Abstract
Homeostasis of Smad phosphorylation at its C-terminal SXS motif is essential for transforming growth factor β (TGFβ) signaling. Whereas it is known that TGFβ signaling can be terminated by phosphatases, which dephosphorylate R-Smads in the nucleus, it is unclear whether there are any cytoplasmic phosphatase(s) that can attenuate R-Smad phosphorylation and nuclear translocation. Here we demonstrate that myotubularin-related protein 4 (MTMR4), a FYVE domain-containing dual-specificity protein phosphatase (DSP), attenuates TGFβ signaling by reducing the phosphorylation level of R-Smads in early endosomes. Co-immunoprecipitation experiments showed that endogenous MTMR4 interacts with phosphorylated R-Smads, and that this interaction is correlated with dephosphorylation of R-Smads. Further analysis showed that overexpression of MTMR4 resulted in the sequestration of activated Smad3 in the early endosomes, thus reducing its nuclear translocation. However, both point mutations at the conserved catalytic site of the phosphatase (MTMR4-C407S) and small interference RNA of endogenous Mtmr4 expression led to sustained Smad3 activation. This work therefore suggests that MTMR4 plays an important role in preventing the overactivation of TGFβ signaling by dephosphorylating activated R-Smads that have been trafficked to early endosomes.
Keywords: Transforming Growth Factor β (TGFβ), SMAD Transcription Factor, Phosphatase, Signal Transduction, Endosomes, MTMR4
Introduction
The transforming growth factor β (TGFβ)3 signaling pathway involves two transmembrane serine/threonine kinases, namely type II (TβRII) and type I TGFβ receptors (TβRI) (1, 2). TβRII, a constitutively active kinase, binds TGFβ to initiate its heterodimerization with TβRI. TβRI then becomes phosphorylated and activates a signaling cascade through a family of intracellular signaling mediators known as Smads. In doing so, TβRI recruits receptor-regulated Smads (R-Smads, including BMP signaling transducers Smad1, -5, and -8 and TGFβ signaling transducers Smad2, -3) and phosphorylates the two conserved C-terminal serines (SXS) of R-Smads. Activated R-Smads form a trimeric complex with a common mediator Smad (Co-Smad, Smad4), and these complexes translocate into the nucleus to regulate the transcription of an array of target genes (3–7). Recent studies have also indicated that Smad Anchor for Receptor Activation (SARA), an early endosomal protein containing a characteristic FYVE domain (Fab1p, YO1B, Vac1p, and EEA1), serves as an anchor protein by directly recruiting Smad2 to the early endosomes (8, 9) and presenting Smad2 to the internalized TβRI/II complex (10, 11). Phosphorylated Smad2 then dissociates from SARA and leaves the early endosome. Subsequently, it forms a Smad2-Smad4 complex for nuclear translocation.
Despite the fact that TGFβ signaling activation is surprisingly simple, there are many layers of negative regulation that fine-tune or terminate the signal. The inhibitory Smads (I-Smads, including Smad6 and -7) competitively inhibit the interaction of R-Smads with Smad4 or receptors to provide a negative checkpoint for TGFβ-signaling activation (12). Ubiquitin-dependent degradation of activated R-Smads was initially thought to cause the negative regulation of TGFβ signaling (13). Further investigation has shown that R-Smads can shuttle between the cytoplasm and the nucleus (3, 14) and that dephosphorylation of activated R-Smads is a prerequisite for their recycling and the down-regulation of TGFβ signaling. Indeed, recent findings have indicated that pyruvate dehydrogenase phosphatase (PDP) in Drosophila (15) and small C-terminal domain phosphatase (SCP2/Os4) in Xenopus (16) act as BMP/Dpp-activated Smad1 phosphatases and that PPM1A/PP2Cα acts as a TGFβ-activated Smad2/3 phosphatase in the nucleus (17). However, whether there are any cytoplasmic phosphatase(s) that dephosphorylate activated Smads and prevent them from being translocated to the nucleus remains unknown.
The myotubularin family is a large group of eukaryotic protein tyrosine/dual-specificity phosphatases (PTP/DSP). The myotubularin family contains 14 genes in humans, named MTM1 and MTM-related 1 to 13 (MTMR1–13). All members share four domains: N-terminal pleckstrin homology/glucosyltransferase Rab-like GTPase activator and myotubularin domain (PH-GRAM), Rac-induced recruitment domain (RID), PTP/DSP active site homology, and SET-interacting domain (SID) (18). Some members also have a protein interaction or phosphoinositide binding domain, such as the coiled-coil domain or FYVE domain at the C terminus. MTMR4 is an intracellular protein (1195 amino acids in length with a calculated molecular mass of 133 kDa) with a unique FYVE domain at its C terminus, which is responsible for binding to PI3P or PI(3,5)P2. Some studies have shown that MTMR4 can dephosphorylate PI3P or PI(3,5)P2 (19), suggesting that MTMR4 is also a lipid phosphatase with important roles in cellular processes. Moreover, recent studies have demonstrated that MTMR4 can homodimerize or heterodimerize with MTMR3, a homolog of MTMR4, to regulate EGF receptor traffic and degradation (20).
In the present study, we demonstrate that MTMR4 negatively regulates TGFβ signaling. We show that MTMR4 localizes to early endosomes via its FYVE domain. MTMR4 interacts with and dephosphorylates activated R-Smads in the early endosomes, and effectively blocks overactivation and nuclear translocation of R-Smads. Our data suggest that MTMR4 might be an endosomal R-Smad phosphatase that attenuates phosphorylation of R-Smads and maintains TGFβ signaling in homeostasis.
