Abstract
The ubiquitin ligase Nedd4 has been proposed to regulate a number of signaling pathways, but its physiological role in mammals has not been characterized. Here we present an analysis of Nedd4-null mice to show that loss of Nedd4 results in reduced insulin-like growth factor 1 (IGF-1) and insulin signaling, delayed embryonic development, reduced growth and body weight, and neonatal lethality. In mouse embryonic fibroblasts, mitogenic activity was reduced, the abundance of the adaptor protein Grb10 was increased, and the IGF-1 receptor, which is normally present on the plasma membrane, was mislocalized. However, surface expression of IGF-1 receptor was restored in homozygous mutant mouse embryonic fibroblasts after knockdown of Grb10, and Nedd4−/− lethality was rescued by maternal inheritance of a disrupted Grb10 allele. Thus, in vivo, Nedd4 appears to positively control IGF-1 and insulin signaling partly through the regulation of Grb10 function.
INTRODUCTION
In mammals, the insulin-like growth factor (IGF) axis is a major regulator of fetal and postnatal growth (1–3). This signaling axis contains three ligands (IGF-1, IGF-2, and insulin, collectively referred to as IGFs/insulin) and two closely-related transmembrane receptor tyrosine kinases, the type 1 insulin-like growth factor receptor (IGF-1R) and the insulin receptor (IR). IGF-1 and IGF-2 promote fetal growth, with the former acting solely through the IGF-1R and the latter signaling through the IGF-1R and IR (1). After birth, growth hormone acts to promote growth independently and by inducing production of systemic and local IGF-1 (3). The third ligand in this axis, insulin, is preeminent for the regulation of whole-body glucose homeostasis and mediates metabolic responses in target tissues by binding to the IR (4). Critical targets for phosphorylation by the IR and IGF-1R are insulin receptor substrates (IRS)-1 and IRS-2, which are docking proteins that, once phosphorylated, provide binding sites for various Src homology 2 (SH2) domain–containing proteins, including Grb2 and the p85 subunit of phosphatidylinositol 3-kinase (PI 3-kinase) (5). Activation of PI 3-kinase and the downstream kinase Akt plays a pivotal role in mediating proliferative, growth, and survival responses to IGFs/insulin as well as metabolic responses to insulin (5, 6). A separate arm of insulin/IGF signaling associated with regulation of proliferation is mediated by tyrosine phosphorylation of Shc, assembly of a complex of Shc and Grb2, and activation of the Ras to extracellular signal–regulated kinase (ERK) pathway (5). The regulation of IGF-1R and IR signaling is complex and involves recruitment of specific protein tyrosine phosphatases, particular SOCS(suppressor of cytokine signaling) family members, the ubiquitin–protein ligase Nedd4 (neuronal precursor cell–expressed developmentally down-regulated gene 4), and the adaptor-type signal modulators Grb10 and Grb14 (5, 7, 8).
Nedd4 is the prototypic member of a highly conserved family of ubiquitin–protein ligases (9–11). The various members of this family have been implicated in controlling several signaling pathways, often by regulating protein trafficking (12, 13). Nedd4 itself is implicated in the regulation of the stability of IGF-1R (7) and vascular endothelial cell growth factor receptor 2 (14), degradation and nuclear localization of the phosphatase PTEN (15), and virus budding (16). Although IGF-1R does not bind Nedd4 directly, the two proteins have been reported to be linked through the adaptor Grb10, leading to IGF-1R ubiquitination, internalization, and degradation (17, 18). In contrast, in this study we provide genetic and cellular evidence to show that Nedd4 is required for efficient cell surface expression of the IGF-1R and IR, and is therefore a positive regulator of IGF-1 and insulin signaling. The abundance of Grb10 is increased in the absence of Nedd4. Furthermore, simultaneous loss of Nedd4 and Grb10 restores cell surface IGF-1R, indicating that Nedd4 acts to oppose the function of the negative regulator Grb10.
RESULTS
Nedd4−/− mice are growth retarded and die perinatally
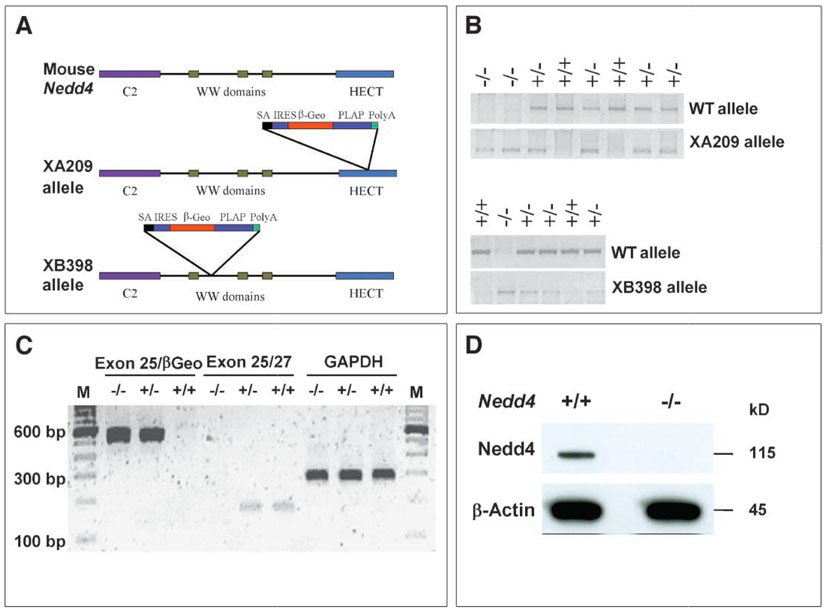
To study the physiological function of Nedd4, we generated knockout (KO) mice with the use of gene-trapped embryonic stem (ES) cell lines (BayGenomics XA209 and XB398) (19), as shown in Fig. 1. In more than 50 offspring generated by intercrossing heterozygotes from each line, genotyping at the time of weaning failed to identify any Nedd4−/− mice, suggesting that the homozygous nullmutation causes either embryonic or perinatal lethality. Because both lines exhibited lethality at similar frequencies, the XA209 line was used for most of the studies. Timed matings revealed the presence of homozygous embryos at the expected ratio at both 12.5 and 18.5 days postcoitum (dpc), suggesting that Nedd4−/− mice die during or shortly after birth. The presence of homozygous embryos was determined by polymerase chain reaction (PCR) and reverse transcription (RT)-PCR with the use of genomic DNA isolated from tail tissue, and the absence of Nedd4 protein in the homozygotes was confirmed by Western blotting (Fig. 1). The homozygous embryos were severely growth retarded at both time points studied (Fig. 2). An additional Nedd4 KO derived from a gene trap line with a retroviral insertion disrupting the Nedd4 gene between exons 17 and 18 was obtained from Mutant Mouse Regional Resource Center (MMRRC) (see Materials and Methods). The phenotype of this KO line was identical to the lines above, with homozygous animals showing severe growth retardation and perinatal lethality. For example, at 13.5 dpc, the body weights of the MMRRC Nedd4 KO animals were significantly lower than the body weights of wild-type littermates (fig. S1).
Fig. 1.
Generation of gene-trapped insertional alleles for the mouse Nedd4 gene. (A) A gene-trapping vector was used to disrupt this gene into two independent cell lines: in the XA209 allele, the gene trap vector is inserted in the middle of the HECT domain (after exon 25), whereas in the XB398 allele, the gene trap vector is inserted after the first WW domain (after exon 12). (B) PCR genotyping of mice potentially harboring one of the alleles. (C) RT-PCR analysis of mice carrying the XA209 allele. Transcript containing exon 25 and the gene trap vector (exon 25-βGeo) was not detected in wild-type mice, whereas the endogenous transcript (exons 25/27) was not detected in mice homozygous for the XA209 allele. GAPDH was used as an internal control. (D) Immunoblot confirming the absence of Nedd4 protein in cells derived from XA209 homozygous mutants. Total cell lysate (50 µg protein) obtained from passage number–matched cultures was loaded in each lane. Primary antibody against the WW2 domain of Nedd4 (Upstate) and goat–anti-rabbit secondary antibody (Super Signal Femto, Pierce) were used.
