Abstract
Lipid signaling pathways define central mechanisms of cellular regulation. Productive lipid signaling requires an orchestrated coupling0020between lipid metabolism, lipid organization, and the action of protein machines that execute appropriate downstream reactions. Using membrane trafficking control as primary context, we explore the idea that the Sec14-protein superfamily defines a set of modules engineered for the sensing of specific aspects of lipid metabolism and subsequent transduction of ‘sensing’ information to a phosphoinositide-driven ‘execution phase’. In this manner, the Sec14–superfamily connects diverse territories of the lipid metabolome with phosphoinositide signaling in a productive ‘crosstalk’ between these two systems. Mechanisms of crosstalk, where non-enzymatic proteins integrate metabolic cues with the action of interfacial enzymes, represent unappreciated regulatory themes in lipid signaling.
Lipids and pathways for membrane trafficking
The identities of proteins that regulate the membrane deformations required for biogenesis and fusion of transport vesicles were discovered by the pioneering studies of Rothman and Schekman some 25 years ago [reviewed in refs 1,2]. Those descriptions of the vesicle cycle described protein-centric points of view. That lipid metabolism must be considered as integral participant in mechanistic descriptions of the vesicle cycle came from studies in permeabilized adrenal chromaffin cells and in yeast [3,4]. In yeast, the phosphatidylinositol (PtdIns)-transfer protein (PITP) Sec14p coordinates multiple lipid metabolic pathways with Golgi function -- a conclusion amply demonstrated by the isolation and characterization of ‘bypass Sec14p’ mutations [5–7; Box 1].
Box 1. ‘Bypass Sec14p’ mutations
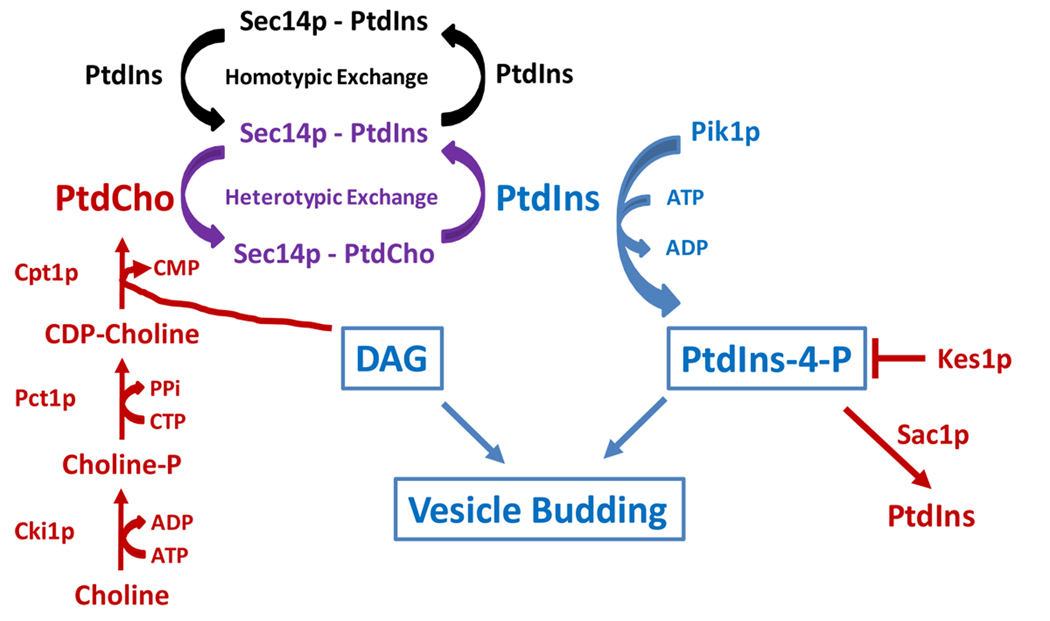
The ‘bypass Sec14p’ mutations uniformly permit cell viability in the complete absence of the normally essential Sec14p. Such mutations are generally interpreted as restoring physiological conditions that are the consequence of Sec14p function in vivo. Analyses of these mutants demonstrated that the cognate mutations represent loss-of-function alleles residing within the coding sequences of seven non-essential genes that cumulatively involve structural genes for enzymes of a specific PtdCho biosynthetic pathway (the CDP-choline pathway) that consumes DAG and represents one of two PtdCho biosynthetic pathways in yeast [5–7], a PtdIns-4-P phosphatase [18–21], and a TGN/endosome-associated PIP-binding protein of the oxysterol-binding protein family [22,23]. Because the ‘bypass Sec14’ gene products identify phospholipid metabolic enzymes or lipid binding proteins, the genetic data identify Sec14p as regulator of an essential functional interface between phospholipid metabolism and TGN/endosomal membrane trafficking activity. Execution points for these gene products in the context of Sec14p-dependent vesicle budding are depicted in Fig. 4.
Subsequently, evidence from numerous laboratories reinforces the concept that lipid metabolism is closely interwoven with the activities of protein machines that regulate membrane trafficking. Pioneering studies to this effect include the purification of mammalian PITP and PtdIns-4-phosphate 5-OH kinase as potentiators of regulated fusion of dense core granules to the neuroendocrine plasma membrane [8,9], and demonstrations that a phosphatidylcholine (PtdCho)-hydrolyzing enzyme (phospholipase D) is activated by PtdIns-4,5-P2 and ADP-ribosylation factor (ARF)-GTP [10,11]. Further demonstrations that phosphoinositides (PIPs) regulate membrane trafficking came from studies of yeast Vps34p, a protein initially described as a novel protein kinase [12], but subsequently shown to be a PtdIns 3-OH kinase [13]. This discovery paved the way for landmark studies that describe roles for 3-phosphorylated PIPs in regulating the multivesicular body pathway for sorting, recycling, and down-regulation of signaling receptors [14,15]. PtdIns-4-phosphate was also shown to be important for Golgi function. Lines of evidence to that effect include functional analyses of the Pik1p PtdIns 4-OH kinase [16,17], demonstrations that the integral membrane multi-specificity PIP-phosphatase Sac1p is the primary conduit for PtdIns-4-phosphate degradation in yeast [18–21], and the description of oxysterol binding proteins as negative regulators trafficking from the trans-Golgi network (TGN) that antagonize PtdIns-4-phosphate signaling [22–25]. Acidic phospholipids (PtdIns-4,5-P2 and phosphatidic acid) are shown to[sc1] influence t-SNARE activities, thereby controlling vesicle fusion to the plasma membrane [26,27]. Similarly, these phospholipids stimulate the activities of specific ARFGAPs [28,29].
Other lipid classes are also intimately involved with the regulation of vesicle trafficking. Indeed, diacylglycerol (DAG) regulates vesicle budding throughout the exocytic pathway [30–34], and regulates protein components of the membrane trafficking machinery [29,35]. Protein kinase D (PKD) isoforms have emerged as regulatory hubs that translate DAG signaling to lipid metabolic events that promote trafficking from the TGN [36,37]. In addition, the roster of lipids that regulate membrane trafficking has expanded to include sterols [38] and sphingolipids [39], and lipid flippases were shown to control the formation of specific TGN-derived vesicles, indicating that regulated transformations in lipid topology also contribute to vesicle formation [40].
Many of the involvements of lipid metabolism in membrane trafficking control employ PIP metabolism as direct component of the regulatory circuit. What is the nature of the interface that connects lipid metabolism to PIP signaling and, ultimately, to proper regulation of membrane trafficking reactions? In exploring this major unresolved question, we first briefly summarize current ideas regarding lipid-mediated regulation of membrane trafficking.
Lipid platforms
Present discussions of the roles for lipids in control of membrane trafficking are dominated by the well-established concepts that lipids form binding platforms for proteins with quasi-specific lipid-binding motifs [reviewed in 41]. Particular attention in this context has been paid to PIPs, whose binding motifs, including pleckstrin-homology (PH)-domains, exhibit rather promiscuous preferences for PtdIns-4,5-P2, PtdIns-4-P and 3-OH PIPs. In addition, the structurally unrelated FYVE-domains, which display pronounced specificities for 3-OH PIPs, and phox-homology (PX)-domains, which also prefer 3-OH PIPs, are well described [41]. Because high chemical specificities for PIP binding are the exception rather than the rule, the lipid-binding properties of these various modules typically cooperate with additional binding elements to impose suitably strict targeting specificities for individual proteins [41,42].
Mammalian cells produce 7 PIPs that, when combined with ancillary targeting signals, allow for creation of a highly diverse set of binding platforms [42]. This diversification strategy demands coordinated activities of PtdIns-kinases and PIP phosphatases, and both classes of enzymes are well-represented in the list of membrane trafficking regulators. Excellent reviews focusing on the roles of PIPs (and their downstream effectors) in controlling membrane trafficking treat the various topics summarized above in detail [43–45]. These discussions focus on product-centric models, however, and fail to capture dynamic involvements of broader aspects of lipid metabolism in regulating PIP signaling and membrane trafficking.
Lipid transfer proteins: In vitro veritas?
Lipid transfer proteins (LTPs) are not enzymes; therefore operational assays for their activities are denied the clarity of interpreting biological outcomes for chemical conversion of defined substrate to specific product. This presents conceptual difficulties in connecting LTP biochemical assays with functional insights. Nonetheless, mechanisms for LTP (and PITP) function are interpreted in the historical context by which LTPs were first identified in vitro. For PITPs, in vitro transfer assays are interpreted to reflect delivery of PtdIns from the endoplasmic reticulum (ER) to membranes engaged in PIP signaling [46,47]. The cycle is closed by vectorial delivery of PtdCho from ‘signaling’ membranes back to the ER. Other recent reviews broadly summarize the involvements of PITPs (and other LTPs) in connecting lipid metabolic pathways with membrane trafficking [48–50]. Most of these interpret LTP and PITP function in the context of lipid transfer between the membranes of distinct organelles.
