Abstract
The herpes simplex virus type 1 (HSV-1) UL25 protein is one of seven viral proteins that are required for DNA cleavage and packaging. Together with UL17, UL25 forms part of an elongated molecule referred to as the C-capsid-specific component or CCSC. Five copies of the CCSC are located at each of the capsid vertices on DNA-containing capsids. To study the conformation of UL25 as it is folded on the capsid surface, we identified the sequence recognized by a UL25-specific monoclonal antibody and localized the epitope on the capsid surface by immunogold electron microscopy. The epitope mapped to amino acids 99-111 adjacent to the region of the protein (amino acids 1-50) that is required for capsid binding. In addition, cryo-EM reconstructions of C-capsids in which the green fluorescent protein (GFP) was fused within the N-terminus of UL25 localized the point of contact between UL25 and GFP. The result confirmed the modeled location of the UL25 protein in the CCSC density as the region that is distal to the penton with the N-terminus of UL25 making contact with the triplex one removed from the penton. Immunofluorescence experiments at early times during infection demonstrated that UL25-GFP was present on capsids located within the cytoplasm and adjacent to the nucleus. These results support the view that UL25 is present on incoming capsids with the capsid binding domain of UL25 located on the surface of the mature DNA-containing capsid.
Keywords: herpes simplex virus, capsid, DNA packaging, UL25, Cryo electron microscopy (cryo EM)
Introduction
The herpes simplex virus virion is bounded by a membrane envelope in which a proteinaceous layer called the tegument surrounds the icosahedral capsid that in turn contains the double-stranded linear DNA viral genome. The capsid plays critical roles in virion assembly and maturation, DNA packaging, and delivery of the genome into a host nucleus. Herpesvirus capsids are assembled in the infected cell nucleus where a closed DNA-free procapsid is first formed and later filled with DNA; capsid assembly involves interaction of the major capsid protein with a scaffolding protein and DNA is packaged by an ATP-driven pump located at a specialized vertex of the capsid containing a dodecameric portal structure; and packaging triggers conformational changes in the capsid.
HSV-1 infected cells contain three distinct types of capsids called A-, B- and C-capsids, respectively 1;2. The three are distinguishable morphologically in electron micrographs and can be separated from each other preparatively by sucrose density gradient ultracentrifugation.3. All have the same shell structure but they differ primarily in the material present inside the capsid cavity. C-capsids contain the viral DNA and can mature into infectious virions. They are closely similar, if not identical, in structure and composition to the capsids present in infectious virions 1. A- and B-capsids lack DNA. The B-capsid cavity has a scaffold remnant, VP22a, the cleaved form of the scaffolding protein, while in A-capsids the cavity lacks significant amounts of either DNA or protein.4. Studies with HSV mutants demonstrate that procapsids are able to package DNA and mature into C-capsids that exit the nucleus and are found in infectious virions. A-capsids are considered to be abortive forms that result from failed attempts to package DNA and B-capsids are angularized capsids that never enter the packaging pathway.
Electron microscopy reveals the capsid to be 125 nm in diameter with walls 15 nm thick comprised of tall capsomers arranged on a T=16 icosahedral lattice 5. Three-dimensional reconstructions of the capsid have been calculated from cryo-electron microscopy (cryoEM) data to resolutions of 30–8.4 Å allowing visualization of the major capsid protein, VP5, as pentameric capsomers (pentons) at the icosahedral vertices and hexameric capsomers (hexons) elsewhere; triplex molecules composed of VP19C and VP23 at sites of local 3-fold symmetry; and the small 12 kDa VP26 protein bound to the tips of hexons 6; 7; 8; 9; 10; 11; 12; 13; 14. In addition to these four major structural proteins, the HSV-1 capsid contains a number of minor capsid proteins, including the gene products of UL6, UL17 and UL25, as well as several (UL15, UL28 and UL33) that are only transiently associated with the capsid. All of these minor proteins are required for the processing and packaging of replicated viral DNA into preformed capsid shells 15; 16; 17; 18; 19; 20; 21; 22. With sound architectural details established to better than 1 nm resolution for the four major capsid proteins, structural data for these essential minor components are necessary for understanding their function in the DNA packaging reaction, and for developing new targets for anti-viral therapeutics. In this report, we focus on the UL25 protein.
The HSV-1 UL25 gene encodes a 580 amino acid (62-KDa) protein that acts to maintain stable incorporation of DNA in capsids. In cells infected with a UL25 null virus, DNA appears to enter the capsid and is cleaved by the cleavage/packaging machinery but the DNA is subsequently lost resulting in the accumulation of A-capsids 17. The crystal structure of the UL25 gene product [PDB ID: 2F5U] showed that it contains a flexible N- terminal tail and a more rigid core of packed α-helices 23. UL25 is a capsid-associated protein present on all capsid forms in increasing amounts from procapsid to B-, A-, C-capsids, and virions 24; 25. The UL25 protein forms part of an elongated molecule referred to as the C-capsid-specific component or CCSC 26. The CCSC has only been observed on the outer surface of C-capsids and is proposed to be a heterodimer of UL25 and UL17 with five copies surrounding each of the capsid vertices. Conformational changes of the capsid proteins induced by DNA packaging are proposed to expose UL25 binding sites accounting for the 2-3 fold higher amount of UL25 on C capsids 26. The C-capsid bound UL25 appears to be a multi-functional protein that serves to: stabilize the capsid after DNA is packaged; trigger nuclear egress of the capsid once a stable DNA-containing capsid is formed; anchor VP1/2, the major tegument protein to capsids; and is required early in the virus growth cycle in uncoating of the viral genome 17; 24; 27; 28; 29. Although the location of the CCSC on the capsid has been determined from cryoEM studies, neither the precise location of UL25 in the CCSC nor the regions of UL25 that are required for interaction with its multiple protein partners have been identified. Our progress to date has demonstrated that amino acids 1-50 of the UL25 protein are required for binding of UL25 to capsids and for successful DNA packaging 27.
Here we present the coordinated application of immuno-gold EM and epitope mapping of a UL25-specific monoclonal antibody to examine UL25 binding to the HSV capsid. In addition, we have definitively localized the UL25 protein within the HSV-1 CCSC by cryoEM reconstruction of HSV-1 C-capsids containing a UL25-GFP fusion protein. The results confirm the modeled location of the UL25 protein in the CCSC as the fragment in the lobe that is distal to the icosahedral 5-fold axis with the N-terminus of UL25 making contact with the triplex (Tc triplex) that is one removed from the penton.
Results
The epitope for mAb 25E10 maps to the N-terminal domain of UL25
Recently, we reported that amino acids 1-50 of the UL25 protein are necessary and sufficient for binding of UL25 to capsids and for successful DNA packaging 27. The N-terminal 133 amino acids are missing from the UL25 crystal structure which has precluded direct modeling this region in the cryo EM reconstructions of the CCSC component of HSV-1 C-capsids. In order to determine the position of the N-terminal region of the UL25 protein within the CCSC we labeled the capsid bound UL25 with antibody and gold particles and then determined if this region of the protein was exposed on the surface of the capsid. A UL25 monoclonal antibody was used in this study and we mapped the epitope of this antibody (25E10) to a small region within the N-terminus of the protein. Recombinant proteins expressing segments of the 580 amino acid UL25 protein were created to map the binding region for mAb 25E10 (Fig.1). Regions of the UL25 gene corresponding to amino acids 1-580, 1-560, 51-580, and 1-212 were expressed from the mammalian cell expression vector, pcDNA, while amino acids 1-99, 1-120 and 1-155 were expressed as UL25-TAP fusion proteins from the pcDNA-cTAP vector. The 177 amino acid TAP domain consists of a protein A IgG binding domain and a calmodulin binding peptide. Immunoblot analysis demonstrated that the two C-terminal truncated UL25 constructs expressing amino acids 1-560 (pFH249) and 1-212 (pFH352) along with the N-terminal truncated UL25 construct expressing amino acids 51-580 (pFH380) all reacted with mAb 25E10 (Fig. 1a,c). These results mapped the mAb 25E10 epitope to the region between amino acids 51 and 211 of the UL25 protein. The recombinant UL25-TAP fusion proteins were examined to more precisely map the epitope within this region of UL25. The ability of the plasmids to express all of the recombinant fusion proteins was confirmed by probing the blots with a rabbit anti-mouse HRP conjugated secondary antibody (Fig. 1b). This antibody reacted with the three UL25-TAP fusion proteins but not with the wild type UL25 protein expressed from the pFH283 plasmid. The rabbit antibodies will bind to the protein A domain of the TAP protein. In contrast, goat antibodies do not bind protein A. A goat anti-mouse HRP conjugated secondary antibody was therefore used in order to detect binding of mAb 25E10 to the UL25 portion of TAP fusion proteins (Fig. 1c). The fusion protein that contained UL25 amino acids 1-99 (pFH374) did not react with mAb 25E10, while the constructs expressing amino acids 1-120 (pFH376) and 1-155 (pFH375) specifically reacted with this mAb (Fig.1c). These results further defined the antibody binding domain to amino acids 99-120. In order to further map the epitope for mAb 25E10, a set of overlapping 19-mer peptides were constructed to span the region from amino acids 95-161 of UL25. Fig. 2 shows the sequence of each peptide along with a dot-immunoblot of each peptide. Only one peptide reacted with m 25E10, mapping the epitope to within amino acids 95-113. When the peptide data is combined with the results of the plasmid expression data the critical residues of the epitope can be narrowed down to amino acids 99-111.