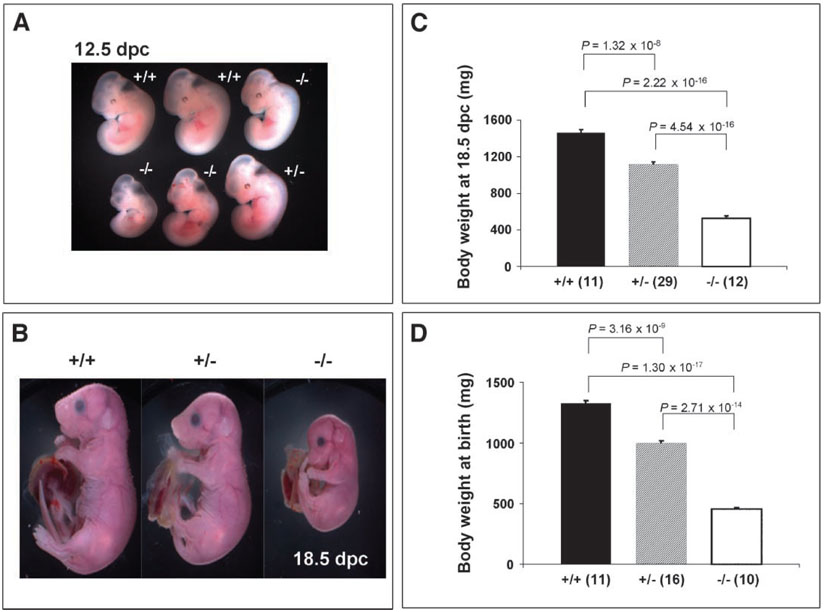
Fig. 2.
Nedd4−/− mice die immediately after birth, and Nedd4+/− and Nedd4−/− mice exhibit intrauterine growth retardation. No mice homozygous for disruption of the Nedd4 gene were found 2 or 3 weeks after birth. Ratios of heterozygotes and homozygous mutants were thus assessed at earlier time points: (A) 12.5 dpc, (B and C) 18.5 dpc, and (D) immediately after birth. Both heterozygotes and homozygous mutants showed signs of intrauterine growth retardation as early as 12.5 dpc (A) and at late gestation [18.5 dpc (B) and (C)]. At the time of birth [post-natal day 1 (D)], the body weights among three genotypes differed significantly:Nedd4−/− body weight averaged 64 to 68% lower relative to that of wild-type littermates; heterozygote body weight averaged about 15 to 20% reduction in body weight relative to that of wild-type littermates. In (C) and (D), the numbers of animals used for the analyses are shown in parentheses; the body weight was significantly different between groups of mice, with P values indicated.
Histological analyses of late-gestation (18.5 dpc) embryos did not reveal gross abnormalities, except membranous ventricular septal defect (VSD) seen in one of three Nedd4−/− embryos and none of five control littermates examined (fig. S2). However, VSD is unlikely to be the cause of perinatal death of Nedd4−/− embryos. Close observation of pregnant females near the term of gestation revealed that Nedd4−/− pups were born at the predicted ratio. Immediately after birth, however, homozygotes turned blue and died. Lungs dissected from Nedd4−/− newborns looked dark and dense and sank when placed in a beaker containing water. In contrast, lungs from heterozygous and wild-type littermates looked red and spongy and floated on water. These observations suggested that Nedd4-null mice die from respiratory distress, probably because of immature lungs, a dysfunctional diaphragm muscle, or both. Lung histology of late-gestation embryos and newborn mice reveals that there is no morphological difference between wild-type and null embryos at late gestation other than the smaller lung size of the null embryos, yet in newborns, the lungs of homozygous animals are relatively unaerated compared with those of wild-type pups, which have expanded alveoli (fig. S3).
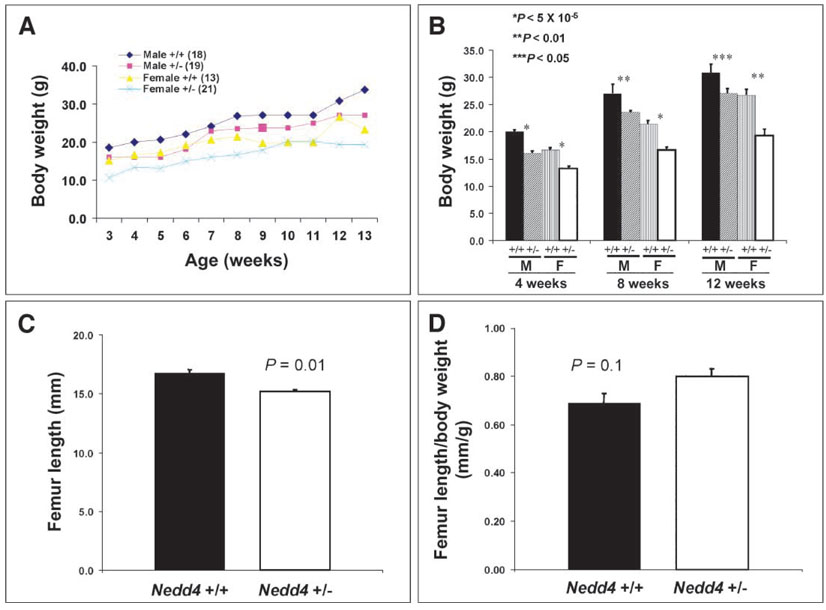
Consistent with the finding of growth retardation during embryo-genesis, Nedd4−/− newborns were small, weighing on average 65% less than their wild-type littermates. Heterozygous littermates were also affected, with body weights reduced by 15 to 20% relative to that of their wild-type littermates (Fig. 2). The difference in body weight between wild-type and heterozygous animals persisted postnatally, until at least 3 months of age, and was paralleled by a reduced femur length in the heterozygous animals (Fig. 3).
Fig. 3.
Mice heterozygous for Nedd4 disruption show significant growth retardation postnatally. Postweaning mice were weighed weekly until they were 13 weeks old. (A and B) Heterozygotes at 3 weeks of age [both male (M) and female (F)] were significantly smaller than control mice, and the body weight difference persisted until at least 3 months of age. The numbers of animals are identical in (A) and (B). The femur length, which was determined by x-ray radiography of 2-month-old mice, showed significant differences between the wild-type and heterozygous mice [(C), P = 0.01]. However, femur length as a function of body weight did not differ significantly between these groups (D). There were 12 mice in each group in (C) and (D).
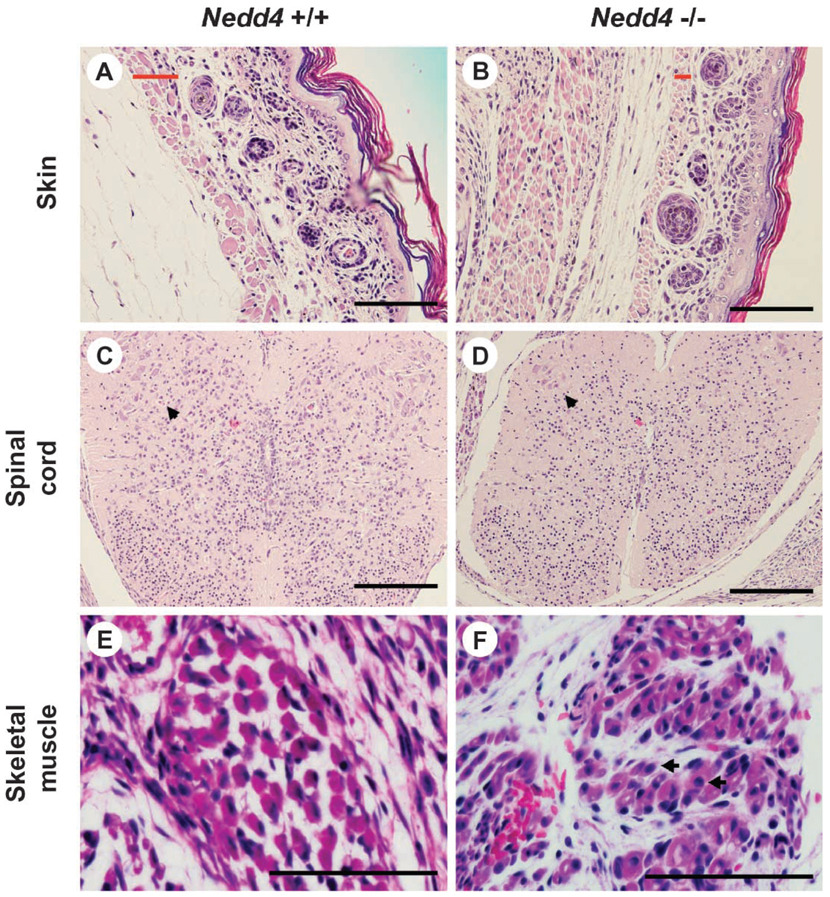
Histological examination by light microscopy failed to reveal any gross malformations in Nedd4−/− embryos or newborn pups. However, signs of immaturity were present in both groups. For example, in skin from these mice, the hair follicles were immature relative to those in the wild-type littermates, and the superficial skeletal muscle (platysma) was underdeveloped. Likewise, the spinal cord was underdeveloped, with few mature ganglion cells present in the anterior horns, consistent with a general tendency toward hypocellularity in this tissue. In the case of skeletal muscle, fiber size was variable in the KO but consistent in the wild-type animals, internal nuclei were common in KO animals, and the interstitium tended to be more immature in the KO than in wild-type mice (Fig. 4).
