Abstract
We have previously demonstrated that the bHLH/PAS transcription factor, singleminded 2s (Sim2s), is required for proper mammary ductal morphogenesis and luminal epithelial differentiation. Furthermore, loss of Sim2s in breast cancer cells resulted in downregulation of epithelial markers and acquisition of a basal-like phenotype. The objective of this study was to further define the role of Sim2s in mammary differentiation. We found that Sim2s is developmentally regulated throughout mammary gland development with highest expression during lactation. Mammary glands from nulliparous mice expressing Sim2s driven by the mouse mammary tumor virus (MMTV) long terminal repeat (LTR) promoter were morphologically indistinguishable from wild-type mice but displayed hallmarks of precocious lactogenic differentiation. These included elevated expression of the milk protein genes Wap and Csn2, and apical localization of the lactation marker Npt2b. Consistent with the in vivo results, Sim2s enhanced prolactin-mediated Csn2 expression in HC11 and CIT3 mouse mammary epithelial cells, and downregulation of Sim2s by shRNA in HC11 cells inhibited Csn2 expression. Chromatin immunoprecipitation (ChIP) analyses of the Csn2 gene found that Sim2s associates with the Csn2 promoter and re-ChIP experiments showed that Sim2s interacted with the RNA II polymerase (RNAPII) complex. Together, these data demonstrate, for the first time, that Sim2s is required for establishing and maintaining mammary gland differentiation.
Keywords: singleminded 2s, Mammary, Lactation, Beta-casein, Mouse
INTRODUCTION
Defining mechanisms that contribute to establishing and maintaining cellular differentiation is crucial for advancing our understanding of cellular development and a novel approach for targeting cancer therapies. A key feature of differentiation involves activation of transcription networks controlling global gene expression to ensure cell commitment to a specific phenotype. In the mammary gland, differentiation of the mammary epithelium takes place throughout the reproductive lifetime of the female. During puberty, terminal end bud (TEB) structures form and are responsible for ductal elongation. As the ducts mature, the cells differentiate into luminal or myoepithelial cells. During pregnancy, alveolar structures form and the luminal epithelium begins to acquire the ability to produce milk. When the offspring are weaned, the majority of alveolar epithelial cells die and the gland regresses, but it maintains the ability to expand in response to subsequent pregnancies. The mammary alveolar epithelial cells respond to the hormone prolactin by initiating a cascade of events leading to activation and nuclear translocation of Stat5a, which binds to regulatory elements of milk protein genes, increasing their expression (Happ and Groner, 1993; Li and Rosen, 1995). Although prolactin-mediated Stat5a activation is central to alveolar epithelial cell function, it is not the only pathway responsible for initiating mammary gland specification towards terminal differentiation. Other transcription factors, including glucocorticoid receptor (GR; Nr3c1 — Mouse Genome Informatics), CCAAT-enhancer binding protein-beta (Cebpb), Elf5 and Gata3 have also been shown to play a role in lactogenic differentiation (Kabotyanski et al., 2006; Kouros-Mehr et al., 2006; Oakes et al., 2008; Seagroves et al., 2000). Thus, commitment to cellular differentiation in the mammary gland appears to require activation and coordination of multiple signaling pathways to insure specific mammary lineage commitment is achieved.
Singleminded 2s (Sim2s) is a member of the bHLH/PAS family of transcription factors and one of two orthologs of Drosophila sim. In fruit flies, sim is considered a ‘master regulator’ of differentiation that is both necessary and sufficient for proper midline cell differentiation during central nervous system development (Chang et al., 2001; Menne et al., 1997; Nambu et al., 1990). In mice, loss of both Sim2s alleles leads to severe developmental abnormalities in the diaphragm, ribs, cartilage and palate, suggesting that Sim2s plays a role in the differentiation of many tissue types (Goshu et al., 2002; Shamblott et al., 2002). We have recently shown that Sim2s is expressed in human breast epithelial cells and is downregulated in primary human breast cancer samples (Kwak et al., 2007). Furthermore, reestablishment of Sim2s in highly invasive cancer cells significantly inhibits growth and motility, suggesting that Sim2s is a breast tumor suppressor (Kwak et al., 2007). In addition, we found that SIM2s expression is suppressed by NOTCH and CEBPβ and that loss of SIM2s in normal human breast and mouse mammary epithelium induces an invasive basal/stem cell-like phenotype (Gustafson et al., 2009; Laffin et al., 2008). Sim2s-null mouse mammary glands do not undergo proper ductal elongation or luminal differentiation (Laffin et al., 2008). These data suggest that Sim2s is required for establishing and/or maintaining differentiation during normal mammary development. As loss of Sim2s results in lethality shortly after birth, transplant studies were used to evaluate the requirement of Sim2s for nulliparous mammary development. This technique did not allow us to evaluate the role of Sim2s in lactogenic differentiation; therefore, the objective of this study was to further define the function of Sim2s in the developing mammary epithelium using alternative models.
Mammary ductal epithelial cells are the primary targets of breast cancer, and much effort has gone into determining the mechanisms governing their development and function. Although it is evident that Sim2s is required for mammary ductal development, and that loss of Sim2s promotes the acquisition of a basal phenotype, it remains unclear what role Sim2s plays in promoting mammary epithelial cell differentiation. Consistent with a role for Sim2s in mammary epithelial differentiation, we report here that Sim2s is developmentally regulated, showing maximal expression during mid-lactation when the mammary gland undergoes terminal differentiation. To further address the role of Sim2s in mammary development, we have generated a transgenic mouse expressing hemagglutinin (HA)-tagged Sim2s targeted to mammary epithelium by the mouse mammary tumor virus (MMTV) long terminal repeat (LTR) promoter. Analysis of MMTV-Sim2s transgenic females revealed precocious lactogenic differentiation, and overexpression of Sim2s in HC11 and CIT3 mouse mammary epithelial cells markedly increased Csn2 expression following exposure to lactogenic hormones. These studies describe, for the first time, a role for Sim2s in regulating mammary differentiation in vitro and in vivo, and might shed light on the nature of the relationship between Sim2s and breast cancer progression.
MATERIALS AND METHODS
Chemicals
Ovine prolactin (Prl) was obtained from the National Hormone and Peptide Program (NHPP), the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK), and Dr Albert F. Parlow (Harbor-UCLA Medical Center).
Animals
Nulliparous females were harvested during estrus as determined by vaginal smears and, unless otherwise indicated, all females were primiparous. All animals were housed three per cage under a standard 12-hour photoperiod. The animals were provided with access to food and water ad libitum. All procedures were approved by the University Laboratory Animal Care Committee at Texas A&M University.