The general embrace of lipid transfer models notwithstanding, the experimental evidence supporting in vivo transfer mechanisms is not compelling for any PITP. In part, this deficiency of evidence reflects the fact that such models are frustratingly difficult to put to an adequate experimental test. Moreover, such models are not easily broken down into testable concepts. It is primarily for these two reasons that our understanding of how PITPs function as molecules has remained rather stagnant. Herein, we explore new ideas for the mechanisms by which PITPs translate lipid binding to biological function and, in so doing, integrate the broader lipid metabolome with PIP signaling and membrane trafficking. This topic is discussed with a focus on Sec14-like PITPs. The unexpected complexities in lipid binding by Sec14-like proteins suggest novel instructive mechanisms for regulating PtdIns-kinases, and provoke new ideas for how biological outcomes for PIP signaling are diversified. Because the quality of experiment directly reflects how good the questions are, we discuss some of the key questions provoked by these new ideas.
The Sec14p tale as told by cells
Genetic studies in yeast have proven invaluable in setting a conceptual framework for what biological activities are responsive to the function of Sec14p, and of other Sec14-like PITPs, and how these PITPs execute biological function. In what is arguably the best understood case for transfer protein function, four lines of evidence are not particularly congruent with classical transfer mechanisms for Sec14p function:
First, Sec14p levels are ca. 100-fold above the cellular threshold for viability [25,51]. Reconciling these data with transfer models requires cells to exhibit extremely low thresholds for PtdIns transfer. This conundrum provides an example for why lipid transfer models are difficult to adequately test. The fervent disciple of transfer models can retreat to interpretations where only minor functional thresholds are required for lipid transfer; i.e. thresholds of insufficient magnitude for experimental test. Second, genetic ablation for a specific pathway for PtdCho biosynthesis, or in specific pathways for PIP metabolism, effect bypass of the normally essential Sec14p requirement for yeast membrane trafficking competence and cell viability [5,21,52]. This discovery argues that Sec14p regulates lipid metabolism, not PtdIns supply/transport. Third, vectorial phospholipid transfer models predict that rank affinity of a PITP for PtdIns vs PtdCho is a critical functional property. Yet, Sec14p activity is surprisingly insensitive to specific reductions in PtdIns binding affinity [25,53]. Fourth, transfer models predict that alternative modes of PtdIns supply to membranes will obviate the essential Sec14p requirement. In contradiction, manipulation of the yeast lipidome such that PtdIns constitutes 40 mole% of total glycerophospholipid mass (which should solve all PtdIns supply demands) fails to relieve cells of the essential Sec14p requirement [5]. This result demonstrates a special relationship between Sec14p and PtdIns that goes beyond supply/transfer. The third and fourth lines of evidence listed above are particularly problematic for transfer models.
Are there alternative possibilities for mechanisms of PITP function that are amenable to direct experimental test[sc2]? In our view, such alternative ideas do indeed exist, but have not been explored. To initiate a discussion of how PITPs actually work, we proffer the hypothesis that Sec14-like PITPs (and perhaps other LTPs), are primarily lipid sensors which instruct specific enzymes when and where to execute biochemical reactions. This regulatory axis incorporates novel mechanisms for coincidence detection in integrating the broader lipid metabolome with the landscape of PIP-driven cell signaling – including the lipid signaling interface with the membrane trafficking machinery. These alternative possibilities suggest concepts that might translate into functional mechanisms for other LTPs. Utilization of non-enzymatic proteins for linking specific channels of metabolic information with the action of interfacial lipid modifying enzymes represents an unappreciated regulatory theme in cell signaling.
The Sec14-superfamily
The Sec14-domain (Smart entry: smart00516), for which the yeast Sec14p is the prototype [54], represents an ancient and versatile structural unit invented by eukaryotes. There are 1551 Sec14 domains, representing 1550 proteins, annotated in the NCBI database (www.ncbi.nlm.nih.gov). Even simple eukaryotes express multiple Sec14-family members. Indeed, Saccharomyces cerevisiae expresses five Sec14-like proteins in addition to Sec14p (55), and Homo sapiens, Mus musculus, Drosophila melanogaster, Caenorhabditis elegans, and Aradidopsis thaliana each invest in greater than twenty individual genes that encode distinct Sec14-superfamily members.
As expected from the diversity of the superfamily, Sec14-domain proteins interface with many cellular activities. Studies in yeast and plants demonstrate multiple roles for Sec14-like PITPs in housekeeping membrane trafficking pathways [52,56,57], developmentally regulated membrane trafficking programs for dimorphic growth and sporulation in yeast [58–60] and root hair biogenesis in plants [61]. Proteins which regulate activities of small GTPases of the Ras, Rho and Rac families often harbor Sec14 domains. Examples include the Ras-GAP neurofibromins NF1 and NF2 [62], Rho- and Cdc42-GAPs of the BCH and BNIP families [63–65], and Rho-GEFs Trio, Dbl and Duo [66,67]. The PTP-MEG2 protein tyrosine phosphatase also harbors a Sec14-domain [68].
The Sec14-domain is also associated with uncharacterized modules that include nodulin domains (in higher plants; 61,69), Golgi dynamics (GOLD) domains in metazoans and higher plants (70), metazoan-specific PRELI-domains, which are thought to represent mitochondrial targeting motifs [70,71], and a GTPase motif of uncertain function [72]. Stand-alone Sec14-domain proteins are well-represented in simpler eukaryotes. By contrast, mammals often incorporate Sec14-modules into more complex arrangements. However, mammalian stand-alone Sec14-domain proteins include cellular retinaldehyde binding protein (CRALBP) [73], caytaxin [74], and α-tocopherol and retinaldehyde transfer proteins [75–77]. The biological importance of Sec14-superfamily members in mammals is demonstrated by the linkage of human diseases to dysfunction of Sec14-like proteins, and of Sec14-domains in more complex modular proteins. Such diseases include autosomal-dominant cancers associated with neurofibromin insufficiencies [78], ataxia with vitamin E deficiency accompanying diminished α-TTP function [79], ataxia associated with caytaxin loss-of-function [74], and retinal degeneration syndromes accompanying CRALBP deficiencies [80,81].
How is the diversity of the Sec14-superfamily employed in the context of the lipid signaling circuitry of eukaryotic cells? We posit that the Sec14-protein superfamily defines a set of modules engineered for sensing specific aspects of lipid metabolism, and for transduction of sensing information to an ‘execution phase’ that employs PIP signaling for action. Below, we describe a physical picture of how the Sec14–superfamily generates a productive ‘crosstalk’ between the larger lipidome, PIP signaling, and membrane trafficking control.
Ligand binding by Sec14-like proteins
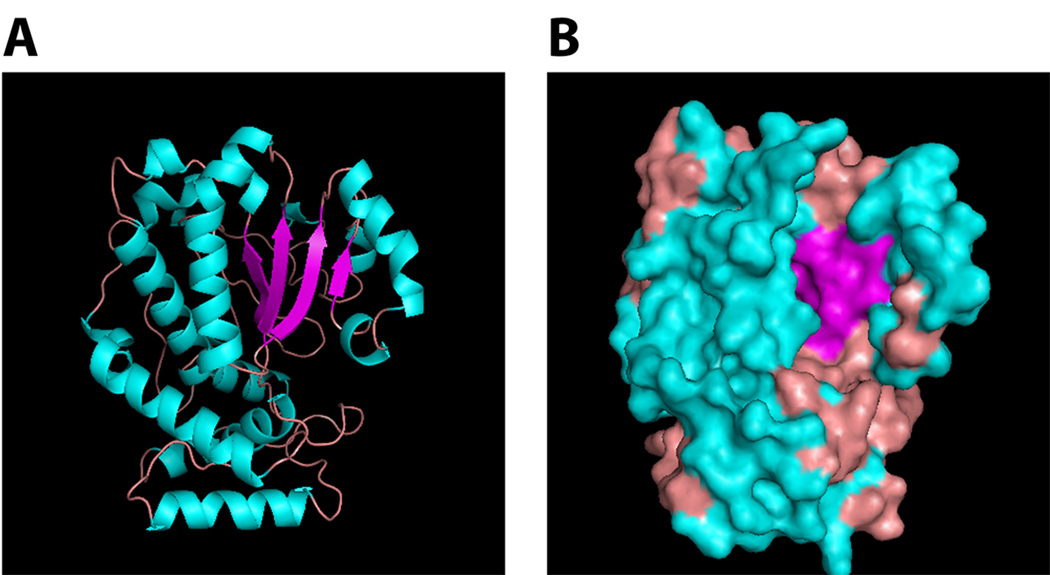
Available crystal structures of Sec14-like proteins include detergent-bound forms of Sec14p (53,54), several phospholipid-bound forms of yeast Sfh1p, the protein most similar to Sec14p [25], ligand-bound and apo-versions of α-tocopherol transfer protein (α-TTP) and the mammalian Sec14-GOLD protein Sec14L2 [82–84], and detergent-bound and phospholipid-bound forms of the neurofibromin Sec14-like domain [85,86]. This ensemble of structures shows that Sec14-domains adopt similar folds – termed the Sec14-fold, an approximately 280-residue two-lobed globular structure. In the case of apo-Sec14p, the amino-terminal lobe consists of four anti-parallel α-helices whereas the carboxy-terminal lobe (also referred to as the CRAL_TRIO-domain) defines the phospholipid binding pocket (Fig. 1). The floor of the Sec14p binding cavity consists of five parallel β-strands comprising a β-sheet that is sandwiched by two long α-helices on one side and two short β-helices plus one 310-helix on the other. The number of β-strands involved in forming the sheet that defines the pocket floor is variable among Sec14-domains, and the numbers and the angles of the helices that form the walls of the corresponding lipid binding cavities also differ [25,54,82–86].
Figure 1.
The Sec14-fold. The apo-Sec14 crystal structure (pdb 1AUA) is rendered as a ribbon diagram (a) and a surface drawing (b). Secondary structure elements are depicted in blue (α-helices), magenta (β-strands) and pink (random coil). The orientation of the molecule in en face with the floor of the lipid binding pocket identified by the β-sheet. This structure highlights not only the essential features of the Sec14-fold, but also the single large hydrophobic pocket in which Sec14-like proteins accommodate one hydrophobic ligand molecule at a time.