EXPERIMENTAL PROCEDURES
Plasmids
The human MTMR4-encoding gene was subcloned from a cDNA clone (hj03819s1, Kazusa DNA Research Institute) by add-on PCR into the pEYFP-C1 and pECFP-C1 vectors (Clontech). Truncation mutants (MTMR4-ΔFYVE1, -ΔFYVE2, -ΔDSP), or fragments (FYVE1 and FYVE2) were similarly fused to YFP or CFP. We designed a point mutation of the catalytic site (MTMR4-C407S) in which the invariant cysteine residue in the CSDGWDR motif (present within all enzymatically active members of the MTMR superfamily (18)) was substituted by a serine residue by mutation using the QuickChange method (Promega). The various truncations of pCMV-Myc-Smad3 (pCMV-Myc-MH1, pCMV-Myc-MH1-Linker, pCMV-Myc-MH2, pCMV-Myc-Linker-MH2), constitutively active Smad3 plasmid (pRK5-HA-Smad3 (SD)), and constitutively active TβRI plasmid (pCS2-GGD-HA) used here have been described previously (21). pDsRed2-N1-EEA1 was constructed by subcloning EEA1 cDNA (provided by Dr. X.-J. Jiang, Institute of Microbiology, CAS) into pDsRed2-N1 vectors (Clontech). Flag-tagged pRK5F-Smad2, pXFIF-Smad3, and pXFIF-Smad4 vectors were kind gifts from Dr. R. Derynck (UCSF). The cDNAs of Smad2, Smad3, and Smad4 were subsequently subcloned into the pECFP-C1 plasmid. All primer sequences used in subcloning are listed in supplemental Table S1. Each derivative clone was verified by DNA sequencing analysis.
Cell Culture, Transfection, and Reporter Assays
Porcine aorta endothelial (PAE) stable cell lines expressing GFP or CFP-MTMR4 (indicated as PAE-GFP or PAE-MTMR4) were established by a standard single cell cloning and G418 selection (400 ng/ml) procedure. PAE cells were routinely maintained in F12 medium (Hyclone) and PAE stable cell lines were maintained in F12 medium with G418 (100 ng/ml). L17 cells (TβRI-deficient Mv1Lu) and HaCaT cells were grown in MEM (Invitrogen), while HeLa and HEK293T cells were cultured in DMEM (Hyclone). All media were supplemented with 10% heat-inactivated FBS (Invitrogen), 1% penicillin and streptomycin (Hyclone). PAE cells were transfected using Effectene (Qiagen), L17, HeLa, and HaCaT cells using Lipofectamine (Invitrogen), and 293T cells by the calcium-phosphate method. Transient cotransfection and Luciferase reporter assays were performed as previously described (22). In brief, 600 ng of 3TP-luciferase reporter plasmid (3TP-lux) and 20 ng of Renilla luciferase plasmid (pCMV-hRL, as the internal control) were cotransfected into PAE-GFP or PAE-MTMR4 cells. Transfectants were starved for 12 h in F12 medium containing 0.2% FBS before being stimulated with TGF-β1 (4 ng/ml, R&D Systems) for 12 h. L17 cells (80% confluent in 24-well plates) were cotransfected with 300 ng of CAGA-luciferase reporter plasmid (23) and 20 ng of pCMV-hRL and either 100 ng of pCMV-TβRI-HA, 60 ng of pCS2-GGD-HA, 100 ng of pCMV-Myc-Smad3, or 100 ng of pRK5-HA-Smad3 (SD). The cells were stimulated with TGF-β1 (2 ng/ml) for 16 h.
PAE cell proliferation assays were performed using a previously published method (22) with a WST-1 kit (Roche Molecular Biochemicals). In brief, PAE stable cell lines (PAE-GFP or PAE-MTMR4) were plated in a 96-well plate (4 × 103 cells/well) in culture medium supplemented with 0.2% FBS for 12 h before TGF-β1 treatment for the times indicated. The cell proliferation rate was calculated using A440 absorbance (CHAMELEON II, Hidex). Data represent the average of triplicate experiments (mean ± S.D.). HaCaT cell proliferation assays using CellTiter 96 One Solution Cell Proliferation Assay were performed according to the manufacturer's instructions (Promega). In brief, cells plated in a 96-well plate (2 × 103 cells/well) were starved in culture medium supplemented with 0.2% FBS for 12 h before treatment for 48 h with different concentrations of TGF-β1 as indicated. Data represent the average of triplicate experiments (mean ± S.D.).
Immunoprecipitation and Western Blotting
Whole cell lysates (300 μg of proteins) were subjected to immunoprecipitation with anti-Flag M2 (2 μg, Sigma) or anti-Myc (2 μg, BD Biosciences) followed by immunoblotting with the antibodies indicated as described previously (22). Antibodies against pSmad2/3 (SXS) were from Cell Signaling (Beverly, MA), Smad2 from BD, Smad3 and GFP from Zymed Laboratories Inc. (San Diego), USF2 from Santa Cruz, and MTMR4 antiserum was provided by Dr. Z. Zhao (University of Oklahoma Health Sciences Center) or from Abgent (San Diego, CA). Nuclear extracts were prepared essentially as described previously (24), and 50 μg of protein were immunoblotted for nuclear translocation assays of Smad2/3.
Fluorescence Microscopy
PAE cells in 35-mm plates were transfected with the plasmids indicated (1 μg each) overnight and then seeded at 10–20% confluence on 20-mm coverslips for an additional 24 h. Cells were then exposed or not to TGF-β1 for 2 h and viewed directly with a fluorescence microscope (Nikon TE2000) for image analysis using a SlideBook 4.1 workstation (Intelligent Imaging Innovations, CO) as previously described (25).
Real-time PCR
Total RNA was extracted with TRIzol reagent (Invitrogen) from PAE-GFP or PAE-MTMR4 cells (1 × 106) previously treated with TGF-β1 for the times indicated. Quantitative RT-PCR (iCycler, Bio-Rad) was carried out as described (26) using 18 S RNA as the internal control. Primer sequences for p15Ink4B, forward: 5′-AGGCCATAGCAGGCACATCACCT-3′ and reverse: 5′-AGCTCACTGCAACCTCTTCCAGCAT-3′; 18 S RNA, forward: 5′-TTGGATGTGGTAGCCGTTTC-3′; reverse: 5′-GATGGTAGTCGCCGTGCC-3′. Data represent the average of three independent experiments (mean ± S.D.).
RNA Interference
The 21-nucleotide siRNA duplexes targeting MTMR4 in the coding region of 2247–2265 (siRNA, 5′-ggagccaccugaacauuguuu-3′) and scrambled siRNA were synthesized and purified by RiboBio Co. (Guangzhou, China). In brief, 200 pmol of siRNA was transfected into 5 × 105 HeLa cells (50% confluence in 60-mm plates). The knockdown efficiency of endogenous MTMR4 was monitored by Western blotting 48-h post-transfection.