Fig. 1.
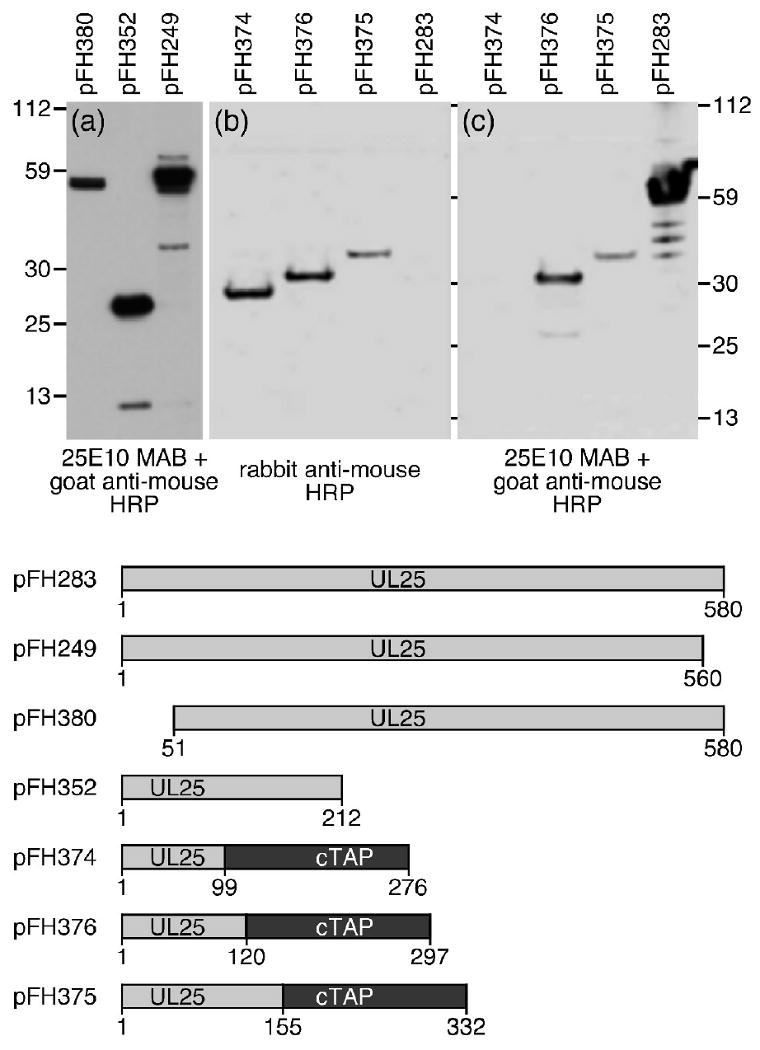
SDS–PAGE and immunoblotting of truncated UL25 proteins or UL25-TAP fusion proteins. Plasmids expressing different UL25 proteins were transfected into Vero cells. Proteins from cell lysates were isolated 48 h post-transfection separated by SDS–PAGE, and then electrophoretically transferred to a nitrocellulose membrane and blotted (a and c) with UL25E10 mAb followed by a goat anti-mouse HRP conjugated antibody or (b) with a rabbit anti-mouse HRP conjugated antibody. Molecular mass standards are shown (kDa) to the left and right.
Fig. 2.
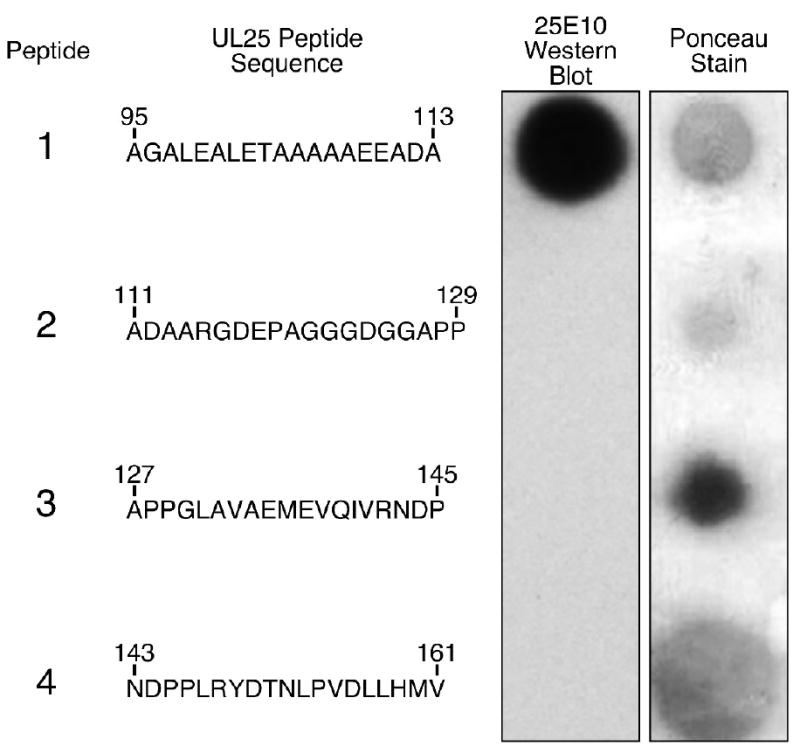
Dot blot analysis of four synthetic UL25 peptides. Peptides were adsorbed onto a nitrocellulose membrane and detected by Ponceau staining or with mAb 25E10. For each peptide, the amino acid sequence and its position in the UL25 molecule are shown.
Immunoelectron microscopy
To observe binding of UL25-specific monoclonal antibody (mAb) 25E10 to the HSV-1 capsid, C-capsids were purified from infected Vero cells by sedimentation on a sucrose gradient. Capsids were incubated with mAb 25E10 followed by a gold bead-conjugated secondary antibody. Preparations were then examined by electron microscopy to identify the location of the gold bead. Micrographs showed that the C-capsids were well labeled with the gold beads (Fig. 3a); most containing more than one bead, and a majority of the beads were located at or near a capsid vertex. In contrast, labeling was not observed with capsids labeled in control experiments performed with nonspecific serum (data not shown) or with B capsids lacking the UL25 protein (Fig. 3b). The location of immunogold label on the capsid was more thoroughly examined by counting capsids that contain a gold particle and individual gold beads were classified as bound at a vertex or bound elsewhere on the capsid. Initial inspection of labeled capsids suggested that a high proportion of gold beads (66%) were located at or near a capsid vertex and 34% elsewhere (Table 1). These results were nearly identical to previous results that we reported using a polyclonal UL25 antibody 24. More importantly, these results demonstrate that he epitope is exposed on the surface of the capsid. These data along with the mapping of the epitope to amino acids 99-111 and the reconstructions of the UL25-GFP capsids localize the capsid binding domain of UL25 on the capsid surface.