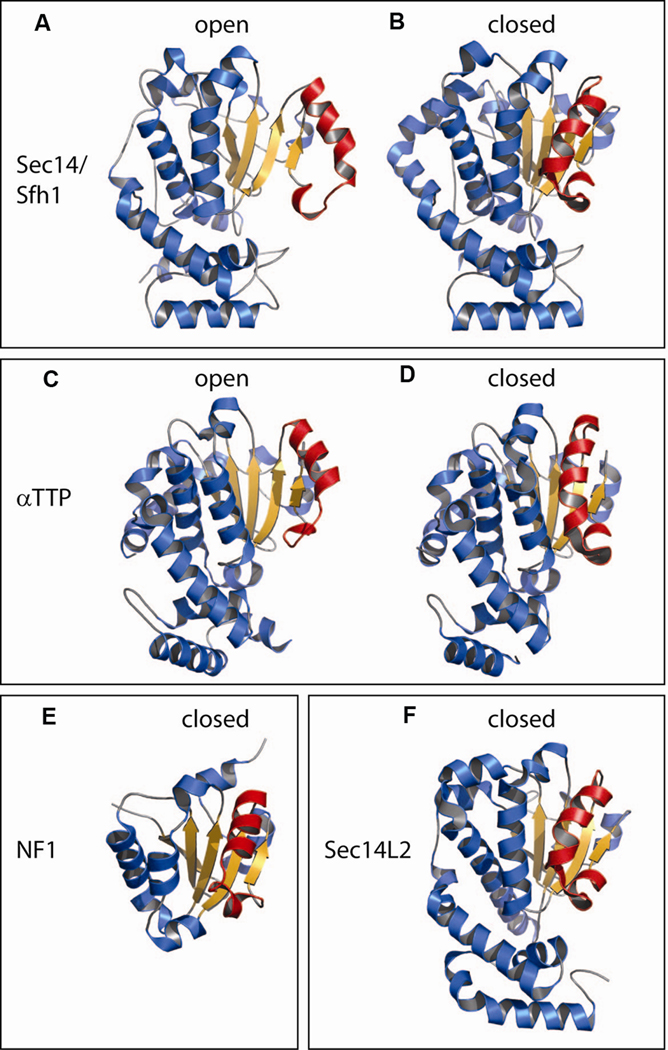
Molecular dynamics simulations [87], comparisons of apo-Sec14p and holo-Sfh1p structures [25,54], the availability of structures for holo-forms of Sec14L2 [83] and α-TTP [84], and a structure of the phosphatidylethanolamine (PtdEtn)-bound form of the neurofibromin Sec14 domain [86], demonstrate that access to the hydrophobic lipid-binding cavity is determined by a helical gate substructure. This gate is flipped open in Sec14-domain apo-structures and is closed in holo-structures (Fig. 2). Transitions from open to closed conformations describe rigid body motions dominated by large displacements of the helical gate. These conformational dynamics accompany interfacial phospholipid exchange reactions on surfaces of biological membranes in the case of Sec14-like PITPs. Other members of the Sec14-superfamily likely undergo similar conformational dynamics during the course of ligand loading/unloading reactions. A regulatory substructure, the approximately 20 residue gating module, regulates an H-bond network that transduces conformational information to the helical gate [87] – likely in response to conformational changes initiated by membrane docking. Examples of human disease missense mutations that map to the gating modules of Sec14-superfamily proteins confirm the functional importance of this substructure [see ref 87].
Figure 2.
Sec14-like proteins gate access to the hydrophobic ligand binding pocket via a mobile helical substructure. Ribbon diagrams of open and closed conformations of a set of Sec14-like proteins or domains (for the NF1 and Sec14L2 structures) are shown. Comparisons of open vs closed conformers highlight the major differences in configuration of the helical substructure (rendered in red) that gates entry into the hydrophobic ligand-binding cavity. α-Helices are depicted in blue and β-strands in orange. The pdb identifiers are 1AUA (A; Sec14p open conformer bound to the detergent β-octylglucoside), 3B7N (B; Sfh1p closed conformer bound to PtdIns), 1OIZ (C; α-TTP open conformer bound to the detergent Triton X-100), 1R5L (D; α-TTP closed conformer bound to α-tocopherol), 2E2X (E; NF-1 Sec14-domain closed conformer bound to PtdEtn), and 1O6U (F; Sec14L2 closed conformer bound to rrr-α-tocopherylquinone), respectively. Neither bound detergent molecules nor bound ligands are represented.
From the perspective of the phospholipid molecule engaged in an exchange reaction, the hydrophobicity gradient within the Sec14p cavity closely matches that found in membrane cytosolic leaflets. Thus, lipid binding/release by Sec14p, and presumably other Sec14-like proteins, is driven by simple substrate partitioning from one aprotic environment to another [88]. How lipid ligands are chosen on the membrane surface, and how these are subsequently configured so as to be offered a choice of partitioning environment, represent mechanistic questions for future analysis. The local membrane deformations that must occur in support of these selection/configuration processes also remain unstudied.
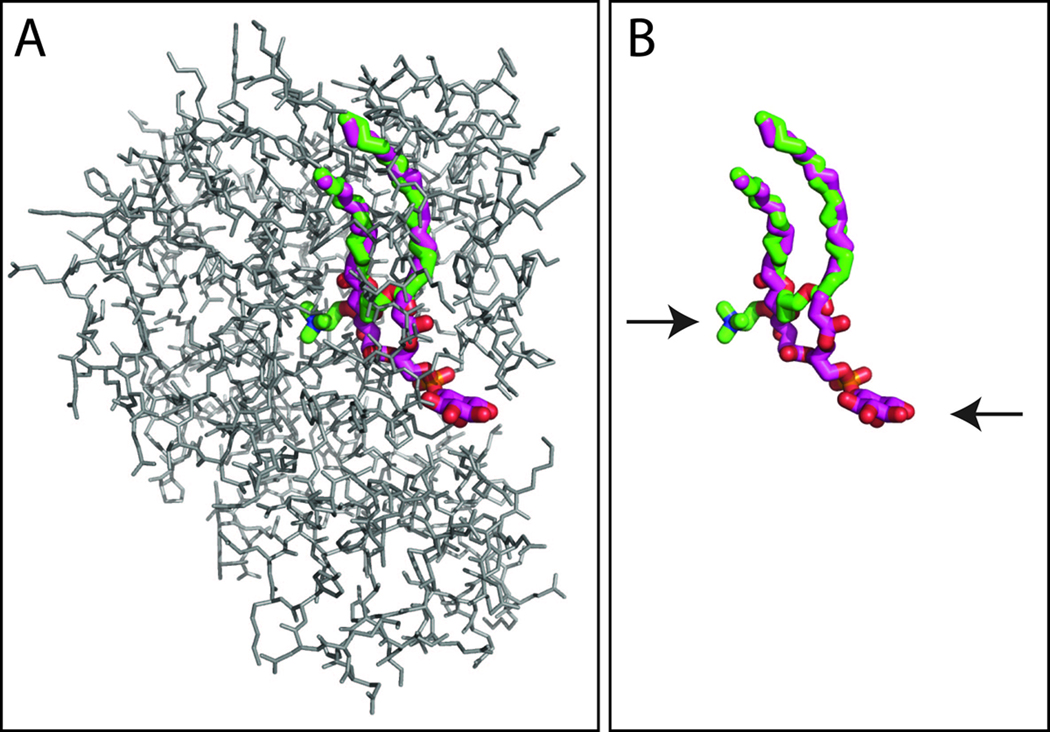
The Sfh1p and Sec14p headgroup binding strategies are surprising as the PtdCho and PtdIns headgroups are bound at distinct sites [25]. The inositol binding site is configured near the protein surface (Fig. 3), consistent with inferences gathered from apo-Sec14p and PtdEtn-bound neurofibromin Sec14-domain structures [53,54,86]. By contrast, the PtdCho headgroup is buried within the protein interior where it is stabilized by tyrosine-mediated cation-π interactions [25; Fig. 3] – a structural theme employed by other alkyl-amine and choline binding proteins [89]. Another unusual feature of Sfh1p and Sec14p is their abilities to accommodate, within the binding cavity, phospholipid molecules with different volumes (cavity volumes for Sfh1p-PtdIns and Sfh1p-PtdCho are 4050.6Å3 and 3068.7Å3, respectively) without significant effect on shape of the protein surface. This is accomplished in part by loading the unoccupied PtdCho headgroup binding site with ordered water molecules in the Sec14-PtdIns complex and, reciprocally, loading the unoccupied PtdIns headgroup binding site with ordered water in the Sec14-PtdCho complex [25]. Flux of water into and from the hydrophobic pocket during heterotypic exchange reactions is a major factor in overcoming the differences in Sec14p relative binding affinities for PtdIns and PtdCho so that heterotypic exchange reactions (PtdIns for PtdCho or PtdCho for PtdIns) can actually take place.
Figure 3.
Sec14-like PITPs exhibit two distinct headgroup binding sites. Bound PtdIns (purple/red) and PtdCho (green/blue) is shown in the context of the Sec14-fold of Sfh1p (gray line drawing; A). (B) The phospholipid configurations are shown with protein electron density subtracted. The positions of the respective headgroups are indicated by arrows. Of note is the remarkable physical separation of the PtdIns and PtdCho headgroup binding regions within the hydrophobic cavity; however, the acyl chain binding regions for the two phospholipid species overlap. We propose that this engineering of the Sec14p molecule with regard to differential phospholipid binding forms the basis for how heterotypic exchange reactions present a PtdIns headgroup to lipid kinases. These depictions are derived from pdb 3B7Z which describes coordinates obtained from crystals composed of a mixture of Sfh1-PtdIns and Sfh1-PtdCho unit cells [25].