Cell culture
HC11 cells were grown in RPMI 1640 media (Gibco) supplemented with 10% calf serum (Atlanta Biologicals), 50 μg/ml gentamycin (Gibco), 5 μg/ml insulin (Sigma) and 10 ng/ml epidermal growth factor (EGF; Sigma). To induce differentiation in HC11 cells, confluent plates were given fresh media for 3 days followed by 24 hours of priming in RPMI 1640 media supplemented with 10% charcoal-stripped horse serum, 50 μg/ml gentamycin, 5 ug/ml insulin and 1 μg/ml hydrocortisone (Sigma). After priming, fresh priming medium containing 1 μg/ml ovine Prl was added with fresh media every 24 hours. CIT3 cells were grown in DMEM/F12 supplemented with 2% calf serum, 1% Pen-Strep (Gibco), 5 μg/ml insulin and 5 ng/ml EGF. Confluent cells were treated with growth media lacking EGF and containing 3 μg/ml hydrocortisone and 3 μg/ml ovine Prl.
RNA isolation and reverse transcription
RNA isolation and reverse transcription (RT) reactions were performed on cultured cells as previously described (Metz et al., 2006). RNA was isolated from tissue using Trizol reagent (Invitrogen), followed by purification using a Qiagen RNEasy Mini Kit as described (Kwak et al., 2007).
In situ hybridization
In situ hybridization was performed as previously described (Wilkensen, 1993). Briefly, 35S-uridine triphosphate (Amersham Pharmacia Biotech, Piscataway, NJ, USA)-labeled antisense and sense probes of mouse Sim2s cDNA were made using the Riboprobe In Vitro Transcription Systems Kit (Promega). The TOPO TA Cloning Kit (Invitrogen) was used to subclone a 437 bp RT-PCR product that was used generate sense and antisense probes using the primers 5′-AAATCAGCTAAACCCAAAAACACAAA-3′ and 5′-AGCATTCACAGGAGAAGGCTCAGAA-3′. Mammary glands from day 7 lactating mice were fixed in 4% paraformaldehyde, embedded in paraffin and cut into 5 μm sections. After deparaffinization and rehydration, sections were pretreated with 20 μg/ml proteinase K and 0.1 mM triethanolamine/acetic anhydride before being coated with labeled probe and incubated overnight at 55°C. Slides were washed with increasing stringency using 5× SSC, followed by 50% formamide/2× SSC/100 mM β-mercaptoethanol for 30 minutes each at 65°C. Slides were then treated with 20 μg/ml ribonuclease A for 30 minutes at 37°C, followed by final washes of 2× SSC and 0.1× SSC for 15 minutes at 65°C, and dehydrated. To visualize the 35S-uridine triphosphate label, slides were dipped in photographic NTB-2 emulsion (Kodak, Rochester, NY, USA) and exposed for 3 days. D-19 developer and fixer (Kodak) were used to develop the slides, followed by hematoxylin counterstain.
PCR
RT-PCR and qRT-PCR were performed as described previously (Kwak et al., 2007). Primers used for qRT-PCR reactions include: Sim2s, sense 5′-AACCAGCTCCCGTGTTTGAC-3′ and antisense 5′-ACTCTGAGGAACGGCGAAAA-3′; Wap, sense 5′-TCAGTCCATGTTCCCAAAAGC-3′ and antisense 5′-CTCGTTGGTTTGGCAGATGA-3′; Csn2, sense 5′-TGTGCTCCAGGCTAAAGTTCACT-3′ and antisense 5′-GGTTTGAGCCTGAGCATATGG-3′; Lalba, sense 5′-GAATGGGCCTGTGTTTTATTTCA-3′ and antisense 5′-TGTGCTGCCGTTGTCGTT-3′; Wdnm1, sense 5′-CGCCCCCACGCAGTT-3′ and antisense 5′-GCCAGAGCACGATGGATCTG-3′; and Cldn7, sense 5′-TCCCTGGTGTTGGGCTTCT-3′ and antisense 5′-ACAGCGTGTGCACTTCATG-3′. Primers used for RT-PCR reactions include: Sim2s transgene, sense 5′-AAGAACCAACCCATATCCCC-3′ and antisense 5′-GGCATAATGCGGCACATCATAAGG-3′; Gapdh, sense 5′-CTAACATCAAATGGGGTGAGG-3′ and antisense 5′-TCATACTTGGCAGGTTTCTCC-3′; and actin exon, sense 5′-TACAGCTTCACCACCACAGC-3′ and antisense 5′-AAGGAAGGCTGGAAAAGAGC-3′. The Csn2 promoter primer sequence has been described elsewhere (Kabotyanski et al., 2006).
Generation of MMTV-Sim2s-HA mice
To generate transgenic mice, the Sim2s coding sequence containing a hemagglutin (HA) tag on the 3′ end was cloned into the MMTV-KCR transgene cassette, pMKBPA (kindly provided by Dr Jeff Rosen, Baylor College of Medicine, Houston, TX, USA). Mice (FVB background) were generated in the Department of Molecular and Cellular Biology Transgenic Mouse Core, Baylor College of Medicine, under the supervision of Dr Francisco DeMayo. Two MMTV-Sim2s lines (3841 and 3829) were generated that transmitted the transgene through the germline as detected by PCR genotyping and immunohistochemistry for the HA epitope tag. However, one line (3829) was found to also have Sim2s expression in tissues outside of the mammary gland, including the hypothalamus and lung, and therefore was not used for these studies. All procedures were approved and followed the guidelines set forth by the Texas A&M Animal Use and Care Committee.
Immunostaining
Immunohistochemical analyses were carried out as previously described (Kwak et al., 2007). Following incubation in blocking solution, primary antibodies were added overnight. Antibodies used included HA (Sigma), Sim2s (Millipore), Npt2b (Alpha Diagnostics), β-casein (Santa Cruz), aquaporin 5 (Alpha Diagnostics), P-Tyr Stat5a/b (Advantex Bioreagents) and normal IgG (Upstate). Tissue processing and Hematoxylin and Eosin (H&E) staining were performed by the Histology Core Facility in the Texas A&M University College of Veterinary Medicine & Biomedical Sciences.
Wholemount analysis
Wholemount analysis was performed as previously described (Laffin et al., 2008). To determine gross ductal branching, wholemount mammary glands were imaged using a stereo microscope equipped with a digital camera and analyzed using NIH Image software. Ductal penetration was defined as the mean average of the length from the point of implantation to the ends of the three longest ducts in each mammary gland. These three ducts were used to calculate the number of branch points, or lateral ducts, per millimeter by averaging the number of unbranched and ramified ducts. The number of branch points directly contacting a vertical reference line, drawn through the center of each mammary gland, were counted directly from pictures of whole mounts.
Western blot
Cells were washed in 10 ml PBS, collected in 2 ml PBS containing complete protease inhibitor (CPI; Roche), pelleted and resuspended in lysis buffer (20 mM tris-Cl pH 8.0, 137 mM NaCl, 10% glycerol, 1% NP-40 and 2 mM EDTA) containing CPI and phosphatase inhibitors (sodium orthovanadate, sodium fluoride and sodium molybdate). Cell lysates were incubated with rocking at 4°C for 30 minutes, centrifuged at 16,000 g for 10 minutes and the supernatant was saved for western blot analysis. Protein concentration was estimated using the RCDC Assay (BioRad). Samples were diluted in water and 6× loading buffer, boiled for 5 minutes and subjected to SDS PAGE. Proteins were transferred to PVDF membranes and blocked for 1 hour to overnight in PBS or TBS containing 0.5% Tween-20 and 5% non-fat milk. Following incubation with primary and secondary antibodies, membranes were developed using the Amersham ECL Plus Kit (Amersham Pharmacia Biotech).