Fig. 3.
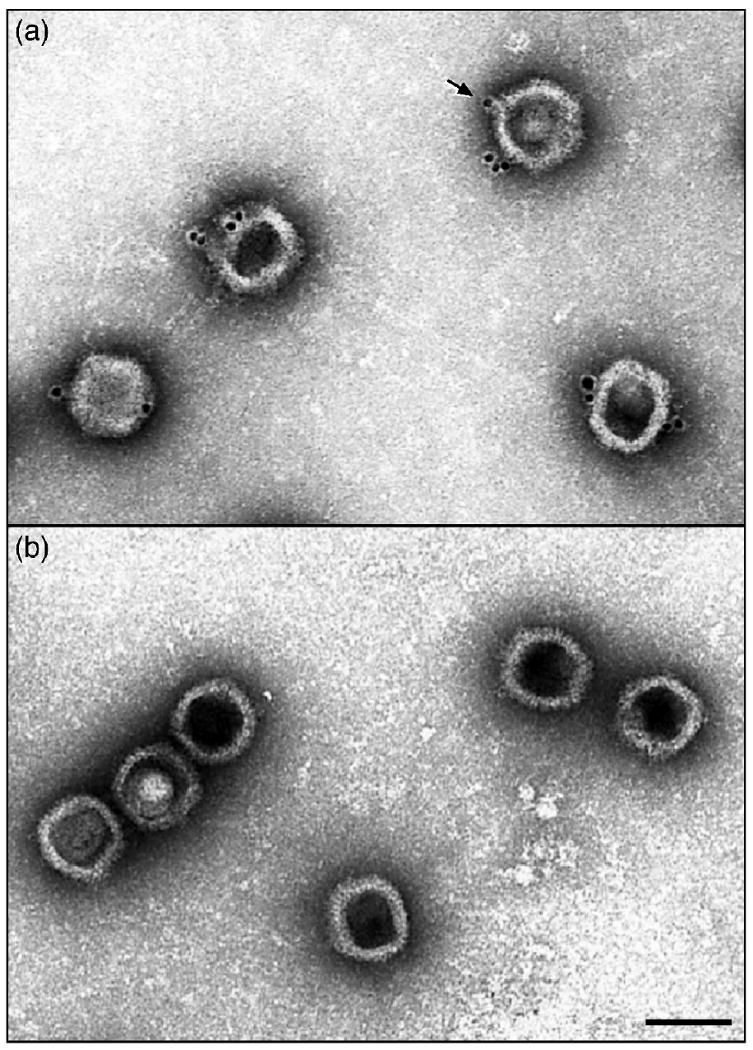
Electron micrographs showing (a) C capsids isolated from KOS infected cells or (b) B capsids isolated from UL25 null virus infected cells after staining with the UL25 mAb 25E10 followed by an anti-antibody conjugated to gold beads. Note that the gold beads are found only on the C capsids and appear to be located at the capsid vertices (one is indicated by an arrow) and the majority of the C capsids have labels at more than one site. Bar = 1000Å.
Table 1. Immunoelectron microscopic localization of the UL25 N-terminal epitope on the surface of HSV-1 capsids.
| Gold Bead Label | Wild Type C-capsids | UL25 Null B-capsids1 |
|---|---|---|
| Label2 | 317 (59.5%) | 8 (0.2%) |
| No Label | 216 (40.5%) | 406 (99.8%) |
| Capsid Location | ||
| Vertex | 210 (66.3%) | ND3 |
| Other location | 107 (33.7%) | ND |
Capsids isolated from cells infected with KUL25NS3.
One or more gold beads located anywhere on the capsid.
Not determined.
Analysis of HSV UL25-GFP virus
The HSV-1 mutant vUL25-GFP encodes UL25-GFP fusion protein where the 726 bp GFP gene was inserted in-frame between codons 50 and 51 of the UL25 gene 27 (Fig. 4a). The UL25-GFP gene was introduced into the viral genome through genetic manipulation of an HSV-1 (KOS) genome maintained in a recombinant bacterial artificial chromosome (BAC). The UL25-GFP BAC was transfected onto UL25-complementing 8-1 cells, and the recovered virus was plaque purified on Vero cell and titrated on Vero and 8-1 cells. The vUL25-GFP mutant yielded identical titers on Vero cells and 8-1 cells 27. In one-step growth analyses after infection at a MOI of 5, maximum titers of vUL25-GFP on Vero cells were only slightly lower than those of wild type HSV-1 KOS; indicating expression of the UL25-GFP fusion protein did not significantly alter virus replication (Fig. 4b). Intranuclear capsids were purified from Vero cells infected with vUL25-GFP and the presence of the UL25 protein in A, B, and C capsids was examined by Western blot analysis (Fig. 5). Identical blots were probed with either the UL25 mAb 25E10 or with a rabbit GFP polyclonal antibody (Fig. 5). The UL25-GFP fusion protein which is approximately 25 kDa larger then the wild type UL25 protein was detected in A, B, and C capsids with both antibodies while the UL25 present in C capsids isolated from KOS infected cells was only detected with the 25E10 mAb. Using the HSV major capsid protein (VP5) as a loading control, the amount of UL25 was observed to be higher in C-capsids as compared to A and B capsids (Fig. 5). These results are in agreement with previous studies indicating that the UL25 content is higher in C capsids than in A and B capsids 24; 26.
Fig. 4.
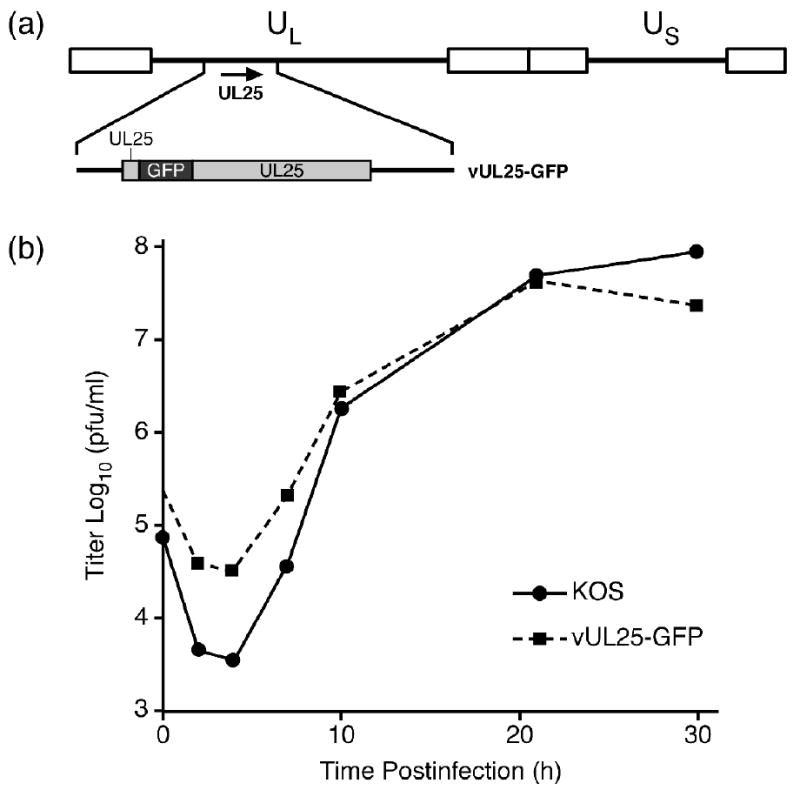
Single-step growth of KOS and vUL25-GFP. (a) The HSV-1 genome is represented at the top; UL and US refer to the long and short unique region sequences, respectively. The region of the HSV-1 genome that contains the UL25 gene is expanded below. The UL25-GFP fusion protein that is expressed from the vUL25-GFP virus is shown. (b) Replicate cultures of Vero cells were infected with KOS or vUL25-GFP at an MOI of 5. The cultures were harvested at the indicated times postinfection and freeze-thawed three times, and the yield of virus at each time point was determined by plaque titer determination on Vero cells.
Fig. 5.