RESULTS
Overexpression of MTMR4 Inhibits TGFβ Signaling
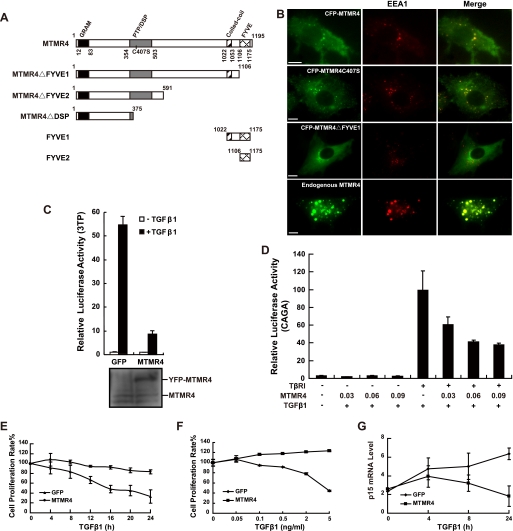
We first showed that MTMR4 was up-regulated and critically involved in B lymphocyte terminal differentiation.4 In agreement with recent findings (20) and consistent with a function in endosomal targeting, we found that the conserved FYVE domain (schematic presentation in Fig. 1A) is responsible for MTMR4 localization in early endosomes (based on its co-localization with early endosome antigen 1 (EEA1)), because MTMR4 without the FYVE domain (MTMR4ΔFYVE1) exhibited a diffused distribution in the cytoplasm (Fig. 1B). Because previous reports indicate that the FYVE domain-containing protein, SARA, is a Smad2 anchor protein required for the activation of Smad2 by TβRI in early endosomes (10), we wondered whether MTMR4 is also involved in TGFβ signaling. To test this hypothesis, we performed TGFβ-responsive promoter reporter (3TP-lux) assays (27) in an MTMR4-expressing PAE stable cell line. Results showed that the ∼55-fold transactivation of the luciferase reporter in PAE-GFP cells, which was induced by TGFβ1 stimulation was reduced to around a 7-fold transactivation in PAE-MTMR4 cells (Fig. 1C). To exclude any nonspecific effects, we repeated the assay using the TβRI-deficient cell line L17. Overexpression of MTMR4 in L17 had no effect on the TGF-β1-driven Smad3-responsive reporter (CAGA-luciferase), but we found that MTMR4 reduced reporter activity in a dose-dependent manner after L17 cells were reconstituted with TβRI (Fig. 1D). These results suggest that, unlike SARA, MTMR4 inhibits TGF-β1-induced transcriptional activation. Ectopic MTMR4 overexpressed at the concentrations shown in Fig. 1D apparently did not disturb endosome membrane lipid metabolism as basal levels of 3TP-lux and CAGA-luc activity were not affected. It remains unclear, however, whether MTMR4 inhibits TGFβ signaling by competing with endogenous SARA for association with PI3P in endosomes, resulting in redistribution of SARA or related proteins (28).
FIGURE 1.
Endosomally localized MTMR4 suppresses TGFβ signaling. A, domain organization of MTMR4. The PH-GRAM (black), PTP/DSP (gray), coiled-coil (slash), and FYVE (grid) domains are shown, including the various truncations (MTMR4ΔFYVE1, MTMR4ΔFYVE2, MTMR4ΔDSP, FYVE1, and FYVE2) and the phosphatase-dead mutant (MTMR4-C407S) used in this study. B, FYVE domain is required for MTMR4 localization in early endosomes. PAE cells were transiently cotransfected with various MTMR4 constructs (green) and pDsRed2-N1-EEA1 (red). Fluorescence images were captured and merged for colocalization analysis. The scale bar indicates 10 μm. C, MTMR4 suppresses TGFβ-induced 3TP-luciferase reporter transcriptional activation. PAE-GFP or PAE-MTMR4 cells were cotransfected with 3TP-lux and pCMV-hRL for 36 h. Cells were then treated with (black) or without (white) TGF-β1 and harvested for assaying luciferase activities. The experiment was performed in triplicate, and the data (relative luciferase activities) represent the means and standard deviations of three independent experiments after normalization to Renilla luciferase activity. D, MTMR4 inhibits the TGF-β1-induced expression of CAGA-luciferase in a dose-dependent manner. L17 cells were cotransfected with CAGA reporter, pCMV-hRL, pCMV-HA-TβRI, and pYFP-MTMR4 for 30 h before being stimulated with TGF-β1. Luciferase assays were performed as in C. E, MTMR4 suppresses TGF-β1-induced cell growth inhibition in PAE cells. PAE-GFP or PAE-MTMR4 cells were treated with TGF-β1 for the times indicated, and cell proliferation was measured using the WST-1 assay. F, MTMR4 suppresses the TGFβ-induced cell arrest in HaCaT cells. Control or overexpressed MTMR4 HaCaT cells were stimulated with various concentrations of TGF-β1 as indicated. Cell proliferation rates were assessed using the CellTiter 96 One Solution Cell Proliferation Assay. G, MTMR4 inhibits TGFβ-induced p15 Ink4B gene expression. PAE-GFP or PAE-MTMR4 cells were treated with TGF-β1 for the times indicated. 2 μg of total RNA was used for qPCR analysis. p15Ink4B mRNA levels were normalized against 18 S RNA.
TGFβ is a potent growth inhibitor and induces apoptosis in a variety of cell types (29, 30). To test whether MTMR4 can antagonize the growth inhibition effect of TGFβ, we performed cell proliferation assays in PAE-GFP and PAE-MTMR4 cells. Whereas TGF-β1 efficiently inhibited cell proliferation in PAE-GFP cells as expected, PAE-MTMR4 cells were resistant to TGF-β1-induced growth inhibition (Fig. 1E). To exclude potential bias from using one particular cell line, we then repeated the assay in a classic TGFβ-responsive cell line, HaCaT. MTMR4 overexpression efficiently overcame growth inhibition induced by increasing concentrations of TGF-β1 (Fig. 1F). Transcriptional activation of the p15Ink4B gene is under the control of Smad2/3 and is abrogated by c-Myc gene involved in oncogenic proliferation (31). Pro-proliferative activity of MTMR4 correlated with its inhibition of p15Ink4B gene expression in PAE-MTMR4 cells as shown by quantitative PCR analysis (Fig. 1G). These results suggest that overexpression of MTMR4-rendered cells resistant to the growth arrest caused by TGF-β1 stimulation.