Retroviral transduction
Lentiviral transduction of HC11 and CIT3 cells was performed as previously described (Kwak et al., 2007).
Chromatin immunoprecipitation assays
Crosslinking was achieved by the addition of 1% formaldehyde directly to culture media for 10 minutes at room temperature with gentle agitation, followed by the addition of glycine at 1.25 mM. Monolayers were washed twice with ice-cold PBS and collected in 2 ml PBS plus CPI. Cell pellets were resuspended in SDS lysis buffer containing CPI and incubated for 10 minutes on ice. Lysates were sonicated 10×10 seconds, which resulted in fragments of 200-1200 kb. Chromatin was diluted 5-fold in chromatin immunoprecipitation (ChIP) dilution buffer containing CPI and pre-cleared with 80 μl salmon sperm DNA/protein A/G agarose beads (Upstate) 2×30 minutes at 4°C with rocking. Antibodies were added overnight followed by the addition of 60 μl beads for one hour. Beads were washed for 10 minutes at 4°C with each of the following solutions: low salt buffer, high salt buffer, LiCl buffer and twice in TE buffer (Tris, EDTA) at room temperature. Complexes were eluted at room temperature in 10% SDS and 0.1 M sodium bicarbonate. Crosslinks were reversed by addition of 0.3 M NaCl and 1 μl 10 mg/ml RNase A and incubated at 65°C for 5 hours. Two and a half volumes of 100% EtOH were added and samples were placed at −20°C overnight. Following centrifugation for 10 minutes at 16,000 g, pellets were resuspended in 100 μl water containing tris-HCl pH 6.5, EDTA and 1 μl 20 mg/ml proteinase K. Samples were incubated at 45°C for 1.5 hours and DNA was purified using Qiagen PCR Purification Kit (Qiagen). Two microliters were used for subsequent PCR analysis. For re-ChIP experiments, chromatin was incubated with antibody overnight at 4°C after pre-clearing. Eighty microliters of protein A or protein G agarose beads were added for 1 hour at 4°C and were washed according to standard ChIP protocol. Following TE washes, beads were incubated with 1 volume of 20 mM dithiothreitol (DTT) for 30 minutes at 37°C with agitation. Supernatants were collected and diluted to 1000 μl with re-ChIP Dilution Buffer (Upstate). Ten percent of the eluate was reserved for input and the remaining eluate was divided in half for no antibody control and immunoprecipitation with a secondary antibody, respectively. After overnight incubation with the antibody, immune complexes were precipitated with agarose beads and washed/eluted according to standard ChIP protocol. Antibodies used in ChIP analyses include AcH3 (Upstate), Sim2s (Millipore), RNAPII (Abcam), Stat5 (Millipore) and normal IgG (Upstate).
RESULTS
Sim2s is differentially expressed during mammary gland development
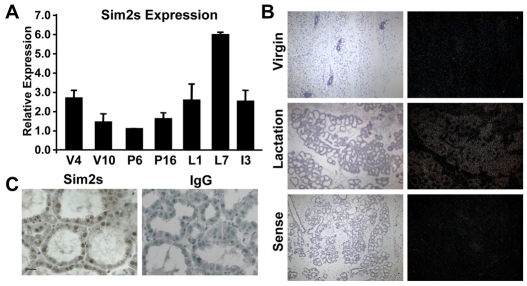
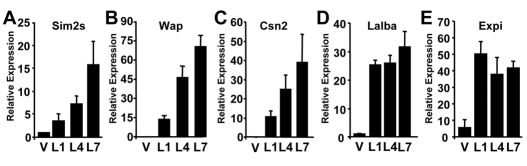
Based on our previous studies, we hypothesized that Sim2s plays a role in establishing and/or maintaining mammary epithelial cell phenotypes. Initially, we investigated Sim2s expression during mouse mammary gland development by qRT-PCR. Total RNA isolated from mouse mammary tissues at various developmental time points was analyzed for the expression of the Sim2s isoform as we have previously shown that mouse mammary epithelial cells do not express full-length Sim2s (Laffin et al., 2008). Sim2s expression is developmentally regulated. It is first detected in the mammary glands of four-week-old pre-pubertal mice and then decreases in mature virgin and early pregnant glands, which is consistent with Sim2s playing a role in the differentiation of the luminal epithelial cells as the TEBs progress through the gland (Laffin et al., 2008). Sim2s expression increases during late pregnancy and throughout lactation, peaking on day seven (L7) when the gland is fully differentiated (Fig. 1A). In situ hybridization confirmed that Sim2s mRNA increases during lactation in the luminal alveolar mammary epithelial cells (Fig. 1B). Further, immunohistochemical analysis revealed that the Sim2s protein is present in the nuclei of alveolar epithelial cells in L7 mammary glands (Fig. 1C). To better define the role of Sim2s during this process, additional samples obtained from lactation days one and four were analyzed by qRT-PCR for Sim2s and several milk protein markers of mammary differentiation. These data show that Sim2s expression progressively increases during the first week of lactation (Fig. 2A), coordinate with the milk protein genes encoding whey acidic protein (Wap, Fig. 2B) and β-casein (Csn2, Fig. 2C). Interestingly, two additional mammary differentiation markers, α-lactalbumin (Lalba) and Wdnm1 (encoded by Expi), were expressed at high levels on lactation day 1 and remained relatively unchanged over the course of lactation (Fig. 2D,E).
Fig. 1.
Sim2s expression is developmentally regulated in mouse mammary tissues. (A) Total RNA isolated from C57 mouse mammary glands (n=3 animals per group) at weeks 4 and 10 of virgin development (V4 and V10), days 6 and 16 of pregnancy (P6 and P16), days 1 and 7 of lactation (L1 and L7) and day 3 of involution (I3) was used for qRT-PCR analysis of Sim2s expression. (B) In situ hybridization analysis of Sim2s mRNA from 5-week-old virgin (top) and L7 (middle) mouse mammary glands. The sense control is shown in the bottom panel. Left images are Hematoxylin and Eosin (H&E)-stained adjacent sections. (C) Immunohistochemical (ICH) staining of L7 FVB mouse mammary glands for Sim2s (left panel) showing nuclear localization in the mammary epithelium. Normal rabbit IgG (right panel) was used as a negative control.
Fig. 2.
Sim2s expression patterns correlate with a subset of milk protein genes during lactation. (A-E) Total RNA isolated from V8, L1, L4 and L7 FVB mammary glands (n=5 animals per group) was used for qRT-PCR analysis of (A) Sim2s, (B) Wap, (C) Csn2, (D) Lalba and (E) Expi expression. Cldn7 was used to normalize all mammary qRT-PCR data.
