Abstract
The reaction cycle of the double-ring chaperonin GroEL is driven by ATP binding that takes place with positive cooperativity within each seven-membered ring and negative cooperativity between rings. The positive cooperativity within rings is due to ATP binding-induced conformational changes that are fully concerted. Herein, it is shown that the mutation Asp-155 → Ala leads to an ATP-induced break in intra-ring and inter-ring symmetry. Electron microscopy analysis of single-ring GroEL particles containing the Asp-155 → Ala mutation shows that the break in intra-ring symmetry is due to stabilization of allosteric intermediates such as one in which three subunits have switched their conformation while the other four have not. Our results show that eliminating an intra-subunit interaction between Asp-155 and Arg-395 results in conversion of the allosteric switch of GroEL from concerted to sequential, thus demonstrating that its allosteric behavior arises from coupled tertiary conformational changes.
Keywords: allostery, chaperonin, protein folding, cooperativity, electron microscopy
Allosteric regulation of oligomeric protein function is often achieved by ligand-induced conformational changes that give rise to cooperativity in binding of the same ligand (homotropic cooperativity) or other ligands (heterotropic cooperativity) (1). Several models, in particular the Monod–Wyman–Changeux (MWC) model (2) and the Koshland–Némethy–Filmer (KNF) model (3), have been developed to describe cooperativity in ligand binding. In both models, cooperativity is due to ligand binding-induced conformational changes that may be either concerted (MWC), sequential (KNF), or a combination of both (4).
A striking allosteric system is the chaperonin GroEL, which promotes protein folding in vivo and in vitro in an ATP-dependent manner (for reviews, see refs. 5–7). GroEL consists of 14 identical subunits that form two stacked back-to-back heptameric rings (8), the cavities of which provide a protective environment for protein folding. It undergoes ATP-induced conformational changes (9, 10) that are responsible for the alternation (between protein substrate-binding and -release states) that is crucial for its folding function (11–13). ATP-induced conformational changes may also provide the energy for forced unfolding of bound misfolded protein substrates (14). Steady-state measurements of initial rates of ATP hydrolysis by GroEL at different concentrations of ATP showed that it undergoes two ATP-induced allosteric transitions: one with a midpoint at relatively low ATP concentrations and the second at higher ATP concentrations (15). Each of the allosteric transitions is reflected in intra-ring positive cooperativity. The higher ATP concentration required to effect the second allosteric transition reflects the inter-ring negative cooperativity. A nested model for cooperativity in ATP binding by GroEL that describes these findings was proposed (12, 15) in which, in accordance with the Monod–Wyman–Changeux representation (2), each ring is in equilibrium between two states that interconvert in a concerted manner: a T state, with low affinity for ATP and high affinity for nonfolded protein substrates, and an R state with high affinity for ATP and low affinity for nonfolded protein substrates. The GroEL double-ring undergoes sequential ATP-induced Koshland–Némethy–Filmer-type (3) transitions from the TT state via the TR state to the RR state.
Several lines of evidence indicate that the allosteric transition of each GroEL ring is indeed concerted. It was shown by generating the Asp-83 → Cys, Lys-327 → Cys double mutant of GroEL that a disulphide cross-link at these positions in only one subunit of each ring is sufficient to block its T → R allosteric transition (ref. 16 and G. Curien and G. H. Lorimer, unpublished results). In addition, a value of one for the ratio between the Hill coefficient values determined from steady-state data and transient kinetic data was observed for a series of GroEL mutants (17), indicating that the allosteric transition of each GroEL ring is concerted (18). Finally, computer simulations have shown that steric repulsions would arise if one subunit changed its conformation while its neighbors did not (19, 20). Herein, it is shown that the allosteric transition of a GroEL ring is converted from concerted to sequential by introducing the single mutation Asp-155 → Ala.