Fig. 4.
Nedd4−/− mice are developmentally delayed. Tissue samples were taken from mice on postnatal day 1 and processed for histology. Wild-type littermates were used as controls. More than six mice were used for each genotype, and typical histology is shown. (A and B) Skin: In the mutant mouse, the hair follicles are immature and the superficial skeletal muscle (platysma) is underdeveloped. The width of the platysma muscle layer is indicated by a red bar in each panel. (C and D) Spinal cord: In the mutant mouse, the spinal cord tends to be hypocellular, with fewer mature ganglion cells in the anterior horns than in the control (see arrowhead). (E and F) Skeletalmuscle: In the mutant mouse, fiber size varies significantly and internal nuclei are common [arrows in (F)]. Also, the interstitium tends to be more immature. Scale bar, 100 µm.
Nedd4−/− mouse embryonic fibroblasts exhibit reduced mitogenic activity
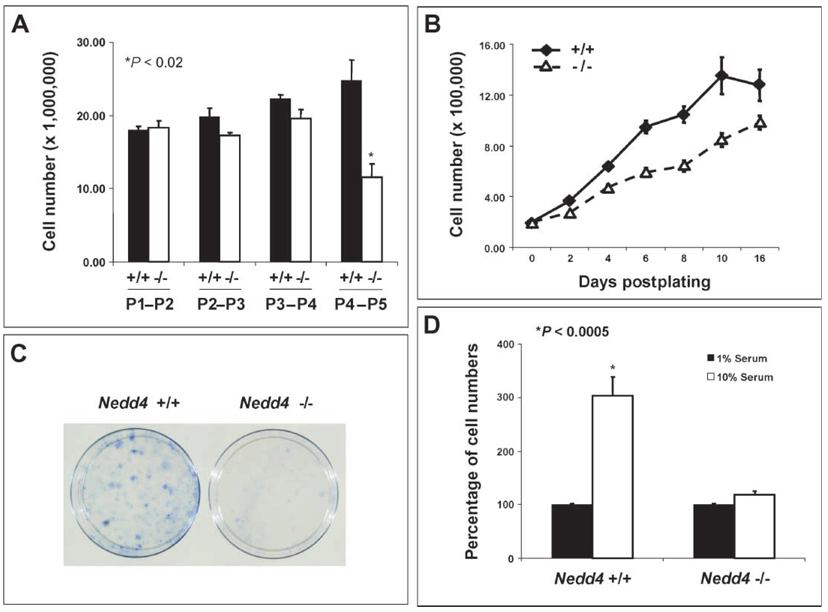
To understand the cellular basis of growth retardation of Nedd4−/− animals, we used mouse embryonic fibroblasts (MEFs) isolated from mid-gestation embryos (13.5 dpc). At early passages (P1 to P4), Nedd4−/− and wild-type MEFs exhibited no significant differences in growth in medium containing 10% serum (Fig. 5A). At later passages, however, proliferation of the Nedd4−/− MEFs under these conditions was greatly reduced compared with that of the wild-type MEFs. From P4, the number of Nedd4−/− MEFs did not increase measurably over a 24-hour period, suggesting that the doubling time was reduced compared with that of the Nedd4+/+ MEFs. To investigate this further, P4 Nedd4+/+ and Nedd4−/− MEFs were seeded at moderate density and their growth properties examined. The Nedd4−/− MEFs proliferated more slowly in 10% serum than did wild-type MEFs (Fig. 5B). Also, after low-density seeding at P2, colony formation was reduced in the case of the Nedd4−/− MEFs (Fig. 5C). This was the earliest in vitro readout of differential growth among the Nedd4 genotypes.
Fig. 5.
MEFs isolated from Nedd4−/− embryos show reduced mitogenic activity. MEFs were isolated from mid-gestation embryos (13.5 dpc) and passage number–matched MEFs were used for each experiment. (A) MEFs from Nedd4−/− embryos exhibit passage number–dependent decreases in growth rate, whereas Nedd4+/+ MEFs do not. (B) Passage 4 Nedd4−/− MEFs grew more slowly than did Nedd4+/+ MEFs. (C) Nedd4−/− MEFs exhibit reduced colony-forming activity after low-density seeding at passage 2.Data shown are representative of three independent experiments. (D) Wild-type MEFs require high concentrations of exogenous growth factors from serum for Nedd4-dependent stimulation of cell growth to take place.
As expected, P4 MEFs from both Nedd4+/+ and Nedd4−/− mice failed to proliferate under low (1%) serum conditions. However, when switched to normal serum (10%)–containing media, the wild-type MEFs proliferated much faster than the KO MEFs (Fig. 5D). These results suggest that Nedd4 regulates MEF growth and proliferation and that these parameters require serum-derived growth factors. This prompted us to analyze the cell cycle in passage-matched (P2–P5) asynchronous MEFs grown under normal culture conditions. This analysis showed that both Nedd4+/− and Nedd4−/− cells accumulated in the G0 and G1 phases, and the differences between the wild-type and heterozygous as well as between the wild-type and KO MEFs were significant (fig. S4A). As expected, the percentages of heterozygous and homozygous KO cells in the G2/M and S phases decreased in parallel (fig. S4, B and C). Growth factors, including IGF-1, are responsible for promoting progression from the G0 into the G1 phase and ultimately through the rest of the cell cycle (20). A reduction in IGF-1 signaling in Nedd4-deficient cells may be responsible for the decreased progression through the cell cycle.
IGF-1 and insulin signaling is reduced in Nedd4−/− MEFs
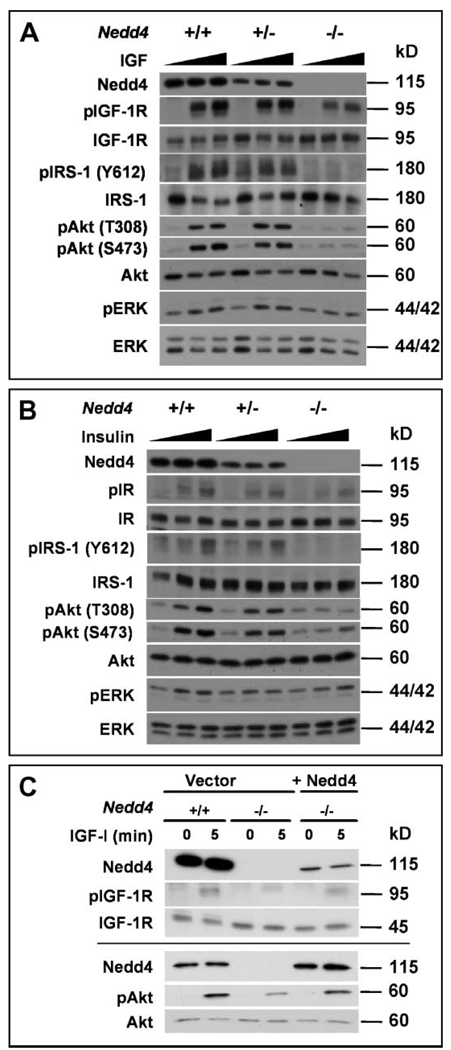
Mice homozygous for Igf1r deletion are severely growth retarded, with a birth weight 55% lower than that of normal littermates, and they die immediately after birth (2). Similarly, double mutants lacking the IGF-1 and IGF-2 ligands, or lacking the IGF-2 ligand and IGF-1R, show a 70% reduction of the normal body weight at birth (1, 2). Thus, our observation of a 65% reduction in normal birth weight in Nedd4−/− embryos and newborn pups is consistent with the absence of Nedd4 leading to an inhibition of IGF signaling during fetal growth. Immunoblotting of MEF lysates showed that Nedd4 deficiency did not significantly affect the total cellular amount of either the IGF-1R precursor or the β-subunit (Fig. 6A and fig. S5). However, IGF-1–mediated signaling was substantially reduced in Nedd4−/− MEFs and to a lesser extent in Nedd4+/− cells (Fig. 6A). As shown in Fig. 6A, IGF-1–induced tyrosine phosphorylation of IGF-1R and IRS-1, as well as activation of Akt and ERK, was reduced in Nedd4-deficient cells, whereas the total abundance of these proteins was not altered. Prolonged incubation with IGF-1 did not overcome this defect in Nedd4−/− MEFs, indicating that signaling was not delayed. Insulin-induced signaling was also decreased in the Nedd4+/− and Nedd4−/− cells compared to the wild-type cells, and similar to the IGF-1 results, ligand-stimulated receptor phosphorylation was reduced and the Akt and ERK signaling pathways were affected, although the effect on ERK phosphorylation was modest (Fig. 6B). The defect in IGF-1 signaling in Nedd4−/− cells was due specifically to the loss of Nedd4, because expression of Nedd4 in Nedd4−/− MEFs restored IGF-1 signaling, as seen by IGF-1R and Akt phosphorylation (Fig. 6C). These data indicate that Nedd4 regulates IGF-1R and IR signaling.