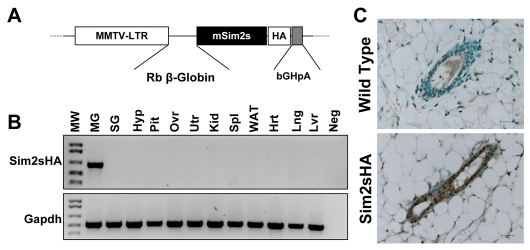
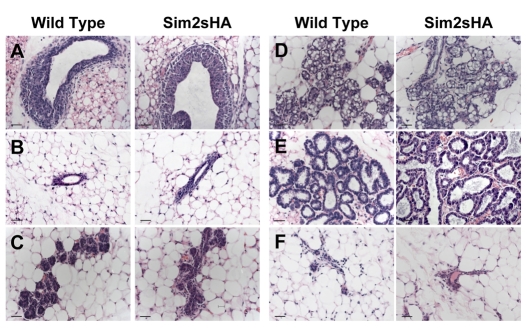
The observation that Sim2s expression coincides with mammary lactogenic differentiation led us to ask if Sim2s was sufficient to regulate this process. As a first step, we generated transgenic mice expressing an HA-tagged full-length Sim2s under control of the MMTV LTR promoter (Fig. 3A). Two separate lines of MMTV-Sim2s-HA mice were generated; however, PCR analyses revealed that one of the lines displayed inappropriate transgene expression with low mammary Sim2s-HA mRNA and high levels of transgene message in the hypothalamus, spleen and lung (see Fig. S1 in the supplementary material). In the second line, transgene expression was detectable exclusively in mammary tissues (Fig. 3B); therefore, further analyses were carried out using this line. Immunohistochemical analysis of wild-type (WT) and MMTV-Sim2s-HA mouse mammary sections using an HA antibody demonstrated that the Sim2s transgene was specifically found in the nucleus of mammary ductal epithelial cells (Fig. 3C; see also Fig. S1 in the supplementary material). Morphologically, MMTV-Sim2s-HA mammary glands were indistinguishable from WT glands throughout development. During early virgin development, TEB structures formed appropriately (Fig. 4A) and mature epithelial ducts were seen throughout the mammary fat pad by 8 weeks of age in transgenic females (Fig. 4B). During pregnancy, alveolar structures expanded and the ability of alveolar epithelial cells to undergo secretory differentiation was not impaired in glands from MMTV-Sim2s-HA females (Fig. 4C,D). Mammary glands of transgenic females were indistinguishable from WT glands during lactation (Fig. 4E), and MMTV-Sim2s-HA females were able to nurse large litters to weaning age. Following pup removal, the mammary epithelium from MMTV-Sim2s-HA females fully involuted and did not differ from that of WT females (Fig. 4E).
Fig. 3.
Generation of MMTV-Sim2s-HA transgenic mice. (A) Schematic diagram of the MMTV-Sim2s-HA targeting construct. The MMTV-LTR construct was provided by Dr Jeffery Rosen at Baylor College of Medicine. Exons 2 and 3 (E2, E3) and Intron 2 (I2) are derived from the rabbit β-globin gene (RβG). The poly-A signal is derived from the bovine growth hormone gene (bGHpA). (B)RT-PCR analysis of pooled samples from three transgenic females showing expression of the Sim2s-HA transgene exclusively in the mammary gland. Hrt, heart; Hyp, hypothalamus; Kid, kidney; Lng, lung; Lvr, liver; Neg, negative control (water); MG, mammary gland; Pit, pituitary; Ovr, ovary; SG, salivary gland; Spl, spleen; Utr, uterus; WAT, white adipose tissue. Gapdh was used as a loading control. (C) IHC analysis of HA localization in mammary glands from wild-type (WT) and transgenic (Sim2s-HA) V8 females.
Fig. 4.
Assessment of mammary morphology in Sim2s-HA transgenic and non-transgenic female mice. (A-F) Mammary glands were isolated from (A) V5, (B) V8, (C) P6, (D) P16, (E) L2 and (F) I14 FVB females, fixed in paraformaldehyde, sectioned and stained with H&E. Scale bars: 100 μm.
Transgenic MMTV-Sim2s nulliparous mice display hallmarks of precocious lactogenic differentiation
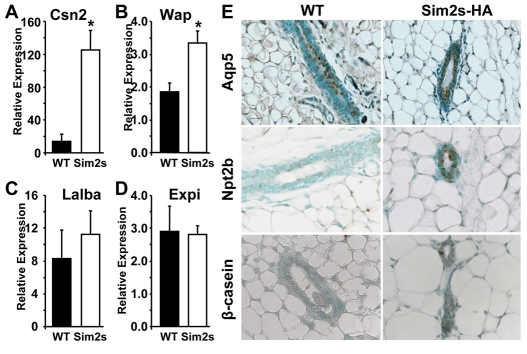
The observation that Sim2s levels increase during lactation, following the same pattern as a subset of milk proteins, led us to hypothesize that Sim2s plays a role in gene regulation during alveolar epithelial cell differentiation. Using qRT-PCR, we analyzed mature virgin WT and MMTV-Sim2s-HA mouse mammary tissues for Csn2, Wap, Lalba and Expi expression. Interestingly, both Csn2 and Wap mRNA levels were significantly higher in eight-week-old virgin transgenic mouse mammary tissues in comparison with WT glands (Fig. 5A,B). However, expression of both Lalba (Fig. 5C) and Expi (Fig. 5D) did not change as a result of Sim2s expression. Although it has been reported that Csn2 expression changes during the estrus cycle (Robinson et al., 1995), mice used in these studies cycled normally (data not shown) and all of the nulliparous females were sacrificed at estrus. Together, these data imply that Sim2s enhances alveolar differentiation at the molecular level and might play a role in regulating the expression of a subset of milk protein genes. To further characterize the effects of Sim2s on mammary cell differentiation, we analyzed sections of WT and MMTV-Sim2s-HA mouse mammary glands for specific epithelial cell type cell markers. aquaporin 5 (Aqp5) is an epithelial cell marker expressed on the luminal surface of ductal mammary epithelial cells in non-pregnant females (Shillingford et al., 2003). As mammary ductal epithelium differentiates into alveolar cells, Aqp5 is lost and various luminal surface solute transporters, such as Slc12a2 (Nkcc1) and Slc34a2 (Npt2b), are upregulated (Shillingford et al., 2003). Analysis of eight-week-old virgin WT and transgenic females revealed no differences in levels or localization of Aqp5 owing to Sim2s expression (Fig. 5E, top). However, apical localization of Npt2b was detected in approximately 70% of MMTV-Sim2s-HA glands (Fig. 5E, middle), suggesting that Sim2s expression is inducing a partial alveolar phenotype in these cells. In support of this, and in confirmation of the effects of Sim2s on Csn2 mRNA expression, immunohistochemical analysis of WT and MMTV-Sim2s-HA mammary sections revealed an increase in β-casein-positive epithelial cells in virgin glands (Fig. 5E, bottom). Furthermore, we found no differences in staining for phospho-Stat5a in WT and Sim2s transgenics compared with glands from L7 mice (data not shown), suggesting that this phenotype is not mediated by activation of Stat5a. Together, these data indicate that Sim2s expression in ductal epithelium induces a gene expression profile characteristic of alveolar epithelial cells.
