Abstract
Background
Gene expression profiling has successfully identified the prognostic significance of the host response in lymphomas. The aggressive T-cell/histiocyte-rich large B-cell lymphoma and the indolent nodular lymphocyte-predominant Hodgkin’s lymphoma are both characterized by a paucity of tumor cells embedded in an overwhelming background. The tumor cells of both lymphomas share several characteristics, while the cellular composition of their microenvironment is clearly different.
Design and Methods
We collected 33 cases of T-cell/histiocyte-rich large B-cell lymphoma and 56 cases of nodular lymphocyte-predominant Hodgkin’s lymphoma and performed microarray gene expression profiling on ten cases of each lymphoma, to obtain a better understanding of the lymphoma host response. By quantitative reverse transcriptase polymerase chain reaction we verified that these 20 selected cases were representative of the entire population of T-cell/histiocyte-rich large B-cell and nodular lymphocyte-predominant Hodgkin’s lymphomas.
Results
We observed that the microenvironment in nodular lymphocyte-predominant Hodgkin’s lymphoma is molecularly very similar to a lymph node characterized by follicular hyperplasia, while the microenvironment in T-cell/histiocyte-rich large B-cell lymphoma is clearly different. The T-cell/histiocyte-rich large B-cell lymphoma signature is hallmarked by up-regulation of CCL8, interferon-γ, indoleamine 2,3 dioxygenase, VSIG4 and Toll-like receptors. These features may be responsible for the recruitment and activation of T cells, macrophages and dendritic cells, characterizing the stromal component of this lymphoma, and may point towards innate immunity and a tumor tolerogenic immune response in T-cell/histiocyte-rich large B-cell lymphoma.
Conclusions
The gene expression profile of T-cell/histiocyte-rich large B-cell lymphoma, in comparison with that of nodular lymphocyte-predominant Hodgkin’s lymphoma, shows features suggestive of a distinct tolerogenic host immune response that may play a key role in the aggressive behavior of this lymphoma, and that may serve as a potential target for future therapy.
Keywords: gene expression profiling, Hodgkin’s lymphoma, THRLBCL
Introduction
B-cell lymphomas with a high content of T cells, occasionally misinterpreted as T-cell lymphomas in the past, have been recognized as a peculiarity by pathologists and were, therefore, indicated as “T-cell-rich B-cell lymphoma”.1 Initial studies demonstrated that a particular subgroup of T-cell-rich B-cell lymphomas mirror nodular lymphocyte-predominant Hodgkin’s lymphoma (NLPHL) and are characterized by a T-cell and histiocyte-rich stroma.2,3 These lymphomas have a distinct clinical behavior and a bad prognosis.4 In the World Health Organization (WHO) classification of 2001, T-cell/histiocyte-rich large B-cell lymphoma (THRLBCL) is defined by the presence of a limited number of scattered large B cells in a background rich in T cells, with or without histiocytes.5 When following this definition, THRLBCL is indeed a heterogeneous disease.6 However, using a stricter definition requiring the presence of a prominent histiocytic component as a major feature and almost complete absence of small B lymphocytes, the unique character of this lymphoma has been demonstrated.7,8
The precise relationship between THRLBCL and NLPHL remains unclear.2,3,9 Indeed, the atypical B cells of NLPHL and THRLBCL share many characteristics, including expression of pan-B-cell markers, germinal center B-cell origin and common chromosomal imbalances.8,10–12 Recently, genome-wide analysis of isolated tumor cells from NLPHL and THRLBCL revealed further similarities between the tumors cells of the two lymphomas.13 Despite the similarities of their malignant cells, an important difference between the two lymphomas lies in their clinical presentation and prognosis. THRLBCL is a very aggressive disorder, which often does not respond to therapy.7 Patients with THRLBCL frequently present with stage III or IV disease, splenomegaly, hepatomegaly and bone marrow involvement. In contrast, NLPHL is an indolent disorder. Most patients are diagnosed at an early stage of disease and have a good prognosis.14
Gene expression profiling of lymphomas clearly illustrated that apart from the characteristics of the tumor cells, the microenvironment of the tumor also defines the profile of the lymphoma, and, more importantly, plays a role in predicting the prognosis.15,16 In the present study, we used gene expression profiling on full tissue sections to evaluate the profile of the microenvironment as a marker that identifies THRLBCL and NLPHL as two distinct entities.
Design and Methods
Patients
A series of 98 cases, all documented by frozen material, were retrieved from the files of the Department of Pathology of the University Hospitals of K.U. Leuven. The series includes all cases recorded over the last 25 years (i) as Hodgkin’s lymphoma rich in lymphocytes (NLPHL or lymphocyte-rich classical Hodgkin’s lymphoma) or (ii) as diffuse large B-cell lymphoma (DLBCL) with a prominent T-cell-rich stromal component. As an additional and external control series, 26 similarly selected cases recorded at the Department of Pathology of the Rikshospitalet-Radiumhospitalet HF Oslo were added to the study material. All cases were reviewed and the 2008 version of the WHO criteria were applied to assign cases to the different categories.17 Thirty-one cases were excluded from the study because the frozen material was not representative, additional material for review or further immunostains was not available, or because upon review they were diagnosed as DLBCL rich in T cells but lacking a prominent histiocytic stromal component or fulfilled the criteria for diagnosis as classical lymphocyte-rich Hodgkin’s lymphoma. A diagnosis of THRLBCL and NLPHL was confirmed in 34 and 57 cases, respectively. In all these cases, the atypical cells represented less than 10% of the tumor mass. Finally, two cases were excluded from the study because an unambiguous diagnosis of NLPHL or THRLBCL could not be made; their morphological features resembled those of the cases described by Boudova et al.18
From the Leuven series, we randomly selected ten typical NLPHL and ten typical THRLBCL cases for microarray expression profiling. Finally, a pool of five reactive lymph node biopsies, characterized by follicular hyperplasia, was constructed for use as reference tissue. Most lymphoma cases were included in one of our previous studies on NLPHL and/or THRLBCL.3,7,8,12,19
This study was approved by the local ethical commissions of the University Hospitals K.U. Leuven.