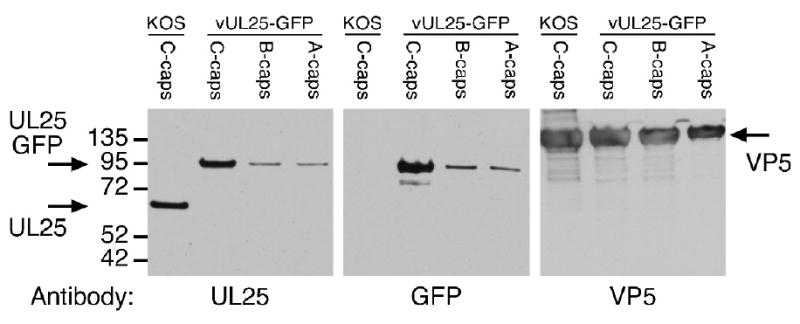
UL25 content in capsids isolated from KOS- or vUL25-GFP-infected Vero cells. Equivalent amounts of KOS C-capsids or vUL25-GFP C-, B-, or A-capsids were separated by SDS-PAGE, and the gels was immunoblotted with either the UL25 antibody 25E10, the GFP antibody A11122, or the VP5 antibody NC1. Molecular mass standards are visible on the left (kDa).
The UL25-GFP protein is present on incoming capsids
Immunofluorescence from the UL25-GFP protein was examined to determine whether this protein was present on incoming capsids. To verify that the GFP signal was associated with HSV capsids, cells were infected with vUL25-GFP and at 3 h postinfection, fixed, permeabilized and probed for the triplex protein VP23 (Fig. 6). The UL25-GFP signal was found to colocalize with the triplex protein signal and was observed with cytoplasmic capsids and also with capsids at the nuclear surface (Fig. 6, arrows). The coincidence of the two fluorescent signals indicated that the UL25GFP protein was bound to the incoming capsids.
Fig. 6.
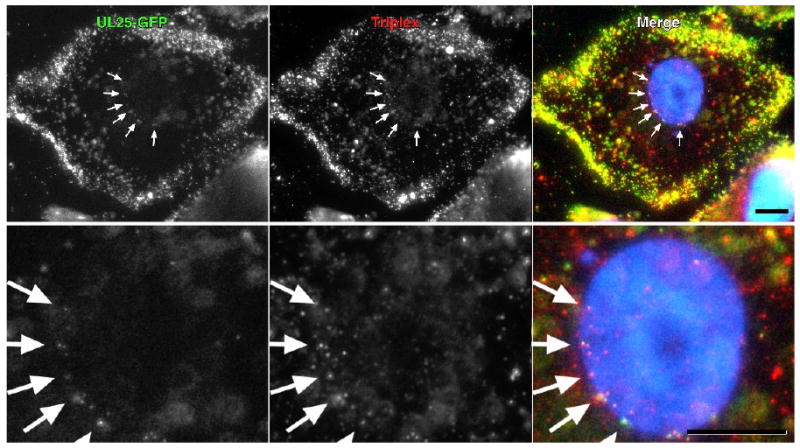
Colocalization of UL25-GFP and capsid triplex protein VP23 by fluorescence microscopy. Vero cells were infected for 3 h with vUL25-GFP and the colocalization of the capsid associated UL25-GFP with the capsid triplex protein VP23 was examined in cells that were fixed and immunostained for VP23 (red). Nuclei were DAPI stained. Note that the UL25-GFP and VP23 protein labels occur coincidently in the cytoplasm and at the nuclear surface (arrows). Bar = 10 um. A close-up image of the nuclei for each of the upper panels is shown in the lower panel.
Structural analysis of HSV UL25-GFP capsids
The location of the UL25 protein has previously been assigned in cryoEM-based reconstruction to a region termed the CCSC that is arrayed around pentons on C-capsids of HSV-1 26. This assignment was based on SDS-PAGE and mass spectrometry analysis of wild-type and UL25 null capsids which demonstrated that the presence of the CCSC density was directly correlated with detection of the UL25 protein. In addition, fitting of the UL25 crystal structure within the CCSC density demonstrated that the best fit for UL25 was in the region of the CCSC distal to the penton. With our UL25-GFP capsids we are in a position to conclusively localize UL25 by visualizing the GFP label in cryoEM reconstructions, and comparing the structure with capsids containing the wild type UL25 protein and capsids lacking UL25.
We calculated density maps of HSV-1 capsids with the UL25-GFP protein to a resolution estimated at 13.7Å for C-capsids and to 16.5Å resolution for B-capsids (see Materials and Methods). We also used the HSV-1 C-capsid map with wild type UL25 for comparison 26. Central sections through the capsids and exterior surface views are shown in Fig. 7. Both C-capsid reconstructions (UL25-GFP and w.t. UL25) show density ascribed to the CCSC that appears to make contact with the two triplexes closest to the penton (triplexes Ta and Tc, in the nomenclature of Zhou et al. see inset Fig.7), and contacts one of two adjacent hexons (Figs. 7a,b & 8a,b). In contrast, CCSC density is absent from the HSV-1 B-capsid reconstruction (Fig. 7c). Compared to the wild-type UL25 C-capsid reconstruction, the UL25-GFP C-capsid shows extra density just above the end of the CCSC distal to the penton. We interpret the extra density to the GFP insertion in UL25 between residues 50 and 51 (Fig. 7a). This provides a definitive identification of UL25 with the CCSC and its location in the distal region.
Figure 7.
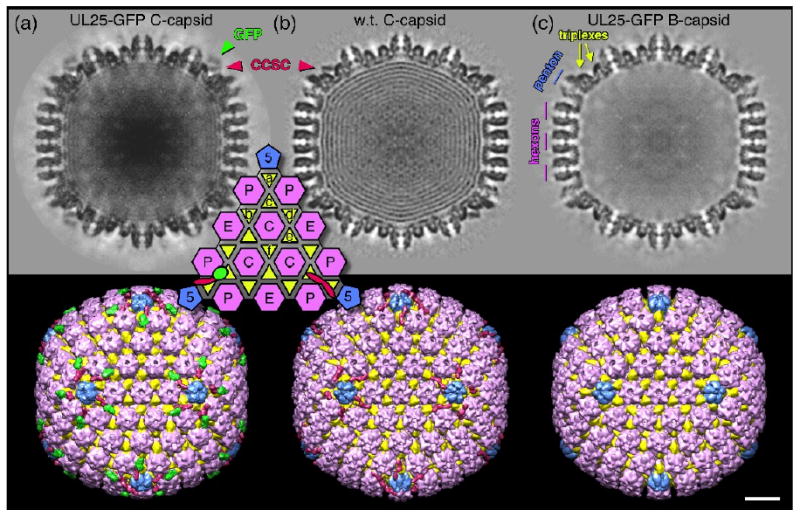
Density maps calculated from (a) C-capsids with UL25-GFP, (b) C-capsids with w.t. UL25 26, and (c) B-capsids that appear to lack the CCSC density as well as packaged DNA. Central sections are shown at top, and surface views along the 2-fold axis below. Capsomers, triplexes and the CCSC molecules are colored as indicated although the exact boundaries are not known in detail. The inset shows the positions of the four kinds of capsomer and six kinds of triplex on one triangular facet. The CCSC density common to both C-capsid samples (marked in red in a, b, and the inset) binds both the triplex (Ta) adjacent to the penton and the next closest (Tc) that are indicated in (c), as well as appearing to contact one of the two adjacent hexons. We attribute the additional density above the CCSC (green in a and the inset lower left) to the GFP protein inserted between residues 50 and 51 of UL25. Bar = 200Å.
Fig. 8.