Fig. 6.
Reduced IGF-1–induced and insulin-induced signaling in Nedd4-deficient MEFs. (A) IGF-1 signaling: MEFs of the indicated genotypes were serum-starved for 2 hours and then left untreated or stimulated with either 10 or 100 ng IGF-1/ml for 5 min. Cell lysates were then immunoblotted as indicated. (B) Insulin signaling: As in (A), except that cells were stimulated with either 10 or 100 ng insulin/ml. (C) Restoration of IGF-1 signaling in Nedd4−/− MEFs by reintroduction of Nedd4. A Nedd4 expression vector was transiently transfected in MEFs as indicated, cells were treated with IGF-1, and lysates were blotted as in (A). The results from two separate transfection experiments are shown. “p,” phospho-.
Previous studies have implicated Nedd4 in proteasomal degradation of PTEN tumor suppressor protein (15); however, the abundance of PTEN in Nedd4+/+ and Nedd4−/−cells was similar (fig. S6A). We also failed to see any changes in PTEN localization in Nedd4+/+ and Nedd4−/− MEFs.
Cell surface expression of the IGF-1R and IR is reduced in Nedd4−/− MEFs
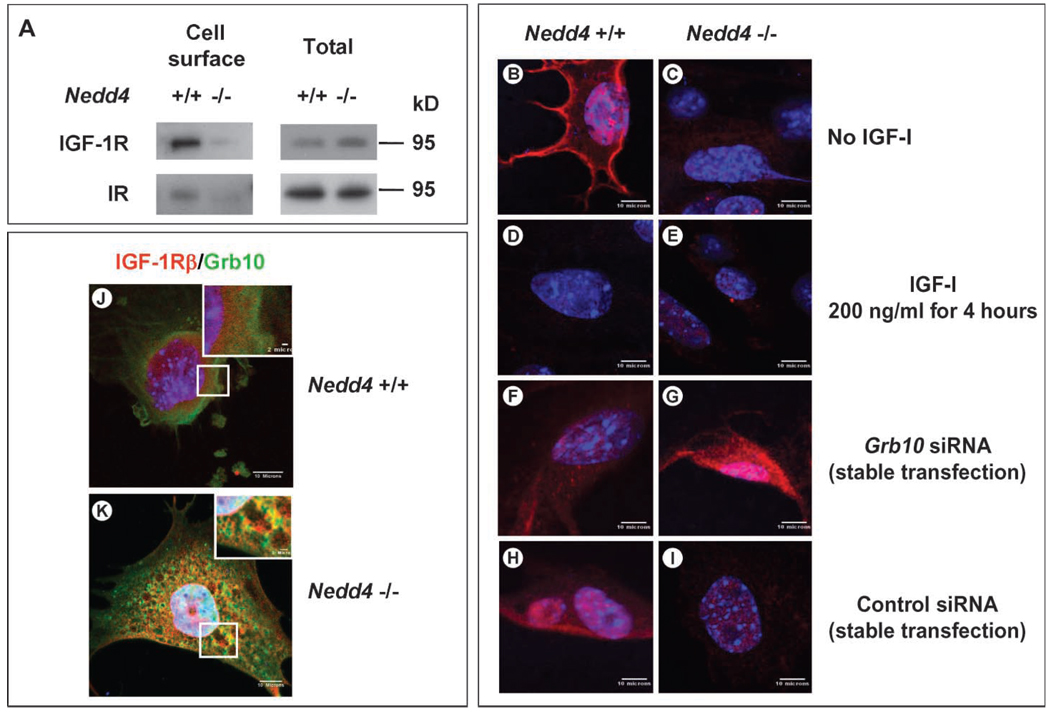
The reduction in IGF-1– and insulin-induced signaling as a result of Nedd4 ablation could be explained if the abundance of the corresponding receptors at the cell surface were lower in the Nedd4−/− MEFs. To test this hypothesis, we performed biotin labeling of cell surface proteins followed by isolation with streptavidin-agarose beads (Fig. 7A) and determined the subcellular localization of the IGF-1R by confocal microscopy (Fig. 7, B and C). Using the former approach, we found that the abundance of both the IGF-1R and IR at the cell surface was substantially lower in the Nedd4−/− cells compared to the wild-type cells. Consistent with these data, the IGF-1Rα was readily detected on the cell surface of Nedd4+/+ MEFs before ligand stimulation, and treatment with IGF-1 for 4 hours stimulated receptor internalization, such that no surface staining remained (Fig. 7, B and D). In contrast, in the Nedd4−/− MEFs, immunofluorescent labeling did not reveal appreciable amounts of IGF-1R on the cell surface (Fig. 7,C and E). Expression ofNedd4 in Nedd4−/−MEFs restored IGF-1R to the cell surface as determined by flow cytometry (fig. S7).
Fig. 7.
Cell surface IGF-1R concentrations change when Nedd4 and Grb10 protein expression are altered. (A) Decreased cell surface IGF-1R and IR in Nedd4-deficient MEFs. Cell surface proteins were isolated by biotin labeling and affinity purification and detected by immunoblotting. (B to I) IGF-1R concentrations at the plasma membranes of Nedd4+/+ and Nedd4−/− MEFs were determined in the absence or presence of an siRNA against Grb10 (rabbit anti–IGF-1Rα antibody was used at 1:100, Santa Cruz Biotechnology). (B and C) No IGF-1 stimulation. (D and E) Cells stimulated with recombinant IGF-1 (at 200 ng/ml, Sigma) for 4 hours. (F and G) Cells stably transfected with siRNA targeting Grb10. (H and I) Cells stably transfected with control siRNA. IGF-1R was not detectable on Nedd4−/− cells, but Grb10 knockdown restored IGF-1R to the plasma membrane. (J and K) Grb10 and IGF-1R are partially colocalized in Nedd4−/− MEF cells. MEFs grown in DMEM + 10% fetal bovine serum were fixed with 2% paraformaldehyde for 30 min at room temperature. Grb10 (green) and IGF-1R (red) were detected with monoclonal antibody against Grb10 and rabbit polyclonal antibody against IGF-1Rβ (Santa Cruz Biotechnology). Scale bar, 10 µm in the main figures and 2 µm in the insets.
Grb10 is involved in IGF-1R regulation by Nedd4
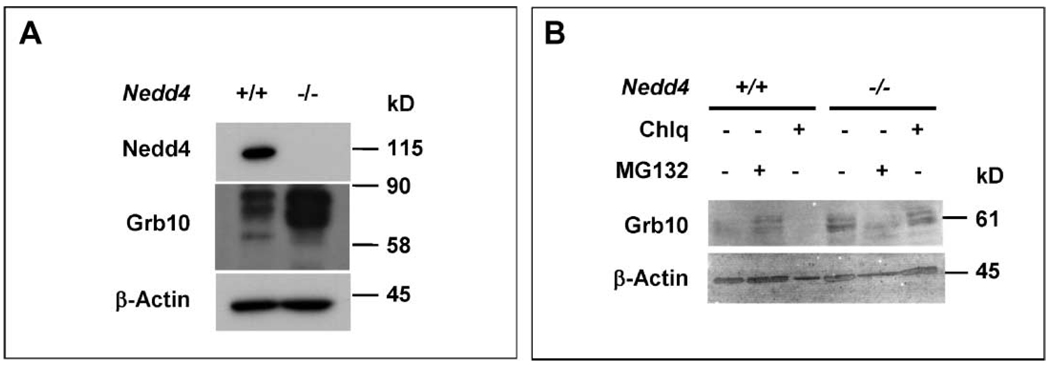
Nedd4 interacts with the adaptor Grb10 (7), and this has been proposed to result in IGF-1R ubiquitination and degradation (21). Previous studies have also shown that Grb10 is an inhibitor of both IGF-1R and IR signaling (17). We found that a portion of intracellular Grb10 and IGF-1Rβ proteins colocalized in Nedd4−/− MEFs (Fig. 7K). Given these prior reports and results with Nedd4−/− MEFs, we analyzed the possible role of Grb10 in the functional interaction between IGF-1R and Nedd4. The abundance of Grb10 protein was substantially increased in Nedd4−/− MEFs (Fig. 8A), although the abundance of Grb10 mRNA was similar in Nedd4+/+ and Nedd4−/− MEFs (fig. S8).