Coincidence sensors that couple lipid metabolic inputs to PIP synthesis
The differential PtdIns vs PtdCho binding strategies of Sec14p and Sfh1p are revealing. Both PtdIns- and PtdCho-binding activities must reside on the same Sec14p molecule to generate a biologically functional protein that productively stimulates PtdIns 4-OH kinase activity [25]. Thus, heterotypic exchange reactions are required for Sec14p-mediated stimulation of PtdIns kinases (and PIP synthesis) in vivo -- indicating that Sec14p cannot stimulate PtdIns 4-OH kinases in cells unless sufficient amounts of ‘accessible’ PtdCho are present to drive the heterotypic exchange reactions which activate PtdIns kinases (Fig. 4). This concept assigns Sec14p a dual role as a PtdCho sensor, and as a PtdIns-presenting nanoreactor, that transmits PtdCho metabolic information to PIP synthesis. Such a definition is congruent with demonstrations of an intimate coupling between the CDP-choline pathway for PtdCho biosynthesis and membrane trafficking control [5,6].
Figure 4.
Sec14 and coordination of lipid metabolism with membrane trafficking. A DAG-requiring vesicle formation pathway is sensitive to flux through the cytidine-diphosphate (CDP)-choline pathway activity because PtdCho production via this mechanism occurs at the expense of DAG. Sec14 surveys flux by binding (sensing) the newly synthesized PtdCho (i.e. the accessible PtdCho pool). Sec14p initiates heterotypic exchange reactions and stimulates PtdIns-4-phosphate production by the Pik1p PtdIns 4-OH kinase. In a single vesicle budding pathway model, PIP substitutes for DAG in promoting vesicle budding. Alternatively, in a model where there are two pathways for vesicle budding, the two modes are distinguished by their threshold requirements for DAG and PtdIns-4-phosphate. Sec14p inactivation compromises both vesicle budding pathways. The activity of multiple pathways for vesicle formation in the yeast TGN/endosomal system is well-established [40]. Negative regulators of Sec14p-dependent vesicle budding pathways are highlighted in red -- including the proteins identified by loss-of-function ‘bypass Sec14p’ mutations (choline kinase, Cki1p; the choline-phosphate cytidylyltransferase, Pct1p; choline phosphotransferase, Cpt1p, the PtdIns-4-phosphatase Sac1p, the oxysterol binding protein Kes1p). Positive regulators of Sec14p-dependent vesicle budding pathways are highlighted in blue. The interface between these two opposing is regulated by Sec14p-mediated heterotypic phopsholipid exchange (purple).
The Sec14p requirement for coordinating the PtdCho biosynthesis/membrane trafficking interface is suggested as a mechanism for ensuring that DAG pools critical for TGN/endosomal trafficking are not exhausted by the CDP-choline pathway for PtdCho biosynthesis – i.e. Sec14p senses PtdCho as a readout for DAG consumption [56]. As PtdCho-levels rise in response to surges in flux through the CDP-choline pathway, Sec14p is activated for heterotypic PtdIns/PtdCho exchange and, in turn, stimulates PIP production by PtdIns 4-OH kinases (Fig. 4). One possibility is that newly synthesized PtdCho represents the ‘accessible’ PtdCho bound by Sec14p. Consequent elevations in PtdIns-4-phosphate activate downstream effectors that promote vesicle budding from TGN/endosomes – perhaps via alternative budding pathways. PtdIns-4-phosphate might also inhibit the cholinephosphate cytidylyltransferase, the rate-determining enzyme of the CDP-choline pathway (Fig. 4).
These concepts hold implications for other Sec14-superfamily members. Structure-based predictive bioinformatic approaches allow the assignment of primary sequence ‘bar codes’ for PtdIns- and PtdCho-binding [25]. These signatures forecast that PtdCho-binding is not a conserved feature of Sec14-like proteins; the PtdCho-binding bar code is recognized in some Sec14-like proteins, but is contra-indicated in others. The holo-Sec14L2 and α-TTP structures, and the biochemical properties of CRALBP and related proteins, demonstrate that Sec14-superfamily proteins lacking PtdCho-binding signatures bind hydrophobic ligands with diverse chemical properties [25, 83–86]. Predictive bioinformatics recognize the PtdIns-binding bar code across the superfamily, however – suggesting the Sec14-like PITP cohort is much larger than presently appreciated. Thus, a two-ligand PITP-mediated mechanism for PtdIns kinase activation (analogous to that described for Sec14p and Sfh1p) might be broadly utilized across the eukaryotic signaling landscape via the action of Sec14-superfamily members, thereby linking diverse territories of the lipid metabolome with PIP signaling.
Instructive mechanisms for regulating PtdIns-kinase activity
Can a physical picture to be drawn to illustrate how Sec14-like PITPs potentiate PtdIns kinase activities? Such a description requires an understanding of how PtdIns kinases register substrate lipids. In vitro PtdIns kinase assays commonly present PtdIns to the kinase in the context of mixed phospholipid-detergent micelles. Although such presentation schemes support kinase activity, it is remarkable how poorly these enzymes register PtdIns residing in membrane bilayers [25]. One factor that no doubt contributes to such behavior is the use of isolated recombinant kinase catalytic subunits in such measurements; PtdIns 3-OH and 4-OH kinases are equipped with accessory subunits essential for enzyme activity in vivo [90–93]. Yet, requirements for Sec14p and Sec14-like PITPs for physiologically sufficient activities of the yeast PtdIns 4-OH kinases report fundamental inadequacies of these enzymes in engaging membrane-incorporated PtdIns.
We entertain the idea that PtdIns 4-OH kinases have evolved to be inefficient interfacial enzymes because such an intrinsic inefficiency provides substantial advantages for regulating lipid signaling. The inadequacies of PtdIns 4-OH kinases in engaging microsomal PtdIns provide opportunities for ‘front-loaded’ regulation of PIP synthesis by Sec14-like PITPs. That is, Sec14-like PITPs solve the substrate accessibility problem for PtdIns-kinases and, in the process, impose exquisite channels for coincidence detection at the level of PtdIns kinase activity. We suggest that such front-loaded strategies define significant mechanisms for how the lipid metabolome is interfaced with membrane trafficking and with other intracellular signaling pathways. The idea that Sec14-like PITPs mediate instructive regulation of PtdIns 4-OH kinases by executing dual functions as coincidence detection sensors, and as non-catalytic PtdIns-presenting scaffolds, justifies their description as components of nanoreactors for PIP synthesis [25,94].
Heterotypic phospholipid exchange and stimulation of PtdIns-kinases
The ability of a primed Sec14-like PITP (i.e. one associated with sensor ligand; such as PtdCho in the case of Sec14p) to switch into substrate-presentation mode via heterotypic exchange reactions raises interesting questions. These data suggest that primed Sec14-like PITPs generate a temporally and spatially-regulated flurry of ‘presented’ PtdIns headgroups now susceptible to modification by PtdIns 4-OH kinases (Fig. 5). The window of opportunity for a PtdIns kinase to modify the ‘presented’ PtdIns monomer is certainly short. However, a transient vulnerability for any single PtdIns molecule might be stochastically compensated by frequency of the ‘presented state’ when local populations of PtdIns molecules are incorporated into PITP-mediated heterotypic exchange reactions. The efficiency with which a PtdIns kinase productively modifies ‘presented’ PtdIns monomers is also low. Yet, as exchange reactions occur rapidly, and at no energy cost, such a simple and inefficient system can be gainfully employed for regulated presentation of PtdIns headgroups to neighboring kinases.
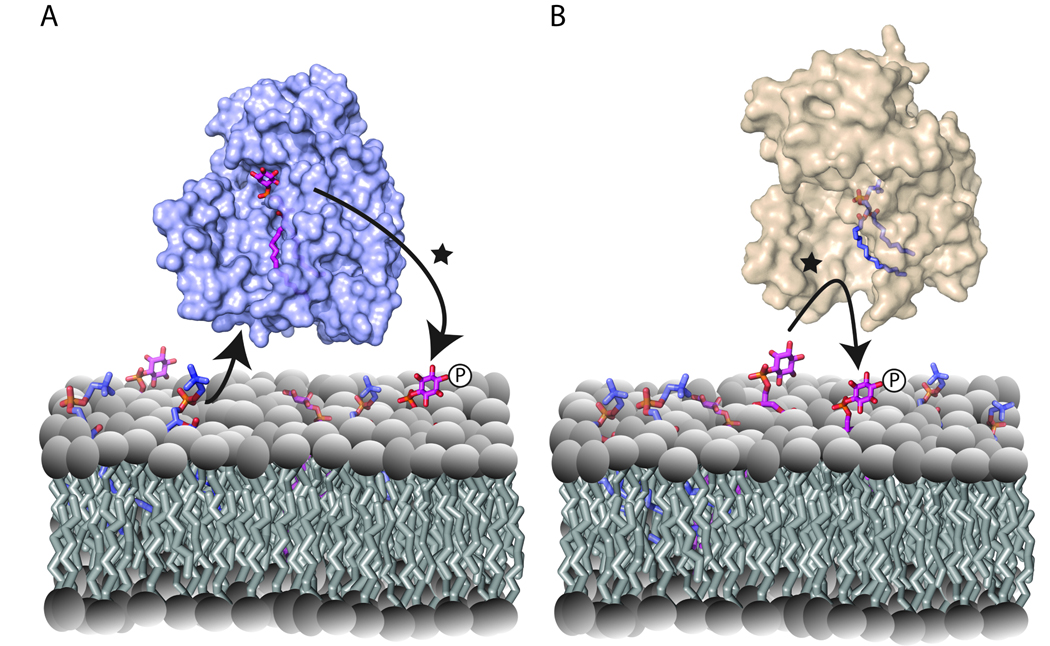
Figure 5.