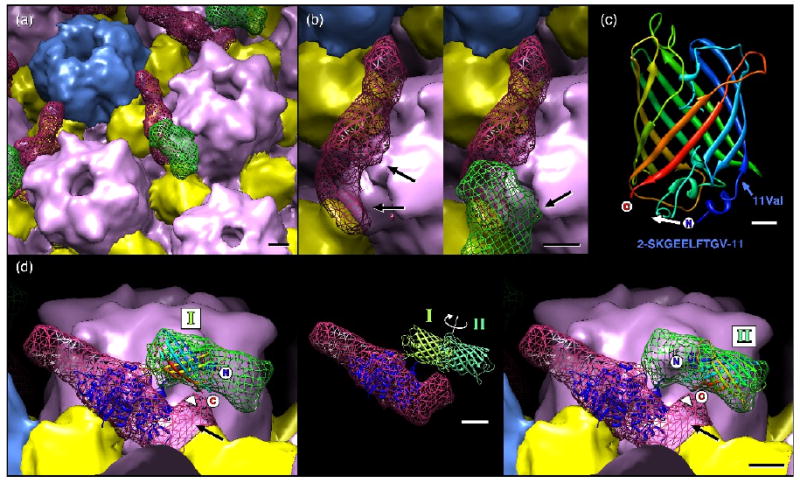
Close-up views of the UL25-GFP density. (a) Subunit boundaries have been estimated and colored as for Figure 7 with UL25 and GFP densities shown as a red and a green mesh, respectively. Bar = 20Å. (b) An alternative view revealing contacts between a hexon and the CCSC density (with GFP removed to aid visibility), at left, and the GFP density, at right. Bar = 20Å. (c) Ribbons representation of the GFP atomic model with termini and amino acid 11 (valine) marked, along with the sequence of GFP amino acids 2-11 (below). When inserted into UL25, the N- and C-terminus of GFP may be closer together, possibly by refolding the N-terminal 11 amino acids moving the N-terminus as indicated (white arrow). Bar = 5Å. (d) Atomic models for UL25 (PDB ID: 2F5U – dark blue ribbons) and GFP (PDB ID: 1EMA rainbow and green ribbons) positioned within the cryoEM density. The UL25 atomic model (residues 134-580) is positioned so that the missing N-terminal 133 residues would occupy the distal region (black arrow) where contact with GFP appears most likely. The GFP model may be placed in two non-overlapping orientations to fill the green mesh, as indicated at left (position I) and at right (position II). These locations place the termini midway along the GFP density that closely approaches the CCSC density (arrowhead). Bar = 20Å. The inset shows the two GFP positions, represented by light and dark green models, superimposed on the CCSC density. The C-termini of the GFP are placed in the GFP density where it most closely approaches the UL25 density (arrowhead).
A closer examination of the UL25-GFP C-capsid density map suggests two likely locations for the GFP insertion (Fig. 8). Low occupancy of the CCSC and GFP densities is the primary cause of uncertainty, but nonetheless the positions of UL25 residues 50-51 are restricted to a small area of the surface. There is, however, no atomic model of the 133 N-terminal portion of UL25 that may be used to constrain any fit of UL25 in the relatively weak CCSC volume. Further, the UL25 crystal structure lacks surface loops useful for guiding any fitting procedure, including the following residues: 249-254 (6 amino acids); 335-345 (11); 417-425 (9); 479-483 (5); 511-513 (3), as well as the C-terminal tri-peptide 578-580. Nonetheless, based on the shape of the CCSC density, the incomplete atomic model of UL25 and the positions revealed for residues 50-51, we propose a likely location of UL25 in the cryoEM density map, as shown in Fig. 8, where the missing N-terminal region is in the CCSC volume most distal to the adjacent penton (black arrow in Fig. 8d) where it makes contact with the Tc triplex. This largely confirms the previously proposed location for UL25 within the CCSC 26.
Likewise, an atomic model of GFP may be positioned into the density ascribed to it. This region of the cryoEM density is sufficient to accommodate two copies of GFP, and although weaker than the CCSC volume, it is rather well defined. Since we do not observe a smeared volume in the reconstruction, which would represent a range of positions adopted by GFP, we infer that the well defined density instead represents an average between sites where GFP is in one of only two positions, and these are related by a ∼180° twist about a central point where GFP inserts into UL25, as indicted in Fig. 8d. The termini of GFP must be in close proximity to make the insertion, and examination of the GFP atomic model shows the termini to be just 19Å apart and with several turns at the N-terminus that could well be folded differently in the UL25-GFP protein (Fig. 8c). This model places the UL25 insertion residues, 50 and 51, at the surface of an unoccupied region of CCSC that we assign to the 133 N-terminal residues of UL25 missing from the crystal structure. Since flexibility at the GFP termini is necessary to accommodate the insertion, the constraints limiting GFP to two positions may be due to non-specific binding to the capsid and/or to some specific low-energy arrangement of the residues at the point of insertion.
Discussion
The herpesvirus capsid is relatively large and complex, constructed from a number of different proteins following an elaborate assembly pathway that is not understood in detail 1; 2. Electron microscopy studies has located the major components of the capsid including; the major capsid protein VP5, the two triplex proteins VP19C and VP23, the hexon decoration protein VP26, and the portal UL6. Unfortunately the resolutions so far obtained from these EM studies have not yet allowed subunit boundaries to be delimited, much less the complete folds of any of these subunits. Similarly, while crystallographic analysis has revealed atomic detail for some portions of VP5, no high resolution data are available on subunit interfaces from which the processes of assembly and maturation of the capsid could be understood 30. Functional studies have highlighted the essential roles of minor capsid proteins, such as UL25 for which a substantial atomic model has been determined for the C-terminal residues 134-580 17; 23; 27. However, the capsid-binding domain (residues 1-50) that we have previously determined, and confirm here, is not included in that model. Nonetheless, because of its requirement for several steps in the infectious cycle including; (i) stabilizing the capsid on completion of DNA packaging signaling, (ii) nuclear exit, (iii) anchoring the major tegument protein to capsids, and (iv) uncoating of the viral genome, UL25 is an interesting subject for structural and functional studies as well as a potential new target for interfering with virus replication.
In this study, we described several approaches to extend our understanding of the location and structure of HSV-1 UL25. Immuno-gold EM analysis and epitope mapping of a UL25-specific monoclonal antibody demonstrated the binding of UL25 to the vertices of the HSV-1 C-capsid, in agreement with previous immuno-electron microscopy data 24, while labeling was not observed with control B capsids that lack UL25. The epitope for the UL25 MAB was mapped to UL25 amino acids 99-111, which are located adjacent to the capsid binding region of the protein (amino acids 1-50) 27. The fact that an antibody can be used to bind a gold-labeled secondary antibody to intact capsids clearly demonstrates that the capsid binding domain of UL25 is on the surface of the mature DNA-containing capsids.
Utilizing cryoEM and image reconstruction we precisely localized the UL25 protein on the surface of the HSV-1 C-capsids containing a UL25-GFP fusion protein. The GFP density is clearly visible in the density map, despite the ∼50% occupancy of UL25-GFP on the capsid, and serves as a definitive marker for the location of UL25 as well as to pinpoint the location of the UL25 residues 50 and 51 between which GFP was inserted. The low occupancy of the CCSC containing the wild type UL25 protein has also been reported in a previous cryoEM study26. Our EM results confirm the modeled location of the UL25 protein as a component of the five CCSC's that surround the pentons located at each of the capsid vertices. The earlier study26 comparing wild type C capsids to B capsids isolated from a UL25 null virus suggested that the UL25 protein was situated in the lobe of the CCSC that is distal to the icosahedral 5-fold axis. We also positioned the atomic model of UL25 in the CCSC density and similarly found it to be most likely located in the volume distal to the adjacent penton. However, since the UL25 model is missing the 133 N-terminal amino acids, and the CCSC density is not fully occupied in the cryoEM density map, precise fitting of the model is not yet possible.
The CCSC lobe attributed to UL25 appears to be anchored to both the penton-proximal triplex Ta and the adjacent next-most proximal triplex Tc and also contacts one of the neighboring hexon subunits. The region we attribute to the 133-residue N-terminal domain of UL25 appears likely to bind the Tc triplex, and this may well involve the 50 N-terminal portion of UL25 that is essential for capsid binding. UL25 further appears to provide part or all of the support for the UL17 protein, the second viral protein that is presumed to be part of the CCSC structure occupying the region nearest to the penton. Thus we conclude that UL25 makes contact with one or both subunits of two differently positioned triplex molecules, at least one VP5 subunit from a hexon, and UL17, suggesting that its essential role in capsid maturation may be based on this multi-subunit contact.