RNA extraction
Total RNA was extracted from 20 micron sections of each frozen tissue sample using the TriZol reagent (Invitrogen, Merelbeke, Belgium), followed by purification using an RNeasy mini kit (Qiagen, Venlo, The Netherlands), according to the manufacturers’ recommendations. RNA quality and concentration were measured using a Nanodrop spectrophotometer (Nanodrop Technologies, Wilmington, DE, USA).
Gene expression profiling
Five micrograms of RNA were biotin-labeled and hybridized onto human oligonucleotide microarrays (Affymetrix HG-U133 Plus 2.0; Affymetrix, High Wycombe, UK). The resulting data are available online at the Gene Expression Omnibus (www.ncbi.nlm.nih.gov/projects/geo/), accession number GSE7788. These data were analyzed using Bioconductor software.20 Statistical testing for genes differentially expressed between the two types of lymphomas was done by a t-test. Corrections for multiple testing were made using a step-down maxT procedure.21
The statistical significance of overlap with other expression profiling studies was calculated using hypergeometric statistics.
Immunohistochemistry
Apart from the immunohistochemical stains used for diagnostic purposes, including CD20, CD3, CD4, CD8, CD23 and CD57 stain, paraffin-embedded sections were stained with a commercially available mouse anti-indoleamine 2,3 dioxygenase (IDO) monoclonal antibody (Chemicon International) and a rabbit anti-STAT1 polyclonal antibody (STAT1 p84/p91, Santa Cruz Biotechnology), following the manufacturers’ recommendations.
Results and Discussion
Clinical data
The clinical characteristics of the patients are summarized in Table 1. There is a clear male predominance in both THRLBCL and NLPHL. Ann Arbor staging, the International Prognostic Index (IPI) and the initial response to treatment confirm that THRLBCL is a very aggressive disease, while NLPHL is an indolent disorder. These results were further strengthened by the Kaplan-Meier estimates of overall survival (Online Supplementary Figure S1).
Table 1.
Clinical data.
Expression profiling of T-cell/histiocyte-rich large B-cell lymphoma compared to nodular lymphocyte-predominant Hodgkin’s lymphoma
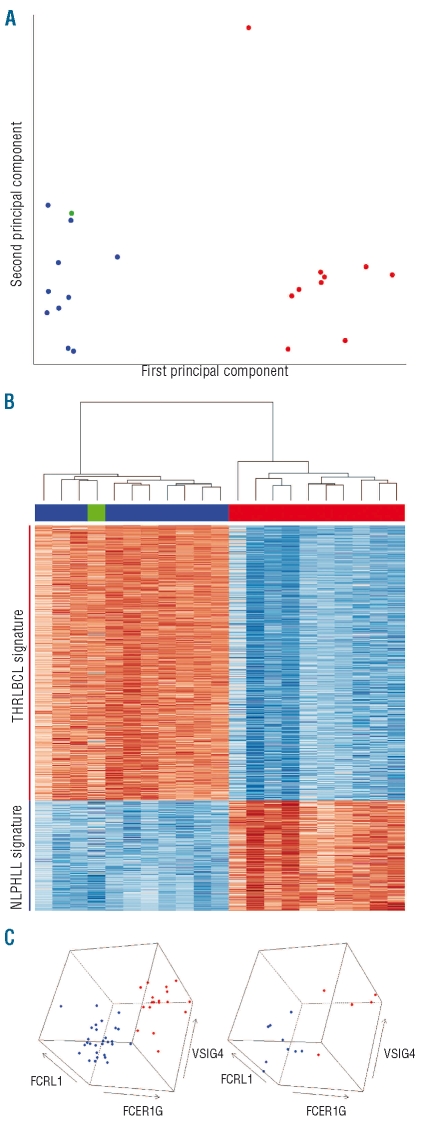
Since malignant cells in both conditions are rare (i.e. comprising considerably less than 10% of all cells on the tissue sections) and share many features,13 we assumed that the gene expression profiling of the entire tissue sections represented predominantly the profile of the microenvironment. Principal component analysis revealed a clear distinction between these two lymphomas (Figure 1A). One THRLB-CL was clearly separated from the other THRLBCL cases. Interestingly, this was the only sample taken from a spleen, while all other samples originated from lymph nodes. As the separation of this sample from the other THRLBCL tumors was in a direction perpendicular to the direction of separation of THRLBCL and NLPHL (Figure 1A), this sample was not removed in subsequent analyses. However, as a control, all subsequent analyses were repeated leaving out this aberrant sample, revealing similar results (data not shown). The reactive lymph node pool was located near the NLPHL samples, in agreement with expectations. Indeed, the microenvironment in NLPHL comprises components of the B follicle (with numerous small B cells, follicular dendritic cells and follicular T cells) as well as adjacent T-cell areas (with numerous T cells), while remnants of B follicles or their components are mostly absent in THRLBCL.
Figure 1.