Fig. 8.
The amount of cellular Grb10 changes in response to amount of Nedd4. (A) The abundance of Grb10 in Nedd4−/− MEFs is much higher than in Nedd4+/+ MEFs. (B) Pretreatment of Nedd4+/+ MEFs with the proteasome inhibitor MG132 increases the abundance of Grb10. Cells were treated with chloroquine (Chlq) (40 µM) or MG132 (20 µM) for 4 hours before harvesting of cells and preparation of cell lysates. The β-actin immunoblot signals serve as protein loading controls.
The role of Grb10 was further investigated by small interfering RNA (siRNA)–mediated knockdown of Grb10 in MEFs. In the case of wild-type MEFs, the abundance of IGF-1R at the cell surface was slightly decreased by reduction of Grb10 through knockdown (compare panels F and B, Fig. 7), but there was no difference between cells transfected with siRNA and a control siRNA (compare panels F and H, Fig. 7). In contrast, IGF-1R was detected at the cell surface when Grb10 was knocked down in the Nedd4−/− cells (compare panels G and C, Fig. 7), whereas the presence of the control siRNA had no effect (compare panels I and C, Fig. 7).
To test if the increased abundance of Grb10 in Nedd4−/− MEFs was a result of increased protein stability, we treated Nedd4+/+ and Nedd4−/− MEFs withMG132 (proteasomal inhibitor) or chloroquine (lysosomal inhibitor) and analyzed Grb10 protein by immunoblotting. When normalized against the control (β-actin) signals, the results showed that treatment with MG132, but not with chloroquine, resulted in increased accumulation of Grb10 in Nedd4+/+ MEFs, whereas in Nedd4−/− MEFs this was not evident (Fig. 8B). These data suggest that proteasomal degradation controls Grb10 abundance and that Nedd4 plays a role in this process.
Inactivating Grb10 partially rescues lethality of Nedd4−/− mice
Next we took a genetic approach to confirm the functional interaction among Nedd4, Grb10, and IGF-1R observed in vitro. Mice carrying a disrupted Grb10 allele were generated from ES cells that harbor a gene-trapping vector in the Grb10 locus (cell line code XC302, obtained from BayGenomics) (fig. S9). Another group has independently generated and characterized mice from the same ES cells, showing that the Grb10-deficient mice have increased insulin signaling, enhanced insulin sensitivity, and increased body weight (22). An additional Grb10 KO mouse has also been described with a similar overgrowth and metabolic phenotype (23, 24). Our Grb10 homozygous null mice were not viable after birth, and Grb10 heterozygotes were slightly larger (~5%) than wild-type littermates at birth.
Mice doubly heterozygous for Nedd4 and Grb10 alleles were then generated and intercrossed so that any genetic interactions could be investigated. The genotypes of the offspring are listed in Table 1. Genotyping nearly 100 offspring revealed that the distribution of mice of various genotypes was significantly different from the expected Mendelian distribution (χ2 = 5.7 × 10−10). We did not obtain any mice that were doubly homozygous for the disruptions of Nedd4 and Grb10. However, we obtained six viable Nedd4−/− mice on the Grb10+/− background. The Grb10 gene is imprinted and expressed predominantly from the maternally transmitted allele, although the paternally transmitted Grb10 allele is expressed at low concentrations in cartilage, bones, heart, lungs, and gut during embryonic development (at both 12.5 and 14.5 dpc) (23). To test which allele contributed to the rescue of the perinatal lethality, mating was carried out between male Nedd4+/−;Grb10+/− mice and female Nedd4+/−;Grb10+/+ mice (mating no. 1), and between female Nedd4+/−;Grb10+/− mice and male Nedd4+/−;Grb10+/+ mice (mating no. 2). In this type of mating, we expected to observe 12.5%of the offspring carrying Nedd4−/−;Grb10+/− genotypes. The male Nedd4+/−;Grb10+/− mice (in mating no. 1) did not produce any Nedd4−/−;Grb10+/− mice in five litters (of a total of 39 mice), whereas the female Nedd4+/−;Grb10+/− mice (in mating no. 2) produced one Nedd4−/−;Grb10+/− animal out of 23 mice in three litters. Because males used in mating no. 2 carried only the paternal wild-type Grb10 allele (+p), the surviving Nedd4−/− mice must be Grb10−m/+p, and thus would have a lower amount of Grb10 than those from a maternally derived Grb10 allele. These data indicate that the imprinted paternally inherited allele rescues the perinatal lethal phenotype of the Nedd4−/− mice. Consistent with this prediction, although staining was much less intense than that seen in age-matched wild-type muscle, Grb10 was detected by immunohistochemistry in Nedd4−/−;Grb10−m/+p adult skeletal muscle from animals at 5 months of age (fig. S10, A and B).At the time of weaning, there was no longer a body weight difference between the Nedd4−/−;Grb10−m/+p mice and the rest of the littermates.
Table 1.
Genotypes of offspring from intercrosses of Nedd4 and Grb10 double heterozygotes.
| Nedd4 locus | +/+ | +/+ | +/+ | +/− | +/− | +/− | −/− | −/− | −/− |
|---|---|---|---|---|---|---|---|---|---|
| Grb10 locus | +/+ | +/− | −/− | +/+ | +/− | −/− | +/+ | +/− | −/− |
| Predicted (%) | 6.3 | 12.5 | 6.3 | 12.5 | 25.0 | 12.5 | 6.3 | 12.5 | 6.3 |
| No. of mice | 13 | 18 | 0 | 22 | 37 | 0 | 0 | 6 | 0 |
| Observed (%) | 13.5 | 18.8 | 0.0 | 22.9 | 38.5 | 0.0 | 0.0 | 6.0 | 0.0 |
DISCUSSION
A major conclusion of our data is that IGF-1 and insulin signaling are positively regulated by Nedd4. We also present both genetic and biochemical data indicating that Nedd4 and Grb10 have opposing roles in regulating IGF-1R function and that loss of Nedd4 leads to increased abundance of Grb10 in MEFs. Grb10, an adaptor protein, binds various receptor tyrosine kinases, including IGF-1R and IR, and may link the receptors with downstream signaling proteins or may play a role in receptor internalization and inactivation. A protein complex consisting of Grb10, Nedd4, and IGF-1R had been proposed to regulate IGF-1R internalization, with the indirect interaction between Nedd4 and IGF-1R through Grb10 promoting IGF-1R ubiquitination and degradation (21). Consistent with this model, inactivation of Grb10, either through paternal uniparental disomy (25) or maternal transmission of the Grb10 deletion (22, 23), resulted in the enhancement of both placental and embryonic growth. However, this model of interaction among Nedd4, Grb10, and IGF-1R cannot explain the growth deficiency seen in the Nedd4−/− mice.
Our data are consistent with a model in which Nedd4 binds and regulates the abundance of Grb10, thus affecting its ability to inhibit IGF-1R signaling. Grb10 may regulate both the proportion of receptor at the membrane and receptor interactions with substrates (26, 27); thus, the abundance and localization of Grb10 may be crucial to signaling by IGF-1R and IR. Several lines of evidence support this model. Grb10 is recruited to the activated IGF-1R (17); it negatively regulates IGF-1R signaling (24, 28), promotes internalization and degradation of the IGF-1R when overexpressed (17), and directly binds Nedd4 (7). RNA interference–mediated knockdown of Grb10 increased IGF-1–stimulated phosphorylation of IRS and Akt (29). Consistent with these published observations, Nedd4−/− MEFs exhibit decreased IGF-1R signaling and increased abundance of Grb10.
