Abstract
The identification of tumor antigens capable of eliciting an immune response in vivo may be an effective method to identify therapeutic cancer targets. We have developed a method to identify such antigens using frozen tumor-draining lymph node samples from breast cancer patients. Immune responses in tumor-draining lymph nodes were identified by immunostaining lymph node sections for B-cell markers (CD20&CD23) and Ki67 which revealed cell proliferation in germinal center zones. Antigen-dependent somatic hypermutation (SH) and clonal expansion (CE) were present in heavy chain variable (VH) domain cDNA clones obtained from these germinal centers, but not from Ki67 negative germinal centers. Recombinant VH single-domain antibodies were used to screen tumor proteins and affinity select potential tumor antigens. Neuroplastin (NPTN) was identified as a candidate breast tumor antigen using proteomic identification of affinity selected tumor proteins with a recombinant VH single chain antibody. NPTN was found to be highly expressed in approximately 20% of invasive breast carcinomas and 50% of breast carcinomas with distal metastasis using a breast cancer tissue array. Additionally, NPTN over-expression in a breast cancer cell line resulted in a significant increase in tumor growth and angiogenesis in vivo which was related to increased VEGF production in the transfected cells. These results validate NPTN as a tumor-associated antigen which could promote breast tumor growth and metastasis if aberrantly expressed. These studies also demonstrate that humoral immune responses in tumor-draining lymph nodes can provide antibody reagents useful in identifying tumor antigens with applications for biomarker screening, diagnostics and therapeutic interventions.
Keywords: Neuroplastin/SDR-1, Tumor antigen, Germinal center, Immune response, Breast cancer, Sentinel lymph node
Introduction
The identification of novel tumor associated antigens (TAAs) can result in significant improvements in the detection and treatment of malignant tumors. The ideal candidates are antigens selectively expressed by tumor cells and those expressed in excess or in alternative immunogenic forms in tumor cells compared to normal tissue. Proteins that bear these characteristics are capable of eliciting an adaptive immune response and thus evaluating immune responses in cancer patients is an excellent starting point for the discovery of new TAAs.
Humoral immune responses to breast cancer have been extensively documented. Evidence for their existence can be obtained at three levels: responses in tumor draining-lymph nodes, the presence of antibodies in serum, and analysis of tumor infiltrating lymphocytes (TILs). In tumor-draining lymph nodes, histological patterns suggestive of humoral responses, such as germinal center hyperplasia, have been described for decades [19, 42] and a relative abundance of IgG+ B cells, indicative of a mature response, has also been observed [21, 43]. Tumor reactive serum antibodies are present in more than half of breast cancer patients analyzed and react with several TAAs such as Her-2/Neu, MUC1 and p53 [7]. Finally, although T lymphocytes are the predominant component of TILs in most breast cancers, significant B cell clusters can be present in a small subset of cases [6, 13]. Taken together, these observations indicate that the humoral arm of the immune system can react to breast tumors. However, it has been hard to prove that the immune responses are protective, as only a few are associated with a significantly better prognosis and tumor regression is infrequent [7, 14].
In spite of this, the study of humoral responses in individual patients may significantly improve the discovery of TAAs which may serve as therapeutic targets. Tumor draining-lymph nodes, the primary location for lymphocyte priming, represent a potential source of anti-tumor antibodies. Accordingly, hybridomas generated with B cells extracted from axillary lymph nodes have given rise to several antibodies that react with malignant tissue and/or breast cancer-derived cell lines [2, 9, 12, 21, 22, 30, 35]. In these studies, cell suspensions from whole lymph node sections were used and thus no selection criteria for tumor-responsive B cells were applied prior to hybridoma fusion. Recent studies have provided evidence for clonal expansion (CE) and somatic hypermutation (SH) in tumor infiltrating-B cells from breast cancer patients by amplification and sequencing of rearranged immunoglobulin genes [8, 27, 38]. As these parameters are reliable indicators of antigen-driven humoral responses, their identification in tumor draining-lymph node samples can reveal antibodies that have reacted to tumor antigens in vivo and can provide an effective methodology to identify antigen-driven humoral responses and a technology for tumor antigen identification.
Here, we generated Ig heavy chain variable domain (VH) gene cDNA libraries to evaluate the presence of SH and CE in distinct lymph node zones defined by the presence of B cell activation and cellular proliferation markers. This analysis guided the selection of VH sequences to be used as recombinant single-domain antibodies (sdAbs) in an antigen-trap assay that led to the discovery of a candidate TAA, neuroplastin (NPTN). Finally, we conducted experiments to evaluate NPTN expression in breast cancer specimens and its effect on tumor biology that validate it as a new TAA for breast cancer.
Materials and methods
Patient samples
Samples of primary breast tumors (>1.5 cm diameter) and medial sections of local draining lymph nodes were obtained within 5–10 min of resection by a staff pathologist. The samples were placed into optimal cutting temperature (OCT) media (Shandon Lipshaw, Pittsburgh, PA, USA) and immediately frozen in liquid nitrogen. All samples were obtained under an Institutional Review Board approved protocol and devoid of any personal identification information.
Immunohistochemistry, tissue coring and RNA isolation
Lymph node cryosections (7–10 μm in thickness) were obtained and post-fixed with methanol:acetone (1:1), dried and rehydrated in phosphate buffered saline (PBS). Immuno-staining was performed on serial sections with anti-CD20 (Dako, Carpinteria, CA, USA), anti-CD23 (BD Pharmingen, San Jose, CA, USA) or anti-Ki67 (Dako) and detected with Vector ABC kit using DAB substrate according to the manufacturer’s protocol (Vector Laboratories, Burlingame, CA, USA). Controls without addition of primary antibody were performed to assure signal specificity. Immuno-stained slides were imaged with a color digital camera (Zeiss AxioCam HRc) and serial slide overlays were produced using Adobe Photoshop (Adobe Systems, Inc., San Jose, CA, USA) and pixel-by-pixel color masks with Image-Pro Plus (Media Cybernetics, Inc., Silver Spring, MD, USA) software. Selected areas of interest were identified with a marker pen on the primary hematoxylin and eosin (H&E) stained slide and aligned with the coordinate frozen tissue block. The area of interest was cored out of the block using a 1.0 mm copper core tube (Beecher Tissue Arrayer acceptor core, Beecher Instruments, Silver Spring, MD, USA) sterilized with 0.5% SDS and 70% ethanol and the cored tissue was transferred into RNA lysis buffer and total RNA isolated with the RNAeasy kit (Qiagen, Valencia, CA, USA) according to the manufacturer’s protocol.
cDNA synthesis, V gene amplification and generation of VH libraries
RNA yields from isolated cores were typically 2–6 μg. Two micrograms of RNA were subjected to reverse transcription (RT) using Superscript II (Invitrogen, Carlsbad, CA, USA) with random hexamer primers in a total reaction volume of 20 μl as described in the manufacturer’s protocol. One microliter of the RT reaction was amplified via PCR with Platinum Pfx DNA Polymerase (Invitrogen) using a modification of degenerate primers described previously [34]: forward primer 5′-ATGCAGGTGCAGCTGGTGSAGTCTGG-3′; reverse primer: 5′-TGAGGAGACGGTGACCAKGGT-3′. Amplifications were initiated with a 5 min hot start at 94°C followed by 30 cycles at 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min each, and a terminal extension at 72°C for 10 min. PCR products were agarose gel purified and terminal adenosine was added to purified fragments with Taq polymerase (Invitrogen) and 200 μM ATP for 10 min at 72°C. Fragments were ligated into pCR-T7TOPO expression vector (Invitrogen) and resultant transformants were screened by PCR using Vent DNA Polymerase (New England Biolabs, Beverly, MA, USA), forward primer: 5′-CGCGAAATTAATACGACTCACTATAGGG-3′, and 3′ primer: 5′-CCTAAATTGTAAGCGTTAATCCGG-3′. A positive screen resulted in a ~730 bp PCR fragment (~410 bp VH sequence + ~320 bp vector sequence). PCR fragments that screened positive were separated from their primer precursors on a Qiagen MinElute 96 UF PCR purification plate (Qiagen).