Expression profiling of NLPHL and THRLBCL. (A) Principal component analysis, performed on the complete microarray data (54675 probe sets) of ten NLPHL cases, ten THRLBCL cases, and a reference pool of lymph nodes with follicular hyperplasia. Blue: NLPHL; red: THRLBCL; green: reactive lymph node pool. The first principal component (separating NLPHL from THRLBCL) captured 42% of the total variance. The second principal component captured 11% of the total variance. (B) Heat map of the 874 differentially expressed probesets (527 unique genes, Online Supplementary Table S1). Top: cluster dendrogram, showing the a priori expected separation between the THRLBCL and NLPHL samples, and confirming the similarity between NLPHL and the reactive lymph node reference; middle: identity of samples (colors as in A); bottom: graphical representation of gene expression (blue: high expression; red: low expression). (C) Expression of three selected genes, measured by real-time quantitative PCR, in 55 in-house (left) and 14 external (right) THRLBCL and NLPHL cases. Colors as in A.
Using highly significant differentially expressed genes, we constructed expression signatures for THRLBCL and NLPHL (Online Supplementary Figure S2). The THRLBCL signature comprised 392 genes, while the NLPHL signature contained 135 genes (Figure 1B, Online Supplementary Table S1). Consistent with the principal component analysis above, the reactive lymph node reference sample clustered together with the ten NLPHL cases, when the 21 microarray profiles were clustered using only these two gene expression signatures (Figure 1B). Finally, the sample originating from a THRLBCL located in the spleen again clustered together with the other THRLBCL cases. This result persisted even when only the other nine THRLBCL samples were used for building the gene expression signatures (data not shown).
Finally, we validated the obtained gene expression data by real-time quantitative reverse transcriptase polymerase chain reaction (RT-PCR) (Online Supplementary Design and Methods), obtaining similar results (Online Supplementary Table S2).
T-cell/histiocyte-rich large B-cell lymphoma and nodular lymphocyte-predominant Hodgkin’s lymphoma gene expression signatures in additional cases
As we observed large differences between the expression profiles of NLPHL and THRLBCL, we wondered whether a simple and intuitive view of gene expression in these lymphomas, based on real-time quantitative RT-PCR measurements of a very limited number of genes, would be able to discriminate additional cases of the two entities. We, therefore, selected three genes from the gene expression signatures and assayed these in 69 additional cases of NLPHL and THRLBCL (both in-house and external), based on quantitative RT-PCR measurements (see Online Supplementary Design and Methods and Online Supplementary Table S3). Using only the expression values for these three selected genes, THRLBCL and NLPHL presented as two distinct groups (Figure 1C). These results further confirmed that the NLPHL and THRLBCL cases selected for microarray expression analysis were representative of the two lymphoma entities.
The gene expression signature of nodular lymphocyte-predominant Hodgkin’s lymphoma: a predominance of B-cell genes
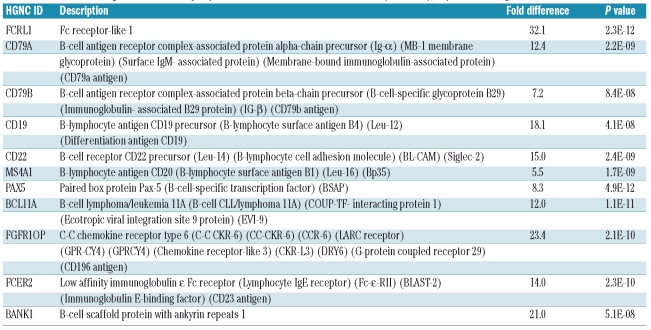
In comparison with the gene expression profile of THRL-BCL, the expression signature of NLPHL comprises mainly genes characteristic of B cells (Table 2A, Online Supplementary Table S1A), in line with the morphological findings. Moreover, the observed similarities between the expression profiles of NLPHL and the reactive lymph nodes, characterized by follicular hyperplasia, suggest that the components of the B follicle play a major role in both profiles. This predominance of B cells in the NLPHL microenvironment was confirmed by immunohistochemical staining for CD20, a B-cell marker (Figure 2 and Table 3). In line with these findings, the NLPHL signature shows significant overlap with the signature Monti et al.16 found to be related to B-cell receptor/proliferation in a subgroup of DLBCL (of the 43 genes in the B-cell receptor/proliferation signature present on our microarray platform, 7 were a part of the NLPHL signature, P=4.4×10−8; Online Supplementary Table S4A). In contrast, the overlap with the oxidative phosphorylation signature and host response signature of Monti et al.16 was not more than would be randomly expected (2 genes and 0 genes, respectively).
Table 2A.
A selection of genes differentially expressed between NLPHL and THRLBCL (P<0.001), expressed at higher levels in NLPHL.
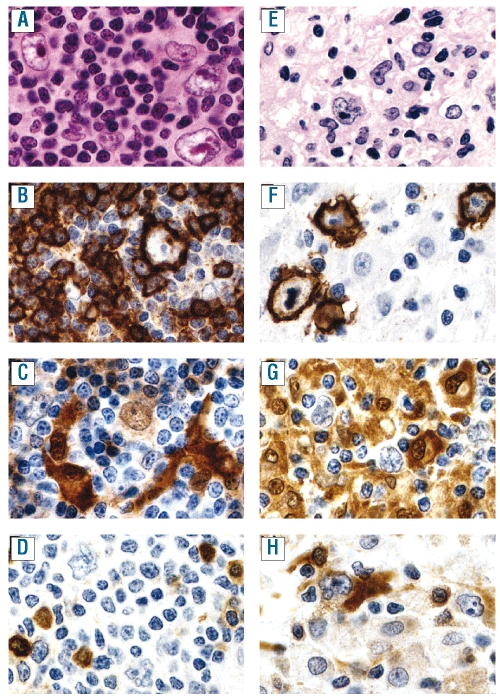
Figure 2.
