Abstract
Anthranilate synthase in Pseudomonas putida is a two component enzyme system. The proteins, termed AS I and AS II, have respective molecular weights of 65,000 and 18,000. Five additional Pseudomonas species, both tryptophan requiring and independent strains, were examined and all were shown to contain similar two component systems. Anthranilate formation by „amide transfer,” with L-glutamine as nitrogen donor, requires both proteins; „amination,” utilizing ammonium ion, proceeds at pH 9 with only the larger component, AS I. The product of the P. putida trpA gene, AS I, carries the chorismate binding and tryptophan feedback inhibition sites whereas the smaller component, AS II, functions in glutamine binding. We have not been able to prepare mutants lacking AS II activity nor have other catalytic activities been detected for this protein.
The second step unique to tryptophan biosynthesis is catalyzed by phosphoribosyl transferase, in P. putida the trpB gene product. Phosphoribosyl transferase, EC 2.4.2.14, is separable from both AS I and II, and is not required for anthranilate synthesis. This chromosomal and protein organization differs from the array found in the enteric bacteria where phosphoribosyl transferase carries also the AS CoII enzymic activity.
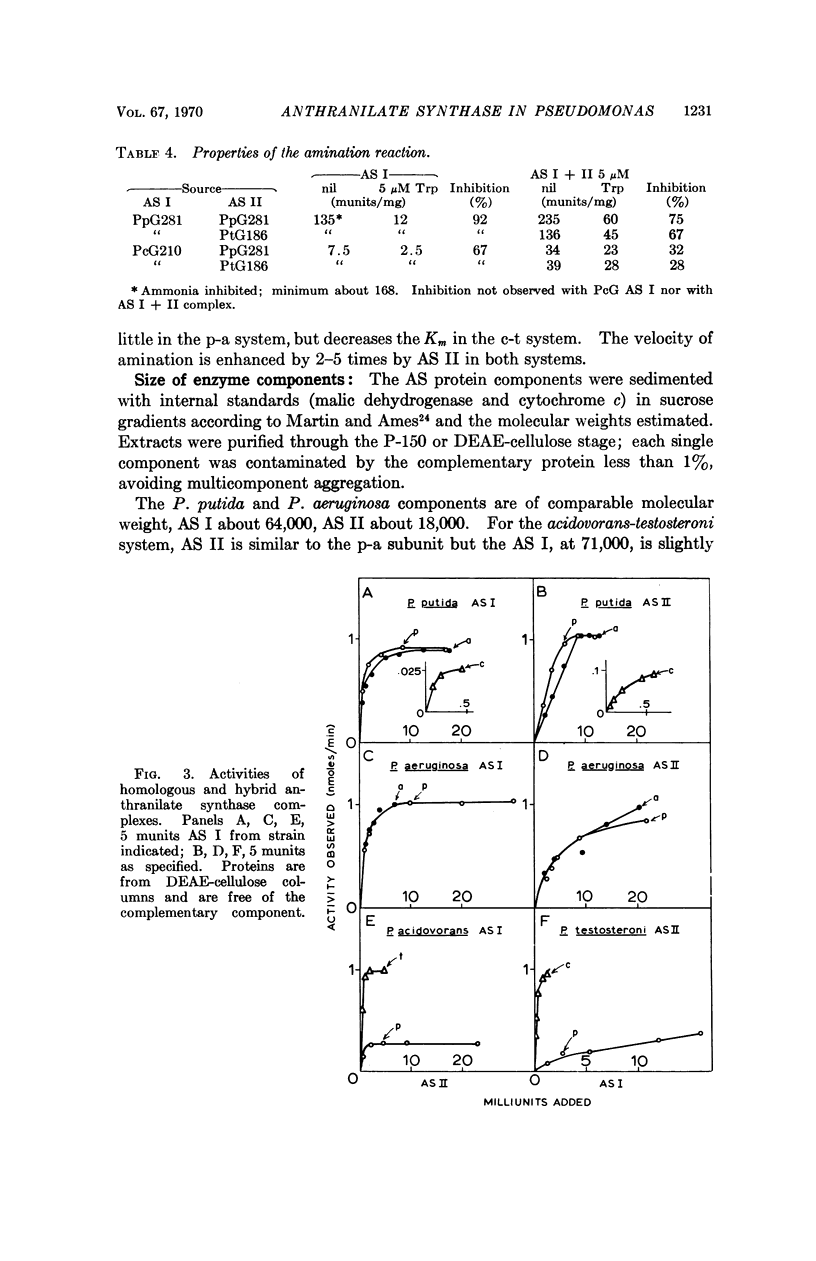
Among the six Pseudomonas species examined, two groups may be distinguished on the basis of subunit complementation, the putida-aeruginosa (p-a), and the acidovorans-testosteroni (c-t). Within these groups the hybrid enzymes are equivalent in activity to the native enzyme and the level of subunits required is comparable. Between the groups the hybrid enzymes are lower in activity than either native form. The c-t components separate with difficulty and the aggregate appears to be larger than the more freely dissociable p-a complex. P. stutzeri resembles the p-a class and P. multivorans the c-t class.