Although our data show that Nedd4 controls the abundance of Grb10, there is currently no evidence that Grb10 is ubiquitinated by Nedd4 (17). Our results suggest that the abundance of Grb10 may be controlled by proteasomal degradation, as MG132 treatment stabilizes Grb10 in the wild-type MEFs without further increasing the amount of Grb10 in the Nedd4−/− MEFs. Although the mechanism of this regulation remains to be determined, the data suggest that Nedd4 directly or through other proteins controls the ubiquitination, or stability, or both, of Grb10.
Because IGF-1R is known to be ubiquitinated after activation and this modification can be inhibited by expression of a dominant-negative Grb10 mutant (17), it is possible that another ubiquitin ligase is recruited by Grb10 and participates in the ubiquitination and down-regulation of IGF-1R. This model predicts that the Nedd4-null phenotype would be alleviated or rescued by a reduction in Grb10, because the unknown ubiquitin ligase could not negatively regulate IGF-1R in the absence of Grb10. Indeed, we have shown that, first, reduction of Grb10 through siRNA knockdown resulted in increased cell surface IGF-1R in the Nedd4−/− MEFs; second, in the presence of a maternally transmitted Grb10 null allele and paternally transmitted wild-type allele, Nedd4−/− mice are able not only to maintain normal growth during embryonic development, but also to survive the neonatal period.
The identity of the proposed ubiquitin ligase responsible for ubiquitinating IGF-1R as part of this regulatory complex remains unknown. Two ubiquitin ligases, Mdm2 and c-Cbl, have been shown to be involved in the polyubiquitination of IGF-1R upon ligand stimulation (29, 30). There is no reported evidence of an interaction among the Nedd4, Grb10, and Mdm2 proteins, and the study of potential interactions at the genetic level is complicated by the fact that Mdm2-null mice die early in gestation (31). Mice deficient in c-Cbl exhibit hyperplasia in specific tissues, indicating that c-Cbl negatively regulates signaling events that control cell proliferation (32). The second member of the Cbl family, Cbl-b, controls immune cell function (33, 34). Cbl-b has been shown to be a target of Nedd4-mediated ubiquitination and degradation (35). In Nedd4−/− MEFs, however, we do not see any appreciable change in the concentrations of either c-Cbl or Cbl-b (fig. S6B).
In the absence of Nedd4, Grb10 concentration is elevated and Grb10 and IGF-1R partially colocalize internally. By modulating Grb10 abundance, Nedd4 tunes IGF-1R/IR signaling by regulating the abundance of the receptors at the cell surface and their signal output. In vitro data suggest that Nedd4 regulates many cellular processes (12, 13). The data presented here provide evidence that in vivo one of its main functions is to regulate fetal growth, most likely by regulating IGF-1 signaling through Grb10.
MATERIALS AND METHODS
Generation of Nedd4- and Grb10-null mice
Targeted ES cell lines with the mouse Nedd4 and Grb10 gene disrupted (XA209 and XB398, and XC302, respectively) were obtained from BayGenomics, and the locations of the gene trapping vector inserted in an intron were determined according to the sequences obtained from rapid amplification of 5′ complementary DNA ends (19). ES cells were injected into mouse blastocysts for the generation of chimeras as described previously (36, 37). Several chimeras were generated, and germline transmission occurred in all three cell lines. The following primers were used to genotype mice with the Nedd4 gene disruption. XA209f (5′-AGG TCA TCC ACT GGT TCT GG-3′) and XA209r (5′-TCT GAG AGC TCT GCA CAG GA-3′) amplify a 930-bp fragment from the wild-type allele, whereas XA209f andXA209r2 (5′-TGT CCT CCA GTC TCC TCC AC-3′) amplify a 400-bp fragment from the gene-trapped allele.
An additional Nedd4 KO line (Nedd4Gt(IRESBetageo)249Lex) was obtained from the MMRRC. This KO line is derived from a gene trap line with a retroviral insertion disrupting the Nedd4 gene between exons 17 and 18 (www.mmrrc.org/strains/11742/011742.html). For this line, the following primers were used to genotype mice with the Nedd4 gene disruption. Primers genoN4 WT5′ (5′-GGA GTC TTT GGA TAT TGT AAG AGC-3′) and genoN4 WTandKO 3′ (5′-GAG CGT GCG CCT CAC AAG TAT GA-3′) amplify a 226-bp fragment from the wild-type allele, whereas genoN4 KO5′ (5′-AAA TGG CGT TAC TTA AGC TAG CTT GC-3′) and genoN4 WTandKO 3′ amplify a 137-bp fragment from the gene-trapped allele.
The following primers were used to genotype mice with Grb10 gene disruption. XC302f4 (5′-CTT GCT GTC TTT GCC TTT CC-3′) and XCWTr (5′-TCA CTT GGC ATG GTA CTC TCC-3′) amplify a 320-bp fragment from the wild-type allele, whereas XC302f4 and XC302r (5′-CAA CAC TTG TAT GGC CTT GG-3′) amplify a 1200-bp fragment from the gene-trapped allele.
All animal procedures were approved by the University of Iowa Animal Care and Use Committee or the Institute of Medical and Veterinary Science Animal Ethics Committee.
RT-PCR
Total RNA was isolated from whole late-gestation embryos (18.5 dpc) and from MEFs generated from these animals with the use of Tri-Reagent (Sigma Chemical Company, St. Louis, MO) according to the manufacturer’s instructions. Two sets of primers were designed to identify the endogenous and gene-trapped Nedd4 transcripts. XAf2 (5′-TGA TAC CAC AGG ATC TCA TCA AG-3′) and WTR2 (5′-ATG ACC TGG TGG TTC ATG C-3′) amplify a 180-bp fragment that spans exons 25 to 27 of the endogenous allele; XAf2 and KOR2 (5′-GCG GAT TGA CCG TAA TGG-3′) amplify a 600-bp fragment from the gene trapped allele. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as internal control. The forward (5′-CGT CTT CAC CAC CAT GGA GA-3′) and reverse (5′-CGG CCA TCA CGC CAC AGT TT-3′) primers amplify a 300-bp fragment, as previously described (36).
Immunoblot analysis
MEF lysates were used for immunoblotting as previously described (38). Antibodies were purchased from the following suppliers: Cell Signalling Technology (phospho- and total ERK and Akt); Millipore (IRS-1); BD Biosciences (IR β-subunit, c-Cbl and Nedd4); Upstate (Nedd4); Santa Cruz Biotechnology Inc. (Grb10, IGF-1R β-subunit, Cbl-b); and Biosource (phosphorylated IGF-1R/IR and IRS-1). Where indicated, MEFs were transfected with a Nedd4 expression construct (11) with Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol. In one experiment (Fig. 6C, lower panel), the Nedd4 expression vector was cotransfected with a green fluorescent protein (GFP) vector (10:1) and cells were sorted for GFP expression. The day after transfection, MEFs were serum starved for 2 hours, stimulated with 100 ng IGF-1/ml for 5 min where indicated, and lysates were prepared for blotting.
Biotinylation of cell surface proteins
Cell monolayers were washed with phosphate-buffered saline (PBS) and then incubated with sulfo-NHS-LC-Biotin (1 mg/ml in PBS) for 20 min at 4°C. The cells were then washed twice with ice-cold PBS containing 100 mM glycine, once with PBS, and then lysed. The cleared lysate was then incubated with steptavidin-agarose beads for 1 h. The beads were washed in lysis buffer, then resuspended in SDS-PAGE sample buffer.
Cell-cycle study
Cell-cycle information was obtained with passage number–matched asynchronous MEFs (P2 to P4) under normal culture conditions [Dulbecco’s modified Eagle’smedium(DMEM) + 10% fetal bovine serum]. Cells were stained with propidium iodide and treated with RNase A to remove cytoplasmic RNA as previously described (39). Sorting of cells with different DNA content was performed with an LSR flow cytometer and analyzed with CELLQUEST software (BD Biosciences, San Jose, CA).
Cell proliferation studies
MEFs were isolated from mid-gestation embryos (13.5 dpc) and passage number–matched MEFs were used for each experiment. Experiments for the analyses of passage-dependent cell growth (cells were seeded at 200,000 cells per 6-cm dish), low-density seeding (cells were seeded at 100,000 cells per 10-cm dish), and cell growth in 1% versus 10% serum were performed.