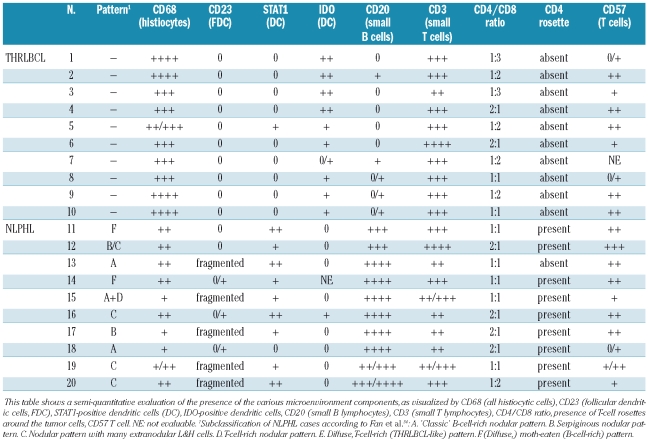
Representative pictures of the hematoxylin and eosin (A, E), CD20 (B, F), STAT1 (C, G) and IDO (D, H) stains on NLPHL (A–D) and THRLBCL (E–H), all taken with a 40X objective. (A) Atypical cells (popcorn cells) in NLPHL embedded in a background rich in small lymphocytes. (B) CD20 expression by a popcorn cell surrounded by CD20-negative lymphocytes, resulting in a T-cell rosette embedded in CD20-positive small B cells. (C) STAT1 is weakly expressed by most popcorn cells in the nucleus and in the cytoplasm, in some of the small lymphocytes and in large dendritic cells adjacent to the T-cell rosettes. (D) Popcorn cells, as well as surrounding lymphocytes do not express IDO, while some plasma cell-like, non-dendritic cells express IDO in their cytoplasm and in their nucleus. These IDO-positive cells might correspond to plasmacytoid monocytes/plasmacytoid dendritic cells. (E) The atypical cells of THRLBCL, embedded in a stroma rich in histiocytes and a limited number of small lymphocytes. (F) CD20 expression by the tumor cells, and absence of small CD20+ lymphocytes in the surrounding stroma. (G) Most of the stromal components of THRLBCL express STAT1, while the tumor cells are clearly negative. (H) IDO is expressed in small plasma cell-like cells in THRLBCL, assumed to correspond to plasmacytoid monocytes/plasmacytoid dendritic cells (as in NLPHL). In addition, large dendritic cells frequently located nearby tumor cells also clearly express IDO in all of the THRLBCL (and none of the NLPHL) cases.
Table 3.
Summary of the immunohistochemical findings of the composition of the microenvironement within the tumor areas in the ten cases each of THRLBCL and NLPHL used for gene expression profiling.
Three genes that were expressed in our NLPHL signature overlapped with the signature Brune et al.13 found in microdissected NLPHL cells compared to microdissected THRLBCL cells (Online Supplementary Table S4B). This finding may suggest that some genes are expressed by the lymphoma cells as well as the cells in the microenvironment. However, high expression of these genes only in the NLPHL tumor cells, combined with lack of expression in THRLBCL tumor cells and in the cells of the microenvironment of both entities, would explain this result as well. Although this overlap strongly suggests that our gene expression signatures are not solely those of the cells in the microenvironment, the limited degree of this overlap (39 of the 42 genes of the NLPHL tumor cell signature determined by Brune et al. were not present in our 135 gene NLPHL signature) confirms that the major part of our NLPHL signature is not determined by the tumor cells.
The gene expression signature of T-cell/histiocyte-rich large B-cell lymphoma: a crucial role for interferon-γ and innate immune responses
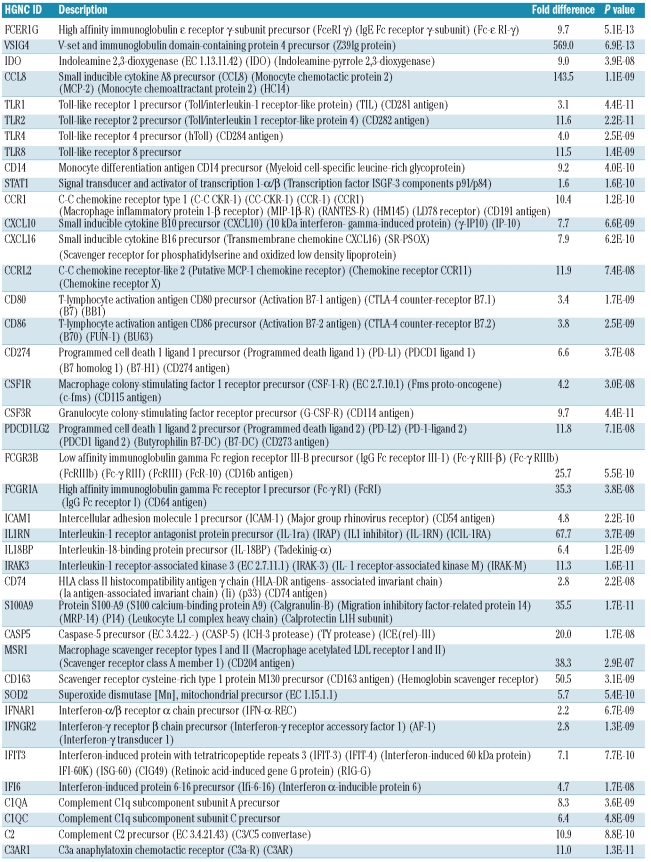
The THRLBCL signature underlines the crucial role of an IFN-γ regulated and tolerogenic pathway within the microenvironment. Indeed, IFN-γ is up-regulated in THRL-BCL (Online Supplementary Table S2), as are several genes encoding for proteins that are up-regulated in macrophages and dendritic cells upon treatment with IFN-γ.22,23 (Table 2B, Online Supplementary Table S1B), indicative of an activated macrophage status. These genes include those encoding for STAT1, Fc-γ receptor I (FcRI or CD64), ICAM-1, IFN-γ induced protein (IP-10/CXCL10), CXCL16 and, in particular, CCL8 and IDO (Table 2B, Online Supplementary Table S1B). CCL8, also designated as monocyte chemotactic protein 2 (MCP2), belongs to the CC chemokines. It is strongly induced by IFN-γ24 and is one of the most potent chemoattractants for mononuclear cells, including monocytes and T cells.22 Thus, CCL8 may contribute to the histiocyte-rich (and T-cell rich) composition of the microenvironment in THRLBCL. IFN-γ also promotes, in a STAT1-dependent way, the induction of the tryptophan-degrading enzyme IDO in monocytes, macrophages and dendritic cells.25,26 Interestingly, both B7-1 (CD80) and B7-2 (CD86) were part of this signature, and through a reverse