DNA sequencing and analysis
Purified VH PCR fragments were sequenced using a standard primer to the T7 promoter with the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA), and an ABI automated cycle sequencer (Applied Biosystems). To determine V, D and J gene segment usage the sequences were aligned to sequences from the V BASE database using DNAPLOT software at http://vbase.mrc-cpe.cam.ac.uk/. Each sequence was compared to human germline sequence with the highest matching score to identify mutations in framework regions (FRs) 1, 2 and 3 and complementarity-determining regions (CDRs) 1 and 2. To establish if mutations in the CDRs were caused by antigen pressure, the multinomial distribution model described by Lossos et al. [26] was applied using the applet provided at http://www-stat.stanford.edu/immunoglobulin/.
Recombinant VH synthesis and purification
The sdAb proteins were synthesized in bacteria by amplifying clones of interest with in-frame oligos for cloning into the pTrcHis2 C vector (Invitrogen) providing a fusion with C-terminal c-myc and 6× His affinity purification tags. The fusion plasmid and induction of expression by IPTG and purification from bacterial extracts was performed using histidine tag affinity chromatography with Ni-NTA Columns (Clontech) according to the manufacturer’s protocol. Purified proteins were dialyzed overnight in 10 mM Tris–HCl, pH = 7.5, 150 mM NaCl, 0.5 mM EDTA at 4°C. VH fusion sdAB proteins were analyzed for protein recovery using the BioRad DC reagent kit and immunoblot with anti-c-myc antibody (Invitrogen).
Breast tumor extracts and sdAB binding/LC-MS/MS identification
Primary breast tumor samples were obtained and immediately frozen. Frozen tissue was directly homogenized in 5 volumes of lysis buffer (1% Triton X-100, 50 mM Hepes, pH 7.5, 10 mM sodium pyrophosphate, 150 mM NaCl, 100 mM NaF, 0.2 mM sodium orthovanadate, 1 mM EGTA, pH 7.5, 1.5 mM MgCl2, 10% glycerol, and fresh Protease Inhibitor Cocktail (10 μl/ml) (Sigma–Aldrich)), using a Polytron Homogenizer with 5 high-power burst to completely homogenize the tissue. The tissue was centrifuged at 4°C once at 800×g and supernatant centrifuged at 14,000×g to remove insoluble debris. Protein concentration was determined by BioRad DC reagent using bovine serum albumin (BSA) as a standard. Purified sdAB recombinant proteins (10 μg/ml) were allowed to bind to high capacity nickel assay plates (Sigma–Aldrich) at 200 μl/well and washed 5× with phosphate binding buffer (PBS pH-7.5, Tween-20 (0.01%), 5 mM imidazole) using an automated plate washer. Each antibody was bound in triplicate for each condition and each lysate. Antibody plates were blocked with charcoal-treated ovalbumin (1%w/v in PBS) for 1 h at RT. Tumor or cell line protein Triton X-100 lysates were diluted with PBS (1:3) and 200 μl added with 80 μg of equivalent protein for each sample tested. The plates were incubated on an orbital shaking platform overnight at 4°C and washed 5× with binding buffer and 1× with PBS, aspirated and air-dried. One well of each antibody was eluted with SDS sample buffer (30 μl) and tested on SDS-PAGE and immunobloted for sdAb recovery using anti-c-myc antibodies or silver stained. All wells recovered the same qualitative amounts of primary sdAb being tested and no immuno-selected proteins could be detected with silver stain. The duplicate remaining wells were treated in situ with sequencing grade porcine trypsin (Princeton Separations, Adelphi, NJ, USA) at 37°C for 3 h. The samples were transferred to a microfuge tube, dried by evaporation under vacuum with centrifugation and resuspended in LC-MS/MS loading buffer [5% Acetic Acid, 5% acetonitrile (ACN)]. The samples were then loaded individually onto a 100 μm C18 column using a Famos Autosampler. Samples were then analyzed by on-line LC-MS/MS on a Finnigan LTQ Iontrap mass spectrometer in a data-dependent MS/MS mode. The MS/MS spectra generated from the 90 min gradient (5–80% ACN) were searched against NCBI-human proteome database using the SEQUEST algorithm [45]. Stringent filtering criteria was used to select for correct assignment with less than 1% false positive rates (Xcorr cutoff of 1.9 for 1+, 2.2 for 2+, and 3.7 for 3+ peptides, delta Cn of 0.1, minimum of two unique peptide identification for each protein). Only proteins selected with multiple or repeat peptides and in duplicate for each sdAb tested were considered specific.
Stromal derived receptor-1/neuroplastin cDNA isolation and expression vector cloning
Human breast cancer cell lines, MDA-MB-231 and MCF-7 cells were obtained from ATCC and cultured as recommended. Total RNA was isolated from cells using the Qiagen RNAeasy kit. Total RNA (2 μg) was used in reverse transcription using Superscript II (Invitrogen) and random hexamer primers as described above. Neuroplastin cDNA was amplified from 1/20th of the RT reaction using coding region primers; ATG start codon: 5′-CAT GTC GGG TTC GTC GCT GCC CAG-3′ and Termination: 5′-TTA ATT TGT GTT TCT CTG GCG CAA-3′. Amplified sequences were cloned into pCR-T7/TOPO and sequenced to assure neuroplastin clones. The full length cDNA was cloned into the pcDNA/V5/GW/D-TOPO expression vector (Invitrogen) by amplifying the 5′-Kozak sequence and an oligonucleotide primer including the last amino acid prior to the termination codon. 5′ primer sequence—5′-CAC CGA ATT CGC CAT GTC GGG TTC GTC GCT GCC CAG-3′ and 3′ primer sequence—5′-ATT TGT GTT TCT CTG GCG CAA-3′, human neuroplastin, beta isoform, NM_012428 (NPTN_036560.1). Individual clones were sequenced to completion with three overlapping primers to assure no mutations had occurred in the clones used.
Synthesis and analysis of human breast tumor tissue arrays
Human breast tumor tissue arrays were developed in house using a Beecher Instruments tissue Arrayer (Beecher Instruments, Silver Spring, MD, USA). Forty de-identified breast tumor formalin-fixed paraffin embedded tissue blocks were obtained from clinical pathology under an approved IRB protocol. Two tumor arrays were established with duplicate cores from each case consisting of 20 estrogen receptor alpha (ERα) negative and 20 ERα positive tumor samples. Cores consisted of 1.0 mm cores from the most dense tumor areas as determined from the H&E stained slide for each tumor block. A second breast tumor tissue array was obtained from the NCI biological repository resource (NCINBR) which contained 280 individual cases from normal ductal tissue to advanced metastatic lesions. Immunohistochemistry was performed using anti-neuroplastin antibodies described previously [39]. Immunostaining was developed with biotinylated secondary antibodies (Dako) and the Vector ABC kit (Vector Laboratories), according to the manufacturers’ instructions. Stained cores were assessed by two independent observers using a scale of 0–4, representing zero to maximal staining specifically in the tumor cells within each core.
Cell culture and transfection
MDA-MB-435S cells were obtained from ATCC (Bethesda, MD, USA) and cultured in Dulbecco’s modified Eagle’s medium (Gibco) supplemented with 10% fetal bovine serum, 2 mM l-glutamine, 10 U/ml penicillin, and 10 μg/ml streptomycin under either normoxic (5% CO2, 21% O2, 74% N2) or hypoxic conditions (5% CO2, 1.5% O2, 93.5% N2) in humidified triple gas incubators at 37°C. Cells were stably transfected with a pcDNA 3.1 vector (Invitrogen) containing NPTN-beta isoform or eGFP cDNA using Lipofectamine™ Reagent (Invitrogen) and selected with neomycin or geneticin as described previously [1].