Materials and Methods
Mutant Construction and Purification. The Phe-44 → Trp, Asp-155 → Ala GroEL double mutant was generated as before (21) by using single-stranded DNA of the plasmid pOA (21) containing the gene for the Phe-44 → Trp GroEL mutant (22) and the mutagenic oligonucleotide (Asp-155 → Ala): 5′-ACCTACGGTTTCGGCGGAGTTAGCGGA-3′. Construction of the Phe-44 → Trp GroEL mutant has been described (22). A single-ring version of GroEL containing the single mutation Phe-44 → Trp was generated by PCR by using double-stranded DNA of the SR1 plasmid (23) and the following oligonucleotides: 5′-GGATAAATCTTGGGGTGCACCGA-3′ (Phe-44 → Trp FOR); and 5′-TCGGTGCACCCCAAGATTTATCC-3′ (Phe-44 → Trp BACK). The single-ring version of GroEL containing the mutations Phe-44 → Trp and Asp-155 → Ala was generated by digesting the Phe-44 → Trp, Asp-155 → Ala pOA plasmid with ClaI and SacII and subcloning the resulting fragment into the SR1 plasmid. Protein expression was carried out as before (21, 23), and purification was achieved as described (22).
Kinetic Experiments. All of the steady-state and transient kinetic experiments were carried out at 25°C in 50 mM Tris·HCl buffer (pH 7.5) containing 10 mM MgCl2, 10 mM KCl, and 1 mM DTT (buffer A). The ATPase activity of GroEL was measured as described (21) by using a final GroEL oligomer concentration of 25 nM. ATP-induced conformational changes in GroEL were initiated by rapid mixing of equal volumes of different nucleotide concentrations and GroEL by using an Applied Photophysics (Leatherhead, U.K.) SX.17MV stopped-flow apparatus. The final GroEL oligomer concentration in these experiments was 0.25 μM. The conformational changes were followed by excitation at 295 nm and monitoring the fluorescence at wavelengths longer than 320 nm by using a cutoff filter. A 0.2-cm pathlength was used, and both the entrance and exit monochromator wavelength band passes were set to 7 nm. Six or more traces (each with 4,000 data points) were collected by using a split time base and averaged for each concentration of ATP. The data were fitted to a triple-exponential equation with a floating end point yielding estimates for the amplitudes and apparent rate constants. Plots of residuals with random deviations about zero were obtained for all of the kinetic traces.
Kinetic Data Analysis. Data of initial ATPase velocities by the Phe-44 → Trp, Asp-155 → Ala GroEL double mutant at different ATP concentrations were fitted to:
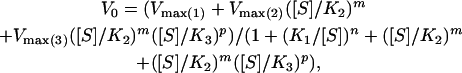 |
[1] |
where V0 is the observed initial rate of ATP hydrolysis; [S] is the substrate (ATP) concentration; Vmax(1), Vmax(2), and Vmax(3) are the respective maximal initial rates of ATP hydrolysis corresponding to the three allosteric transitions; n, m, and p are the respective Hill coefficients for the three allosteric transitions; and K1, K2, and K3 are the respective apparent binding constants of ATP for the three allosteric transitions. Data of initial ATPase velocities by the Phe-44 → Trp mutant at different ATP concentrations were fitted to an equation similar to Eq. 1 for two allosteric transitions (24). Data of observed rate constants of the T → R conformational change of the Phe-44 → Trp, Asp-155 → Ala GroEL double mutant at different ATP concentrations were fitted to:
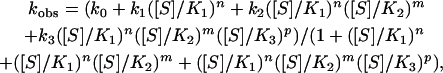 |
[2] |
where kobs is the observed forward rate constant of the conformational change; k0, k1, k2, and k3 are the respective forward rate constants of conformational change in the absence of ATP and at saturating ATP concentrations required to effect the three allosteric transitions; and [S], n, m, p, K1, K2, and K3 are defined as before. Data of observed rate constants of the T → R conformational change of the Phe-44 → Trp mutant at different ATP concentrations were fitted to an equation similar to Eq. 2 for two allosteric transitions (22, 25). All data fitting was carried out by using origin 7 (OriginLab, Northhampton, MA). Estimates of parameters (± SE) are reported.