Fig. 5.
Sim2s expression results in precocious expression of a subset of milk proteins and alveolar epithelial cell surface markers. (A-D) qRT-PCR was used to evaluate gene expression in samples from nulliparous wild type (WT) and transgenic (Sim2s) females (n=5 animals per group). (A) Csn2 expression. (B) Wap expression. (C) Lalba expression. (D) Expi expression. *, P<0.05. Cldn7 expression was used to normalize qRT-PCR data. (E) Immunohistochemical analysis of aquaporin 5 (Aqp5, top), the sodium-inorganic phosphate co-transport protein Npt2b (middle) and β-casein (bottom) in mammary glands from V8 WT and transgenic females.
Sim2s is upregulated in prolactin-treated HC11 and CIT3 mouse mammary epithelial cells and is required for Csn2 (β-casein) gene expression
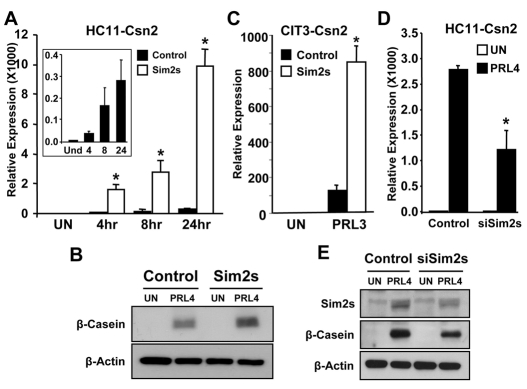
To further analyze the molecular effects of Sim2s on mammary epithelial cell differentiation, we moved to an in vitro model system. The mouse mammary epithelial cell line HC11, derived from the COMMA-D cell line that was isolated from a mid-pregnant Balb/c mouse, has been widely used to study gene expression during lactogenic differentiation in a relatively homogeneous environment (Ball et al., 1988). Following confluent growth arrest, HC11 cells can be treated with hydrocortisone and prolactin (HC + PRL) to induce Csn2 expression. Sim2s expression was analyzed in undifferentiated HC11 cells (UN) and in cells treated with HC + PRL for one or four days (Fig. 6A). We found that Sim2s mRNA levels began to increase within 24 hours after hormone stimulation and were elevated further by day four, which is consistent with our in vivo observations. Western blot analyses confirmed that both Sim2s and β-casein levels increased in HC11 cells after four days of hormone treatment (Fig. 6B). CIT3 cells, another COMMA-D-derived line, were also found to upregulate Sim2s following treatment with HC + PRL (Fig. 6C). These results indicate that these cells represent an appropriate model to study Sim2s regulation of mammary differentiation, and adequately recapitulate the effect on Sim2s seen during lactogenic differentiation in vivo.
Fig. 6.
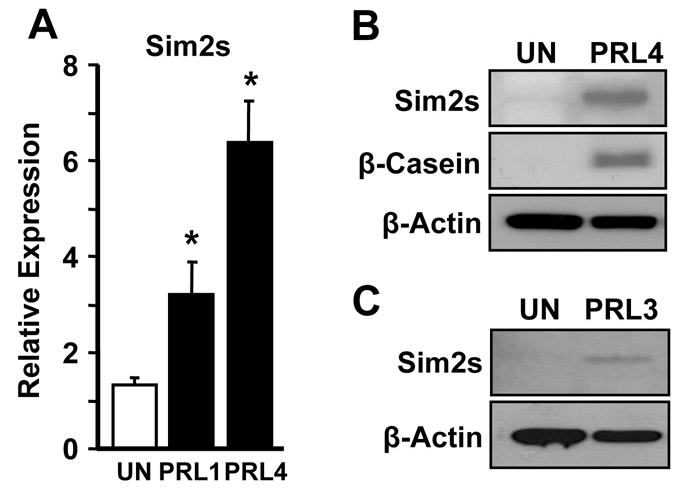
Sim2s expression is upregulated in differentiated HC11 and CIT3 cells. (A) qRT-PCR was used to detect Sim2s in undifferentiated HC11 cells (UN) and cells treated with HC + Prl for 1 or 4 days (PRL1 and PRL4). (B) Western blot analysis of Sim2s and β-casein protein levels in undifferentiated (UN) HC11 cells and PRL4 cells. (C) Western blot analysis of Sim2s levels in UN CIT3 cells and PRL3 cells. β-actin was used as a loading control for western analyses.
A lentiviral transduction system was used to overexpress Sim2s in both HC11 and CIT3 cells. Using qRT-PCR, we determined that Csn2 expression in cells expressing Sim2s, was significantly upregulated following hormone stimulation in both HC11 and CIT3 cells (Fig. 7A,C, respectively). In HC11 cells, the increase in Csn2 was detectable in as little as four hours after treatment with HC + PRL, and continued through 24 hours (Fig. 7A). Western blot analyses confirmed increased β-casein protein in Sim2s-overexpressing cells (Fig. 7B), in agreement with the in vivo data. With the observation that Sim2s enhances β-casein, we sought to determine whether Sim2s is required for Csn2 expression in vitro. To do this, we transduced HC11 cells with an shRNA construct targeting exon 6 of the Sim2 gene. Following selection, colonies were pooled and cells were treated for 4 days with HC + PRL and analyzed for Csn2 expression. Our results show that Csn2 induction was reduced by approximately 50% in HC11 cells targeted with the Sim2s shRNA construct (siSim2s) in comparison with scrambled control cells (Fig. 7D). Western blot analyses revealed that although Sim2s levels were not completely abrogated in siSim2s cells, the reduction in protein was associated with a decrease in β-Casein protein in this cell line (Fig. 7E). These data suggest that Sim2s expression is necessary for the robust hormone-mediated induction of Csn2 in this culture system.
Fig. 7.
Sim2s is necessary and sufficient for robust prolactin-mediated Csn2 expression. (A) qRT-PCR analysis of Csn2 expression in control and Sim2s-overexpressing HC11 cells. Total RNA was isolated from UN cells or cells treated for 4, 8 or 24 hours with HC + Prl. Csn2 expression in control cells is shown on a smaller scale in the inset panel. (B) Western blot analysis of β-casein production in UN and HC + Prl treated control and Sim2s-overexpressing HC11 cells. (C) qRT-PCR analysis of Csn2 expression in control and Sim2s-overexpressing CIT3 cells. Total RNA was isolated from UN cells or PRL3 cells. (D)qRT-PCR analysis of Csn2 expression in HC11 cells stably expressing a scrambled shRNA sequence (control) or expressing shRNA targeting Sim2s (siSim2s). (E) Western blot analysis of Sim2s and β-casein in control and stable shSim2s UN cells or PRL4 cells. *, P<0.05.