Xenograft tumor model
MDA-MB-435S cells selected for NPTN or GFP expression were grown to 70–90% confluence, trypsinized, washed with PBS twice, and resuspended in Hank’s Balanced Salt Solution to a final concentration of 1 × 107 cells/ml. Two hundred microliter (2 × 106cells) of cell suspension was injected into the third axial mammary fat pad of 4 week old, female nude mice using a 30 g needle affixed to a 1 cc tuberculin syringe. Tumor size was measured twice per week and tumor volume calculated with the formula: Volume (mm3) = Length (mm) × Width (mm)2 × 0.52. Ten weeks after injection, the mice were euthanized, tumors removed, weighed and split into thirds, with one-third frozen in optimal cutting temperature cryosectioning media (OCT), one-third flash frozen in liquid nitrogen and a third formalin fixed and paraffin embedded for histological analysis.
Immunohistochemistry and histological quantification
Tumor cryo-sections were obtained (7 mm) and immunostained for vessels using the anti-human CD31 monoclonal antibody (Santa Cruz Biotech, Santa Cruz, CA, USA) at a 1:100 dilution. Positive staining was detected as described above. Tumor microvascular density was obtained by imaging three high power fields in areas of viable tumor and capturing digital color images. A pixel-by-pixel quantification of DAB staining was performed using Image Pro Plus. Pixels were normalized to the average pixel per vessel as defined by the value obtained from ten independent microvessels in cross section/10 to estimate total vessels per area.
Protein extraction and VEGF ELISA
MDA-MB-435 cells or tumors from the xenograft experiment were lysed in Triton X-100 lysis buffer (1% Triton X-100, 50 mM Hepes, pH 7.5, 10 mM sodium pyrophosphate, 150 mM NaCl, 100 mM NaF, 0.2 mM sodium orthovanadate, 1 mM EGTA, pH 7.5, 1.5 mM MgCl2, 10% glycerol, and fresh protease inhibitor cocktail (10 μl/ml) (Sigma)). Protein concentrations were determined using Biorad DC protein assay (BioRad Inc.) according to the manufacturer’s instructions. A human VEGF sandwich ELISA was performed as described previously [37].
RNA Isolation and qRT-PCR
Cores (2 mm) were removed from human breast tumor samples and RNA was isolated with the RNeasy kit (Qiagen) according to the manufacturer’s protocol. Reverse transcription reactions and quantitative PCR was performed as previously described [29]. NPTN-beta (NM_012428) expression was normalized to the ribosomal transcript RPL0 (NM_532753). Primers used for qRT-PCR were: NPTN-beta (NM_012428) forward 5′-TGTGCTGAGAATAACCCGGCT-3′, NPTN-beta reverse 5′-TTGGCTTCTGAAGGACGCTTA-3′, RPL0 (NM_053275.3) forward 5′-TCCTCGTGGAAGTGACATCGT-3′, RPL0 reverse 5′-CTGTCTTCCCTGGGCATC-3′.
Statistical analysis
Data from individual experiments were represented as mean ± standard error unless otherwise stated. Statistical comparison of groups was performed using 2-tailed Student’s t test, ANOVA, or Chi-square with appropriate tests for equal variances. Statistical significance was defined and indicated as P ≤ 0.05 (*) or P ≤ 0.01 (**).
Results
B cell activation and proliferation zones can be identified in tumor-draining lymph nodes from breast cancer patients by multiplex immunostaining
Our first objective was to establish if B cell activation and proliferation, both hallmarks of antigen-driven immune responses, can be detected in tumor-draining lymph nodes from breast cancer patients. To achieve this, 32 sentinel tumor-draining lymph nodes were stained for three markers: CD20, a B cell-specific cell surface molecule; CD23, an indicator of B cell activation, which is not expressed on plasma cells [33]; and Ki67, a proliferation marker [36]. As shown in Fig. 1, multiple CD20-positive zones were found in each lymph node (average 42.5 zones per node), but only 27% of these were positive for CD23. Interestingly, clusters of Ki67+ cells within the CD20+CD23+ zones were rare (1.13 clusters per lymph node on average), indicating that only a small fraction of B cell clusters were proliferating at the time of surgical resection. These results indicate that zones of B cell activation and proliferation can be identified in frozen lymph node samples and that they account for just a small proportion of the total area occupied by B cells in each sentinel lymph node. These zones may represent active germinal centers where antigen-driven B cell proliferation was taking place.
Fig. 1.
Immunohistological analysis of B cell zones in tumor-draining lymph nodes from breast cancer patients. a Immunohistochemistry staining for CD20, CD23 and Ki67 in serial sections of a tumor-draining lymph node (bar = 1,000 μm). b Merged image of the three stains in the zone highlighted in a. Masks of different colors were applied to each marker: CD20 yellow, CD23 blue, and Ki67 pink (bar = 1,000 μm). c Number of zones positive for CD20, CD20 and CD23 or a combination of the three markers in each lymph node (average and standard deviations are shown, n = 32)
Clonal expansion and somatic hypermutation events can be identified in proliferative B cell zones
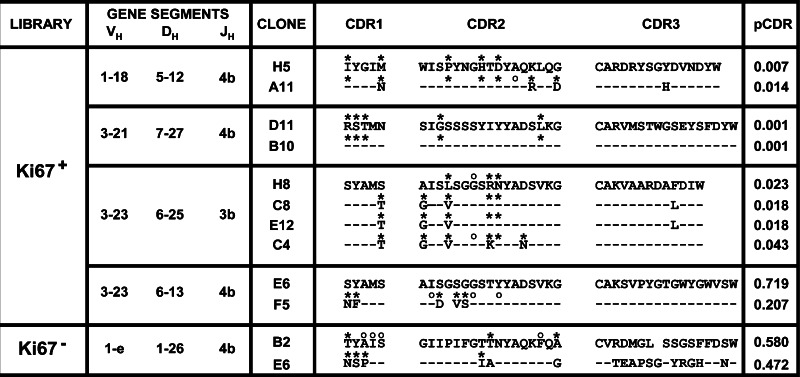
Next, we wanted to determine if signs of antigen-driven immune responses could be detected in activated B cell zones that were defined by immunohistochemistry with activation and proliferation markers. Our approach was to core either proliferative (Ki67+) CD20+CD23+ or non-proliferative (Ki67−) zones from the same lymph node and isolate total RNA for the generation of VH cDNA sequence libraries. Forty-six complete open reading frame sequences were obtained from the Ki67+ B cell zone and 50 from its Ki67− counterpart. These sequences were analyzed to determine V, D and J gene segment usage and to establish if mutations in the complementary determining regions (CDRs) were caused by antigen pressure. Four groups of sequences that shared the same VDJ rearrangement were found in the Ki67+ library, Table 1. These groups included three pairs and a group of four for a total of ten sequences, which represented 21.7% of the library. Conversely, the Ki67− library had a single pair of sequences with matching VDJ segments, representing only 4% of the sequences recovered, Table 1. Significant clustering of replacement mutations in CDR1&2 was found in 52.5% of the sequences from the Ki67+ library, while 22% of the sequences showed this characteristic in the Ki67− pool, Table 1. When considering only sequences that where part of clonal groups, three of the four groups in the Ki67+ library had significant replacement mutations in their CDRs as determined by the algorithm of Lossos et al. [26]. None of the sequences in the Ki67− group had significant mutational events. In total, eight out of ten clonal sequences (80%) had signs of somatic hypermutation (SH). As shown in Fig. 2, alignment of the CDR sequences from the groups sharing VDJ rearrangements show that several mutations were shared, although most clones had different sequences. Two pairs of sequences (clones D11/B10 and C8/E12) had identical CDRs, although their sequences were not identical overall. This distribution strongly suggests that the sequences that shared the same VDJ rearrangement originated from one clone and that their mutations were caused by antigen pressure. In summary, clonal groups with mutations indicative of antigen selection were found only in clones sequenced from the proliferative B cell zone library. These results indicate that both CE and SH were present in the VH sequences obtained from the CD20+CD23+Ki-67+ zone and, consequently, that antigen-driven immune responses could be identified in tumor-draining lymph nodes based upon B-cell activation and proliferation markers.