The recent report of an interaction between UL25 and the HSV portal protein UL6 31 suggests that the UL25 protein is present on all twelve of the capsid vertices including the portal vertex – the site where DNA enters and exits the capsid. The portal is not visualized in our cryoEM reconstructions that have icosahedral symmetry imposed because the single portal vertex is averaged with the eleven other penton vertices. Since the capsid subunits UL25 contacts around pentons are also in place at the portal vertex, we may expect that UL25 binds in a similar fashion precluding direct contact with the encapsidated dsDNA. The role of UL25 in retaining the viral chromosome may then hinge on it providing a support for UL17, which appears more likely to make direct contact with the portal. Visualizing the portal vertex separately from the pentons will be addressed in future by collecting sufficient capsid images to offset the reduction in symmetry that is entailed.
The UL25-GFP was demonstrated to be present on incoming capsids since its fluorescence signal co-localized with the red immunostained triplex protein, VP23 (Fig. 6). The capsid bound UL25 appears to be a multi-functional protein that serves several functions during the virus life cycle, suggesting that interfering with the UL25 capsid interaction may be a unique mechanism for blocking infection.
An interesting aspect of the UL25-GFP mutant is that the GFP insertion between residues 50 and 51 was not detrimental to growth. Single-step growth studies demonstrated that the UL25-GFP virus replicated similar to wild type HSV-1. UL25 is an essential protein that is required to retain viral DNA in mature capsids. Since the GFP fluorescence was readily observed, the insertion of GFP into the UL25 protein clearly did not alter the function of either protein. Comparing reconstructions of capsids containing either the wild type UL25 protein or the UL25-GFP fusion protein showed that the occupancy of UL25 as part of the CCSC structure was similar. These results suggest that there was no change in the binding interface of UL25 to the rest of the HSV-1 capsid, nor to UL17 or any other of its putative partners. Further, the morphology of the CCSC molecule containing either form of the UL25 protein was similar, with the difference limited to addition of GFP density above the CCSC in the UL25-GFP map where it does not appear to alter the capsid during assembly, DNA packaging, tegumentation or envelopment. Finally, the point of insertion in the UL25 protein must be flexible enough to accommodate the N- and C-termini of the GFP label, which are significantly more distant from each other than the consecutive residues 50 and 51 of UL25.
Since the GFP domain of the fusion protein appears to be completely exposed on the surface of the capsid in the cryo-EM reconstructions (Figs. 7, 8) it may be possible to use this insertion site for studying the structure of other viral and cellular proteins. By exploiting the icosahedral symmetry of the capsid to which the UL25 fusion protein is bound the structure of the exposed fusion protein could be determined by conventional cryoEM reconstruction of the capsids. Currently the occupancy of UL25 on HSV-1 C-capsid preparations is somewhat limited, impacting interpretation of this part of the density maps, although cryoEM reconstruction of the pseudorabies virus C capsid has proven more successful in our hands both in resolution (10 Å) and in UL25 occupancy (100%) (Homa and Conway unpublished data). Phage display technology, with the coat or tail proteins of bacteriophages, has been used to display both small peptides and large proteins, and then employed as a tool to study protein interactions and as a means to present antigens 32. In a similar manner the UL25 protein may provide a platform for the surface display of proteins or peptides on HSV-1 capsids.
Further structural work is also indicated on orienting the UL25 structure within the CCSC density, such as by visualizing GFP inserted into regions of the UL25 protein that are part of the crystal structure, i.e. the C-terminal residues from 134-580. Localizing surface residues of the atomic model would constrain the position and orientation of the model within the cryoEM envelope and would additionally reveal the regions of UL25 that interact with other capsid subunits such as VP5 and the triplexes as well as the other putative CCSC component, UL17. Previous studies from our lab using transposon mutagenesis identified several regions of the UL25 protein, in addition to residues 50-51, where amino acid insertions did not affect the function of the protein 27. Labeling with antibodies, gold particles, and terminal sequence extensions to determine positions of specific amino acids or peptides has been successful in a number of cases, including our experiences with the hepatitis B virus capsid 33; 34; 35.
Materials and Methods
Cells, viruses and antibodies
African green monkey kidney cells (Vero; American Type Culture Collection, Rockville, MD.) and UL25 transformed 8-1 cells were propagated as previously described 17. HSV wild type KOS, UL25 null viruses, KUL25NS and vΔUL25, and the vUL25-GFP virus have been previously described 17; 27. Western blots were done using rabbit polyclonal antibodies NC1 36 to detect the HSV-1 major capsid protein VP5, rabbit polyclonal antibody A11122 from Molecular Probes (Eugene, OR) to detect the GFP protein and mouse monoclonal antibody 25E10 to detect the UL25 protein 27.
Capsid purification
Vero cells (1.5 × 108) were infected overnight (18 h at 37°C) at an MOI of 5 PFU/cell. Infected cells were harvested, rinsed with PBS and resuspended in 20mM Tris pH 7.5 plus protease inhibitors (Roche), adjusted to 1% Triton X-100 and incubated for 30 minutes on ice. The resulting nuclei were harvested by low-speed centrifugation, resuspended in 10 ml TNE (500mM NaCl, 10mM Tris, 1mM EDTA, pH 7.5), and then sonicated to lyse the nuclei. The nuclear lysate was adjusted to 20 mM MgCl2, and incubated with DNaseI (100 μg/mL) at room temperature for 30 min. The lysate was then cleared by low speed centrifugation and the resulting supernatant was layered on top of a 5-ml cushion of 35% sucrose (SW28 rotor; 23,000 rpm for 1 h). The pellets were suspended in 3 ml TNE and adjusted to 1 mM dithiothreitol and capsids were separated by centrifugation on 20-50% sucrose (in TNE) gradients (SW41 rotor at 24000 rpm for 1hr). The position of A, B, or C capsids were observed as light scattering bands with A capsids being found highest (least dense) on the gradient and C capsids found lowest (dense fractions) on the gradients. The different capsid fractions can also be identified based on the presence or absence of the scaffold protein, VP22a, since only B capsids contain the scaffold protein. The fractions were collected using a Beckman Fraction Recovery system (Beckman catalog # 34890). The apparatus has a mechanism to puncture the bottoms of the tubes and fractions are collected from the bottom to top. Fractions (0.75ml) were collected, and protein was precipitated by adding an equal volume of 16% trichloroacetic acid. Pellets were resuspended in 35 μL of 2x PAGE loading buffer (Invitrogen) supplemented with 0.4M Tris-base. Gradient fractions were run on a 5-12% SDS-PAGE and the gels were either stained with Imperial Blue (Pierce) to visualize capsid proteins or analyzed by immunoblotting.
Immunoblotting
Protein extracts were separated on a 5-12% SDS-polyacrylamide gel, and proteins were transferred to nitrocellulose. The nitrocellulose was washed twice in TBS and blocked overnight in TBS-T (20mM Tris, 0.5M NaCl pH 7.5 plus 0.5% Tween-20) supplemented with 10% nonfat dry milk. UL25 monoclonal antibody, 25E10, and the VP5 rabbit polyclonal antibody, NC1, were diluted to 1:5000 in blocking buffer; the GFP polyclonal antibody A11122 was diluted 1:1000. The diluted antibodies were reacted with the blocked nitrocellulose for 2 h at room temperature, washed, and reacted with horseradish peroxidase-conjugated anti-goat or anti-mouse IgG diluted at 1:5000 dilution in TBS-T plus 10% nonfat dry milk. The bound immunoglobulins were revealed by enhanced chemiluminescence (Pierce ECL kit).
Capsid labeling with UL25 monoclonal antibody 25E10
Immunoelectron microscopy was performed with capsids adsorbed to carbon-Formvar-coated copper grids. The procedure used for labeling was the same as that described previously 37 except that (i) the primary antibody was the UL25 mAb 25E10 and (ii) the secondary antibody was conjugated to 10-nm-diameter gold beads (EY Laboratories, San Mateo, CA). Images were recorded on film as described previously 37, scanned on a flatbed scanner and displayed for counting with Photoshop CS3.