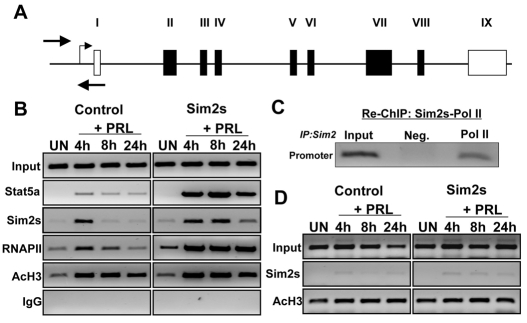
Based on the data indicating a direct relationship between Sim2s and Csn2 expression, and because Sim2s is a transcription factor, we hypothesized that it is directly involved in regulating Csn2 gene expression. Mechanisms governing Csn2 transcription have been well documented, and within 1 hour of prolactin treatment, increased association of Stat5a, GR, Cebpb and RNAPII can be detected at the Csn2 proximal promoter (Kabotyanski et al., 2006). Although this region of the Csn2 promoter does not contain consensus Sim2s response elements, we monitored the occupancy of proteins around the Csn2 transcription start site in control and Sim2s-overexpressing HC11 cells (Fig. 8A, solid arrows). Undifferentiated cells (UN) or cells treated for 4, 8 or 24 hours with HC + PRL were analyzed for the presence of various transcription factors using ChIP (Fig. 8B). In control cells, increased Stat5a binding to the Csn2 promoter was observed 4 hours after HC + PRL treatment and slowly decreased thereafter. In Sim2s cells, Stat5a binding to the promoter region was also apparent 4 hours after HC + PRL treatment; however, the relative level of bound Stat5a was much greater in Sim2s HC11 cells, peaking 8 hours after hormone treatment and remaining high even at 24 hours post-induction (Fig. 8B). Sim2s binding to the Csn2 promoter was also detected within 4 hours in both control and Sim2s HC11 cells, the latter of which maintained Sim2s occupancy even at 24 hours post-treatment. These changes correlated with enhanced RNAPII association at the Csn2 promoter, and Sim2s cells showed both increased RNAPII binding and duration of association in comparison with control HC11 cells (Fig. 8B). Assessment of acetylated histone 3 (AcH3) binding, indicative of active transcription, did not reveal significant differences between control and Sim2s cells. To further examine the association pattern of Sim2s and the RNAPII complex, we performed re-ChIP analyses using chromatin from HC + PRL-treated WT HC11 cells with a Sim2s antibody. Before crosslink reversal, this chromatin was subjected to a second ChIP using an RNAPII-specific antibody. The resulting chromatin was analyzed for Csn2 promoter and exon 7 DNA by PCR (Fig. 8C). These results show that Sim2s is associated with the RNAPII complex at the promoter region of the Csn2 gene. The absence of Sim2s at the Actb gene supports that this is gene-specific and not a general effect of Sim2s on transcription (Fig. 8D). These results suggest that, rather than altering the accessibility of Csn2 promoters in the population of HC11 cells, Sim2s is enhancing recruitment of Stat5a and RNAPII to the Csn2 promoter, which might result in higher levels of mRNA transcription.
Fig. 8.
Sim2s alters the binding of regulatory factors to the Csn2 promoter region. (A) Schematic diagram of the Csn2 gene. White boxes represent non-coding exons and black boxes represent coding exons (II-VIII). Arrows indicate the location of proximal promoter primers. (B) ChIP analyses of the proximal promoter were performed on chromatin isolated from UN HC11 cells or cells treated for 4, 8 or 24 hours with HC + Prl using the antibodies indicated to the left. (C) Re-ChIP analysis of Sim2s and RNAPII at the proximal promoter of Csn2 in control cells at PRL4. Following an initial pull down with Sim2s antibody, chromatin was re-immunoprecipitated with RNAPII antibody and the Csn2 promoter sequence was amplified using PCR. (D) ChIP analysis of Sim2s and AcH3 occupancy of Actnb coding region in control and Sim2s-overexpressing HC11 cells is included as a control.
DISCUSSION
This study demonstrates, for the first time, a role for Sim2s in promoting differentiation of mammary alveolar epithelial cells. We have previously shown that SIM2s is expressed in human breast luminal epithelial cells and loss of SIM2s in normal human breast cell lines and mouse mammary epithelium during virgin development induces an invasive basal/stem cell-like phenotype (Laffin et al., 2008; Gustafson et al., 2009). To further evaluate the role of Sim2s in mammary gland development, we provide new data demonstrating that Sim2s is upregulated in a stepwise manner during early lactation, similar to that of Csn2 and Wap, but not Lalba and Expi. Furthermore, overexpression of Sim2s in the mouse mammary epithelium leads to an increase of a subset of differentiation markers. In nulliparous transgenic female mice, mRNA levels of Csn2 and Wap, but not Expi and Lalba, were significantly increased, revealing slight differences in the mechanisms governing regulation of these genes. Similarly, Sim2s expression increased in HC11 and CIT3 mouse mammary epithelial cells with lactogenic hormone treatment, and overexpressing Sim2s in vitro resulted in enhanced Csn2 expression and association of Stat5a and RNAPII with the Csn2 promoter. These results suggest that Sim2s is required to establish and maintain mammary ductal epithelial character in the developing virgin gland and promote lactation-dependent gene expression.
The observation that Sim2s expression peaks after parturition points toward Sim2s playing a role in the acquisition and maintenance of alveolar differentiation. Analysis of milk protein gene expression in whole tissue lysates from mammary glands of nulliparous transgenic and WT females showed that only Csn2 and Wap were significantly upregulated as a result of Sim2s overexpression. Taken with the data showing differential patterns of milk protein gene expression during the first week of lactation, these results suggest that distinct mechanisms govern the regulation of genes in the mammary alveolar epithelium, and that Sim2s might participate in regulating a subset of lactation specific genes. This is consistent with previous studies that looked at milk protein gene expression in mammary glands from nulliparous mice and during pregnancy and lactation (Robinson et al., 1995). The authors showed that, during pregnancy, Csn2 and Wap were detectable at different times, but in the same cell type. The expression of Lalba and Expi were temporally regulated during pregnancy as well, but were not expressed in the same cells as Csn2 and Wap. During lactation, all four milk protein genes were highly expressed, but expression levels were different between cells contained within an alveolar structure. Recently, it was reported that Wap expression in vivo is regulated by the demand of the nursing offspring (Creamer et al., 2009). Using a Wap-luciferase reporter transgenic mouse model, the results show that females nursing litters of three pups produced less Wap than those nursing eight pups. The increase in Sim2s expression during the first week of lactation might reflect a role in modulating milk protein gene transcription in response to increasing pup demand. The ChIP data obtained from HC11 cells showing enhanced recruitment of RNAPII to the Csn2 promoter as a result of Sim2s overexpression supports this theory. Based on these studies, it is probable that specific factors expressed in the mammary epithelium are responsible for the variability in milk protein gene expression and for the coordinated regulation of Csn2 and Wap in the same cells.