Table 1.
Gene clonal expansion and mutational analysis of sequences sharing V and J segments

|
A list of clones with V/J shared segments that were identified from the Ki67 + and Ki67- libraries are shown. Clonal expanded groups sharing V/J and D segments are identified by bars (|) and numbered for each library. A dash (–) in the D segment column indicates there was no matching D segment from the database. Significant difference from germline sequence was determined by the method of Lossos et al. [26], and P values are presented for each clone (pCDR) and asterisks (*) indicate P ≤ 0.05
Fig. 2.
Alignment of amino acid sequences of the CDRs of VH clones that shared VDJ rearrangements. The VH, DH and JH gene segments used by each clonal group from a Ki67 + and Ki67 − libraries are indicated. Amino acid sequences are provided for each clone and aligned with similar VH, DH, and JH clones. Dashes indicate same amino acid sequence and asterisks (*) above each amino acid indicates difference from germline sequence (not shown). Locations of silent mutations are indicated by open circles above their respective amino acids. CDR3 sequences could not be compared to germline through direct database matching thus differences from germline are not reported for CDR3. The p value for clustering of replacement mutations in CDR1&2 (pCDR) according to the method described by Lossos et al. [19] are provided for each clone, P ≤ 0.05 is considered significant
Single domain antibodies extracted from tumor-draining lymph nodes can be used to identify new tumor antigens
Our analysis of several tumor-draining lymph node sections and two VH sequence libraries obtained from one of them showed that antigen driven humoral immune responses can be detected in these tissues. However, the origin and identity of the antigens could not be determined with this type of analysis. Our possession of VH sequences with evidence of antigen-driven mutations prompted us to devise a method to screen for, and to identify potential tumor antigens. There is ample evidence that VH domains alone (termed sdAbs) retain antigen-binding properties and can be used to screen protein samples [17, 18, 46]. Therefore, we developed an assay to trap tumor antigens with recombinant sdAbs synthesized with the VH sequences retrieved from the tumor-draining lymph node. Since the accumulation of replacement mutations in CDRs indicates antigen pressure, we selected a sequence from the Ki67+ library with high CDR1&2 mutational events (clone A3; P = 0.005). As a control, a sequence from the Ki67− library with the same V and J segments, but with no significant clustering of replacement mutations in the CDRs was chosen (clone H1, P = 0.746). As shown in Fig. 3a, these sequences differ mainly by their variation in CDR domains.
Fig. 3.
Identification of NPTN in tumor protein extracts. a Alignment of amino acid sequences of A3 and H1 sdAbs. Dashes indicate same amino acid between A3 and H1 clones. Replacement mutations in FRs 1, 2 and 3 and CDRs 1 and 2 are indicated by an asterisk (*), CDR3 was not directly compared to germline sequences (germline sequence not shown). Gray boxes indicate CDRs 1–3. b Amino acid sequence of NPTN beta. The peptide identified by LC-MS/MS is boxed. c Immunoblot of 5 primary breast tumor samples using anti-NPTN (NP) (top panel) or A3 sdAb (tagged with c-myc) and detected with anti-c-myc antibody (bottom panel)
The sequences were cloned into a bacterial expression vector which provides a C-terminal myc-epitope tag and a 6× His purification tag. Both proteins were synthesized in bacteria and the purified recombinant sdAb proteins dialyzed to promote refolding. The protein production and purification was followed by anti-myc immunoblots to normalize the amounts of purified sdAb proteins obtained (data not shown). The purified sdAbs were allowed to bind to nickel-coated plates and used to select proteins from tumor extracts obtained from the same patient as the lymph node VH library. Proteins trapped by sdAbs were subjected to in-solution direct trypsin protease digestion and peptides released were analyzed by LC-MS/MS. One peptide (IVTSEEVIIR) was identified multiple times in the tumor protein selection and peptide analysis by LC-MS/MS with the A3 sdAb but not in the control H1 protein trap. A protein database search revealed that this peptide matched with a member of the Ig superfamily of proteins, neuroplastin (NPTN), also called Stromal Derived Receptor-1 (SDR-1), Fig. 3b. The H1 sdAb did not trap and identify proteins with high confidence by LC-MS/MS from the same extract performed in the same plate and at the same time.
NPTN is a transmembrane glycoprotein originally described as a component of synaptic membranes in rat brain [39]. Two alternatively spliced isoforms exist: NPTN-65, a brain-specific isoform with three extracellular Ig-like domains and NPTN-55, a widely expressed isoform that has only two Ig-like domains [24, 25]. To confirm the specificity of the A3 sdAb, we tested it as a detection reagent in immunoblots and compared it to the signal obtained with an antibody raised against the NPTN-65 protein. Five breast tumor protein extracts were assayed, including the sample in which the NPTN protein was identified. The anti-NPTN antibody revealed the presence of a 55 kDa isoform in all tumors tested and an alternative 45 kDa band that varied in intensity among samples, three of them having a stronger signal, Fig. 3c. Probing with the A3 sdAb resulted in the presence of the 45 kDa bands, closely resembling the pattern found with the anti-NPTN antibody, albeit with a weaker signal. The A3 sdAb also detected bands at 55 kDa. These results suggest that A3 is a sdAb specific for NPTN and confirms the presence of NPTN in the tumor sample used for the antigen-trapping assay.
Neuroplastin is expressed in a subset of human breast cancers
To better understand the expression of NPTN within a population of breast cancers, we evaluated a human breast tumor tissue array containing cores of normal breast tissue and invasive breast carcinoma using immunohistochemistry using a polyclonal anti-NPTN antibody that had been previously used to characterize NPTN expression in rodent and human tissues [5, 39]. A subset of invasive carcinoma samples had significant positive staining in tumor cells but not in the adjacent stroma, while normal breast tissue was negative, Fig. 4a–d. To quantify the data, the extent of NPTN-immunostaining was scored from 0 to 4 by two independent observers. While only one out of 40 (2.5%) normal breast tissue samples had a score of 3 or higher, 38 out of 179 (21.2%) invasive carcinoma specimens showed a positive staining for NPTN using the same threshold. Interestingly, when grouping the cases according to the presence of lymph node or distant metastasis, a larger percentage of NPTN-positive tumors was found in the group that had distant metastasis, Fig. 4e, indicating that NPTN expression may promote tumor invasion and/or metastatic potential.
Fig. 4.
Neuroplastin is expressed in a subset of breast tumors. A tissue array containing normal breast and breast cancer tissue cores was stained by immunohistochemistry with an anti-NPTN antibody. Representative cases are shown: a Normal breast, b Invasive ductal breast carcinoma, node negative. c Invasive ductal breast carcinoma, node positive. d Invasive ductal breast carcinoma, distant metastasis (bar = 100 μm). e Percentage of cases from each group that had a score of 3 or higher for NPTN staining; N0 node negative, N ≥ 1 node positive, M1 distant metastasis
Over-expression of NPTN in MDA-MB-435 breast carcinoma cells confers a growth advantage in vivo
Like other members of the Ig superfamily, NPTN participates in cell adhesion phenomena [10, 39] and could have profound effects on tumor biology. To test this hypothesis, we transfected the breast cancer-derived cell line MDA-MB-435 with a vector expressing the NPTN beta protein tagged with the V5 peptide or EGFP as a control. Cell pools were selected with neomycin resistance using geneticin-sulfate and evaluated for protein expression by immunoblot, Fig. 5a. Expression of a ~49 kDa protein was detected by anti-V5 Western blot in two MDA-MB-435 pools (1&2) stably expressing NPTN. MDA-MB-435 cells transfected with GFP were used as controls for background staining. To assess whether the NPTN isoform was highly glycosylated as reported in the literature, extracts were exposed to both N- and O-linked glycosidases and evaluated by immunoblot. The main 49 kDa isoform was observed to be reduced to an apparent molecular weight of 37 kDa after glycosidase treatment. Some of the glycosylated form was still present indicating that de-glycosidation was not quite complete. Immunoblot for β-actin showed equal loading of extracts in each lane.