Heterotypic exchange and PtdIns presentation. Two possibilities for how heterotypic exchange reactions support PtdIns presentation are shown. (A) Sec14p-PtdIns represents the primed intermediate and PtdCho entry displaces bound PtdIns from the open Sec14p conformer in a vectorial head-first manner. The PtdIns 4-OH kinase (not shown) executes the modification on the leaving PtdIns substrate that exits through a portal (depicted by a star) distinct from that occupied by the invading PtdCho. (B) Sec14p-PtdCho represents the primed intermediate and the sequestered PtdCho molecule frustrates entry of the invading PtdIns into the hydrophobic pocket. The frustrated PtdIns is a superior substrate for PtdIns 4-OH kinase (not shown), which executes modification of the invading PtdIns substrate at its of entry portal (depicted by a star). Both models satisfy the requirement that nascent PtdIns-4-phosphate not collapse back into the hydrophobic pocket as that circumstance results in a dead-end Sec14p-PIP complex that cannot be further resolved by phospholipid exchange [26].
The term ‘heterotypic exchange’ implies complete transition, in one cycle, of a holo-Sec14p-PtdCho complex to a holo-Sec14p-PtdIns complex, or vice versa. This definition might be too strict. The effect of heterotypic exchange reactions on interfacial presentation of PtdIns to PtdIns 4-OH kinases could reflect consequences of abortive heterotypic exchange reactions [25]. Assuming that the Sec14p-PtdCho state represents the ‘primed’ Sec14p, sequestration of PtdCho in the hydrophobic pocket might result in a ‘slow’ exit of this phospholipid from the pocket, thereby occluding the pocket and frustrating full accommodation of an invading PtdIns. The invading molecule, neither membrane incorporated nor sequestered by the engaged Sec14p, is now susceptible to modification by PtdIns 4-OH kinase. Multiple abortive PtdIns invasion events could accompany a single round of PtdCho clearance from the Sec14p hydrophobic pocket if PtdIns invasion is fast relative to PtdCho egress. In support of this proposition, rates of PtdIns-transfer are 20-fold faster than those of PtdCho-transfer in vitro. Such abortive PtdIns entry mechanisms identify PtdCho binding as prerequisite to PtdIns presentation events. However, alternative scenarios where PtdIns presentation is described as a PtdCho-driven ejection of PtdIns from the lipid-binding cavity also remain plausible (Fig. 5). These alternative mechanisms might not be mutually exclusive. These conjectures make distinguishing experimental predictions, but the test highlights the need for detailed descriptions of lipid trajectories during heterotypic exchange. Computationally-intensive molecular dynamics approaches will be required for such efforts.
Definition of sensing territories
Spatial and temporal restriction of PITP activity can be achieved by multiple paths. Protein-protein or protein-lipid interactions that involve the Sec14-like PITP surface (as opposed to the hydrophobic binding pocket) can restrict localization to membrane subdomains. Indeed, several yeast Sec14-like PITPs stably target to specific intracellular locations [55,95]. The identities of proteins (or lipids) that impart specific localization to PITPs have yet to be described. PITP-receptors are of interest as such proteins (or lipid platforms) define ‘sensing’ territories. In that regard, the PtdIns kinases themselves represent obvious candidates for PITP receptors. Is there a requirement for dedicated physical interactions between PITPs and the kinases? This is unlikely to be the case as Sec14p defects are rescued by the expression in yeast of mammalian class 1 PITPs (i.e. proteins with no primary sequence homology or structural similarity to Sec14p) [96].
Highly transient membrane associations can also be productively employed to stimulate PtdIns kinases. Sec14p provides a case in point as this PITP stimulates both the plasma membrane-localized Stt4p and the TGN/endosomal Pik1p PtdIns 4-OH kinases in vivo, even though Sec14p does not exhibit stable membrane associations. A nanoreactor-type mechanism accounts for such stimulation provided the heterotypic exchange cycle is much faster than the koff for Sec14p (or any other PITP) from membrane surfaces. By example, Sec14p could execute as many as ten exchange cycles on a membrane surface in a two second residence window if an exchange reaction runs in a 200 msec cycle (our calculations suggest both values are reasonable estimates). Direct measurements of the time scales for lipid exchange by individual PITP molecules, especially when considered relative to PITP membrane residence times, define important future directions essential for assessing nanoreactor vs lipid transfer models.
A fast exchange cycle relative to membrane residence time also solves a problem with system noise – i.e. what prevents priming of Sec14p at irrelevant sites and transmission of inappropriate information to Stt4p or Pik1p kinases by nomadic Sec14p molecules? So long as a Sec14p cycles through multiple rounds of phospholipid exchange on the membrane surface upon which it is docked, the ‘memory’ of previous membrane associations will be erased (i.e. Sec14p ‘reboots’ its hard drive). PtdIns kinase inefficiencies in registering PtdIns monomers ‘presented’ by Sec14p molecules engaged in heterotypic exchange reactions would synergize with such rebooting in limiting translation of inappropriate sensor information from Sec14p to PtdIns 4-OH kinases. In this regard, Sec14-domains in modular proteins (e.g. Ras/Rho GAPs and GEFs and the MEG2 PTP; 62–67,97) are well-engineered to act as intrinsic lipid sensing units for instructing local changes in PIP environment in response to metabolic cues (Box 2).
Box 2. Multidomain Sec14 proteins
Both GEF and GAP activities are responsive to PIPs, thereby raising the attractive prospect that the Sec14-domain senses the lipid environment, transduces the signal to PtdIns kinases for ‘on-demand’ PIP synthesis, and that the GEF/GAP/PTP catalytic domains juxtaposed to Sec14 modules (e.g. as in Dbl and Dbs), are the immediate downstream PIP effectors. Some studies are interpreted as suggesting that lipid binding by the Sec14-domain localizes Ras-GEFs to membrane surfaces [65,66,97]. Such membrane-targeting activities most likely involve protein-lipid interactions on the Sec14-domain surface, however, and not the hydrophobic pocket.
Similar designs might also apply to Sec14-nodulin proteins of higher plants [61,69]. The AtSfh1 Sec14-nodulin protein expressed in Arabidopsis root hair cells both stimulates synthesis of PtdIns-4,5-P2 synthesis and organizes distribution of this PIP in growing root hairs. In this fashion, an AtSfh1-PtdIns-kinase-PIP-kinase axis supports polarized programs for membrane trafficking, actin organization, and calcium signaling in growing root hairs [61]. AtSfh1 might help pattern the PIP landscape via a sensing role for the Sec14 domain, and consequent translation of a sensing signal to stimulated PIP synthesis catalyzed by neighboring PtdIns 4-OH and PtdIns4-phosphate 5-OH kinases [102,103].
Nodulin-domains represent membrane association elements, and the chemical properties of the AtSfh1 nodulin-domain, and of other nodulin-domains, suggest additional layers of regulation. These units present basic carboxy-terminal tails [61,69] -- thereby resembling known PIP-binding motifs that operate via membrane surface electrostatics (e.g. the MARCKS peptide; 104). It is a testable proposition that AtSfh1 functionally specifies dedicated PIP pools at the levels of both production and organization. Regulated electrostatic interactions suggest mechanisms for imprinting ‘caged’ PIP patterns for subsequent and regulated PIP release in a spatially organized program of downstream signaling [104]. The multiplicity of plant Sec14-nodulin proteins forecasts a large diversity in such a strategy for coupling distinct Sec14-like modules of multidomain proteins to developmental pathways for membrane morphogenesis [61]. Sec14-GOLD domain proteins that bind PIPs (e.g. PATELLIN 1) might also operate similarly [105].
Concluding remarks
Recent progress, from various lines of research, is providing unprecedented insights into mechanisms for how lipid metabolism is incorporated into the engineering diagram for membrane trafficking and signaling. Realizations that Sec14-like PITPs integrate diverse aspects of lipid metabolism with PIP signaling, and the recent developments that contribute to a physical appreciation of how such integration could work, introduce a number of new questions for experimental address. Although we highlight the idea that PITPs primarily function as nanoreactors for regulating lipid metabolism, rather than as lipid carriers, the mechanism will vary from case to case. For instance, the metazoan-specific PITPs (e.g. fly RdgBα and mammalian NIR2) are structurally unrelated to the Sec14-like PITPs, yet bind PtdIns and PtdCho in the same binding site [98,99]. Are these nanoreactors for PIP signaling or transfer proteins? Similar issues confront interpretation of how other putative transfer proteins operate in cells [e.g. oxysterol binding proteins and ceramide transfer protein; 100–101]. The competing conceptual frameworks can now be experimentally examined, thereby providing an escape from the circular arguments that have plagued the lipid transfer protein field since its inception.
‘Front-loaded’ strategies that employ non-enzymatic proteins, such as Sec14-like PITPs, for the coupling of metabolic cues to the action of interfacial lipid modifying enzymes may represent a common theme in regulation of signal transduction. Indeed, Sec14p itself might be more versatile in this regard than what we portray here, given that PtdIns might not be the only inositol-phospholipid binding substrate for Sec14p. The yeast sphingolipid inositol-phosphoceramide is a close molecular cousin to PtdIns. There is no structural reason to exclude the possibility that Sec14p also binds shorter chain inositol-phosphoceramides in vivo, a property that could additionally impart on Sec14p the ability to transmit PtdCho metabolic information to inositol sphingolipid metabolism.
From a biological perspective, our understanding of the spatial and temporal regulation of lipid metabolic flux is inadequate. Navigating the technical obstacles that confound acquisition of such information presents major challenges for the future. Our understanding of when and where Sec14-like PITPs are active in living cells is also rudimentary. The engineering of reliable conformational biosensors is required to address these questions. When these goals are met, the biochemical properties of Sec14-like PITPs will establish these proteins as unique tools for studying the dynamic interface between lipid metabolism and signaling in living cells.
Acknowledgements
We thank Fred Maxfield, Anant Menon, Todd Graham, Chris Burd, Will Prinz, Pete Downes, Minoru Nakano, and Eric Ortlund for useful discussions regarding the issues highlighted in this review. This work was supported by grants from the National Institutes of Health to V.A.B. G.S. is supported by grants from the Deutsche Forschungsgemeinschaft.
Glossary
- ARF
(ADP-ribosylation factor): a group of highly conserved GTPases of the Ras-superfamily originally identified as cofactors in the cholera toxin-stimulated ADP ribosylation of Gs subunits of heterotrimeric G-proteins. ARFs are master regulators of membrane trafficking and do so in part by mediating the recruitment of proteins that form vesicle coats.