Immunofluorescence
vUL25-GFP was added to Vero cells and allowed to attach for 1hr at 4°C. The medium was then aspirated to remove any unattached virus. The medium was replaced with fresh medium and cells were incubated at 37°C with 7.5% CO2 for 3hrs. Cells were then washed one time with 1X PBS and fixed by submersion in 4% paraformaldehyde for 5 minutes. Cells were washed three times with 1X PBS and permeabilized by submersion in ice-cold methanol for 4 minutes. Cells were then washed once in 1X PBS followed by blocking with 5% goat serum in water at room temperature for 1h. Primary VP23 monoclonal antibody 1D2 was diluted 1:500 in 5% goat serum and added to cells for 1hr at room temperature. Cells were washed three times in 1X PBS. Secondary antibody (Alexa conjugated goat anti-mouse (Molecular Probes)) was added at a dilution of 1:2000 for 1hr at room temperature. Cells were washed three times in 1X PBS and DAPI stain (0.5μg/ml in 150mM Tris-Cl pH 7.5) was added for 5 minutes. Cells were washed three times in 1X PBS and viewed with a Nikon Eclipse TE2000-E fluorescence microscope using a 63x oil immersion lens (Improvision Open Lab software).
Cryo-electron microscopy and image reconstructions
Purified capsid samples were placed on regular lacey carbon grids for blotting and plunge-freezing in an FEI Vitrobot (FEI, Hillsboro, OR). The cryogen used was a 50:50 mix of ethane and propane, which maintains liquid nitrogen temperature without the cryogen freezing as may happen with ethane alone 38. Grids were transferred to a Gatan 626 cryoholder (Gatan, Pleasanton, CA) for entry into an FEI Tecnai TF20 electron microscope, maintaining the liquid nitrogen temperature throughout. Fields of capsids were imaged at 200kV and 29,000x magnification (30,000x calibrated) onto film using standard low-dose techniques. Micrographs were developed with full-strength Kodak D19 for 12 minutes and digitized on a Nikon Super CoolScan 9000 scanner at 6.35Å/pixel, yielding an effective sampling at the specimen of 2.12 Ångstroms/pixel. Images of C-capsids and B-capsids were selected separately, assessed for contrast transfer function parameters (defocus and astigmatism) by micrograph 39, and each population was submitted independently to the Random Model method 40 for arriving at a first 3d model in each case. The first models started the orientation search and refinement procedure using AUTO3DEM 41. Details of the data sets are listed in Table 1. UCSF Chimera 42 was used to visualize density maps and to position the atomic models of UL25 (residues 134-580, PDB ID: 2F5U) 23 and green fluorescent protein (PDB ID: 1EMA) 43. Occupancy of the CCSC volume was estimated at ∼50% by comparison of the peak density within it to peak density in the capsid (100% occupancy) and background (0% occupancy). The HSV-1 C-capsid density map with w.t. UL25 26 is available in the Macromolecular Structure Database of the European Bioinformatics Institute with accession code EMD-1354 at the following link: http://www.ebi.ac.uk/pdbe-srv/emsearch/atlas/1354_summary.html
Table 2. Details of CryoEM reconstructions of HSV-1 capsids.
| Sample | Resolution1 (Å) | Particle Count2 | Micrograph Count3 |
|---|---|---|---|
| UL25-GFP C-capsids |
13.7 | 10583/14515 | 112 |
| UL25-GFP B-capsids |
16.5 | 12721/15333 | 47 |
| UL25 C-capsids4 |
19.9 | 643 pairs | - |
Resolution is estimated by Fourier Shell Correlation 41 calculated over the radial zone occupied by the capsid density (∼ 430 to 670Å) and dropping to a correlation coefficient of 0.5.
Number of particle images used in the final reconstruction compared to the number of particles collected in total from scanned electron micrographs.
Number of scanned electron micrographs.
The UL25 C-capsids density map is from the EMI database (entry 1354), which was calculated from pairs of images taken at different defocus settings 19. The number of micrographs was not reported.
Acknowledgments
We thank D. Anjum and A. Makhov for their excellent support in electron microscopy and J. Huffman for technical support. This work was supported by NIH awards AI060836 (F.L.H.), AI041644 (J.C.B.), training grant T32 AI049820 (S.K.C.), and the Commonwealth of Pennsylvania SAP 4100031302 (J.F.C.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Homa FL, Brown JC. Capsid assembly and DNA packaging in herpes simplex virus. Rev Med Virol. 1997;7:107–122. doi: 10.1002/(sici)1099-1654(199707)7:2<107::aid-rmv191>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 2.Brown J, McVoy MA, Homa FL. Packaging DNA into herpesvirus capsids. In: Bogner E, H A, editors. Structure-function relationships of human pathogenic viruses. Kluwer Acacdemic/Plenum Publishers; New York: 2002. pp. 111–155. [Google Scholar]
- 3.Gibson W, Roizman B. Proteins specified by herpes simplex virus. 8. Characterization and composition of multiple capsid forms of subtypes 1 and 2. J Virol. 1972;10:1044–52. doi: 10.1128/jvi.10.5.1044-1052.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Newcomb WW, Brown JC. Structure of the herpes simplex virus capsid: effects of extraction with guanidine hydrochloride and partial reconstitution of extracted capsids. J Virol. 1991;65:613–20. doi: 10.1128/jvi.65.2.613-620.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caspar DL, Klug A. Physical principles in the construction of regular viruses. Cold Spring Harb Symp Quant Biol. 1962;27:1–24. doi: 10.1101/sqb.1962.027.001.005. [DOI] [PubMed] [Google Scholar]
- 6.Newcomb WW, Thomsen DR, Homa FL, Brown JC. Assembly of the herpes simplex virus capsid: identification of soluble scaffold-portal complexes and their role in formation of portal-containing capsids. J Virol. 2003;77:9862–71. doi: 10.1128/JVI.77.18.9862-9871.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spencer JV, Newcomb WW, Thomsen DR, Homa FL, Brown JC. Assembly of the herpes simplex virus capsid: preformed triplexes bind to the nascent capsid. J Virol. 1998;72:3944–51. doi: 10.1128/jvi.72.5.3944-3951.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grunewald K, Desai P, Winkler DC, Heymann JB, Belnap DM, Baumeister W, Steven AC. Three-dimensional structure of herpes simplex virus from cryo-electron tomography. Science. 2003;302:1396–8. doi: 10.1126/science.1090284. [DOI] [PubMed] [Google Scholar]
- 9.Booy FP, Trus BL, Newcomb WW, Brown JC, Conway JF, Steven AC. Finding a needle in a haystack: detection of a small protein (the 12-kDa VP26) in a large complex (the 200-MDa capsid of herpes simplex virus) Proc Natl Acad Sci U S A. 1994;91:5652–6. doi: 10.1073/pnas.91.12.5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Trus BL, Homa FL, Booy FP, Newcomb WW, Thomsen DR, Cheng N, Brown JC, Steven AC. Herpes simplex virus capsids assembled in insect cells infected with recombinant baculoviruses: structural authenticity and localization of VP26. J Virol. 1995;69:7362–6. doi: 10.1128/jvi.69.11.7362-7366.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou ZH, Dougherty M, Jakana J, He J, Rixon FJ, Chiu W. Seeing the herpesvirus capsid at 8.5 A. Science. 2000;288:877–80. doi: 10.1126/science.288.5467.877. [DOI] [PubMed] [Google Scholar]
- 12.Zhou ZH, He J, Jakana J, Tatman JD, Rixon FJ, Chiu W. Assembly of VP26 in herpes simplex virus-1 inferred from structures of wild-type and recombinant capsids. Nat Struct Biol. 1995;2:1026–30. doi: 10.1038/nsb1195-1026. [DOI] [PubMed] [Google Scholar]
- 13.Zhou ZH, Prasad BV, Jakana J, Rixon FJ, Chiu W. Protein subunit structures in the herpes simplex virus A-capsid determined from 400 kV spot-scan electron cryomicroscopy. J Mol Biol. 1994;242:456–69. doi: 10.1006/jmbi.1994.1594. [DOI] [PubMed] [Google Scholar]