Fig. 5.
NPTN expression in MDA-MB-435 cells does not affect in vitro cell growth. a The stable expression of NPTN in MDA-MB-435 cells by immunoblot. Top panel Detection of V5 epitope tag in extracts from parental MDA-MB-435 (435) cells, cells expressing EGFP (MDA-MB-435-GFP) (GFP), and two lines expressing NPTN-V5 (NP-1, NP-2), middle panel Detection of V5-epitope after treatment of extracts with N- and O-glycosidases. Bottom panel Control for extract loading with β-actin immunoblot. b MTT viability assay of cells plated in triplicate after overnight seeding (time 0), 48 and 72 h later. MDA-MB-435-GFP (GFP), MDA-MB-435-NPTN (NP) and a 1:1 ratio of both (NP:GFP) cells were analyzed. No statistical difference was observed at any time point
To evaluate whether the expression of NPTN in the MDA-MB-435 cell line affected optimal growth in vitro, we plated cells stably expressing EGFP (GFP) and the highest NPTN expressing line, pool 1, and assessed cell viability over time using an MTT assay, Fig. 5b. There were no significant differences in overnight cell seeding (time 0), or proliferation rates over 48–72 h. The cell pools even demonstrated identical growth suppression based upon confluency and media nutrient consumption at 72 h. Analysis of a 1:1 mixture of the GFP cells with the NPTN pool 1 (NPTN:GFP) also showed nearly identical seeding and growth curves over 72 h. NPTN-expressing MDA-MB-435 cells were also assayed for survival during glucose deprivation, anchorage independent growth and invasion through Matrigel matrix barrier, and showed no difference from the GFP control cells in each of these assays (data not shown).
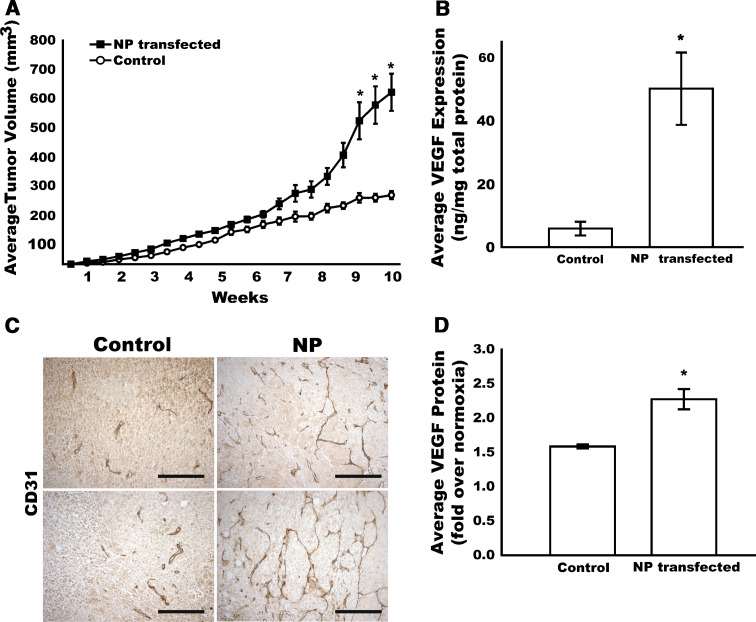
Evaluation of the breast tumor cells in culture suggest that the NPTN expression does not affect growth pathways in the MDA-MB-435 cell line either directly or through secreted mediators. However, it has been demonstrated that Ig proteins with homology to NPTN, such as Basigin/CD147/EMMPRIN have unique effects on stromal cells and within the tumor microenvironment. For example, Basigin/CD147/EMMPRIN demonstrated the ability to be shed from the cell surface and act upon stromal cells to induce migration and VEGF angiogenic gene expression [41, 44, 47]. Thus, to assess whether NPTN over-expression might promote tumor growth or metastasis, cells were implanted into athymic nude mice in the mammary orthotopic site to test tumor growth over time, Fig. 6a. Ten weeks after injection into the mammary fat pad, MDA-MB-435 cells over expressing the NPTN protein showed a significantly improved tumor growth with larger tumor volumes (614.8 ± 132.6 vs. 247.0 ± 29.4 mm3; P = 0.032); as well as tumor weights at harvest (366.7 ± 64.0 vs. 138.3 ± 17.7 mg; P = 0.024) compared to the GFP expressing control cells. Interestingly, vascular staining with anti-CD31, revealed that NPTN-overexpressing tumors had enhanced blood vessel formation and a highly integrated vascular network, Fig. 6b. To test the hypothesis that the NPTN-overexpressing cells could produce this effect through increased vascular endothelial growth factor (VEGF) production, the levels of VEGF were assayed in tumor lysates and in cell culture supernatants of the transfected cells. Tumors derived from the NPTN-transfected cells showed a fivefold increase in VEGF content in vivo, Fig. 6c. Likewise, when the MDA-MB-435 cells were challenged with hypoxia in culture, the NPTN-expressing cells demonstrated increased VEGF production when compared to GFP-expressing controls, Fig. 6d. Taken together, these data support a model for NPTN-enhanced tumor growth through an angiogenic mechanism mediated by increased production of VEGF. This function is likely related to some contribution of NPTN to tumor cells in response to microenvironmental hypoxia, a mechanism that will have to be investigated with more detailed experiments.
Fig. 6.
NPTN expression in MDA-MB-435 cells stimulates tumor growth and angiogenesis in vivo. a Control MDA-MB-435 or NPTN-transfected (NP) cells were injected into the mammary fat pad of athymic nude mice and tumors were measured twice a week. Average tumor volumes (ave. ± SEM) for NPTN-transfected (filled square) and control (open circle) tumors are shown (n = 7). b Vascular immunostaining (CD31) in MDA-MB-435-GFP tumors (Control) or NPTN-overexpressing (NP) tumors. Two representative cases for each group are shown (bar = 100 μm). c VEGF protein expression in NPTN-overexpressing and control tumor lysates normalized to the total amount of protein in each sample (n = 7). d Hypoxia induced VEGF secretion into media by MDA-MB-435 cells (control) or with overexpression of NPTN (NP) over 24 h (n = 3) (*P ≤ 0.05)
Neuroplastin beta correlates with angiogenic phenotype in primary human breast cancer
Immunological detection of NPTN isoforms and the complexity of post-translational modification by glycosylation cannot ascertain the NPTN isoform(s) that are prevalent in human breast cancer. To address which NPTN isoform is expressed in human breast cancers we performed qRT-PCR for NPTN-alpha and NPTN-beta on mRNA isolation from estrogen receptor alpha (ERα) positive, MCF-7 breast carcinoma cells and the ERα negative MDA-MB-231 cells. The data indicated that the NPTN-beta isoform was expressed and was the dominant isoform in MDA-MB-231 cells and very little amplification of NPTN-alpha or beta was observed in MCF-7 cells (data not shown).