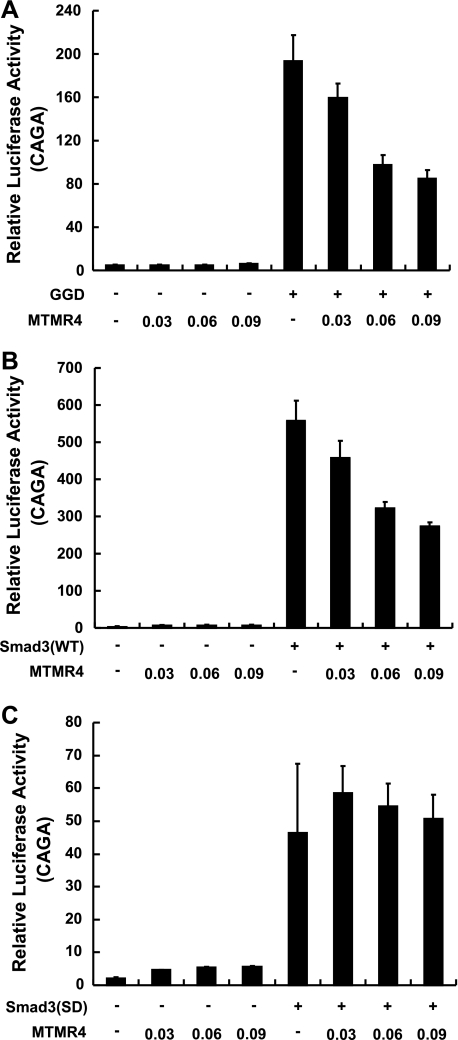
Negative regulation of TGFβ signaling by MTMR4 prompted us to determine whether it functions by either inhibiting TβRI or R-Smad activation. To do this, we reconstituted L17 cells with GGD, a dominant active form of TβRI. We then assessed whether MTMR4 still inhibited TβRI activation-independent signaling. Overexpression of MTMR4 effectively reduced Smad3-responsive CAGA reporter activities in a dose-dependent manner in L17/GGD cells (Fig. 2A), suggesting that MTMR4 blocked signaling downstream of TβRI. On the other hand, overexpressed MTMR4 in L17 cells effectively inhibited Smad3-driven CAGA reporter activities in a dose-dependent manner (Fig. 2B), but failed to do so in the presence of ectopically expressed Smad3-SD, a constitutively active Smad3 mutant, which is activated in a phosphorylation-independent manner (Fig. 2C). These results therefore indicate that MTMR4 acts downstream of TβRI in inhibition of TGFβ signaling and most likely acts on Smad3.
FIGURE 2.
MTMR4 functions downstream of TβRI. A, MTMR4 inhibits constitutively active TβRI (GGD)-induced CAGA-luciferase reporter transcriptional activation in a dose-dependent manner. L17 cells were cotransfected with CAGA reporter, pCMV-hRL, pCS2-GGD-HA and increasing amounts of pYFP-MTMR4. Luciferase assays were performed as in Fig. 1C. B, MTMR4 inhibits Smad3-induced CAGA-luciferase reporter transcriptional activation in a dose-dependent manner. L17 cells were co-transfected with CAGA reporter, pCMV-hRL, pCMV-Myc-Smad3, and increasing amounts of pYFP-MTMR4. Luciferase assays were performed as in Fig. 1C. C, MTMR4 fails to inhibit constitutively active Smad3 (SD)-induced CAGA-luciferase reporter transcriptional activation. L17 cells were co-transfected with CAGA reporter, pCMV-hRL, pRK5-HA-Smad3 (SD) and increasing amounts of pYFP-MTMR4. Luciferase assays were performed as in Fig. 1C.
MTMR4 Interacts with Smad2/3
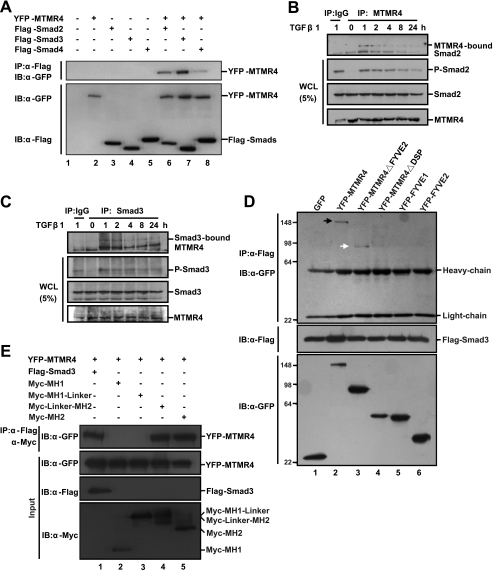
Because FYVE-domain-containing proteins usually regulate TGFβ signaling through Smads (10, 32) and the data above show that MTMR4 most likely targets R-Smads, we examined whether MTMR4 can bind to Smads using co-immunoprecipitation experiments. We transiently co-expressed YFP-MTMR4 with Flag-tagged Smad2, -3, or -4 in HEK293T cells. Results showed that MTMR4 could bind to Smad2, -3, and -4 (Fig. 3A) whereas the interaction between MTMR4 and Smad4 was relatively weaker than that with Smad2/3. We speculated that co-Smad bound to MTMR4 indirectly, most likely through a trimeric complex of Smad2/3/4. Because MTMR4 resides in early endosomes, whereas Smad2/3 has to be phosphorylated in the early endosome, we wondered whether the intermolecular interaction between MTMR4 and R-Smads could actually happen under physiological conditions. Indeed, co-immunoprecipitation assays showed that endogenous MTMR4 and endogenous Smad2 (Fig. 3B) or Smad3 (Fig. 3C) in HaCaT cells interact with each other, and such interaction only occurred after TGF-β1 stimulation. Interestingly, the amount of Smad2/3 associated with MTMR4 was reduced in proportion with its decreasing phosphorylation levels (Fig. 3, B and C), suggesting that MTMR4 interacts with phosphorylated R-Smads.
FIGURE 3.