interaction with CTLA4, these membrane proteins have been shown to activate IDO expression, as reviewed by Munn and Mellor.27 IDO has been described to promote tumor immune tolerance by suppressing local T-cell responses and by altering the conversion of effector T cells into T regulatory cells.28,29 Intriguingly, VSIG4 (V-set and Ig domain-containing 4, also known as Z39Ig), one of the most significant and strongly up-regulated genes of the THRLBCL signature, is a B7 family-related protein expressed by macrophages and dendritic cells and acts as a strong negative regulator of CD4 and CD8 T-cell activation in vitro and in vivo.30 Thus, together with IDO, VSIG4 may contribute to a state of immune suppression and tumor tolerance. Of interest, aside from its suppressive properties on T cells, VSIG4 has also been recognized as a new complement receptor of the immunoglobulin superfamily (CRIg), required for phagocytosis of circulating pathogens.31 In line with this finding, the THRLBCL signature includes scavenger receptors (CXCL16, MSR1, CD163) and Toll-like receptors (TLR1, TLR2, TLR4 and TLR8). These data are indicative of innate immune responses in THRLBCL. A possible involvement of pathogens in the initiation or propagation of the disease cannot, therefore, be excluded and should be further investigated.
Table 2B.
A selection of genes differentially expressed between NLPHL and THRLBCL (P<0.001), expressed at higher levels in THRLBCL.
The THRLBCL signature shows significant overlap with the signature Dave et al.15 found to be related to an unfavorable immune response in some follicular lymphomas (9 of 23 genes, P=8.7×10−10, Online Supplementary Table S4C). In addition, the signature Monti et al.16 found to be related to host response in a subgroup of DLBCL, enriched in THRL-BCL cases, was also significantly overrepresented in our THRLBCL signature (14 of 59 genes, P=3.8×10−11, Online Supplementary Table S4D). In contrast, the favorable immune response signature found by Dave et al.,15 as well as the oxidative phosphorylation and B-cell receptor/proliferation signatures of Monti et al.,16 did not overlap with our THRLBCL signature more than would have been randomly expected.
Remarkably, nine genes of our THRLBCL signature were also found to be up-regulated in microdissected lymphoma cells in NLPHL compared to in normal B cells and in B-cell non-Hodgkin’s lymphoma (P=8.5×10−11, Online Supplementary Table S4E), and four genes of our THRLBCL signature were found to be up-regulated in microdissected lymphoma cells in NLPHL versus classical Hodgkin’s lymphoma (and germinal center B cells)13 (P=5.4×10−5, Online Supplementary Table S4F). One of these genes is STAT1, a signal transducer and activator of transcription factors in response to interferons. In order to find an explanation for this apparent discrepancy, we performed immunohistochemistry for STAT1 (and other markers) on THRLBCL and NLPHL tissue sections. A representative stain is shown in Figure 2 and a summary is listed in Table 3. Staining for STAT1 revealed that in THRLBCL almost all stromal cells were positive, whereas in NLPHL STAT1 staining was restricted to dendritric cells (in close vicinity to the T-cell rosettes around the tumor cells) and to the tumor cells itself. The STAT1-positive cells in NLPHL may correspond to the STAT1-positive cells described in a subpopulation of follicular lymphomas.32 The differences in staining pattern that we found between THRLBCL and NLPHL may explain the increased STAT1 gene expression in THRLBCL versus NLPHL, as well as the upregulation of the STAT1 gene in NLPHL in the study by Brune et al.13
When sections were stained for IDO, we observed strongly IDO-positive dendritic-like cells in THRLBCL, in close contact with small lymphocytes and/or tumor cells (Figure 2). These IDO-positive dendritic cells are not present in NLPHL. IDO-positive, round, medium-sized cells are found in both NLPHL and THRLBCL sections (Table 3). Based on their phenotype (CD68+, IDO+) these cells might be considered plasmacytoid monocytes or plasmacytoid dendritic cells.