- BCH
(BNIP-2 and Cdc42GAP homology) domain: an approximately 145 amino acid domain first found in BNIP-2 and Cdc42GAP, also known as p50-RhoGAP, that is a GTPase-activating protein for the small GTPase Cdc42. In the cases studied, the BCH domain negatively regulates the GTPase-activating protein activities of the p50-RhoGAP.
- BNIP
A BCH-domain protein originally identified as binding partner for the anti-apoptotic proteins of the Bcl2 family and the 19-kDa adenovirus E1B protein.
- CRALBP
(cellular retinaldehyde binding protein): a water-soluble 36 kDa protein that binds 11-cis-retinol and acts as substrate carrier for 11-cis-retinol dehydrogenase which converts 11-cis-retinol to 11-cis-retinal in the isomerization step of the rod visual cycle.
- FYVE
an approximately 60 residue motif conserved in Fab1,YOTB, Vac1 and EEA1 Zn2+-finger domains.
- GAP
(GTPase activating protein): regulatory proteins that bind specifically to GTP-bound forms of G-proteins and stimulate the intrinsic GTPase activities of the G-proteins for the purpose of terminating signaling (or promoting cycling between GTP and GDP-bound states). ARF-GAPs provide an arginine finger that contributes to the GTPase active site by stabilizing the positions of the nucleophilic water and the γ-phosphate of GTP bound to ARF. GAPs oppose GEFs in regulating the binary switch of G-proteins between active and inactive states.
- GEFs
(guanine nucleotide exchange factors): proteins that accelerate exchange of GDP from a G-protein for GTP, thereby activating G-proteins for interaction with downstream effectors. GEFs oppose GAPs in regulating the binary switch of G-proteins between active and inactive states.
- MARCKS
(myristoylated alanine-rich C-kinase substrate): a PtdIns-4,5-P2 binding protein whose association to membranes requires both N-terminal myristoylation and an extended cluster of basic amino acids. Association of MARCKS with membranes is regulated by phosphorylation of Ser and Thr residues within the basic cluster by protein kinase C.
- PH-domain
(pleckstrin homology domain): a β-sandwich structural module of approximately 120 residues that binds phosphoinositides and phosphorylated proteins and is a feature of many proteins involved in signal transduction where it acts to recruit these proteins to sites of signaling.
- PTPs
(protein tyrosine phosphatases): a large group of enzymes that hydrolyze the tyrosine-linked phosphomonoester bond in tyrosine-phosphorylated proteins.
- PX
(phox-homology domain): a phosphoinositide-binding unit of approximately 120 amino acids which is found in >100 proteins involved in signal transduction or membrane trafficking. Like PH-domains, these modules primarily serve to recruit proteins to membrane sites of signaling.
- t-SNAREs
(soluble n-ethylmaleimide sensitive factor receptors): are a large family of short-range tethers that mediate docking and fusion of transport vesicles with target membranes. Whereas t-SNAREs represent the target membrane-associated proteins in this process, v-SNARES are associated with vesicles.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Rothman JE. The protein machinery of vesicle budding and fusion. Pro. Science. 1996;5:185–194. doi: 10.1002/pro.5560050201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schekman R, Orci L. Coat proteins and vesicle budding. Science. 1996;271:1526–1533. doi: 10.1126/science.271.5255.1526. [DOI] [PubMed] [Google Scholar]
- 3.Eberhardt DA, et al. Evidence that the inositol phospholipids are necessary for exocytosis: loss of inositol phospholipids and inhibition of secretion in permeabilized cells caused by bacterial phospholipase C and removal of ATP. Biochem. J. 1990;268:15–25. doi: 10.1042/bj2680015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bankaitis VA, et al. An essential role for a phospholipid transfer protein in yeast Golgi function. Nature. 1990;347:561–562. doi: 10.1038/347561a0. [DOI] [PubMed] [Google Scholar]
- 5.Cleves AE, et al. Mutations in the CDP-choline pathway for phospholipid biosynthesis bypass the requirement for an essential phospholipid transfer protein. Cell. 1991;64:789–800. doi: 10.1016/0092-8674(91)90508-v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cleves AE, et al. Phospholipid transfer proteins: a biological debut. Trends Cell Biol. 1991;1:30–34. doi: 10.1016/0962-8924(91)90067-j. [DOI] [PubMed] [Google Scholar]
- 7.McGee TP, et al. A phosphatidylinositol transfer protein controls the phosphatidylcholine content of yeast Golgi membranes. J. Cell Biol. 1994;124:273–287. doi: 10.1083/jcb.124.3.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hay JC, Martin TFJ. Phosphatidylinositol transfer protein required for ATP-dependent priming of Ca2+-activated secretion. Nature. 1993;366:572–575. doi: 10.1038/366572a0. [DOI] [PubMed] [Google Scholar]
- 9.Hay JC, et al. ATP-dependent inositide phosphorylation required for Ca(2+)-activated secretion. Nature. 1995;374:173–177. doi: 10.1038/374173a0. [DOI] [PubMed] [Google Scholar]
- 10.Brown HA, et al. ADP-ribosylation factor (ARF), a small GTP-dependent regulatory protein, stimulates phospholipase D activity. Cell. 1993;75:1137–1144. doi: 10.1016/0092-8674(93)90323-i. [DOI] [PubMed] [Google Scholar]
- 11.Ktistakis N, et al. Phospholipase D is present on Golgi-enriched membranes and its activation by ADP ribosylation factor is sensitive to brefeldin A. Proc. Natl. Acad. Sci. USA. 1995;92:4952–4956. doi: 10.1073/pnas.92.11.4952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Herman PK, et al. A novel protein kinase homolog essential for protein sorting to the yeast lysosome-like vacuole. Cell. 1991;64:425–437. doi: 10.1016/0092-8674(91)90650-n. [DOI] [PubMed] [Google Scholar]
- 13.Schu PV, et al. Phosphatidylinositol 3-kinase encoded by yeast VPS34 gene essential for protein sorting. Science. 1993;260:88–91. doi: 10.1126/science.8385367. [DOI] [PubMed] [Google Scholar]
- 14.Katzmann DJ, et al. Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex ESCRT-I. Cell. 2001;106:145–155. doi: 10.1016/s0092-8674(01)00434-2. [DOI] [PubMed] [Google Scholar]
- 15.Babst M. A protein’s final ESCRT. Traffic. 2005;6:2–9. doi: 10.1111/j.1600-0854.2004.00246.x. [DOI] [PubMed] [Google Scholar]
- 16.Walch-Solimena C, Novick P. The yeast phosphatidylinositol-4-OH kinase Pik1 regulates secretion at the Golgi. Nature Cell Biol. 1999;1:523–555. doi: 10.1038/70319. [DOI] [PubMed] [Google Scholar]
- 17.Hama H, et al. Direct involvement of phosphatidylinositol-4-phosphate in secretion in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 1999;274:34294–34301. doi: 10.1074/jbc.274.48.34294. [DOI] [PubMed] [Google Scholar]
- 18.Cleves AE, et al. Mutations in the SAC1 gene suppress defects in yeast Golgi and yeast actin function. J. Cell Biol. 1989;109:2939–2950. doi: 10.1083/jcb.109.6.2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Whitters EA, et al. SAC1p is an integral membrane protein that influences the cellular requirement for phospholipid transfer protein function and inositol in yeast. J Cell Biol. 1993;122:79–94. doi: 10.1083/jcb.122.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo S, et al. SAC1-like domains of yeast SAC1, INP52 and INP53, and human synaptojanin encode polyphosphoinositide phosphatases. J. Biol. Chem. 1999;274:12260–12265. doi: 10.1074/jbc.274.19.12990. [DOI] [PubMed] [Google Scholar]
- 21.Rivass MP, et al. Relationship between altered phospholipid metabolism, DAG, 'bypass Sec14p', and the inositol auxotrophy of yeast sac1 mutants. Mol. Biol. Cell. 1999;10:2235–2250. doi: 10.1091/mbc.10.7.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fang M, et al. Kes1p shares homology with human oxysterol binding protein and participates in a novel regulatory pathway for yeast Golgi-derived transport vesicle biogenesis. EMBO J. 1996;15:6447–6459. [PMC free article] [PubMed] [Google Scholar]
- 23.Li X, et al. Analysis of oxysterol binding protein homologue Kes1p function in regulation of Sec14p-dependent protein transport from the yeast Golgi complex. J Cell Biol. 2002;157:63–77. doi: 10.1083/jcb.200201037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fairn GD, et al. The oxysterol binding protein Kes1p regulates Golgi apparatus phosphatidylinositol-4-phosphate function. Proc Natl Acad Sci U S A. 2007;104:15352–15357. doi: 10.1073/pnas.0705571104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schaaf G, et al. The functional anatomy of phospholipid binding and regulation of phosphoinositide homeostasis by proteins of the Sec14-superfamily. Molecular Cell. 2008;29:191–206. doi: 10.1016/j.molcel.2007.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.James DJ, et al. Phosphatidylinositol 4,5-bisphosphate regulates SNARE-dependent membrane fusion. J Cell Biol. 2008;182:355–366. doi: 10.1083/jcb.200801056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murray DH, Tamm LK. Clustering of syntaxin-1A in model membranes is modulated by phosphatidylinositol 4,5-bisphosphate and cholesterol. Biochemistry. 2009;48:4617–4625. doi: 10.1021/bi9003217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brown MT, et al. ASAP1, a phospholipid-dependent ARF GTPase-activating protein that associates with and is activated by Src. Mol. Cell. Biol. 1998;18:7038–7051. doi: 10.1128/mcb.18.12.7038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yanagisawa L, et al. Activity of specific lipid-regulated ARFGAPs is required for Sec14p-dependent Golgi secretory function in yeast. Mol. Biol. Cell. 2002;13:2193–2206. doi: 10.1091/mbc.01-11-0563.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kearns BG, et al. Essential role for diacylglycerol in protein transport from the yeast Golgi complex. Nature. 1997;387:101–105. doi: 10.1038/387101a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baron CL, Malhotra V. Role of diacylglycerol in PKD recruitment to the TGN and protein transport to the plasma membrane. Science. 2002;295:325–328. doi: 10.1126/science.1066759. [DOI] [PubMed] [Google Scholar]
