rpn11-1. RPN11, rpn11-1, and rpn11AXA were transformed with plasmids expressing FLAG-Rad23, FLAG-Ddi1, and FLAG-Dsk2. Protein extracts were incubated with FLAG-agarose, and the bound proteins were resolved by SDS-PAGE and transferred to nitrocellulose. Incubation of the filter with anti-ubiquitin antibodies revealed strong interaction with shuttle factors in rpn11-1 extracts (Fig. 7A, lanes 5, 8, and 11).
To further test these findings, we measured peptidase activity that was purified with the shuttle factors as this would provide an independent gauge of their ability to bind proteasomes. Protein extracts were prepared in the presence of ATP to minimize proteasome disassembly. Shuttle factors were recovered on FLAG-agarose, and the affinity beads were incubated with LLVY-AMC to measure chymotryptic activity. In the absence of SDS, peptidase activity in total extract was lower in rpn11-1 but significantly higher in rpn11AXA (Fig. 7B). However, the addition of SDS stimulated the chymotryptic activity in all three strains and was particularly robust in rpn11-1 (consistent with Fig. 3C). This result supports the idea that higher levels of free 20 S catalytic particles accumulate in rpn11-1 as its activity can be stimulated by SDS. Protein extracts were applied to FLAG-agarose to measure the peptidase activity in proteasomes that were co-purified with the shuttle factors (Fig. 7C). As expected, no activity was detected in a strain that did not express a FLAG-tagged protein (vector). Significant chymotryptic activity was immunopurified with FLAG-Rad23 from RPN11 and rpn11AXA in the absence of SDS. In contrast, very low peptidase activity was immunopurified with FLAG-Rad23 from rpn11-1. The addition of SDS resulted in a similar trend in chymotryptic activity, although the magnitude of hydrolysis was reduced. We believe that this might be due to an adverse effect of detergent on the affinity matrix. Purification of FLAG-Ddi1 yielded very low activity, consistent with the absence of significant interaction with proteasomes (Fig. 7A). In contrast, FLAG-Dsk2 gave qualitatively similar results to FLAG-Rad23, suggesting that the stability defect of rpn11-1 proteasomes affects its interactions with multiple UbL/UBA shuttle factors.
DISCUSSION
The role of chaperones in promoting the assembly of the 20 S catalytic particle has been characterized extensively (2–5). In contrast, knowledge of 19 S complex assembly (6–12) and 26 S maturation is only now emerging. The 19 S base subcomplex was recently reported to occur through a succession of intermediate structures, with the 20 S particle serving as a platform for assembly (8). The ensuing recruitment of a preformed lid complex could yield a complete proteasome.
Mutation in the Rpn11 deubiquitinating enzyme can affect proteasome assembly (21, 23) and interaction with multiubiquitinated proteins. The rpn11-1 (mpr1-1) mutant contains a frameshift near the carboxyl terminus (40), which results in the replacement of the terminal 31 amino acids with nine non-native residues. This well characterized temperature-sensitive mutant (20, 40) displays growth and proteolytic deficiencies (21, 40), as well as a mitochondrial defect (20). The rpn11AXA mutant contains changes in the MPR1-PAD1-N terminus (MPN) domain and does not support viability, although its biochemical properties can be examined in an rpn11-1 mutant (21).
The significant proteolytic and growth defects of rpn11-1 led us to examine proteasome interaction with ubiquitinated proteins and shuttle factors. Proteasomes are structurally defective in rpn11-1 and show a specific loss of 19 S subunits. Consequently, the low peptidase activity of proteasomes in rpn11-1 may be due to the diminished stimulatory effect that the 19 S regulatory particle is known to exert. These proteasomes also showed significantly reduced interaction with multiubiquitinated proteins. In contrast, proteasomes containing the catalytically inactive rpn11AXA protein are structurally sound and bind very high levels of multiubiquitinated proteins due to defective deubiquitination (21). These results show that the interaction between ubiquitinated proteins and the proteasome can be uncoupled from the subsequent steps involving deubiquitination and degradation.
The carboxyl-terminal 39 residues of Rpn11 can function in trans to partially restore proteasome assembly. GST-CRpn11 improved growth of rpn11-1 at the non-permissive temperature and increased survival in the presence of translational inhibitors. Additionally, normal resistance to UV light was restored, and the degradation of a proteolytic substrate was improved. Direct binding studies failed to reveal any interaction between FLAG-rpn11-1 and GST-CRpn11, indicating that suppression is not achieved by dimerization of these protein sequences (data not shown and Ref. 22). Because rpn11-1 protein is catalytically active but destabilizes proteasomes, we propose that the carboxyl terminus promotes proteasome stability, consistent with previous reports showing its requirement for Rpn11 function (21–23).
Multiubiquitinated proteins accumulated in rpn11-1 but were not detected at correspondingly high levels in the proteasome. However, high levels of ubiquitinated proteins were co-purified with shuttle factors. Proteasomes purified from rpn11-1 contained reduced levels of the 19 S subunit Rpn1, which forms the primary interaction with shuttle factors (38). This provides an explanation for the reduced interaction between shuttle factors and proteasomes in rpn11-1. In contrast, the high levels of multiubiquitinated proteins in proteasomes in rpn11AXA indicate that shuttle factors can bind intact proteasomes, even if they are catalytically inactive for deubiquitination.