- 14.Newcomb WW, Trus BL, Booy FP, Steven AC, Wall JS, Brown JC. Structure of the herpes simplex virus capsid. Molecular composition of the pentons and the triplexes. J Mol Biol. 1993;232:499–511. doi: 10.1006/jmbi.1993.1406. [DOI] [PubMed] [Google Scholar]
- 15.Adelman K, Salmon B, Baines JD. Herpes simplex virus DNA packaging sequences adopt novel structures that are specifically recognized by a component of the cleavage and packaging machinery. Proc Natl Acad Sci U S A. 2001;98:3086–91. doi: 10.1073/pnas.061555698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.al-Kobaisi MF, Rixon FJ, McDougall I, Preston VG. The herpes simplex virus UL33 gene product is required for the assembly of full capsids. Virology. 1991;180:380–8. doi: 10.1016/0042-6822(91)90043-b. [DOI] [PubMed] [Google Scholar]
- 17.McNab AR, Desai P, Person S, Roof LL, Thomsen DR, Newcomb WW, Brown JC, Homa FL. The product of the herpes simplex virus type 1 UL25 gene is required for encapsidation but not for cleavage of replicated viral DNA. J Virol. 1998;72:1060–70. doi: 10.1128/jvi.72.2.1060-1070.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Patel AH, MacLean JB. The product of the UL6 gene of herpes simplex virus type 1 is associated with virus capsids. Virology. 1995;206:465–78. doi: 10.1016/s0042-6822(95)80062-x. [DOI] [PubMed] [Google Scholar]
- 19.Salmon B, Cunningham C, Davison AJ, Harris WJ, Baines JD. The herpes simplex virus type 1 U(L)17 gene encodes virion tegument proteins that are required for cleavage and packaging of viral DNA. J Virol. 1998;72:3779–88. doi: 10.1128/jvi.72.5.3779-3788.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tengelsen LA, Pederson NE, Shaver PR, Wathen MW, Homa FL. Herpes simplex virus type 1 DNA cleavage and encapsidation require the product of the UL28 gene: isolation and characterization of two UL28 deletion mutants. J Virol. 1993;67:3470–80. doi: 10.1128/jvi.67.6.3470-3480.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lamberti C, Weller SK. The herpes simplex virus type 1 UL6 protein is essential for cleavage and packaging but not for genomic inversion. Virology. 1996;226:403–7. doi: 10.1006/viro.1996.0668. [DOI] [PubMed] [Google Scholar]
- 22.Lamberti C, Weller SK. The herpes simplex virus type 1 cleavage/packaging protein, UL32, is involved in efficient localization of capsids to replication compartments. J Virol. 1998;72:2463–73. doi: 10.1128/jvi.72.3.2463-2473.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bowman BR, Welschhans RL, Jayaram H, Stow ND, Preston VG, Quiocho FA. Structural characterization of the UL25 DNA-packaging protein from herpes simplex virus type 1. J Virol. 2006;80:2309–17. doi: 10.1128/JVI.80.5.2309-2317.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Newcomb WW, Homa FL, Brown JC. Herpes simplex virus capsid structure: DNA packaging protein UL25 is located on the external surface of the capsid near the vertices. J Virol. 2006;80:6286–94. doi: 10.1128/JVI.02648-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sheaffer AK, Newcomb WW, Gao M, Yu D, Weller SK, Brown JC, Tenney DJ. Herpes simplex virus DNA cleavage and packaging proteins associate with the procapsid prior to its maturation. J Virol. 2001;75:687–98. doi: 10.1128/JVI.75.2.687-698.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trus BL, Newcomb WW, Cheng N, Cardone G, Marekov L, Homa FL, Brown JC, Steven AC. Allosteric signaling and a nuclear exit strategy: binding of UL25/UL17 heterodimers to DNA-Filled HSV-1 capsids. Mol Cell. 2007;26:479–89. doi: 10.1016/j.molcel.2007.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cockrell SK, Sanchez ME, Erazo A, Homa FL. Role of the UL25 protein in herpes simplex virus DNA encapsidation. J Virol. 2009;83:47–57. doi: 10.1128/JVI.01889-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Coller KE, Lee JI, Ueda A, Smith GA. The capsid and tegument of the alphaherpesviruses are linked by an interaction between the UL25 and VP1/2 proteins. J Virol. 2007;81:11790–7. doi: 10.1128/JVI.01113-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Preston VG, Murray J, Preston CM, McDougall IM, Stow ND. The UL25 gene product of herpes simplex virus type 1 is involved in uncoating of the viral genome. J Virol. 2008;82:6654–66. doi: 10.1128/JVI.00257-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bowman BR, Baker ML, Rixon FJ, Chiu W, Quiocho FA. Structure of the herpesvirus major capsid protein. Embo J. 2003;22:757–65. doi: 10.1093/emboj/cdg086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pasdeloup D, Blondel D, Isidro AL, Rixon FJ. Herpesvirus capsid association with the nuclear pore complex and viral DNA release involve the nucleoporin CAN/Nup214 and the capsid protein pUL25. J Virol. 2009;83:6610–23. doi: 10.1128/JVI.02655-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mullen LM, Nair SP, Ward JM, Rycroft AN, Henderson B. Phage display in the study of infectious diseases. Trends Microbiol. 2006;14:141–7. doi: 10.1016/j.tim.2006.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zlotnick A, Cheng N, Stahl SJ, Conway JF, Steven AC, Wingfield PT. Localization of the C terminus of the assembly domain of hepatitis B virus capsid protein: implications for morphogenesis and organization of encapsidated RNA. Proc Natl Acad Sci U S A. 1997;94:9556–61. doi: 10.1073/pnas.94.18.9556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Conway JF, Cheng N, Zlotnick A, Stahl SJ, Wingfield PT, Steven AC. Localization of the N terminus of hepatitis B virus capsid protein by peptide-based difference mapping from cryoelectron microscopy. Proc Natl Acad Sci U S A. 1998;95:14622–7. doi: 10.1073/pnas.95.25.14622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Conway JF, Cheng N, Zlotnick A, Stahl SJ, Wingfield PT, Belnap DM, Kanngiesser U, Noah M, Steven AC. Hepatitis B virus capsid: localization of the putative immunodominant loop (residues 78 to 83) on the capsid surface, and implications for the distinction between c and e-antigens. J Mol Biol. 1998;279:1111–21. doi: 10.1006/jmbi.1998.1845. [DOI] [PubMed] [Google Scholar]
- 36.Cohen GH, Ponce de Leon M, Diggelmann H, Lawrence WC, Vernon SK, Eisenberg RJ. Structural analysis of the capsid polypeptides of herpes simplex virus types 1 and 2. J Virol. 1980;34:521–31. doi: 10.1128/jvi.34.2.521-531.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Newcomb WW, Juhas RM, Thomsen DR, Homa FL, Burch AD, Weller SK, Brown JC. The UL6 gene product forms the portal for entry of DNA into the herpes simplex virus capsid. J Virol. 2001;75:10923–32. doi: 10.1128/JVI.75.22.10923-10932.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tivol WF, Briegel A, Jensen GJ. An improved cryogen for plunge freezing. Microsc Microanal. 2008;14:375–9. doi: 10.1017/S1431927608080781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Conway JF, Steven AC. Methods for reconstructing density maps of “single” particles from cryoelectron micrographs to subnanometer resolution. J Struct Biol. 1999;128:106–18. doi: 10.1006/jsbi.1999.4168. [DOI] [PubMed] [Google Scholar]
- 40.Yan X, Dryden KA, Tang J, Baker TS. Ab initio random model method facilitates 3D reconstruction of icosahedral particles. J Struct Biol. 2007;157:211–25. doi: 10.1016/j.jsb.2006.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yan X, Sinkovits RS, Baker TS. AUTO3DEM--an automated and high throughput program for image reconstruction of icosahedral particles. J Struct Biol. 2007;157:73–82. doi: 10.1016/j.jsb.2006.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–12. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 43.Ormo M, Cubitt AB, Kallio K, Gross LA, Tsien RY, Remington SJ. Crystal structure of the Aequorea victoria green fluorescent protein. Science. 1996;273:1392–5. doi: 10.1126/science.273.5280.1392. [DOI] [PubMed] [Google Scholar]