- 32.Litvak V, et al. Maintenance of the diacylglycerol level in the Golgi apparatus by the Nir2 protein is critical for Golgi secretory function. Nat Cell Biol. 2005;7:225–234. doi: 10.1038/ncb1221. [DOI] [PubMed] [Google Scholar]
- 33.Fernandez-Ulibarri I, et al. Diacylglycerol is required for the formation of COPI vesicles in the Golgi-to-ER transport pathway. Mol Biol Cell. 2007;18:3250–3263. doi: 10.1091/mbc.E07-04-0334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Asp L, et al. Early stages of Golgi vesicle and tubule formation require diacylglycerol. Mol Biol Cell. 2009;20:780–790. doi: 10.1091/mbc.E08-03-0256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Antonny B, et al. Activation of ADP-ribosylation factor 1 GTPase-activating protein by phosphatidylcholine-derived diacylglycerols. J. Biol. Chem. 1997;272:30848–30851. doi: 10.1074/jbc.272.49.30848. [DOI] [PubMed] [Google Scholar]
- 36.Baron CL, Malhotra V. Role of diacylglycerol in PKD recruitment to the TGN and protein transport to the plasma membrane. Science. 2002;295:325–328. doi: 10.1126/science.1066759. [DOI] [PubMed] [Google Scholar]
- 37.Bard F, Malhotra V. The formation of TGN-to-plasma-membrane transport carriers. Annu Rev Cell Dev Biol. 2006;22:439–455. doi: 10.1146/annurev.cellbio.21.012704.133126. [DOI] [PubMed] [Google Scholar]; 37 Bossard C, et al. Dimeric PKD regulates membrane fission to form transport carriers at the TGN. J. Cell Biol. 2007;179:1123–1131. doi: 10.1083/jcb.200703166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Proszynski TJ, et al. A genome-wide visual screen reveals a role for SLs and ergosterol in cell surface delivery in yeast. Proc. Natl. Acad. Sci. 2005;102:17981–17986. doi: 10.1073/pnas.0509107102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Klemm RW, et al. Segregation of sphingolipids and sterols during formation of secretory vesicles at the trans-Golgi network. J. Cell Biol. 2009;186:601–612. doi: 10.1083/jcb.200901145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muthusamy B-P, et al. Linking phospholipid flippases to vesicle-mediated transport. Biochim. Biophys. Acta. 2009 doi: 10.1016/j.bbalip.2009.03.004. (In Press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lemmon MA. Membrane recognition by phospholipid-binding domains. Nat Rev Mol Cell Biol. 2008;9:26–111. doi: 10.1038/nrm2328. [DOI] [PubMed] [Google Scholar]
- 42.Balla T. Inositol-lipid binding motifs: signal integrators through protein–lipid and protein–protein interactions. J. Cell Sci. 2005;118:2093–2104. doi: 10.1242/jcs.02387. [DOI] [PubMed] [Google Scholar]
- 43.Roth MG. Phosphoinositides in constitutive membrane traffic. Physiol Rev. 2004;84:699–730. doi: 10.1152/physrev.00033.2003. [DOI] [PubMed] [Google Scholar]
- 44.Di Paolo G, De Camilli P. Phosphoinositides in cell regulation and membrane dynamics. Nature. 2006;443:651–657. doi: 10.1038/nature05185. [DOI] [PubMed] [Google Scholar]
- 45.D’Angelo G, et al. The multiple roles of PtdIns(4)P – not just the precursor of PtdIns(4,5)P2. J. Cell Science. 2008;121:1955–1963. doi: 10.1242/jcs.023630. [DOI] [PubMed] [Google Scholar]
- 46.Allen-Baume V, et al. Current thoughts on the phosphatidylinositol transfer protein family. FEBS Lett. 2002;531:74–80. doi: 10.1016/s0014-5793(02)03412-9. [DOI] [PubMed] [Google Scholar]
- 47.Cockcroft S, Carvou N. Biochemical and biological functions of class I phosphatidylinositol transfer proteins. Biochim Biophys Acta. 2007;1771:677–691. doi: 10.1016/j.bbalip.2007.03.009. [DOI] [PubMed] [Google Scholar]
- 48.Phillips SE, et al. The diverse biological functions of phosphatidylinositol transfer proteins in eukaryotes. Crit. Rev. in Bioch. & Mol. Biol. 2006;41:1–28. doi: 10.1080/10409230500519573. [DOI] [PubMed] [Google Scholar]
- 49.De Matteis MA, et al. Lipid-transfer proteins in membrane trafficking at the Golgi complex. Biochim. Biophys. Acta. 2008;1771:761–768. doi: 10.1016/j.bbalip.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 50.Neumann S, van Meer G. Sphingolipid management by an orchestra of lipid transfer proteins. Biol. Chem. 2008;389:1349–1360. doi: 10.1515/BC.2008.154. [DOI] [PubMed] [Google Scholar]
- 51.Salama SR, et al. Cloning and characterization of the Kluyveromyces lactis SEC14: A gene whose product stimulates Golgi secretory function in S. cerevisiae. J. Bacteriol. 1990;172:4510–4521. doi: 10.1128/jb.172.8.4510-4521.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xie Z, et al. Phospholipase D activity is required for suppression of yeast phosphatidylinositol transfer protein defects. Proc. Natl. Acad. Sci. U.S.A. 1998;95:12346–12351. doi: 10.1073/pnas.95.21.12346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Phillips SE, et al. Yeast Sec14p deficient in phosphatidylinositol transfer activity is functional in vivo. Mol. Cell. 1999;4:187–197. doi: 10.1016/s1097-2765(00)80366-4. [DOI] [PubMed] [Google Scholar]
- 54.Sha B, et al. Crystal structure of the Saccharomyces cerevisiae phosphatidylinositol transfer protein Sec14. Nature. 1998;391:506–510. doi: 10.1038/35179. [DOI] [PubMed] [Google Scholar]
- 55.Li X, et al. Identification of a novel family of nonclassic yeast phosphatidylinositol transfer proteins whose function modulates phospholipase D activity and Sec14p-independent cell growth. Mol Biol Cell. 2002;11:1989–2005. doi: 10.1091/mbc.11.6.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Skinner HB, et al. Phosphatidylinositol transfer protein stimulates yeast Golgi secretory function by inhibiting choline-phosphate cytidylyltransferase activity. Proc. Natl. Acad. Sci. U.S.A. 1265;92:112–116. doi: 10.1073/pnas.92.1.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu WI, et al. A new gene involved in the transport-dependent metabolism of phosphatidylserine, PSTB2/PDR17, shares sequence similarity with the gene encoding the phosphatidylinositol/phosphatidylcholine transfer protein, SEC14. J Biol Chem. 2000;275:14446–14456. doi: 10.1074/jbc.275.19.14446. [DOI] [PubMed] [Google Scholar]
- 58.Carmen-Lopez M, et al. A phosphatidylinositol/phosphatidylcholine transfer protein is required for differentiation of the dimorphic yeast Yarrowia lipolytica from the yeast to the mycelial form. J. Cell Biol. 1994;124:113–127. doi: 10.1083/jcb.125.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nakase Y, et al. The Schizosaccharomyces pombe spo20(+) gene encoding a homologue of Saccharomyces cerevisiae Sec14 plays an important role in forespore membrane formation. Mol. Biol Cell. 2001;4:901–917. doi: 10.1091/mbc.12.4.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rudge SA, et al. Roles of phosphoinositides and of Spo14p (phospholipase D)-generated phosphatidic acid during yeast sporulation. Mol Biol Cell. 2004;15:207–218. doi: 10.1091/mbc.E03-04-0245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vincent P, et al. A Sec14p-nodulin domain phosphatidylinositol transfer protein polarizes membrane growth of Arabidopsis thaliana root hairs. J. Cell Biol. 2005;168:801–812. doi: 10.1083/jcb.200412074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Aravind L, et al. Sec14p-like domains in NF1 and Dbl-like proteins indicate lipid regulation of Ras and Rho signaling. Curr. Biol. 1999;9:R195–R197. doi: 10.1016/s0960-9822(99)80127-4. [DOI] [PubMed] [Google Scholar]
- 63.Shang X, et al. Concerted regulation of cell dynamics by BNIP-2 and Cdc42GAP homology/Sec14p-like, proline-rich, and GTPase-activating protein domains of a novel rhoGTPase-activating protein, BPGAP1. J. Biol. Chem. 2003;278:45903–45914. doi: 10.1074/jbc.M304514200. [DOI] [PubMed] [Google Scholar]
- 64.Tcherkezian J, Lamarche-Vane N. Current knowledge of the large RhoGAP family of proteins. Biol. Cell. 2007;26:67–86. doi: 10.1042/BC20060086. [DOI] [PubMed] [Google Scholar]
- 65.Sirokmany G, et al. Sec14 homology domain targets p50RhoGAP to endosomes and provides a link between Rab- and Rho GTPases. J. Biol. Chem. 2005;281:6096–6105. doi: 10.1074/jbc.M510619200. [DOI] [PubMed] [Google Scholar]
- 66.Ueda S, et al. Role of the Sec14-like domain of Dbl family exchange factors in the regulation of Rho family GTPases in different subcellular sites. Cell Signal. 2004;16:826–906. doi: 10.1016/j.cellsig.2004.01.007. [DOI] [PubMed] [Google Scholar]