MTMR4 interacts with Smad2/3. A, MTMR4 interacts strongly with Smad2/3. HEK293T cells transiently expressed Flag-tagged Smad2 (0.8 μg), Smad3 (0.8 μg), or Smad4 (0.8 μg) and YFP-MTMR4 (1.5 μg). Whole cell lysates were immunoprecipitated with M2 Flag antibody, and resolved proteins were immunoblotted with anti-GFP antibody (top panel). Protein expression levels were verified by anti-GFP (YFP-MTMR4, middle) or -Flag (Smads, bottom) antibodies, respectively. B and C, endogenous MTMR4 interacts with endogenous Smad2/3 upon TGF-β1 stimulation. Cell lysates were prepared from 90% confluent HaCaT cells after TGF-β1 treatment (2 ng/ml) for different lengths of time as indicated. Endogenous Smad2/3 bound to endogenous MTMR4 was immunoprecipitated with anti-MTMR4 antibody (B) or anti-Smad3 antibody (C) and detected by Western blotting with anti-Smad2 antibody (B) or anti-MTMR4 antibody (C). D, PTP/DSP domain of MTMR4 is required for interaction with Smad3. 293T cells were cotransfected with Flag-tagged Smad3 (0.8 μg) and pEYFP-MTMR4 or various mutant derivatives (1.5 μg) as indicated for 48 h. Cell lysates were immunoprecipitated and blotted as in A. Expression levels of Flag-tagged Smad3 (middle) and MTMR4 (bottom) are also shown. Arrows indicate positive interactions. E, MH2 domain of Smad3 is required for interaction with MTMR4. 293T cells were cotransfected for 48 h with pEYFP-MTMR4 (1.5 μg) and Flag-tagged Smad3 or various Myc-tagged mutant derivatives (0.8 μg) as indicated. Cell lysates were immunoprecipitated with M2 Flag antibody or anti-Myc antibody and blotted with anti-GFP antibody (top panel). Expression levels of YFP-MTMR4 (middle) and Flag-Smad3 and various mutants (two bottom panels) are also shown. WCL, whole cell lysates.
To map the region of contact between MTMR4 and Smad3 in greater detail, we performed co-immunoprecipitation of various MTMR4 truncation mutants with Flag-tagged Smad3 overexpressed in 293T cells. We found that Smad3 interacted with MTMR4 between amino acid residues 375 and 591 (Fig. 3D), a region that overlaps with the conserved PTP/DSP phosphatase domain (Fig. 1A). Reciprocal co-immunoprecipitation showed that the MH2 domain of Smad3, but not its MH1 domain is in contact with MTMR4 (Fig. 3E). These results therefore suggest that the PTP/DSP domain of MTMR4 and the MH2 domain of Smad3 are required for their interaction. This mode of interaction seems reasonable because it locates the DSP domain of MTMR4 adjacent to the C-terminal SXS motif of Smad2/3, which is convenient for its dephosphorylation.
MTMR4 Blocks R-Smad Phosphorylation and Nuclear Translocation
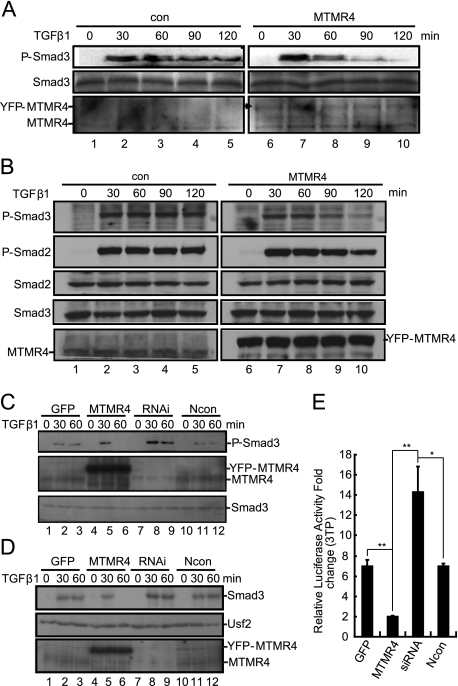
Interestingly, overexpression of MTMR4 appeared to reduce Smad3-Smad4 complex formation after TGF-β1 stimulation (supplemental Fig. S1). R-Smad3 can interact with Smad4 in the cytoplasm or nucleus only after it has been phosphorylated and has left the early endosome (12). Therefore, the reduced level of interaction between Smad3 and Smad4 observed here suggested that MTMR4 might be able to dephosphorylate activated Smad3 in early endosomes. To test this hypothesis, we measured the phosphorylation levels of endogenous Smad3 in PAE-MTMR4 cells (Fig. 4A) and in HaCaT cells transiently expressing MTMR4 (Fig. 4B). Immunoblotting with anti-phosphor-Smad3-specific antibodies showed that the SXS motif of Smad3 was rapidly phosphorylated (within 30 min after TGF-β1 treatment), and slowly dephosphorylated ∼1.5 h post-TGF-β1 treatment. The onset of Smad3 phosphorylation in PAE-MTMR4 or HaCaT cells overexpressing MTMR4 did not alter during the early phase of TGF-β1 stimulation. However, Smad3 was dephosphorylated at an accelerated rate after 1 h of TGF-β1 treatment. The reduced phosphorylation level of Smad3 was not due to its proteasomal degradation, because the level of Smad3 protein remained the same in the presence or absence of overexpressed MTMR4.
FIGURE 4.