Knockdown of Grb10 expression by RNA interference
Grb10 protein was reduced by transfecting cells with siRNA (purchased from Open Biosystems, Huntsville, AL) according to the manufacturer’s protocols. Nedd4+/+ and Nedd4−/− cells were plated in six-well plates and allowed to grow to 60 to 80% confluence. Nedd4+/+ and Nedd4−/− cells were transfected with siRNAs specific for Grb10 (Open Biosystems) with the use of the Arrest-In transfection reagent (Open Biosystems). An siRNA against an unrelated gene (eGFP) was used as a control. After 48 hours of transfection, the cells were transferred to a medium containing 3.5 µg/ml puromycin (InvivoGen, San Diego, CA) for selection. All cells were harvested after 14 days of selection and were then processed for immunostaining and Western blotting. All siRNA experiments were done in triplicate for each condition. Similar results were obtained in all experiments.
Immunofluorescence
Untransfected and stably transfected (14 days after transfection and selection, see above protocol) Nedd4+/+ and Nedd4−/−MEFs were grown on a permeable supported membrane with pore sizes of 0.4-µm Cell Culture Insert (BD Labware, Franklin Lakes, NJ). MEFs grown to 50 to 70% confluence were fixed with 4% paraformaldehyde for 10 min at room temperature, treated with 0.2% Triton X-100 for 10 min, blocked with 3% bovine serum albumin for 30 min, and incubated with primary antibodies overnight at 4°C. Antibodies against IGF-1Rα (rabbit), IGF-1Rβ (rabbit), and Grb10 (mouse) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA), and antibody against Nedd4 (mouse) was purchased from BD Biosciences (Franklin Lakes, NJ). The next day, the cells were washed with PBS and incubated with an Alexa 568–labeled goat anti-rabbit secondary antibody or streptavidin–Alexa 488 for 30 min. The nucleus was washed and stained with ToPro-3 (Molecular Probes, Eugene, OR). Stained sections were mounted in a mounting medium (Vectashield Mounting medium, Vector Laboratories, Burlingame, CA). Images were obtained with a Bio-Rad MRC-1024 Confocal Microscope (Bio-Rad Laboratories, Hercules, CA) with a 60× oil objective.
Statistics and data analysis
Data analysis between different groups of animals was performed by unpaired Student’s t test. We considered P values <0.05 statistically significant. All values are expressed as the mean ± SE.
Supplementary Material
Footnotes
SUPPLEMENTARY MATERIALS
www.sciencesignaling.org/cgi/content/full/1/38/ra5/DC1
Fig. S1. MMRRC Nedd4 KO animals are growth retarded.
Fig. S2. Some Nedd4−/− embryos have ventricular septal defect (VSD).
Fig. S3. Normal lung development in Nedd4−/− embryos.
Fig. S4. Mouse embryonic fibroblasts isolated from both Nedd4+/− and Nedd4−/− embryos show growth arrest at G0–G1 phase of the cell cycle.
Fig. S5. Total cellular levels of the IGF-1R precursor and the processed β subunit do not vary consistently with Nedd4 gene dosage.
Fig. S6. Cellular PTEN, c-Cbl, and Cbl-b levels are not significantly different in Nedd4−/− MEFs.
Fig. S7. Reexpression of Nedd4 in Nedd4−/− MEFs restores IGF-1R to the cell surface.
Fig. S8. Similar levels of Grb10 transcript in Nedd4+/+ and Nedd4−/− MEFs.
Fig. S9. Generation and genotyping of Grb10-deficient mice.
Fig. S10. Paternally inherited Grb10 allele is expressed at low levels in muscle.
REFERENCES AND NOTES
- 1.Baker J, Liu JP, Robertson EJ, Efstratiadis A. Role of insulin-like growth factors in embryonic and postnatal growth. Cell. 1993;75:73–82. [PubMed] [Google Scholar]
- 2.Liu JP, Baker J, Perkins AS, Robertson EJ, Efstratiadis A. Mice carrying null mutations of the genes encoding insulin-like growth factor I (Igf-1) and type 1 IGF receptor (Igf1r) Cell. 1993;75:59–72. [PubMed] [Google Scholar]
- 3.Lupu F, Terwilliger JD, Lee K, Segre GV, Efstratiadis A. Roles of growth hormone and insulin-like growth factor 1 in mouse postnatal growth. Dev. Biol. 2001;229:141–162. doi: 10.1006/dbio.2000.9975. [DOI] [PubMed] [Google Scholar]
- 4.Saltiel AR, Kahn CR. Insulin signalling and the regulation of glucose and lipid metabolism. Nature. 2001;414:799–806. doi: 10.1038/414799a. [DOI] [PubMed] [Google Scholar]
- 5.Taniguchi CM, Emanuelli B, Kahn CR. Critical nodes in signalling pathways: Insights into insulin action. Nat. Rev. Mol. Cell Biol. 2006;7:85–96. doi: 10.1038/nrm1837. [DOI] [PubMed] [Google Scholar]
- 6.Glass DJ. Skeletal muscle hypertrophy and atrophy signaling pathways. Int. J. Biochem. Cell Biol. 2005;37:1974–1984. doi: 10.1016/j.biocel.2005.04.018. [DOI] [PubMed] [Google Scholar]
- 7.Morrione A, Plant P, Valentinis B, Staub O, Kumar S, Rotin D, Baserga R. mGrb10 interacts with Nedd4. J. Biol. Chem. 1999;274:24094–24099. doi: 10.1074/jbc.274.34.24094. [DOI] [PubMed] [Google Scholar]
- 8.Holt LJ, Daly RJ. Adapter protein connections: The MRL and Grb7 protein families. Growth Factors. 2005;23:193–201. doi: 10.1080/08977190500196267. [DOI] [PubMed] [Google Scholar]
- 9.Kumar S, Tomooka Y, Noda M. Identification of a set of genes with developmentally down-regulated expression in the mouse brain. Biochem. Biophys. Res. Commun. 1992;185:1155–1161. doi: 10.1016/0006-291x(92)91747-e. [DOI] [PubMed] [Google Scholar]
- 10.Staub O, Dho S, Henry P, Correa J, Ishikawa T, McGlade J, Rotin D. WW domains of Nedd4 bind to the proline-rich PY motifs in the epithelial Na+ channel deleted in Liddle’s syndrome. EMBO J. 1996;15:2371–2380. [PMC free article] [PubMed] [Google Scholar]
- 11.Kumar S, Harvey KF, Kinoshita M, Copeland NG, Noda M, Jenkins NA. cDNA cloning, expression analysis, and mapping of the mouse Nedd4 gene. Genomics. 1997;40:435–443. doi: 10.1006/geno.1996.4582. [DOI] [PubMed] [Google Scholar]
- 12.Harvey KF, Kumar S. Nedd4-like proteins: An emerging family of ubiquitin-protein ligases implicated in diverse cellular functions. Trends Cell Biol. 1999;9:166–169. doi: 10.1016/s0962-8924(99)01541-x. [DOI] [PubMed] [Google Scholar]
- 13.Shearwin-Whyatt L, Dalton HE, Foot N, Kumar S. Regulation of functional diversity within the Nedd4 family by accessory and adaptor proteins. Bioessays. 2006;28:617–628. doi: 10.1002/bies.20422. [DOI] [PubMed] [Google Scholar]
- 14.Murdaca J, Treins C, Monthouel-Kartmann MN, Pontier-Bres R, Kumar S, Van Obberghen E, Giorgetti-Peraldi S. Grb10 prevents Nedd4-mediated vascular endothelial growth factor receptor-2 degradation. J. Biol. Chem. 2004;279:26754–26761. doi: 10.1074/jbc.M311802200. [DOI] [PubMed] [Google Scholar]
- 15.Wang X, Trotman LC, Koppie T, Alimonti A, Chen Z, Gao Z, Wang J, Erdjument-Bromage H, Tempst P, Cordon-Cardo C, Pandolfi PP, Jiang X. NEDD4-1 is a proto-oncogenic ubiquitin ligase for PTEN. Cell. 2007;128:129–139. doi: 10.1016/j.cell.2006.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harty RN, Brown ME, Wang G, Huibregtse J, Hayes FP. A PPxY motif within the VP40 protein of Ebola virus interacts physically and functionally with a ubiquitin ligase: Implications for filovirus budding. Proc. Natl. Acad. Sci. U.S.A. 2000;97:13871–13876. doi: 10.1073/pnas.250277297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vecchione A, Marchese A, Henry P, Rotin D, Morrione A. The Grb10/Nedd4 complex regulates ligand-induced ubiquitination and stability of the insulin-like growth factor I receptor. Mol. Cell Biol. 2003;23:3363–3372. doi: 10.1128/MCB.23.9.3363-3372.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Monami G, Emiliozzi V, Morrione A. Grb10/Nedd4-mediated multiubiquitination of the insulin-like growth factor receptor regulates receptor internalization. J. Cell Physiol. 2008;216:426–437. doi: 10.1002/jcp.21405. [DOI] [PubMed] [Google Scholar]