Using HC11 and CIT3 mouse mammary epithelial cell lines, we determined that Sim2s expression increases after stimulation with lactogenic hormones. In HC11 cells, we observed a progressive increase in Sim2s expression over the course of 4 days of hormone treatment, similar to the stepwise increase seen in vivo. When Sim2s was overexpressed in these cell lines, Csn2 expression was significantly higher after hormone treatment, and reduction of Sim2s in HC11 cells revealed a necessity of Sim2s expression for robust Csn2 induction. ChIP analyses in HC11 cells demonstrated increased recruitment of RNAPII and Stat5a to the Csn2 promoter in cells overexpressing Sim2s. In addition, Sim2s itself was detected in association with the Csn2 promoter. Significantly, the changes in RNAPII and Stat5a levels were seen without an increase in AcH3 protein in association with the same region. Re-ChIP analysis of both Sim2s and RNAPII showed that both proteins associated with one another in a complex at the Csn2 promoter. These data indicate that overexpression of Sim2s is not simply facilitating the recruitment of transcription factors to more Csn2 promoters in the population of cells, but rather enhancing the association of these proteins with a particular promoter. However, the mechanism by which Sim2s accomplishes this is unclear, although it might participate in recruiting transcriptional activators, facilitating promoter escape and/or elongation of RNAPII or acting as a transcriptional co-activator through an undefined mechanism.
Although the RNAPII complex is large, and made up of many subunits, it is possible that Sim2s associates with the holoenzyme to target the complex to a particular class of genes during differentiation. The conformation and subunit composition of the general transcription factors can vary depending on the tissue, the cell type within a tissue or the developmental stage of the tissue. In Drosophila spermatocyte development, it has been proposed that transcriptional complexes can vary in the composition of TBP-associated factors (TAFs) that, together with TBP, form TFIID to regulate the expression of testis-specific genes (Hiller et al., 2004). TFIID associates with promoters of genes to recruit RNAPII and promote transcription initiation. In mice, it has been shown that the cellular localization of a TAF is another level of transcriptional regulation during differentiation (Pointud et al., 2003). TAF7L was identified as a germ cell-specific paralog of TAF7. In spermatogonia, TAF7L is localized to the cytoplasm but becomes nuclear and replaces TAF7 in the TFIID complex during spermatogenesis. A study performed in mouse ovaries determined that loss of TAF105 affected expression of a subset of genes associated with follicle development (Freiman et al., 2001). The authors proposed that cell type-specific subunits of TFIID in mammalian cells lead to specific regulation of gene expression programs during differentiation. Cell type-specific replacements for TBP have been identified in spermatocytes and developing myotubules as well (Deato and Tjian, 2007; Zhang et al., 2001). The participation of Sim2s in recruiting RNAPII to specific gene promoters could explain its association with the Csn2 regulatory region in the absence of consensus Sim2s response elements.
In addition to changes in gene expression, mammary epithelial cells from MMTV-Sim2s-HA mice also showed apical localization of Npt2b. The sodium-inorganic phosphate transport protein is normally detected during late pregnancy and lactation and is localized to the luminal surface of alveolar epithelial cells, suggesting that Sim2s promotes a change in cell fate. These results are similar to the transcription factors, Elf5 and Gata3, which have also been shown to play a role in mammary lactogenic differentiation (Choi et al., 2009; Kouros-Mehr et al., 2006; Oakes et al., 2008). Loss of Elf5 in the mammary epithelium led to impaired alveolar formation and Stat5a activation (Choi et al., 2009). In addition, the authors showed that Elf5 was associated with the Csn2 promoter. Our data indicate that Sim2s associates with the Csn2 promoter in vitro and contributes to enhanced transcription of the Csn2 gene. In cultured epithelial cells, Elf5 reduction resulted in a decrease in Stat5a, phosphorylated Stat5a and subsequently, β-casein (Choi et al., 2009). Similarly, shRNA-mediated reduction of Sim2s levels in HC11 cells attenuated Csn2 mRNA and protein levels. Our studies did not investigate the requirement of Sim2s for alveolar expansion during pregnancy; however, overexpression of Sim2s in nulliparous females did not result in precocious morphological changes seen prior to lactation. Gata3 has been demonstrated to play a role in the maintenance of luminal cell fate in vivo (Kouros-Mehr et al., 2006). Loss of Gata3 impaired ductal outgrowth, alveolar formation and milk protein production. Our previous studies with Sim2s-null transplanted mammary epithelium suggest that Sim2s is also required for proper ductal outgrowth and luminal epithelial differentiation during early virgin development, which is taking place while the TEBs progress through the fat pad (Laffin et al., 2008). In the studies herein, we have demonstrated a requirement of Sim2s for robust prolactin-mediated Csn2 expression. Thus, it is possible that Sim2s activation lies either downstream of Gata3 and Elf5 or plays a role in a parallel pathway by not only maintaining ductal epithelial cell character, but also by promoting alveolar differentiation.
A better understanding of how Sim2s governs mammary gland development and the consequences of this regulation might contribute significantly to cancer therapies aimed at inducing tumor cell differentiation. With increasing evidence that breast cancer progression correlates with decreased differentiation and induction of an invasive basal/stem cell-like phenotype (Contesso et al., 1987; Kouros-Mehr et al., 2008; Liu et al., 2007), deciphering signals controlling normal mammary gland development and differentiation will provide much needed insight into treating this disease. As we have shown that loss of Sim2s leads to a basal/stem cell-like phenotype and overexpression promotes differentiation (Laffin et al., 2008; Gustafson et al., 2009), we hypothesize that re-establishing SIM2s expression in highly aggressive human breast cancers will promote tumor cell differentiation and block breast cancer progression and metastasis.
Supplementary Material
Acknowledgements
We acknowledge Dr Jeffery Rosen at Baylor College of Medicine for kindly providing the MMTV-KCR-bGHpA construct. These studies were supported by grants from the National Cancer Institute (NCI) R01CA111551 (W.W.P.) and National Institutes of Environmental Health Sciences (NIEHS) T32ES007273. Deposited in PMC for release after 12 months.
Footnotes
Competing interests statement
The authors declare no competing financial interests.