MTMR4 dephosphorylates activated R-Smads. A, MTMR4 reduces the phosphorylation level of Smad3. PAE-GFP or PAE-MTMR4 cells (1 × 106) were treated with TGF-β1 (4 ng/ml) for the times indicated, and endogenous activated Smad3 was immunoblotted with anti-phosphor-Smad3 antibody (top panel). The protein loading of Smad3 was verified by immunoblotting with an anti-Smad3 antibody (middle panel). Stable expression of YFP-MTMR4 and endogenous MTMR4 was verified with anti-MTMR4 rabbit serum (bottom panel). B, overexpressed MTMR4 reduces the phosphorylation level of R-Smads in HaCaT cells. Control or overexpressed MTMR4 HaCaT cells were treated with TGF-β1 (2 ng/ml) for different lengths of time as indicated. Endogenous activated Smad2/3 was immunoblotted with anti-phosphor-Smad2/3 antibodies (top two panels). The protein loading of Smad2/3 was verified by immunoblotting with anti-Smad2/3 antibodies (middle two panels). Overexpression of YFP-MTMR4 and endogenous MTMR4 was verified with anti-MTMR4 antibody (bottom panel). C, knockdown of endogenous MTMR4 results in sustained Smad3 phosphorylation. HeLa cells were transfected with the control vector (lanes 1–3), pEYFP-MTMR4 (lanes 4–6), MTMR4-specific siRNA (lanes 7–9) or scrambled siRNA (lanes 10–12), and then treated with or without TGF-β1 (4 ng/ml). The phosphorylation level of Smad3 (top) and Smad3 input (bottom) were verified as described in A. Knockdown efficiency was verified with anti-MTMR4 rabbit serum (middle, 50 μg of lysates per lane). D, MTMR4 inhibits Smad3 nuclear translocation. HeLa cells were transfected with different constructs and siRNA as described in C. Nuclear extracts were resolved by SDS-PAGE and Western blotted with anti-Smad3 antibody. Nuclear transcription factor USF2 was immunoblotted as the loading control. MTMR4 knockdown efficiency was verified as described in B. E, knockdown of endogenous MTMR4 up-regulates TGFβ-induced 3TP-luciferase transcriptional activation. HeLa cells were cotransfected for 24 h with different constructs and siRNA as described in C. Cells were subsequently transfected with 3TP-lux and pCMV-hRL for another 36 h and then treated with or without TGF-β1 for 24 h. Luciferase assays were performed as described in Fig. 1C. **, p < 0.01; *, p < 0.05. Ncon, scrambled siRNA control.
On the other hand, siRNA knockdown of the endogenous Mtmr4 gene in HeLa cells (Fig. 4C) not only caused an overall increase in Smad3 phosphorylation (30 min) in response to TGF-β1 (compared with that of the GFP and Ncon controls), but also significantly attenuated Smad3 dephosphorylation (60 min) compared with that of the MTMR4 overexpression control. Western blotting of nuclear proteins showed that enhanced Smad3 phosphorylation by Mtmr4 siRNA correlated with increased nuclear translocation of Smad3 (Fig. 4D, lanes 8 and 9), which was otherwise suppressed by overexpressed MTMR4 after TGF-β1 treatment for 60 min (Fig. 4D, lanes 5 and 6). As expected, siRNA knockdown of endogenous Mtmr4 up-regulated TGFβ reporter activity (3TP-lux), which was inhibited by overexpression of Mtmr4 (Fig. 4E). Therefore, MTMR4 most likely regulates TGFβ signaling through inhibition of R-Smad phosphorylation and nuclear translocation.
The DSP Activity of MTMR4 Inactivates Smad3
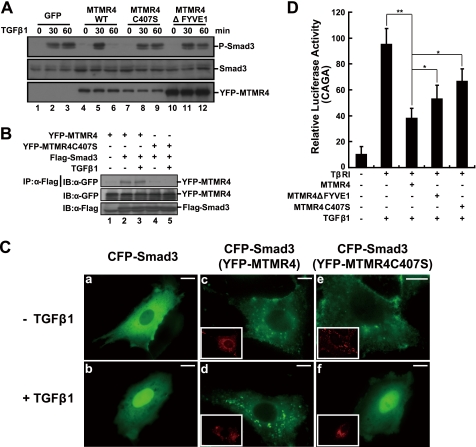
To substantiate the above notion that MTMR4 attenuates TGFβ signaling by dephosphorylating R-Smads, we transiently expressed a DSP-dead mutant (MTMR4-C407S) in HeLa cells. As expected, MTMR4-C407S lost its ability to dephosphorylate Smad3 after TGF-β1 treatment (Fig. 5A, compare lanes 5 and 6 and 8 and 9). Specifically, co-immunoprecipitation experiments showed that MTMR4-C407S failed to interact with Smad3 (Fig. 5B), supporting the notion that MTMR4 used its DSP domain to contact and dephosphorylate the MH2 domain of Smad3 (Fig. 3E). Unexpectedly, however, MTMR4ΔFYVE1 did not dephosphorylate Smad3 as efficiently as wild-type MTMR4, although it possessed an intact DSP domain (Fig. 5A, lanes 11 and 12). This might be due to the fact that cytosol-localized MTMR4ΔFYVE1 (Fig. 1B) was no longer in contact with activated R-Smads in early endosomes for efficient dephosphorylation. Fluorescence microscopy further revealed that overexpression of MTMR4 resulted in the sequestration of Smad3 in early endosomes, thus preventing Smad3 from leaving early endosomes for the nucleus (Fig. 5C, c–d). As expected, MTMR4-C407S, which is deficient for DSP phosphatase activity, was unable to prevent nuclear trafficking of Smad3 (Fig. 5C, e–f). Finally, in comparison with the apparent inhibition of Smad3-responsive CAGA reporter activity by MTMR4, overexpression of MTMR4-C407S resulted in a greatly reduced suppression of the Smad3-dependent transcriptional response to TGF-β1 (Fig. 5D). The less efficient dephosphorylation of Smad3 by the cytosol-localized MTMR4ΔFYVE1 yielded slightly increased CAGA reporter activities compared with wild-type MTMR4. These results further suggest that the intrinsic DSP activity of MTMR4 might be specifically involved in Smad3 dephosphorylation and attenuation of TGFβ signaling.
FIGURE 5.