Our results suggest that the primary defect of rpn11-1 is the instability of the lid complex rather than a failure to properly assemble proteasomes. Although we cannot disregard an assembly defect, our model is supported by the detection of intact proteasomes in rpn11-1 following in vivo cross-linking (Fig. 3). Rad23 and Rpn10, as well as multiubiquitinated proteins, were readily co-purified with proteasomes from rpn11-1 following cross-linking.
Lower amounts of Rad23 and Dsk2 were detected in proteasomes isolated from rpn11-1, suggesting inefficient delivery of shuttle factors. However, high amounts of multiubiquitinated proteins were bound to these shuttle factors. Presumably, multiubiquitinated proteins that are not delivered to the proteasome can remain bound to shuttle factors. The increased amount of fully assembled proteasomes in rpn11AXA might underlie their favorable interaction with multiubiquitinated proteins. Taken together, these findings suggest that shuttle factors and ubiquitinated proteins may bind proteasomes only after the lid subcomplex has assembled with the base-20 S complex. The release of shuttle factors from proteasomes could be linked to the disassembly of multiubiquitin chains, which was shown to be coupled to substrate degradation (21).
The overall abundance of proteasome subunits is not markedly affected in rpn11AXA, as judged by immunoblotting (Fig. 1). However, both in-gel assays and immunoprecipitated proteasomes showed higher chymotryptic activity in rpn11AXA. Because high levels of multiubiquitinated proteins are bound to proteasome in rpn11AXA, we speculate that peptidase activity might be positively affected by interaction with proteolytic substrates, but their degradation requires deubiquitination (21).
The addition of SDS did not stimulate the peptidase activity of proteasomes purified with either FLAG-Rad23 or FLAG-Dsk2 from rpn11-1 (Fig. 7C). Because shuttle factors interact with the 19 S complex, these results support the model that lower amounts of the 20 S particle were co-purified with shuttle factors from rpn11-1. Based on these findings, it will be of interest to determine how the level of proteasome assembly affects substrate turnover and whether deubiquitination regulates the release of shuttle factors from the proteasome.
Supplementary Material
Acknowledgments
Members of the laboratory are thanked for critical review of the manuscript. We thank Drs. Dohmen (University of Cologne), Verma (California Institute of Technology), Deshaies (California Institute of Technology), Enenkel (Humboldt University), Skowyra (St. Louis University), and Peltz (UMDNJ), for plasmids, strains, and antibodies.
This work was supported, in whole or in part, by National Institutes of Health Grants CA83875 and GM083321 (to K. M.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1 and 2.
- GST
- glutathione S-transferase
- CP
- catalytic (20 S) particle
- RP
- regulatory (19 S) particle
- GFP
- green fluorescent protein
- HA
- hemagglutinin
- LLVY-AMC
- leucine-leucine-valine-tyrosine-7-amino-4-methylcoumarin
- UBA
- ubiquitin-associated
- UbL
- ubiquitin-like
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine.
REFERENCES
- 1.Glickman M. H., Ciechanover A. (2002) Physiol. Rev. 82, 373–428 [DOI] [PubMed] [Google Scholar]
- 2.Rosenzweig R., Glickman M. H. (2008) Biochem. Soc. Trans. 36, 807–812 [DOI] [PubMed] [Google Scholar]
- 3.Ramos P. C., Dohmen R. J. (2008) Structure 16, 1296–1304 [DOI] [PubMed] [Google Scholar]
- 4.Le Tallec B., Barrault M. B., Courbeyrette R., Guérois R., Marsolier-Kergoat M. C., Peyroche A. (2007) Mol. Cell 27, 660–674 [DOI] [PubMed] [Google Scholar]
- 5.Kusmierczyk A. R., Kunjappu M. J., Funakoshi M., Hochstrasser M. (2008) Nat. Struct. Mol. Biol. 15, 237–244 [DOI] [PubMed] [Google Scholar]
- 6.Saeki Y., Toh-E. A., Kudo T., Kawamura H., Tanaka K. (2009) Cell 137, 900–913 [DOI] [PubMed] [Google Scholar]
- 7.Roelofs J., Park S., Haas W., Tian G., McAllister F. E., Huo Y., Lee B. H., Zhang F., Shi Y., Gygi S. P., Finley D. (2009) Nature 459, 861–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park S., Roelofs J., Kim W., Robert J., Schmidt M., Gygi S. P., Finley D. (2009) Nature 459, 866–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murata S., Yashiroda H., Tanaka K. (2009) Nat. Rev. Mol. Cell Biol. 10, 104–115 [DOI] [PubMed] [Google Scholar]