In an effort to determine if NPTN-beta expression was related to selective human breast cancer phenotypes, we analyzed its expression in 16 primary breast cancer cases. To avoid excess contamination by non-tumor cell types, tumor samples were obtained by sampling only tumor dense areas from a frozen block using a 2 mm biopsy punch. NPTN-beta mRNA levels, as determined by qRT-PCR, were compared to patient age, tumor size, hormone receptor and Her-2/neu status, as well as angiogenesis, Table 2. Interestingly, the top quartile for NPTN-beta expression was twofold higher than the average of the total and ten-fold higher than the bottom quartile. There appear to be no selectivity or correlation with patient age or tumor size; in fact the tumor sizes were generally smaller in the upper quartile of NPTN-beta expression. Surprisingly, all four samples in the upper quartile were triple negative for ERα, PR, and Her-2/neu. Despite slightly smaller tumor size, the level of angiogenesis in the upper quartile was two-fold higher than the lower quartile and was statistically significant. The microvascular density (MVD) in tumors with high NPTN-beta expression was also higher than the average of the total; however, this did not reach significance due to high variance in the individual tumors. Analysis of whether NPTN-beta expression correlated with tumor angiogenesis was found to be highly significant using Pearson’s correlation statistic (R 2 = 0.7566, P = 0.0243). Thus, even in a small pilot study, the expression of NPTN-beta may have a significant impact upon tumor angiogenesis and subsequently metastatic potential.
Table 2.
Pilot study to evaluate NPTN-β mRNA expression in relation to tumor clinical and vascular phenotypes
| Group | NPTN-b Expression | Age (years) | Tumor size (cm) | ER pos (%) | PR pos (%) | Her-2/neu pos (%) | MVD (vessels/field) |
|---|---|---|---|---|---|---|---|
| Upper 25% (n = 4) | 4.83 ± 0.14 | 45.0 ± 11.0 | 1.4 ± 0.3 | 0% | 0% | 0% | 124.3 ± 32.8 |
| Lower 25% (n = 4) | 0.47 ± 0.49 | 47.0 ± 8.5 | 2.0 ± 0.4 | 75% | 75% | 0% | 61.2 ± 6.04 |
| Total (n = 16) | 2.41 ± 1.47 | 49.5 ± 9.5 | 1.8 ± 1.1 | 62.5% | 50% | 12.5% | 92.7 ± 40.5 |
| Upper vs. lower | **P < 0.0001 | NS P = 0.782 | NS P = 0.0692 | *P = 0.0285 | *P = 0.0285 | NS | *P = 0.31 |
| Upper vs. total | *P = 0.0145 | NS P = 0.421 | NS P = 0.804 | NS P = 0.129 | NS P = 0.172 | NS P = 0.619 | NS P = 0.284 |
Quantitative NPTN-β mRNA expression was determined for 16 primary breast carcinomas and average values provided for top quartile (upper 25%), bottom quartile (lower 25%), and total. Age (years), tumor size as maximum diameter (cm), percent cases positive for estrogen receptor alpha (ERα), progesterone receptor (PR), and Her-2/neu, and average microvascular density (MVD) as determined by quantification of CD31 staining are provided for each group. Values for NPTN-β, age, tumor size, and MVD are average ± standard deviation. Statistical analysis of each category was completed comparing the upper quartile to the lower quartile and the upper quartile to the total. P values are indicated as significant P ≤ 0.05 (*), P ≤ 0.01 (**), or not significant (NS)
Discussion
In the present study, we describe a novel scheme for the identification of TAAs using human tissues. Because it is likely that antigen-driven B cell proliferation in tumor-draining lymph nodes can be activated by TAAs, our first endeavor was to identify areas within the lymph node where such proliferation events were taking place. Immunohistological evaluation of B cell proliferation in tumor-draining lymph nodes using Ki67 as a marker resulted in the identification of very few proliferative zones in each sample (approximately one or two in each lymph node). In germinal centers, Ki67 is expressed in the centroblast-rich dark zone and is lost during B cell differentiation [3, 23, 31]. Thus, when Ki67 signal was overlayed onto the anti-CD20 and -CD23 signals, the triple stain pattern adequately identifies clusters of B cell activation and proliferation, a good indication that these corresponded to germinal centers where an immune response was developing at the time of resection.
To examine this possibility further, Ig VH domain cDNA libraries were generated from either the CD20+CD23+Ki67+ and CD20+CD23+Ki67− zones from one lymph node. Our analysis demonstrated that CE is present to a much larger extent in the Ki67+proliferative zone when compared to Ki67− areas. Furthermore, evaluation of mutations from the closest matching germline genes using a stringent method previously described [26] shows that the distribution and characteristics of SH was present in a significantly higher percentage of sequences from the proliferative zone VH library. These data point toward the possibility of identifying zones with antigen-driven humoral immune responses in frozen tumor-draining lymph node samples by combining immunohistochemistry and Ig VH sequence analysis. Despite a unique and potentially useful approach to identify active germinal centers within a tumor-draining lymph node, these analyses; however, cannot assess whether these events are evoked by the tumor or some basal cycling events within lymph nodes, something that will have to be evaluated in a larger population study.
A significant application of this approach was the ability to utilize the CE and SH VH gene sequences to screen for, and identify potential novel tumor antigens. The discovery of naturally occurring antibodies devoid of light chains in camelids [15] has led to the characterization of sdAbs as highly versatile molecules with potential as diagnostic and therapeutic applications [18]. These sdABs retain antigen-binding capacity and can be synthesized as a stable molecule, and are significantly smaller than full-length antibodies or even scFv recombinant proteins. These characteristics make them uniquely suitable for antigen discovery. A previous study by Zhang et al. [46] used a pentavalent sdAb to identify a candidate TAA from a non-small cell lung carcinoma cell line using proteomic analysis, proving that these reagents can select specific proteins from a complex sample. In this study, we selected a VH sequence that showed a significant SH pattern to synthesize a recombinant sdAb, which could be bound to a solid matrix and served to trap a specific protein from tumor lysates, NPTN a potential breast cancer antigen. Since the VH gene was derived from the same lymph node sample as the patient tumor and, the non-CE/SH control sdAb did not select NPTN, we speculate that an NPTN-specific immune response was detected in the lymph node and served to produce this specific VH sequence. Further evidence for this may have been possible if patient serum was available, however, a matched serum was not available from this patient. These results do however, raise the exciting possibility that this procedure for the discovery of TAAs using antibody sequences selected by immune activation in tumor-draining lymph nodes is entirely possible and in fact may be very efficient and effective.
The identification of NPTN as a potential breast cancer antigen is not surprising since multiple antigens have been identified that are surface molecules with high glycosylation content [4, 11, 16, 28, 32]. NPTN is a surface molecule containing multiple immunoglobulin-like extracellular domains which share homology with those of Basigin/CD147/EMMPRIN gene. The 65 kDa neuronal-specific isoform has been shown to participate in long-term potentiation in neural synapses through homophilic adhesion that triggers p38MAPK activation [10, 39]. Several investigations have indicated that the Basigin/CD147/EMMPRIN family of proteins promote monocarboxylate transporter proteins which are critical for lactate and pyruvate shuttling into and out of cells during metabolic stress [20]. In this study, the overexpression of NPTN beta in a breast cancer cell line significantly enhanced tumor growth and angiogenesis in vivo. The enhanced tumorigenesis correlated with a higher VEGF content in the tumors as well as a more robust production under hypoxic conditions in vitro. Interestingly, CD147 is a molecule which can be highly expressed in tumors and has also been shown to stimulate VEGF production by both tumor and stromal cells when expressed in a breast cancer cell line [40]. In an interesting preliminary screen, it was found that NPTN-beta expression also correlated with primary human breast tumor angiogenesis. Thus, our results implicate NPTN as another member of the Ig superfamily that, when aberrantly expressed by transformed cells, can confer a growth advantage to malignant tumors. The specific mechanism of this response is thus far unclear and may involve tumor microenvironmental signals. Here we determined that environmental hypoxia, when combined with NPTN overexpression, further promotes the expression of VEGF above the level observed in control cells. Thus, NPTN expression may increase the sensitivity or ability to synthesize additional VEGF in tumor cells. A mechanism for this process may be complicated and implications for increased HIF-1 activity, post-transcriptional mRNA stability and even potentiation of translation and vesicular trafficking are all possible and need to be investigated further. The increased VEGF expression may be one explanation of the correlation of NPTN expression with highly invasive and the distally metastatic breast cancers, suggesting that NPTN may promote survival in the circulation and/or invasion and survival at a distal site. Finally, NPTN expression appears to represent multiple glycoforms in human breast cancer suggesting that it may be shed from the cell surface by proteases or there may be alternative patterns of glycosylation in some tumors. Alternative or aberrant glycosylation could be a mechanism that would promote the antigenicity of this protein, especially if shed from the tumor microenvironment at high levels.