The phosphatase catalytic pocket and the FYVE domain of MTMR4 are both critical for TGF-β signaling regulation. A, MTMR4 phosphatase-dead mutant C407S or FYVE domain deletion mutant fails to dephosphorylate activated Smad3. HeLa cells expressing control vector (3.75 μg, lanes 1–3), YFP-MTMR4 (3.75 μg, lanes 4–6), YFP-MTMR4-C407S (3.75 μg, lanes 7–9), or YFP-MTMR4ΔFYVE1 (3.75 μg, lanes 10–12) were treated with or without TGF-β1 for the times indicated. Endogenous activated Smad3 was immunoblotted with an anti-phosphor-Smad3 antibody (top panel). Protein loading of Smad3, YFP-MTMR4, and different mutants was verified by immunoblotting with anti-Smad3 antibody (middle panel) or anti-GFP antibody (bottom panel), respectively. B, phosphatase catalytic pocket of MTMR4 is in contact with Smad3. 293T cells were transiently cotransfected with pYFP-MTMR4 or pYFP-MTMR4-C407S (1.5 μg) and Flag-tagged Smad3 (1 μg) for 36 h. Cell lysates were immunoprecipitated with anti-Flag antibody, and blotted with anti-GFP antibody (top). Expression levels of YFP-MTMR4 (middle) and Smad3 (bottom) were also shown. C, MTMR4 sequesters Smad3 in early endosomes. PAE cells were transiently cotransfected with pCFP-Smad3 (green) and pYFP-MTMR4 or pYFP-MTMR4-C407S (red, insets) for 36 h. The subcellular localization of Smad3 and MTMR4 was detected using GFP and YFP channels, respectively, before and after TGF-β1 treatment. D, MTMR4 phosphatase-dead mutant and the FYVE domain deletion mutant fail to suppress TGFβ-induced 3TP-luciferase reporter activities efficiently. L17 cells reconstituted with TβRI were transiently cotransfected with CAGA reporter plasmid, and pCMV-hRL and either pYFP-MTMR4 30 ng, YFP-MTMR4-C407S 30 ng, or YFP-MTMR4ΔFYVE1 30 ng for 36 h. Luciferase assays were performed as described in Fig. 1C. **, p < 0.01; *, p < 0.05. The scale bar indicates 10 μm.
DISCUSSION
It has become evident that phosphorylation and dephosphorylation of R-Smads is a crucial regulatory mechanism in the control of the activation and termination of TGFβ signaling. The FYVE domain-containing proteins, such as SARA (10), Hgs/Hrs (33) and endofin (34), participate in anchoring Smad2 to early endosomes and present it to TβRI for signal transactivation. In the present work, we have shown for the first time that MTMR4, another FYVE domain-containing protein with a conserved PTP/DSP domain (35), specifically down-regulates the phosphorylation level of activated Smad2/3 in early endosomes to prevent R-Smads from being overactivated. This is particularly interesting because MTMR4 might provide an additional layer for fine-tuning the nucleocytoplasm shuttling of activated R-Smads. PPM1A terminates TGFβ signaling by dephosphorylation of activated Smad2/3 in the nucleus enabling their nuclear export (17). MTMR4, by residing in early endosomes, may help to prevent overactivation of R-Smads and provide the first checkpoint for maintaining the homeostasis of TGFβ signaling.
MTMR4 possesses a few unique features for gate-keeping TGFβ signaling. First, unlike most R-Smad anchors of FYVE-containing molecules that positively regulate TGFβ/BMP signaling (10, 33, 34, 36), MTMR4 negatively regulates TGFβ signaling through R-Smads dephosphorylation. Second, unlike most transcription co-factors (12) or phosphatases (17) that interact with Smads in the nucleus, MTMR4 constitutively resides in early endosomes (20) and most likely dephosphorylates Smad2/3 in situ. This explains why dislocated MTMR4-ΔFYVE1, with its intact PTP/DSP catalytic domain, can no longer efficiently dephosphorylate p-Smad2/3, and hence only weakly inhibited CAGA-luciferase activity. On the other hand, because MTMR4 forms a homodimer (20), that overexpression of the phosphatase-dead mutant MTMR4C407S antagonized endogenous MTMR4 DSP activity on R-Smads seems reasonable.
Intriguingly, MTMR4 most likely comes into play at a relatively late phase of TGFβ signaling, as demonstrated by the normal Smad3 activation observed within 30 min of TGF-β1 stimulation. This can be explained by the requirement of phosphorylated R-Smads for contact with MTMR4 and the physical separation of inactivated R-Smads from MTMR4. In the first 5–15 min of TGF-β1 stimulation, we observed a rather swift accumulation of phosphorylated R-Smads in endosomes (data not shown), and it reached a peak around 30 min of TGF-β1 stimulation. Failure of MTMR4 to block the transcriptional response to dominant active Smad3-SD further supports the notion that MTMR4 acts after R-Smads activation.
It is a long time since the suppressive effects of TGFβ signaling in the cytoplasm, such as inhibition of the internalization of TGFβ receptors, and degradation of receptors or R-Smads, have been identified (6, 37), yet to our knowledge there are no previous reports showing that R-Smads is dephosphorylated in the cytoplasm. Here we have demonstrated that MTMR4 gate-keeps the phosphorylation level of R-Smads in the cytoplasm, thus preventing their overactivation, and collaborates with nuclear phosphatase(s) to turn off the TGFβ signal system after it has served its purpose.
Supplementary Material
Acknowledgments
We thank Drs. X. Yang (Military Academy of Medicine, Beijing) and Z.-J. Zhao (University of Oklahoma Health Sciences Center) for kindly providing various constructs and antisera. We thank Drs. R.-W. Xi (National Institute of Biological Sciences, Beijing), J. Zhang (Institute of Genetics and Developmental Biology, CAS), Xin-hua Feng (Baylor College of Medicine), and Ting Xie (Stowers Institute for Medical Research) for technical assistance and stimulating discussions. We are grateful to Dr. Joy Fleming for editing the English in the manuscript and for incisive and helpful comments.
This work was supported in part by grants from the National Basic Research Program of MOST (2006CB504300, 2007DFC30190, 2009CB522506) (to H. T.) and NSFC (30428002) and MOST (2004CB720002, 2006CB910100) (to Y. C.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Fig. S1.
J. Yu, X. Qin, and H. Tang, unpublished results.
- TGFβ
- transforming growth factor β
- MTMR
- myotubularin-related protein
- FBS
- fetal bovine serum
- DSP
- dual-specificity protein phosphatase
- PAE
- porcine aorta endothelial
- YFP
- yellow fluorescent protein
- SARA
- Smad anchor for receptor activation.