- 67.Debant A, et al. The multidomain protein Trio binds the LAR transmembrane tyrosine phosphatase, contains a protein kinase domain, and has separate rac-specific and rho-specific guanine nucleotide exchange factor domains. Proc. Natl. Acad. Sci. U.S.A. 1996;93:5466–5471. doi: 10.1073/pnas.93.11.5466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gu M, et al. Cloning and expression of a cytosolic megakaryocyte protein-tyrosine- phosphatase with sequence homology to retinaldehyde-binding protein and yeast SEC14p. Proc Natl Acad Sci U S A. 1992;89:2980–2984. doi: 10.1073/pnas.89.7.2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kapranov P, et al. Nodule-specific regulation of phosphatidylinositol transfer protein expression in Lotus japonicus. Plant Cell. 2001;13:1369–1382. doi: 10.1105/tpc.13.6.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Anantharaman V, Aravind L. The GOLD domain, a novel protein module involved in Golgi function and secretion. Genome Biol. 2002;3:0023. doi: 10.1186/gb-2002-3-5-research0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dee CT, Moffat KG. A novel family of mitochondrial proteins is represented by the Drosophila genes slmo, preli-like and real-time. Dev. Genes Evol. 2005;215:248–254. doi: 10.1007/s00427-005-0470-4. [DOI] [PubMed] [Google Scholar]
- 72.Habermehl D, et al. Recombinant SEC14-like proteins (TAP) possess GTPase activity. Biochem. Biophys. Res. Comm. 2004;326:254–259. doi: 10.1016/j.bbrc.2004.11.021. [DOI] [PubMed] [Google Scholar]
- 73.Liu T, et al. Structural insights into the cellular retinaldehyde-binding protein (CRALBP) PROTEINS: Structure, Function, and Bioinformatics. 2005;61:412–422. doi: 10.1002/prot.20621. [DOI] [PubMed] [Google Scholar]
- 74.Bomar JM, et al. Mutations in a novel gene encoding a CRAL-TRIO domain cause human Cayman ataxia and ataxia/dystonia in the jittery mouse. Nature Genetics. 2003;35:264–269. doi: 10.1038/ng1255. [DOI] [PubMed] [Google Scholar]
- 75.Gotoda T, et al. Adult-onset spinocerebellar dysfunction caused by a mutation in the gene for alpha-tocopherol transfer protein. N. Engl. J. Med. 1995;333:1313–1318. doi: 10.1056/NEJM199511163332003. [DOI] [PubMed] [Google Scholar]
- 76.Hentati A, et al. Human alpha-tocopherol transfer protein: gene structure and mutations in familial vitamin E deficiency. Ann. Neurol. 1996;39:295–300. doi: 10.1002/ana.410390305. [DOI] [PubMed] [Google Scholar]
- 77.Golovleva I, et al. Disease causing mutations in the cellular retinaldehyde binding protein tighten and abolish ligand interactions. J. Biol. Chem. 2003;278:12397–12402. doi: 10.1074/jbc.M207300200. [DOI] [PubMed] [Google Scholar]
- 78.Cichowski K, Jacks T. NF1 tumor suppressor gene function: narrowing the GAP. Cell. 2001;104:593–604. doi: 10.1016/s0092-8674(01)00245-8. [DOI] [PubMed] [Google Scholar]
- 79.Ouachi K, et al. Ataxia with vitamin E deficiency is caused by mutations in the α-tocopherol transfer protein. Nature Genetics. 1995;9:141–145. doi: 10.1038/ng0295-141. [DOI] [PubMed] [Google Scholar]
- 80.Maw MA, et al. Mutation of the gene encoding cellular retinaldehyde-binding protein in autosomal recessive retinitis pigmentosa. Nat. Genet. 1997;17:198–200. doi: 10.1038/ng1097-198. [DOI] [PubMed] [Google Scholar]
- 81.Fishman GA, et al. Vovel mutations in the cellular retinaldehyde-binding protein gene (RLBP1) associated with retinitis punctata albescens: evidence of interfamilial genetic heterogeneity and fundus changes in heterozygotes. Arch. Opthamol. 2004;122:70–75. doi: 10.1001/archopht.122.1.70. [DOI] [PubMed] [Google Scholar]
- 82.Min KC, et al. Crystal structure of α-tocopherol transfer protein bound to its ligand: Implications for ataxia with vitamin E deficiency. Proc. Natl. Acad. Sci. 2003;100:14713–14718. doi: 10.1073/pnas.2136684100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Meier R, et al. The molecular basis of vitamin E retention: structure of human alpha-tocopherol transfer protein. J Mol Biol. 2003;331:725–734. doi: 10.1016/s0022-2836(03)00724-1. [DOI] [PubMed] [Google Scholar]
- 84.Stocker A, Baumann U. Supernatant protein factor in complex with RRR-alpha-tocopherylquinone: a link between oxidized vitamin E and cholesterol biosynthesis. J. Mol. Biol. 2003;332:759–765. doi: 10.1016/s0022-2836(03)00924-0. [DOI] [PubMed] [Google Scholar]
- 85.D'Angelo I, et al. A novel bipartite phospholipid-binding module in the neurofibromatosis type 1 protein. EMBO Rep. 2006;7:174–179. doi: 10.1038/sj.embor.7400602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Welti S, et al. The sec14 homology module of neurofibromin binds cellular glycerophospholipids: mass spectrometry and structure of a lipid complex. J Mol Biol. 2007;366:551–562. doi: 10.1016/j.jmb.2006.11.055. [DOI] [PubMed] [Google Scholar]
- 87.Ryan MM, et al. Conformational dynamics of the major yeast phosphatidylinositol transfer protein Sec14: Insights into the mechanisms of PL exchange and diseases of Sec14-like protein deficiencies. Mol. Biol. Cell. 2007;18:1928–1942. doi: 10.1091/mbc.E06-11-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Smirnova T, et al. Local polarity and hydrogen bonding inside the Sec14 PL-binding cavity: High-field multifrequency studies. Biophys. J. 2007;92:3686–3695. doi: 10.1529/biophysj.106.097899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Roderick SL, et al. Structure of human phosphatidylcholine transfer protein in complex with its ligand. Nat Struct Biol. 2002;9:507–511. doi: 10.1038/nsb812. [DOI] [PubMed] [Google Scholar]
- 90.Stack JH, et al. A membrane-associated complex containing the Vps15 protein kinase and Vps34 PI 3-kinase is essential for protein sorting to the yeast lysosome-like vacuole. EMBO J. 1993;12:2195–2204. doi: 10.1002/j.1460-2075.1993.tb05867.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hendricks, et al. Yeast homologue of neuronal frequenin is a regulator of phosphatidylinositol-4-OH kinase. Nature Cell Biol. 1999;1:234–241. doi: 10.1038/12058. [DOI] [PubMed] [Google Scholar]
- 92.Baird, et al. Assembly of the PtdIns 4-kinase Stt4 complex requires Ypp1 and Efr3. J. Cell Biol. 2008;183:1061–1074. doi: 10.1083/jcb.200804003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Strahl T, Thorner J. Synthesis and function of membrane phosphoinositides in budding yeast, Saccharomyces cerevisiae. Biochim. Biophys. Acta. 2007;1771:353–404. doi: 10.1016/j.bbalip.2007.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ile KE, et al. Phosphatidylinositol transfer proteins and cellular nanoreactors for lipid signaling. Nature Chem. Biol. 2006;2:576–583. doi: 10.1038/nchembio835. [DOI] [PubMed] [Google Scholar]
- 95.Routt SM, et al. Nonclassical PITPs activate phospholipase D via an Stt4p-dependent pathway and modulate function of late stages of the secretory pathway in vegetative yeast cells. Traffic. 2005;6:1157–1172. doi: 10.1111/j.1600-0854.2005.00350.x. [DOI] [PubMed] [Google Scholar]
- 96.Skinner HB, et al. Phospholipid transfer activity is relevant to but not sufficient for the essential function of the yeast SEC14 gene product. EMBO J. I. 1993:4775–4784. doi: 10.1002/j.1460-2075.1993.tb06166.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kostenko EV, et al. The Sec14 homology domain regulates the cellular distribution and transforming activity of the Rho-specific guanine nucleotide exchange factor Dbs. J. Biol. Chem. 2004;280:2807–2817. doi: 10.1074/jbc.M411139200. [DOI] [PubMed] [Google Scholar]
- 98.Yoder MD, et al. Structure of a multifunctional protein: Mammalian phosphatidylinositol transfer protein complexed with phosphatidylcholine. J. Biol. Chem. 2001;276:9246–9252. doi: 10.1074/jbc.M010131200. [DOI] [PubMed] [Google Scholar]
- 99.Tilley SJ, et al. Structure-function analysis of phosphatidylinositol transfer protein alpha bound to human phosphatidylinositol. Structure. 2004;12:317–326. doi: 10.1016/j.str.2004.01.013. [DOI] [PubMed] [Google Scholar]
- 100.Yan D, Olkkonen VM. OSBP-related proteins: lipid sensors or transporters? Future Lipidology. 2007;2:85–94. [Google Scholar]
- 101.Hanada, et al. CERT and intracellular trafficking of ceramide. Biochim. Biophys. Acta. 2007;1771:644–653. doi: 10.1016/j.bbalip.2007.01.009. [DOI] [PubMed] [Google Scholar]
- 102.Preuss ML, et al. A role for the RabA4b effector protein PI-4Kbeta1 in polarized expansion of root hairs in Arabidopsis thaliana. J. Cell Biol. 2006;172:261–268. doi: 10.1083/jcb.200508116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Stenzel I, et al. The type B phosphatidylinositol-4-phosphate 5-kinase 3 is essential for root hair formation in Arabidopsis thaliana. Plant Cell. 2008;20:124–141. doi: 10.1105/tpc.107.052852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.McLaughlin S, Murray D. Plasma membrane phosphoinositide organization by protein electrostatics. Nature. 2005;438:605–611. doi: 10.1038/nature04398. [DOI] [PubMed] [Google Scholar]
- 105.Peterman TK, et al. Patellin1, a novel Sec14-like protein, localizes to the cell plate and binds phosphoinositides. Plant Physiol. 2004;136:3080–3094. doi: 10.1104/pp.104.045369. [DOI] [PMC free article] [PubMed] [Google Scholar]