- 19.Stryke D, Kawamoto M, Huang CC, Johns SJ, King LA, Harper CA, Meng EC, Lee RE, Yee A, L’Italien L, Chuang PT, Young SG, Skarnes WC, Babbitt PC, Ferrin TE. BayGenomics: A resource of insertional mutations in mouse embryonic stem cells. Nucleic Acids Res. 2003;31:278–281. doi: 10.1093/nar/gkg064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baserga R, Porcu P, Rubini M, Sell C. Cell cycle control by the IGF-1 receptor and its ligands. Adv. Exp. Med. Biol. 1993;343:105–112. doi: 10.1007/978-1-4615-2988-0_11. [DOI] [PubMed] [Google Scholar]
- 21.Sehat B, Andersson S, Vasilcanu R, Girnita L, Larsson O. Role of ubiquitination in IGF-1 receptor signaling and degradation. PLoS ONE. 2007;2:e340. doi: 10.1371/journal.pone.0000340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang L, Balas B, Christ-Roberts CY, Kim RY, Ramos FJ, Kikani CK, Li C, Deng C, Reyna S, Musi N, Dong LQ, DeFronzo RA, Liu F. Peripheral disruption of the Grb10 gene enhances insulin signaling and sensitivity in vivo. Mol. Cell Biol. 2007;27:6497–6505. doi: 10.1128/MCB.00679-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Charalambous M, Smith FM, Bennett WR, Crew TE, Mackenzie F, Ward A. Disruption of the imprinted Grb10 gene leads to disproportionate overgrowth by an Igf2-independent mechanism. Proc. Natl. Acad. Sci. U.S.A. 2003;100:8292–8297. doi: 10.1073/pnas.1532175100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith FM, Holt LJ, Garfield AS, Charalambous M, Koumanov F, Perry M, Bazzani R, Sheardown SA, Hegarty BD, Lyons RJ, Cooney GJ, Daly RJ, Ward A. Mice with a disruption of the imprinted Grb10 gene exhibit altered body composition, glucose homeostasis, and insulin signaling during postnatal life. Mol. Cell Biol. 2007;27:5871–5886. doi: 10.1128/MCB.02087-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cattanach BM, Beechey CV, Rasberry C, Jones J, Papworth D. Time of initiation and site of action of the mouse chromosome 11 imprinting effects. Genet. Res. 1996;68:35–44. doi: 10.1017/s0016672300033863. [DOI] [PubMed] [Google Scholar]
- 26.Dey BR, Frick K, Lopaczynski W, Nissley SP, Furlanetto RW. Evidence for the direct interaction of the insulin-like growth factor I receptor with IRS-1, Shc, and Grb10. Mol Endocrinol. 1996;10:631–641. doi: 10.1210/mend.10.6.8776723. [DOI] [PubMed] [Google Scholar]
- 27.Wick KR, Werner ED, Langlais P, Ramos FJ, Dong LQ, Shoelson SE, Liu F. Grb10 inhibits insulin-stimulated insulin receptor substrate (IRS)-phosphatidylinositol 3-kinase/Akt signaling pathway by disrupting the association of IRS-1/IRS-2 with the insulin receptor. J. Biol. Chem. 2003;278:8460–8467. doi: 10.1074/jbc.M208518200. [DOI] [PubMed] [Google Scholar]
- 28.Dufresne AM, Smith RJ. The adaptor protein GRB10 is an endogenous negative regulator of insulin-like growth factor signaling. Endocrinology. 2005;146:4399–4409. doi: 10.1210/en.2005-0150. [DOI] [PubMed] [Google Scholar]
- 29.Girnita L, Girnita A, Larsson O. Mdm2-dependent ubiquitination and degradation of the insulin-like growth factor 1 receptor. Proc. Natl. Acad. Sci. U.S.A. 2003;100:8247–8252. doi: 10.1073/pnas.1431613100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sehat B, Andersson S, Girnita L, Larsson O. Identification of c-Cbl as a new ligase for insulin-like growth factor-I receptor with distinct roles from Mdm2 in receptor ubiquitination and endocytosis. Cancer Res. 2008;68:5669–5677. doi: 10.1158/0008-5472.CAN-07-6364. [DOI] [PubMed] [Google Scholar]
- 31.Jones SN, Roe AE, Donehower LA, Bradley A. Rescue of embryonic lethality in Mdm2-deficient mice by absence of p53. Nature. 1995;378:206–208. doi: 10.1038/378206a0. [DOI] [PubMed] [Google Scholar]
- 32.Murphy MA, Schnall RG, Venter DJ, Barnett L, Bertoncello I, Thien CB, Langdon WY, Bowtell DD. Tissue hyperplasia and enhanced T-cell signalling via ZAP-70 in c-Cbl-deficient mice. Mol. Cell Biol. 1998;18:4872–4882. doi: 10.1128/mcb.18.8.4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bachmaier K, Krawczyk C, Kozieradzki I, Kong YY, Sasaki T, Oliveira-dos-Santos A, Mariathasan S, Bouchard D, Wakeham A, Itie A, Le J, Ohashi PS, Sarosi I, Nishina H, Lipkowitz S, Penninger JM. Negative regulation of lymphocyte activation and auto-immunity by the molecular adaptor Cbl-b. Nature. 2000;403:211–216. doi: 10.1038/35003228. [DOI] [PubMed] [Google Scholar]
- 34.Chiang YJ, Kole HK, Brown K, Naramura M, Fukuhara S, Hu RJ, Jang IK, Gutkind JS, Shevach E, Gu H. Cbl-b regulates the CD28 dependence of T-cell activation. Nature. 2000;403:216–220. doi: 10.1038/35003235. [DOI] [PubMed] [Google Scholar]
- 35.Magnifico A, Ettenberg S, Yang C, Mariano J, Tiwari S, Fang S, Lipkowitz S, Weissman AM. WW domain HECT E3s target Cbl RING finger E3s for proteasomal degradation. J. Biol. Chem. 2003;278:43169–43177. doi: 10.1074/jbc.M308009200. [DOI] [PubMed] [Google Scholar]
- 36.Oliver PM, Cao X, Worthen GS, Shi P, Briones N, MacLeod M, White J, Kirby P, Kappler J, Marrack P, Yang B. Ndfip1 protein promotes the function of itch ubiquitin ligase to prevent T cell activation and T helper 2 cell-mediated inflammation. Immunity. 2006;25:929–940. doi: 10.1016/j.immuni.2006.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McDonald FJ, Yang B, Hrstka RF, Drummond HA, Tarr DE, McCray PB, Stokes JB, Welsh MJ, Williamson RA. Disruption of the beta subunit of the epithelial Na+ channel in mice: Hyperkalemia and neonatal death associated with a pseudohy-poaldosteronism phenotype. Proc. Natl. Acad. Sci. U.S.A. 1999;96:1727–1731. doi: 10.1073/pnas.96.4.1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cao XR, Shi PP, Sigmund RD, Husted RF, Sigmund CD, Williamson RA, Stokes JB, Yang B. Mice heterozygous for beta-ENaC deletion have defective potassium excretion. Am. J. Physiol. Renal. Physiol. 2006;291:F107–F115. doi: 10.1152/ajprenal.00159.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krishan A. Rapid flow cytofluorometric analysis of mammalian cell cycle by propidium iodide staining. J. Cell Biol. 1975;66:188–193. doi: 10.1083/jcb.66.1.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.We thank J. Snyder, E. Twait, K. Walters, T. Kinney, K. Volk ,and J. Koland for technical support, and J. C. Murray and J. B. Stokes for helpful discussions. This work was supported in part by U.S. NIH grants DK52617 (B.Y.), AR052647 (B.Y.), and DE16215 (B.Y.), American Cancer Society RSG-03-046-01 (N.L.L.), National Health and Medical Research Council (NHMRC Australia) fellowships and grants (S.K. and R.J.D.), and a Cancer Institute of New South Wales fellowship (D.C.). The authors declare that they have no competing financial interests.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.