The identification and evaluation of a large number of potential antigens, particularly alternative forms of proteins normally expressed, may be a useful approach to define novel cancer biomarkers for primary and metastatic cancers. Although this study effectively used patient derived VH domain antibody to define a breast cancer antigen, it remains to be seen whether this is a robust method applicable to breast and other cancers. The use of a patient-activated humoral response to provide the tools for this approach has been demonstrated here and should be applicable to many other human cancers.
Acknowledgments
This work was supported by NIH:NCI IMAT grant, CA114489 (KPC) and The Patrick and Catherine Weldon Donaghue Foundation (KPC). Many thanks to Nancy Ryan for her technical assistance in histology and immunohistochemistry.
References
- 1.Agarwal A, Munoz-Najar U, Klueh U, Shih SC, Claffey KP. N-acetyl-cysteine promotes angiostatin production and vascular collapse in an orthotopic model of breast cancer. Am J Pathol. 2004;164(5):1683–1696. doi: 10.1016/S0002-9440(10)63727-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aihara K, Yamada K, Murakami H, Nomura Y, Omura H. Production of human–human hybridomas secreting monoclonal antibodies reactive to breast cancer cell lines. In Vitro Cell Dev Biol. 1988;24(9):959–962. doi: 10.1007/BF02623911. [DOI] [PubMed] [Google Scholar]
- 3.Angelin-Duclos C, Cattoretti G, Lin KI, Calame K. Commitment of B lymphocytes to a plasma cell fate is associated with Blimp-1 expression in vivo. J Immunol. 2000;165(10):5462–5471. doi: 10.4049/jimmunol.165.10.5462. [DOI] [PubMed] [Google Scholar]
- 4.Annibali NV, Baldi A. Characterization of the ETSA-21 antigen, a glycosylphosphatidyl-inositol anchor glycoprotein identified in breast cancer cells using monoclonal antibody B21. Hybridoma. 1997;16(2):139–145. doi: 10.1089/hyb.1997.16.139. [DOI] [PubMed] [Google Scholar]
- 5.Bernstein HG, Smalla KH, Bogerts B, Gordon-Weeks PR, Beesley PW, Gundelfinger ED, Kreutz MR. The immunolocalization of the synaptic glycoprotein neuroplastin differs substantially between the human and the rodent brain. Brain Res. 2007;1134(1):107–112. doi: 10.1016/j.brainres.2006.11.090. [DOI] [PubMed] [Google Scholar]
- 6.Chin Y, Janseens J, Vandepitte J, Vandenbrande J, Opdebeek L, Raus J. Phenotypic analysis of tumor-infiltrating lymphocytes from human breast cancer. Anticancer Res. 1992;12(5):1463–1466. [PubMed] [Google Scholar]
- 7.Coronella-Wood JA, Hersh EM. Naturally occurring B-cell responses to breast cancer. Cancer Immunol Immunother. 2003;52(12):715–738. doi: 10.1007/s00262-003-0409-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coronella JA, Spier C, Welch M, Trevor KT, Stopeck AT, Villar H, Hersh EM. Antigen-driven oligoclonal expansion of tumor-infiltrating B cells in infiltrating ductal carcinoma of the breast. J Immunol. 2002;169(4):1829–1836. doi: 10.4049/jimmunol.169.4.1829. [DOI] [PubMed] [Google Scholar]
- 9.Cote RJ, Morrissey DM, Houghton AN, Beattie EJ, Jr, Oettgen HF, Old LJ. Generation of human monoclonal antibodies reactive with cellular antigens. Proc Natl Acad Sci USA. 1983;80(7):2026–2030. doi: 10.1073/pnas.80.7.2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Empson RM, Buckby LE, Kraus M, Bates KJ, Crompton MR, Gundelfinger ED, Beesley PW. The cell adhesion molecule neuroplastin-65 inhibits hippocampal long-term potentiation via a mitogen-activated protein kinase p38-dependent reduction in surface expression of GluR1-containing glutamate receptors. J Neurochem. 2006;99(3):850–860. doi: 10.1111/j.1471-4159.2006.04123.x. [DOI] [PubMed] [Google Scholar]
- 11.Freire T, Lo-Man R, Piller F, Piller V, Leclerc C, Bay S. Enzymatic large-scale synthesis of MUC6-Tn glycoconjugates for antitumor vaccination. Glycobiology. 2006;16(5):390–401. doi: 10.1093/glycob/cwj082. [DOI] [PubMed] [Google Scholar]
- 12.Goldstein NI, Nagle R, Villar H, Hersh E, Fisher PB. Isolation and characterization of a human monoclonal antibody which reacts with breast and colorectal carcinoma. Anticancer Res. 1990;10(6):1491–1500. [PubMed] [Google Scholar]
- 13.Grekou AN, Toliou T, Stravoravdi P, Patakiouta F, Tsoukalas T, Pinakidis M, Keramidas G. Correlation of apoptosis with the distribution and composition of lymphocytic infiltrate in human breast carcinomas. Anticancer Res. 1996;16(6C):3991–3995. [PubMed] [Google Scholar]
- 14.Hadden JW. The immunology and immunotherapy of breast cancer: an update. Int J Immunopharmacol. 1999;21(2):79–101. doi: 10.1016/S0192-0561(98)00077-0. [DOI] [PubMed] [Google Scholar]
- 15.Hamers-Casterman C, Atarhouch T, Muyldermans S, Robinson G, Hamers C, Songa EB, Bendahman N, Hamers R. Naturally occurring antibodies devoid of light chains. Nature. 1993;363(6428):446–448. doi: 10.1038/363446a0. [DOI] [PubMed] [Google Scholar]
- 16.Harada Y, Ohuchi N, Masuko T, Funaki Y, Mori S, Satomi S, Hashimoto Y. Characterization of a new breast cancer-associated antigen and its relationship to MUC1 and TAG-72 antigens. Tohoku J Exp Med. 1996;180(3):273–288. doi: 10.1620/tjem.180.273. [DOI] [PubMed] [Google Scholar]
- 17.Holliger P, Hudson PJ. Engineered antibody fragments and the rise of single domains. Nat Biotechnol. 2005;23(9):1126–1136. doi: 10.1038/nbt1142. [DOI] [PubMed] [Google Scholar]
- 18.Holt LJ, Herring C, Jespers LS, Woolven BP, Tomlinson IM. Domain antibodies: proteins for therapy. Trends Biotechnol. 2003;21(11):484–490. doi: 10.1016/j.tibtech.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 19.Hunter RL, Ferguson DJ, Coppleson LW. Survival with mammary cancer related to the interaction of germinal center hyperplasia and sinus histiocytosis in axillary and internal mammary lymph nodes. Cancer. 1975;36(2):528–539. doi: 10.1002/1097-0142(197508)36:2<528::AID-CNCR2820360232>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 20.Iacono KT, Brown AL, Greene MI, Saouaf SJ. CD147 immunoglobulin superfamily receptor function and role in pathology. Exp Mol Pathol. 2007;83(3):283–295. doi: 10.1016/j.yexmp.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Imam A, Drushella MM, Taylor CR, Tokes ZA. Generation and immunohistological characterization of human monoclonal antibodies to mammary carcinoma cells. Cancer Res. 1985;45(1):263–271. [PubMed] [Google Scholar]
- 22.Kjeldsen TB, Rasmussen BB, Rose C, Zeuthen J. Human–human hybridomas and human monoclonal antibodies obtained by fusion of lymph node lymphocytes from breast cancer patients. Cancer Res. 1988;48(11):3208–3214. [PubMed] [Google Scholar]