Supplementary material
Supplementary material for this article is available at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.041657/-/DC1
References
- Ball R. K., Friis R. R., Schoenenberger C. A., Doppler W., Groner B. (1988). Prolactin regulation of beta-casein gene expression and of a cytosolic 120-kd protein in a cloned mouse mammary epithelial cell line. EMBO J. 7, 2089-2095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J., Kim I. O., Ahn J. S., Kim S. H. (2001). The CNS midline cells control the spitz class and Egfr signaling genes to establish the proper cell fate of the Drosophila ventral neuroectoderm. Int. J. Dev. Biol. 45, 715-724 [PubMed] [Google Scholar]
- Choi Y. S., Chakrabarti R., Escamilla-Hernandez R., Sinha S. (2009). Elf5 conditional knockout mice reveal its role as a master regulator in mammary alveolar development: failure of Stat5 activation and functional differentiation in the absence of Elf5. Dev. Biol. 2, 227-241 [DOI] [PubMed] [Google Scholar]
- Contesso G., Mouriesse H., Friedman S., Genin J., Sarrazin D., Rouesse J. (1987). The importance of histologic grade in long-term prognosis of breast cancer: a study of 1,010 patients, uniformly treated at the Institut Gustave-Roussy. J. Clin. Oncol. 5, 1378-1386 [DOI] [PubMed] [Google Scholar]
- Creamer B. A., Triplett A. A., Wagner K. U. (2009). Longitudinal analysis of mammogenesis using a novel tetracycline-inducible mouse model and in vivo imaging. Genesis 4, 234-245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deato M. D., Tjian R. (2007). Switching of the core transcription machinery during myogenesis. Genes Dev. 21, 2137-2149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freiman R. N., Albright S. R., Zheng S., Sha W. C., Hammer R. E., Tjian R. (2001). Requirement of tissue-selective TBP-associated factor TAFII105 in ovarian development. Science 293, 2084-2087 [DOI] [PubMed] [Google Scholar]
- Goshu E., Jin H., Fasnacht R., Sepenski M., Michaud J. L., Fan C. M. (2002). Sim2 mutants have developmental defects not overlapping with those of Sim1 mutants. Mol. Cell. Biol. 22, 4147-4157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafson T. L., Wellberg E., Laffin B., Schilling L., Metz R. P., Zahnow C. A., Porter W. W. (2009). Ha-Ras transformation of MCF10A cells leads to repression of Singleminded-2s through NOTCH and C/EBPbeta. Oncogene 12, 1561-1568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Happ B., Groner B. (1993). The activated mammary gland specific nuclear factor (MGF) enhances in vitro transcription of the beta-casein gene promoter. J. Steroid Biochem. Mol. Biol. 47, 21-30 [DOI] [PubMed] [Google Scholar]
- Hiller M., Chen X., Pringle M. J., Suchorolski M., Sancak Y., Viswanathan S., Bolival B., Lin T. Y., Marino S., Fuller M. T. (2004). Testis-specific TAF homologs collaborate to control a tissue-specific transcription program. Development 131, 5297-5308 [DOI] [PubMed] [Google Scholar]
- Kabotyanski E. B., Huetter M., Xian W., Rijnkels M., Rosen J. M. (2006). Integration of prolactin and glucocorticoid signaling at the beta-casein promoter and enhancer by ordered recruitment of specific transcription factors and chromatin modifiers. Mol. Endocrinol. 20, 2355-2368 [DOI] [PubMed] [Google Scholar]
- Kouros-Mehr H., Slorach E. M., Sternlicht M. D., Werb Z. (2006). GATA-3 maintains the differentiation of the luminal cell fate in the mammary gland. Cell 127, 1041-1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouros-Mehr H., Bechis S. K., Slorach E. M., Littlepage L. E., Egeblad M., Ewald A. J., Pai S. Y., Ho I. C., Werb Z. (2008). GATA-3 links tumor differentiation and dissemination in a luminal breast cancer model. Cancer Cell 13, 141-152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak H. I., Gustafson T., Metz R. P., Laffin B., Schedin P., Porter W. W. (2007). Inhibition of breast cancer growth and invasion by single-minded 2s. Carcinogenesis 28, 259-266 [DOI] [PubMed] [Google Scholar]
- Laffin B., Wellberg E., Kwak H. I., Burghardt R. C., Metz R. P., Gustafson T., Schedin P., Porter W. W. (2008). Loss of singleminded-2s in the mouse mammary gland induces an epithelial-mesenchymal transition associated with up-regulation of slug and matrix metalloprotease 2. Mol. Cell. Biol. 28, 1936-1946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Rosen J. M. (1995). Nuclear factor I and mammary gland factor (STAT5) play a critical role in regulating rat whey acidic protein gene expression in transgenic mice. Mol. Cell. Biol. 15, 2063-2070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R., Wang X., Chen G. Y., Dalerba P., Gurney A., Hoey T., Sherlock G., Lewicki J., Shedden K., Clarke M. F. (2007). The prognostic role of a gene signature from tumorigenic breast-cancer cells. New Engl. J. Med. 356, 217-226 [DOI] [PubMed] [Google Scholar]
- Menne T. V., Luer K., Technau G. M., Klambt C. (1997). CNS midline cells in Drosophila induce the differentiation of lateral neural cells. Development 124, 4949-4958 [DOI] [PubMed] [Google Scholar]
- Metz R. P., Kwak H. I., Gustafson T., Laffin B., Porter W. W. (2006). Differential transcriptional regulation by mouse single-minded 2s. J. Biol. Chem. 281, 10839-10848 [DOI] [PubMed] [Google Scholar]
- Nambu J. R., Franks R. G., Hu S., Crews S. T. (1990). The single-minded gene of Drosophila is required for the expression of genes important for the development of CNS midline cells. Cell 63, 63-75 [DOI] [PubMed] [Google Scholar]
- Oakes S. R., Naylor M. J., Asselin-Labat M. L., Blazek K. D., Gardiner-Garden M., Hilton H. N., Kazlauskas M., Pritchard M. A., Chodosh L. A., Pfeffer P. L., et al. (2008). The Ets transcription factor Elf5 specifies mammary alveolar cell fate. Genes Dev. 22, 581-586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pointud J. C., Mengus G., Brancorsini S., Monaco L., Parvinen M., Sassone-Corsi P., Davidson I. (2003). The intracellular localisation of TAF7L, a paralogue of transcription factor TFIID subunit TAF7, is developmentally regulated during male germ-cell differentiation. J. Cell Sci. 116, 1847-1858 [DOI] [PubMed] [Google Scholar]
- Robinson G. W., McKnight R. A., Smith G. H., Hennighausen L. (1995). Mammary epithelial cells undergo secretory differentiation in cycling virgins but require pregnancy for the establishment of terminal differentiation. Development 121, 2079-2090 [DOI] [PubMed] [Google Scholar]
- Seagroves T. N., Lydon J. P., Hovey R. C., Vonderhaar B. K., Rosen J. M. (2000). C/EBPbeta (CCAAT/enhancer binding protein) controls cell fate determination during mammary gland development. Mol. Endocrinol. 14, 359-368 [DOI] [PubMed] [Google Scholar]
- Shamblott M. J., Bugg E. M., Lawler A. M., Gearhart J. D. (2002). Craniofacial abnormalities resulting from targeted disruption of the murine Sim2 gene. Dev. Dyn. 224, 373-380 [DOI] [PubMed] [Google Scholar]
- Shillingford J. M., Miyoshi K., Robinson G. W., Bierie B., Cao Y., Karin M., Hennighausen L. (2003). Proteotyping of mammary tissue from transgenic and gene knockout mice with immunohistochemical markers: a tool to define developmental lesions. J. Histochem. Cytochem. 51, 555-565 [DOI] [PubMed] [Google Scholar]
- Wilkensen D. G. (1993). In situ hybridization. In Essential Developmental Biology: a Practical Approach (ed. Stern C. D., Holland P. W. H.), pp. 258-263 New York: Oxford University Press; [Google Scholar]
- Zhang D., Penttila T. L., Morris P. L., Roeder R. G. (2001). Cell- and stage-specific high-level expression of TBP-related factor 2 (TRF2) during mouse spermatogenesis. Mech. Dev. 106, 203-205 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.