REFERENCES
- 1.Brummel T. J., Twombly V., Marqués G., Wrana J. L., Newfeld S. J., Attisano L., Massagué J., O'Connor M. B., Gelbart W. M. (1994) Cell 78, 251–261 [DOI] [PubMed] [Google Scholar]
- 2.Massagué J., Attisano L., Wrana J. L. (1994) Trends Cell Biol. 4, 172–178 [DOI] [PubMed] [Google Scholar]
- 3.Inman G. J., Nicolás F. J., Hill C. S. (2002) Mol. Cell 10, 283–294 [DOI] [PubMed] [Google Scholar]
- 4.Shi Y., Massagué J. (2003) Cell 113, 685–700 [DOI] [PubMed] [Google Scholar]
- 5.Xu L., Massagué J. (2004) Nat. Rev. Mol. Cell Biol. 5, 209–219 [DOI] [PubMed] [Google Scholar]
- 6.Massagué J., Chen Y. G. (2000) Genes Dev. 14, 627–644 [PubMed] [Google Scholar]
- 7.Wotton D., Lo R. S., Lee S., Massagué J. (1999) Cell 97, 29–39 [DOI] [PubMed] [Google Scholar]
- 8.Di Guglielmo G. M., Le, Roy C., Goodfellow A. F., Wrana J. L. (2003) Nat. Cell Biol. 5, 410–421 [DOI] [PubMed] [Google Scholar]
- 9.Itoh T., Takenawa T. (2002) Cell Signal. 14, 733–743 [DOI] [PubMed] [Google Scholar]
- 10.Tsukazaki T., Chiang T. A., Davison A. F., Attisano L., Wrana J. L. (1998) Cell 95, 779–791 [DOI] [PubMed] [Google Scholar]
- 11.Hayes S., Chawla A., Corvera S. (2002) J. Cell Biol. 158, 1239–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Massagué J., Seoane J., Wotton D. (2005) Genes Dev. 19, 2783–2810 [DOI] [PubMed] [Google Scholar]
- 13.Lo R. S., Massagué J. (1999) Nat. Cell Biol. 1, 472–478 [DOI] [PubMed] [Google Scholar]
- 14.Pierreux C. E., Nicolás F. J., Hill C. S. (2000) Mol. Cell. Biol. 20, 9041–9054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen H. B., Shen J., Ip Y. T., Xu L. (2006) Genes Dev. 20, 648–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Knockaert M., Sapkota G., Alarcón C., Massagué J., Brivanlou A. H. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 11940–11945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin X., Duan X., Liang Y. Y., Su Y., Wrighton K. H., Long J., Hu M., Davis C. M., Wang J., Brunicardi F. C., Shi Y., Chen Y. G., Meng A., Feng X. H. (2006) Cell 125, 915–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laporte J., Bedez F., Bolino A., Mandel J. L. (2003) Hum. Mol. Genet. 12, R285–R292 [DOI] [PubMed] [Google Scholar]
- 19.Zhao R., Qi Y., Chen J., Zhao Z. J. (2001) Exp. Cell Res. 265, 329–338 [DOI] [PubMed] [Google Scholar]
- 20.Lorenzo O., Urbé S., Clague M. J. (2006) J. Cell Sci. 119, 2953–2959 [DOI] [PubMed] [Google Scholar]
- 21.Zhao X., Nicholls J. M., Chen Y. G. (2008) J. Biol. Chem. 283, 3272–3280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weng C., Li Y., Xu D., Shi Y., Tang H. (2005) J. Biol. Chem. 280, 10491–10500 [DOI] [PubMed] [Google Scholar]
- 23.Dennler S., Itoh S., Vivien D., ten, Dijke P., Huet S., Gauthier J. M. (1998) EMBO J. 17, 3091–3100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tang H., Sharp P. A. (1999) Immunity 11, 517–526 [DOI] [PubMed] [Google Scholar]
- 25.Chen Y., Chen J., Xiong Y., Da Q., Xu Y., Jiang X., Tang H. (2006) Biochem. Biophys. Res. Commun. 345, 106–117 [DOI] [PubMed] [Google Scholar]
- 26.O'Connell R. M., Saha S. K., Vaidya S. A., Bruhn K. W., Miranda G. A., Zarnegar B., Perry A. K., Nguyen B. O., Lane T. F., Taniguchi T., Miller J. F., Cheng G. (2004) J. Exp. Med. 200, 437–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wrana J., Attisano L., Cárcamo J., Zentella A., Doody J., Laiho M., Wang X. F., Massagué J. (1992) Cell 71, 1003–1014 [DOI] [PubMed] [Google Scholar]
- 28.Itoh F., Divecha N., Brocks L., Oomen L., Janssen H., Calafat J., Itoh S., Dijke Pt., P. (2002) Genes Cells 7, 321–331 [DOI] [PubMed] [Google Scholar]
- 29.Aoki C. A., Borchers A. T., Li M., Flavell R. A., Bowlus C. L., Ansari A. A., Gershwin M. E. (2005) Autoimmun Rev. 4, 450–459 [DOI] [PubMed] [Google Scholar]
- 30.Massagué J., Blain S. W., Lo R. S. (2000) Cell 103, 295–309 [DOI] [PubMed] [Google Scholar]
- 31.Feng X. H., Liang Y. Y., Liang M., Zhai W., Lin X. (2002) Mol. Cell 9, 133–143 [DOI] [PubMed] [Google Scholar]
- 32.Kurokawa M., Mitani K., Irie K., Matsuyama T., Takahashi T., Chiba S., Yazaki Y., Matsumoto K., Hirai H. (1998) Nature 394, 92–96 [DOI] [PubMed] [Google Scholar]
- 33.Miura S., Takeshita T., Asao H., Kimura Y., Murata K., Sasaki Y., Hanai J. I., Beppu H., Tsukazaki T., Wrana J. L., Miyazono K., Sugamura K. (2000) Mol. Cell. Biol. 20, 9346–9355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen Y. G., Wang Z., Ma J., Zhang L., Lu Z. (2007) J. Biol. Chem. 282, 9688–9695 [DOI] [PubMed] [Google Scholar]
- 35.Gallego M., Virshup D. M. (2005) Curr. Opin. Cell Biol. 17, 197–202 [DOI] [PubMed] [Google Scholar]
- 36.Shi W., Chang C., Nie S., Xie S., Wan M., Cao X. (2007) J. Cell Sci. 120, 1216–1224 [DOI] [PubMed] [Google Scholar]
- 37.Schilling S. H., Datto M. B., Wang X. F. (2006) Cell 125, 838–840 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.