Absence of T-cell genes in the nodular lymphocyte-predominant Hodgkin’s lymphoma and T-cell/histiocyte-rich large B-cell lymphoma gene expression signatures
Neither the NLPHL nor the THRLBCL gene expression signature contained a significant component of T-cell-associated genes. As shown in Online Supplementary Table S5, the absence of T-cell genes in this comparison is not due to our strict statistical cut-off, as even with a cut-off of P<0.05 after correction for multiple testing, none of the tested T cell-associated genes showed a significant difference. In addition, the ratio between CD4+ and CD8+ T cells, described to change in favor of the CD8+ cells in THRLB-CL8,33 was not reflected in the expression profiles either, although we did observe a (non-significant) tendency towards a higher expression of CD8α (Online Supplementary Table S5). These findings are in line with the immunohistochemical staining data we obtained for CD3, CD4 and CD8 (Table 3). Apart from a difference in the number of T cells within the tumor nodules, as previously described,34,35 all NLPHL cases comprised residual non-neoplastic T-cell areas. Furthermore, CD57 expression is described as a typical feature of the T cells surrounding the tumor cells in NLPHL, but the number of CD57+ T cells present in NLPHL is variable and CD57+ T cells are also found in THRLBCL.8,33 Our immunohistochemical data revealed similar results (Table 3). Thus, the absence of T-cell-associated genes in the THRLBCL expression signature might be regarded as a confirmation that it is not the T cells, but rather the macrophages/histiocytes that represent the functionally important component of the microenvironment in THRLBCL.
A host immune tolerogenic microenvironment in T-cell/histiocyte-rich large B-cell lymphoma as an explanation for the bad prognosis of patients with this lymphoma?
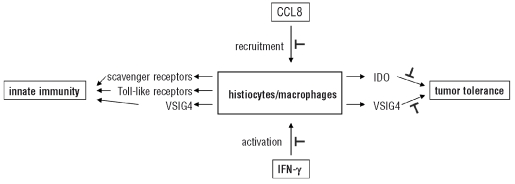
Tumor cells in general have acquired several mechanisms to escape from immune surveillance by immunocompetent cells. In both THRLBCL and NLPHL, the tumor cells produce proteins that counteract the activity of cytotoxic lymphocytes and NK cells.13 Together with their regulation of genes that are involved in inhibition of cell proliferation and programmed cell death, lymphoma cells can, therefore, continue to grow and escape immune attack. However, in addition to such a tumor-induced immune escape, we demonstrate here that the microenvironment of THRLB-CL, in comparison with that of NLPHL, is hallmarked by a distinct tolerogenic host immune response that may play a key role in the aggressive behavior of the former lymphoma. As schematically represented in Figure 3, we speculate that CCL8 and IFN-γ are responsible for, respectively, the recruitment and the activation of monocytes, macrophages and dendritic cells and, in synergy with TLR-ligands, for the production of high levels of IDO and VSIG4. IDO is at least partly produced by dendritic cells, a subpopulation of the numerous histiocytes characterizing the THRLBCL stroma. These dendritic cells are intensely stained by IDO immunohistochemistry, are in proximity of the tumor, and were not found in NLPHL (Figure 2). We speculate that it is this production of IDO and VSIG4 that results in a tolerogenic microenvironment of the tumor cells. This hypothesis, derived from observational data, could explain the bad prognosis of THRLBCL patients, but further investigation is required to pinpoint one of these mediators (CCL8, IFN-γ, IDO, VSIG4) as a novel target for therapy in the aggressive THRLBCL. Such investigations could include analysis of T-cell proliferation (from peripheral bone marrow cells of THRLBCL patients) upon challenge with lymphoma tissue samples, in the presence or absence of IFN-γ, IDO and VSIG4 inhibitors. In addition, a severe combined immunodeficiency xenograft mouse model might be of great value for evaluating the efficacy of these inhibitors on lymphoma tumor growth and lymphadenopathy.
Figure 3.
Schematic proposal of the host immune response in THRLBCL, based on our morphological and gene expression data, and on literature evidence. By morphology, the microenvironment of THRLBCL is hallmarked by the presence of histiocytes/macrophages. Gene expression profiling data confirm the central role of macrophages and/or dendritic cells and suggest that these cells may be recruited by CCL8.22,24 IFN-γ activates these cells to produce IDO.25,26 High levels of IDO, together with VSIG4, suppress the proliferation of effector T cells (such as CD8+ cytotoxic T cells), resulting in tumor tolerance.28,30 Macrophages and dendritic cells also express receptors involved in innate immunity, including scavenger and Toll-like receptors, and VSIG4 as a complement receptor.31 Blocking the production and/or the function of CCL8, IFN-γ, and in particular IDO and VSIG4 may abrogate the induction of tumor tolerance. It is encouraging to note that inhibitors to target IDO are available.36
Supplementary Material
Acknowledgments
The authors thank Andreas Rosenwald for careful review of the manuscript.
Footnotes
The online version of this article has a Supplementary Appendix.
Authorship and Disclosures
All authors meet the criteria for being contributing authors. PVL, PeterM, PatrickM and CDW-P designed the study; PVL, TT, VV, DD, AM, IVB, GV, JD and CDW-P collected data; PVL, TT, PatrickM and CDW-P analyzed and interpreted data; PVL performed the statistical analysis; PVL, TT, VV, PatrickM and CDW-P drafted the manuscript. All authors were involved in the discussion and all approved the final version of the manuscript.
The authors reported no potential conflicts of interest.
Funding: this work was supported by a Concerted Research Action Grant from the K.U. Leuven (Peter M) and grant G.05.28.06 from the Research Foundation – Flanders (FWO) (CDW-P). PVL is supported by a postdoctoral fellowship of the Research Foundation – Flanders (FWO).