- 10.Le Tallec B., Barrault M. B., Guérois R., Carré T., Peyroche A. (2009) Mol. Cell 33, 389–399 [DOI] [PubMed] [Google Scholar]
- 11.Kaneko T., Hamazaki J., Iemura S., Sasaki K., Furuyama K., Natsume T., Tanaka K., Murata S. (2009) Cell 137, 914–925 [DOI] [PubMed] [Google Scholar]
- 12.Funakoshi M., Tomko R. J., Jr., Kobayashi H., Hochstrasser M. (2009) Cell 137, 887–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Glickman M. H., Rubin D. M., Fu H., Larsen C. N., Coux O., Wefes I., Pfeifer G., Cjeka Z., Vierstra R., Baumeister W., Fried V., Finley D. (1999) Mol. Biol. Rep. 26, 21–28 [DOI] [PubMed] [Google Scholar]
- 14.van Nocker S., Sadis S., Rubin D. M., Glickman M., Fu H., Coux O., Wefes I., Finley D., Vierstra R. D. (1996) Mol. Cell Biol. 16, 6020–6028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Díaz-Martínez L. A., Kang Y., Walters K. J., Clarke D. J. (2006) Cell Div. 1, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Funakoshi M., Sasaki T., Nishimoto T., Kobayashi H. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 745–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lambertson D., Chen L., Madura K. (1999) Genetics 153, 69–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elsasser S., Chandler-Militello D., Müller B., Hanna J., Finley D. (2004) J. Biol. Chem. 279, 26817–26822 [DOI] [PubMed] [Google Scholar]
- 19.Verma R., Oania R., Graumann J., Deshaies R. J. (2004) Cell 118, 99–110 [DOI] [PubMed] [Google Scholar]
- 20.Rinaldi T., Ricordy R., Bolotin-Fukuhara M., Frontali L. (2002) Gene 286, 43–51 [DOI] [PubMed] [Google Scholar]
- 21.Verma R., Aravind L., Oania R., McDonald W. H., Yates J. R., 3rd, Koonin E. V., Deshaies R. J. (2002) Science 298, 611–615 [DOI] [PubMed] [Google Scholar]
- 22.Rinaldi T., Hofmann L., Gambadoro A., Cossard R., Livnat-Levanon N., Glickman M. H., Frontali L., Delahodde A. (2008) Mol. Biol. Cell 19, 1022–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rinaldi T., Pick E., Gambadoro A., Zilli S., Maytal-Kivity V., Frontali L., Glickman M. H. (2004) Biochem. J. 381, 275–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gietz R. D., Sugino A. (1988) Gene 74, 527–534 [DOI] [PubMed] [Google Scholar]
- 25.Chuang S. M., Madura K. (2005) Genetics 171, 1477–1484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bachmair A., Finley D., Varshavsky A. (1986) Science 234, 179–186 [DOI] [PubMed] [Google Scholar]
- 27.Ortolan T. G., Tongaonkar P., Lambertson D., Chen L., Schauber C., Madura K. (2000) Nat. Cell Biol. 2, 601–608 [DOI] [PubMed] [Google Scholar]
- 28.Glickman M. H., Rubin D. M., Fried V. A., Finley D. (1998) Mol. Cell Biol. 18, 3149–3162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Groll M., Bajorek M., Köhler A., Moroder L., Rubin D. M., Huber R., Glickman M. H., Finley D. (2000) Nat. Struct. Biol. 7, 1062–1067 [DOI] [PubMed] [Google Scholar]
- 30.Braun B. C., Glickman M., Kraft R., Dahlmann B., Kloetzel P. M., Finley D., Schmidt M. (1999) Nat. Cell Biol. 1, 221–226 [DOI] [PubMed] [Google Scholar]
- 31.Smith D. M., Chang S. C., Park S., Finley D., Cheng Y., Goldberg A. L. (2007) Mol. Cell 27, 731–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Köhler A., Cascio P., Leggett D. S., Woo K. M., Goldberg A. L., Finley D. (2001) Mol. Cell 7, 1143–1152 [DOI] [PubMed] [Google Scholar]
- 33.Chen L., Madura K. (2002) Mol. Cell Biol. 22, 4902–4913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schauber C., Chen L., Tongaonkar P., Vega I., Lambertson D., Potts W., Madura K. (1998) Nature 391, 715–718 [DOI] [PubMed] [Google Scholar]
- 35.Bertolaet B. L., Clarke D. J., Wolff M., Watson M. H., Henze M., Divita G., Reed S. I. (2001) Nat. Struct. Biol. 8, 417–422 [DOI] [PubMed] [Google Scholar]
- 36.Chen L., Shinde U., Ortolan T. G., Madura K. (2001) EMBO Rep. 2, 933–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Biggins S., Ivanovska I., Rose M. D. (1996) J. Cell Biol. 133, 1331–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Elsasser S., Gali R. R., Schwickart M., Larsen C. N., Leggett D. S., Müller B., Feng M. T., Tübing F., Dittmar G. A., Finley D. (2002) Nat. Cell Biol. 4, 725–730 [DOI] [PubMed] [Google Scholar]
- 39.Raasi S., Pickart C. M. (2003) J. Biol. Chem. 278, 8951–8959 [DOI] [PubMed] [Google Scholar]
- 40.Rinaldi T., Ricci C., Porro D., Bolotin-Fukuhara M., Frontali L. (1998) Mol. Biol. Cell 9, 2917–2931 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.