Full text
PDF






Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baker T. I., Crawford I. P. Anthranilate synthetase. Partial purification and some kinetic studies on the enzyme from Escherichia coli. J Biol Chem. 1966 Dec 10;241(23):5577–5584. [PubMed] [Google Scholar]
- Bauerle R. H., Margolin P. A multifunctional enzyme complex in the tryptophan pathway of Salmonella typhimurium: comparison of polarity and pseudopolarity mutations. Cold Spring Harb Symp Quant Biol. 1966;31:203–214. doi: 10.1101/sqb.1966.031.01.028. [DOI] [PubMed] [Google Scholar]
- Chakrabarty A. M., Gunsalus C. F., Gunsalus I. C. Transduction and the clustering of genes in fluorescent Pseudomonads. Proc Natl Acad Sci U S A. 1968 May;60(1):168–175. doi: 10.1073/pnas.60.1.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford I. P., Gunsalus I. C. Inducibility of tryptophan synthetase in Pseudomonas putida. Proc Natl Acad Sci U S A. 1966 Aug;56(2):717–724. doi: 10.1073/pnas.56.2.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson M. I., Gibson F. Preliminary studies on the isolation and metabolism of an intermediate in aromatic biosynthesis: chorismic acid. Biochem J. 1964 Feb;90(2):248–256. doi: 10.1042/bj0900248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunsalus C., Gunsalus C. F., Chakrabarty A. M., Sikes S., Crawford I. P. Fine structure mapping of the tryptophan genes in Pseudomonas putida. Genetics. 1968 Nov;60(3):419–435. doi: 10.1093/genetics/60.3.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HOLLOWAY B. W. Genetic recombination in Pseudomonas aeruginosa. J Gen Microbiol. 1955 Dec;13(3):572–581. doi: 10.1099/00221287-13-3-572. [DOI] [PubMed] [Google Scholar]
- Ito J., Crawford I. P. Regulation of the enzymes of the tryptophan pathway in Escherichia coli. Genetics. 1965 Dec;52(6):1303–1316. doi: 10.1093/genetics/52.6.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MARTIN R. G., AMES B. N. A method for determining the sedimentation behavior of enzymes: application to protein mixtures. J Biol Chem. 1961 May;236:1372–1379. [PubMed] [Google Scholar]
- Ornston L. N., Ornston M. K., Chou G. Isolation of spontaneous mutant strains of Pseudomonas putida. Biochem Biophys Res Commun. 1969 Jul 7;36(1):179–184. doi: 10.1016/0006-291x(69)90666-4. [DOI] [PubMed] [Google Scholar]
- SMITH R. A., GUNSALUS I. C. Distribution and formation of isocitritase. Nature. 1955 Apr 30;175(4461):774–775. doi: 10.1038/175774b0. [DOI] [PubMed] [Google Scholar]
- Stanier R. Y., Palleroni N. J., Doudoroff M. The aerobic pseudomonads: a taxonomic study. J Gen Microbiol. 1966 May;43(2):159–271. doi: 10.1099/00221287-43-2-159. [DOI] [PubMed] [Google Scholar]
- VOGEL H. J., BONNER D. M. Acetylornithinase of Escherichia coli: partial purification and some properties. J Biol Chem. 1956 Jan;218(1):97–106. [PubMed] [Google Scholar]