References
- 1.Ramsay AD, Smith WJ, Isaacson PG. T-cell-rich B-cell lymphoma. Am J Surg Pathol. 1988;12(6):433–43. doi: 10.1097/00000478-198806000-00003. [DOI] [PubMed] [Google Scholar]
- 2.Chittal SM, Brousset P, Voigt JJ, Delsol G. Large B-cell lymphoma rich in T-cells and simulating Hodgkin’s disease. Histopathology. 1991;19(3):211–20. doi: 10.1111/j.1365-2559.1991.tb00024.x. [DOI] [PubMed] [Google Scholar]
- 3.Delabie J, Vandenberghe E, Kennes C, Verhoef G, Foschini MP, Stul M, et al. Histiocyte-rich B-cell lymphoma. A distinct clinicopathologic entity possibly related to lymphocyte predominant Hodgkin’s disease, paragranuloma subtype. Am J Surg Pathol. 1992;16(1):37–48. [PubMed] [Google Scholar]
- 4.Abramson JS. T-cell/histiocyte-rich B-cell lymphoma: biology, diagnosis, and management. Oncologist. 2006;11(4):384–92. doi: 10.1634/theoncologist.11-4-384. [DOI] [PubMed] [Google Scholar]
- 5.World Health Organization. Classification of Tumours Pathology and Genetics of Tumours of Haematopoietic and Lymphoid Tissues. Lyon, France: IARC Press; 2001. [Google Scholar]
- 6.Lim MS, Beaty M, Sorbara L, Cheng RZ, Pittaluga S, Raffeld M, et al. T-cell/histiocyte-rich large B-cell lymphoma: a heterogeneous entity with derivation from germinal center B cells. Am J Surg Pathol. 2002;26(11):1458–66. doi: 10.1097/00000478-200211000-00008. [DOI] [PubMed] [Google Scholar]
- 7.Achten R, Verhoef G, Vanuytsel L, De Wolf-Peeters C. T-cell/histiocyte-rich large B-cell lymphoma: a distinct clinicopathologic entity. J Clin Oncol. 2002;20(5):1269–77. doi: 10.1200/JCO.2002.20.5.1269. [DOI] [PubMed] [Google Scholar]
- 8.Achten R, Verhoef G, Vanuytsel L, De Wolf-Peeters C. Histiocyte-rich, T-cell-rich B-cell lymphoma: a distinct diffuse large B-cell lymphoma subtype showing characteristic morphologic and immunophenotypic features. Histopathology. 2002;40(1):31–45. doi: 10.1046/j.1365-2559.2002.01291.x. [DOI] [PubMed] [Google Scholar]
- 9.Rudiger T, Gascoyne RD, Jaffe ES, de JD, Delabie J, De Wolf-Peeters C, et al. Workshop on the relationship between nodular lymphocyte predominant Hodgkin’s lymphoma and T cell/histiocyte-rich B cell lymphoma. Ann Oncol. 2002;13 (Suppl 1):44–51. doi: 10.1093/annonc/13.s1.44. [DOI] [PubMed] [Google Scholar]
- 10.Brauninger A, Kuppers R, Spieker T, Siebert R, Strickler JG, Schlegelberger B, et al. Molecular analysis of single B cells from T-cell-rich B-cell lymphoma shows the derivation of the tumor cells from mutating germinal center B cells and exemplifies means by which immunoglobulin genes are modified in germinal center B cells. Blood. 1999;93(8):2679–87. [PubMed] [Google Scholar]
- 11.Braeuninger A, Kuppers R, Strickler JG, Wacker HH, Rajewsky K, Hansmann ML. Hodgkin and Reed-Sternberg cells in lymphocyte predominant Hodgkin disease represent clonal populations of germinal center-derived tumor B cells. Proc Natl Acad Sci USA. 1997;94(17):9337–42. doi: 10.1073/pnas.94.17.9337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Franke S, Wlodarska I, Maes B, Vandenberghe P, Achten R, Hagemeijer A, et al. Comparative genomic hybridization pattern distinguishes T-cell/histiocyte-rich B-cell lymphoma from nodular lymphocyte predominance Hodgkin’s lymphoma. Am J Pathol. 2002;161(5):1861–7. doi: 10.1016/S0002-9440(10)64462-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brune V, Tiacci E, Pfeil I, Doring C, Eckerle S, van Noesel CJ, et al. Origin and pathogenesis of nodular lymphocyte-predominant Hodgkin lymphoma as revealed by global gene expression analysis. J Exp Med. 2008;205(10):2251–68. doi: 10.1084/jem.20080809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diehl V, Sextro M, Franklin J, Hansmann ML, Harris N, Jaffe E, et al. Clinical presentation, course, and prognostic factors in lymphocyte-predominant Hodgkin’s disease and lymphocyte-rich classical Hodgkin’s disease: report from the European Task Force on Lymphoma Project on Lymphocyte-Predominant Hodgkin’s Disease. J Clin Oncol. 1999;17(3):776–83. doi: 10.1200/JCO.1999.17.3.776. [DOI] [PubMed] [Google Scholar]
- 15.Dave SS, Wright G, Tan B, Rosenwald A, Gascoyne RD, Chan WC, et al. Prediction of survival in follicular lymphoma based on molecular features of tumor-infiltrating immune cells. N Engl J Med. 2004;351(21):2159–69. doi: 10.1056/NEJMoa041869. [DOI] [PubMed] [Google Scholar]
- 16.Monti S, Savage KJ, Kutok JL, Feuerhake F, Kurtin P, Mihm M, et al. Molecular profiling of diffuse large B-cell lymphoma identifies robust subtypes including one characterized by host inflammatory response. Blood. 2005;105(5):1851–61. doi: 10.1182/blood-2004-07-2947. [DOI] [PubMed] [Google Scholar]
- 17.WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. Lyons, France: IARC Press; 2008. [Google Scholar]