- 23.Lampert IA, Van Noorden S, Wotherspoon AC. Centrocytoid plasma cells of the germinal center. Appl Immunohistochem Mol Morphol. 2005;13(2):124–131. doi: 10.1097/01.pai.0000135614.30196.fb. [DOI] [PubMed] [Google Scholar]
- 24.Langnaese K, Beesley PW, Gundelfinger ED. Synaptic membrane glycoproteins gp65 and gp55 are new members of the immunoglobulin superfamily. J Biol Chem. 1997;272(2):821–827. doi: 10.1074/jbc.272.2.821. [DOI] [PubMed] [Google Scholar]
- 25.Langnaese K, Mummery R, Gundelfinger ED, Beesley PW. Immunoglobulin superfamily members gp65 and gp55: tissue distribution of glycoforms. FEBS Lett. 1998;429(3):284–288. doi: 10.1016/S0014-5793(98)00616-4. [DOI] [PubMed] [Google Scholar]
- 26.Lossos IS, Tibshirani R, Narasimhan B, Levy R. The inference of antigen selection on Ig genes. J Immunol. 2000;165(9):5122–5126. doi: 10.4049/jimmunol.165.9.5122. [DOI] [PubMed] [Google Scholar]
- 27.Nzula S, Going JJ, Stott DI. Antigen-driven clonal proliferation, somatic hypermutation, and selection of B lymphocytes infiltrating human ductal breast carcinomas. Cancer Res. 2003;63(12):3275–3280. [PubMed] [Google Scholar]
- 28.Pancino G, Osinaga E, Charpin C, Mistro D, Barque JP, Roseto A. Purification and characterisation of a breast-cancer-associated glycoprotein not expressed in normal breast and identified by monoclonal antibody 83D4. Br J Cancer. 1991;63(3):390–398. doi: 10.1038/bjc.1991.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Phoenix KN, Vumbaca F, Claffey KP (2008) Therapeutic metformin/AMPK activation promotes the angiogenic phenotype in the ERalpha negative MDA-MB-435 breast cancer model. Breast Cancer Res Treat. doi:10.1007/s10549-008-9916-5 [DOI] [PMC free article] [PubMed]
- 30.Posner MR, Elboim HS, Tumber MB, Wiest PM, Tibbetts LM. An IgG human monoclonal antibody reactive with a surface membrane antigen expressed on malignant breast cancer cells. Hum Antibodies Hybridomas. 1991;2(2):74–83. [PubMed] [Google Scholar]
- 31.Rahman ZS, Rao SP, Kalled SL, Manser T. Normal induction but attenuated progression of germinal center responses in BAFF and BAFF-R signaling-deficient mice. J Exp Med. 2003;198(8):1157–1169. doi: 10.1084/jem.20030495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rughetti A, Turchi V, Ghetti CA, Scambia G, Panici PB, Roncucci G, Mancuso S, Frati L, Nuti M. Human B-cell immune response to the polymorphic epithelial mucin. Cancer Res. 1993;53(11):2457–2459. [PubMed] [Google Scholar]
- 33.Sarfati M. Leucocyte typing VI. New York: Garland Publishing Inc.; 1997. pp. 144–147. [Google Scholar]
- 34.Sblattero D, Bradbury A. A definitive set of oligonucleotide primers for amplifying human V regions. Immunotechnology. 1998;3(4):271–278. doi: 10.1016/S1380-2933(97)10004-5. [DOI] [PubMed] [Google Scholar]
- 35.Schlom J, Wunderlich D, Teramoto YA. Generation of human monoclonal antibodies reactive with human mammary carcinoma cells. Proc Natl Acad Sci USA. 1980;77(11):6841–6845. doi: 10.1073/pnas.77.11.6841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scholzen T, Gerdes J. The Ki-67 protein: from the known and the unknown. J Cell Physiol. 2000;182(3):311–322. doi: 10.1002/(SICI)1097-4652(200003)182:3<311::AID-JCP1>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 37.Shih SC, Mullen A, Abrams K, Mukhopadhyay D, Claffey KP. Role of protein kinase C isoforms in phorbol ester-induced vascular endothelial growth factor expression in human glioblastoma cells. J Biol Chem. 1999;274(22):15407–15414. doi: 10.1074/jbc.274.22.15407. [DOI] [PubMed] [Google Scholar]
- 38.Simsa P, Teillaud JL, Stott DI, Toth J, Kotlan B. Tumor-infiltrating B cell immunoglobulin variable region gene usage in invasive ductal breast carcinoma. Pathol Oncol Res. 2005;11(2):92–97. doi: 10.1007/BF02893374. [DOI] [PubMed] [Google Scholar]
- 39.Smalla KH, Matthies H, Langnase K, Shabir S, Bockers TM, Wyneken U, Staak S, Krug M, Beesley PW, Gundelfinger ED. The synaptic glycoprotein neuroplastin is involved in long-term potentiation at hippocampal CA1 synapses. Proc Natl Acad Sci USA. 2000;97(8):4327–4332. doi: 10.1073/pnas.080389297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tang Y, Nakada MT, Kesavan P, McCabe F, Millar H, Rafferty P, Bugelski P, Yan L. Extracellular matrix metalloproteinase inducer stimulates tumor angiogenesis by elevating vascular endothelial cell growth factor and matrix metalloproteinases. Cancer Res. 2005;65(8):3193–3199. doi: 10.1158/0008-5472.CAN-05-0027. [DOI] [PubMed] [Google Scholar]
- 41.Tang Y, Nakada MT, Rafferty P, Laraio J, McCabe FL, Millar H, Cunningham M, Snyder LA, Bugelski P, Yan L. Regulation of vascular endothelial growth factor expression by EMMPRIN via the PI3 K-Akt signaling pathway. Mol Cancer Res. 2006;4(6):371–377. doi: 10.1158/1541-7786.MCR-06-0042. [DOI] [PubMed] [Google Scholar]
- 42.Tsakraklides V, Olson P, Kersey JH, Good RA. Prognostic significance of the regional lymph node histology in cancer of the breast. Cancer. 1974;34(4):1259–1267. doi: 10.1002/1097-0142(197410)34:4<1259::AID-CNCR2820340436>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 43.Whitford P, George WD, Campbell AM. Flow cytometric analysis of tumour infiltrating lymphocyte activation and tumour cell MHC class I and II expression in breast cancer patients. Cancer Lett. 1992;61(2):157–164. doi: 10.1016/0304-3835(92)90174-T. [DOI] [PubMed] [Google Scholar]
- 44.Yan L, Zucker S, Toole BP. Roles of the multifunctional glycoprotein, emmprin (basigin; CD147), in tumour progression. Thromb Haemost. 2005;93(2):199–204. doi: 10.1160/TH04-08-0536. [DOI] [PubMed] [Google Scholar]
- 45.Yates JR, 3rd, Eng JK, McCormack AL, Schieltz D. Method to correlate tandem mass spectra of modified peptides to amino acid sequences in the protein database. Anal Chem. 1995;67(8):1426–1436. doi: 10.1021/ac00104a020. [DOI] [PubMed] [Google Scholar]
- 46.Zhang J, Li Q, Nguyen TD, Tremblay TL, Stone E, To R, Kelly J, Roger MacKenzie C. A pentavalent single-domain antibody approach to tumor antigen discovery and the development of novel proteomics reagents. J Mol Biol. 2004;341(1):161–169. doi: 10.1016/j.jmb.2004.05.069. [DOI] [PubMed] [Google Scholar]
- 47.Zheng HC, Takahashi H, Murai Y, Cui ZG, Nomoto K, Miwa S, Tsuneyama K, Takano Y. Upregulated EMMPRIN/CD147 might contribute to growth and angiogenesis of gastric carcinoma: a good marker for local invasion and prognosis. Br J Cancer. 2006;95(10):1371–1378. doi: 10.1038/sj.bjc.6603425. [DOI] [PMC free article] [PubMed] [Google Scholar]