- 18.Boudova L, Torlakovic E, Delabie J, Reimer P, Pfistner B, Wiedenmann S, et al. Nodular lymphocyte-predominant Hodgkin lymphoma with nodules resembling T-cell/histiocyte-rich B-cell lymphoma: differential diagnosis between nodular lymphocyte-predominant Hodgkin lymphoma and T-cell/histiocyte-rich B-cell lymphoma. Blood. 2003;102(10):3753–8. doi: 10.1182/blood-2003-02-0626. [DOI] [PubMed] [Google Scholar]
- 19.Franke S, Wlodarska I, Maes B, Vandenberghe P, Delabie J, Hagemeijer A, et al. Lymphocyte predominance Hodgkin disease is characterized by recurrent genomic imbalances. Blood. 2001;97(6):1845–53. doi: 10.1182/blood.v97.6.1845. [DOI] [PubMed] [Google Scholar]
- 20.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5(10):R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dudoit S, Shaffer JP, Boldrick JC. Multiple hypothesis testing in microarray experiments. Statistical Science. 2003;18:71–103. [Google Scholar]
- 22.Mantovani A, Sica A, Sozzani S, Allavena P, Vecchi A, Locati M. The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol. 2004;25(12):677–86. doi: 10.1016/j.it.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 23.Billiau A, Matthys P. Interferon-γ: a historical perspective. Cytokine Growth Factor Rev. 2009;20(2):97–113. doi: 10.1016/j.cytogfr.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 24.Van Damme J, Proost P, Put W, Arens S, Lenaerts JP, Conings R, et al. Induction of monocyte chemotactic proteins MCP-1 and MCP-2 in human fibroblasts and leukocytes by cytokines and cytokine inducers. Chemical synthesis of MCP-2 and development of a specific RIA. J Immunol. 1994;152(11):5495–502. [PubMed] [Google Scholar]
- 25.Taylor MW, Feng GS. Relationship between interferon-gamma, indoleamine 2,3-dioxygenase, and tryptophan catabolism. FASEB J. 1991;5(11):2516–22. [PubMed] [Google Scholar]
- 26.Chon SY, Hassanain HH, Gupta SL. Cooperative role of interferon regulatory factor 1 and p91 (STAT1) response elements in interferon-γ-inducible expression of human indoleamine 2,3-dioxygenase gene. J Biol Chem. 1996;271(29):17247–52. doi: 10.1074/jbc.271.29.17247. [DOI] [PubMed] [Google Scholar]
- 27.Munn DH, Mellor AL. Indoleamine 2,3-dioxygenase and tumor-induced tolerance. J Clin Invest. 2007;117(5):1147–54. doi: 10.1172/JCI31178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mellor AL, Munn DH. IDO expression by dendritic cells: tolerance and tryptophan catabolism. Nat Rev Immunol. 2004;4(10):762–74. doi: 10.1038/nri1457. [DOI] [PubMed] [Google Scholar]
- 29.Curti A, Pandolfi S, Valzasina B, Aluigi M, Isidori A, Ferri E, et al. Modulation of tryptophan catabolism by human leukemic cells results in the conversion of CD25− into CD25+ regulatory T cells. Blood. 2007;109(7):2871–7. doi: 10.1182/blood-2006-07-036863. [DOI] [PubMed] [Google Scholar]
- 30.Vogt L, Schmitz N, Kurrer MO, Bauer M, Hinton HI, Behnke S, et al. VSIG4, a B7 family-related protein, is a negative regulator of T cell activation. J Clin Invest. 2006;116(10):2817–26. doi: 10.1172/JCI25673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Helmy KY, Katschke KJ, Jr, Gorgani NN, Kljavin NM, Elliott JM, Diehl L, et al. CRIg: a macrophage complement receptor required for phagocytosis of circulating pathogens. Cell. 2006;124(5):915–27. doi: 10.1016/j.cell.2005.12.039. [DOI] [PubMed] [Google Scholar]
- 32.Alvaro T, Lejeune M, Camacho FI, Salvado MT, Sanchez L, Garcia JF, et al. The presence of STAT1-positive tumor-associated macrophages and their relation to outcome in patients with follicular lymphoma. Haematologica. 2006;91(12):1605–12. [PubMed] [Google Scholar]
- 33.Fraga M, Sanchez-Verde L, Forteza J, Garcia-Rivero A, Piris MA. T-cell/histiocyte-rich large B-cell lymphoma is a disseminated aggressive neoplasm: differential diagnosis from Hodgkin’s lymphoma. Histopathology. 2002;41(3):216–29. doi: 10.1046/j.1365-2559.2002.01466.x. [DOI] [PubMed] [Google Scholar]
- 34.Fan Z, Natkunam Y, Bair E, Tibshirani R, Warnke RA. Characterization of variant patterns of nodular lymphocyte predominant hodgkin lymphoma with immunohistologic and clinical correlation. Am J Surg Pathol. 2003;27(10):1346–56. doi: 10.1097/00000478-200310000-00007. [DOI] [PubMed] [Google Scholar]
- 35.Nam-Cha SH, Roncador G, Sanchez-Verde L, Montes-Moreno S, Acevedo A, Dominguez-Franjo P, et al. PD-1, a follicular T-cell marker useful for recognizing nodular lymphocyte-predominant Hodgkin lymphoma. Am J Surg Pathol. 2008;32(8):1252–7. doi: 10.1097/PAS.0b013e318165b0d6. [DOI] [PubMed] [Google Scholar]
- 36.Muller AJ, Scherle PA. Targeting the mechanisms of tumoral immune tolerance with small-molecule inhibitors. Nat Rev Cancer. 2006;6(8):613–25. doi: 10.1038/nrc1929. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.