Electron Microscopy. Samples of 50 nM of a single-ring version of GroEL (23) containing the Phe-44 → Trp single mutation or the Phe-44 → Trp, Asp-155 → Ala double mutation were rapidly mixed with buffer A with or without 5 or 100 μM ATP and then fixed within 6–8 s on fresh glow-discharged copper carbon-coated grids with 1% uranyl acetate. Samples were then imaged on an FEI (Eindhoven, The Netherlands) Tecnai F20 transmission electron microscope. Images were directly recorded by using a 1,024 × 1,024 Tietz Video and Image Processing Systems (Gauting, Germany) Biocam charge-coupled device camera at a magnification of ×93,620. Between 6 and 10 tiled images (3 × 3) were recorded for each of the experimental conditions, with defocus values ranging from 1.0 to 2.0 μm in 90% of the cases and from 2.0 to 2.7 μm in the rest. Particle images recognized as top views were manually selected by using boxer (26). The total number of particles for each of the experimental conditions was in the range of 680–800. Contrast transfer-phase flips were corrected for all particles by using ctfit (26). Centered particles were normalized, low-pass filtered to 1.5 nm, aligned, and classified by using the eman software package (26). Initially, a large number of classes were generated for each of the experimental conditions, with ≈50 particles per class. If visual inspection revealed many similar class averages, then the number of particles per class was increased. This process was continued until all class averages seemed unique. Symmetry analysis was carried out by rotating each class average by N (an integer that runs from 1 to 360) × 1° and calculating the cross-correlation coefficients between each of the rotated class averages and the nonrotated class average by using procedures written with spider (27).
Results
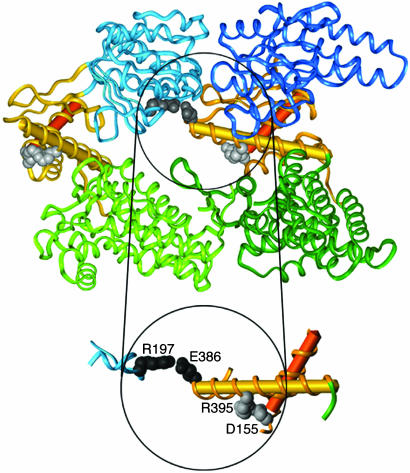
Site of Mutation. Each subunit of GroEL is made up of three domains (8): (i) an apical domain that binds GroES and substrate proteins; (ii) an equatorial domain that contains an ATP binding site and is involved in inter-ring contacts; and (iii) an intermediate domain that connects the apical and equatorial domains (Fig. 1). Asp-155 is a conserved residue at the N-cap of helix G in the intermediate domain. In the apo GroEL structure (8) and in the trans ring of the GroEL-GroES-(ADP)7 complex (28) (without bound ADP), the Oδ1 atom of Asp-155 is hydrogenbonded to the main-chain amides of Thr-l57 and Val-158, and the Oδ2 atom of Asp-155 makes a salt-bridge with NH1 of Arg-395 in helix M in the intermediate domain. In the ADP-bound cis ring of the GroEL–GroES–(ADP)7 complex (28), the Oδ1 atom of Asp-155 is hydrogen bonded to the main-chain amides of Val-155 and Thr-157, but the salt bridge of the Oδ2 atom with NH1 of Arg-395 is broken (the distance changes from 3.19 to 5.41 Å). The break in the Asp-155–Arg-395 salt-bridge accompanies the downward motion of the intermediate domain that occurs at the beginning of the T → R transition. This downward motion brings Asp-398 in helix M into the coordination sphere of the ATP-bound Mg2+, thereby enabling ATP hydrolysis in the R state (29).
Fig. 1.
Ribbon diagram of the structure of two adjacent GroEL subunits (PDB code 1OEL) (33) showing the location of the inter-subunit Arg-197–Glu-386 salt bridge and the intra-subunit Asp-155–Arg-395 salt-bridge. The equatorial, intermediate, and apical domains of the two subunits are shown in different shades of green, gold, and blue, respectively. Helices G and M are shown as red and yellow cylinders, respectively. Single-letter notation for amino acids is used.
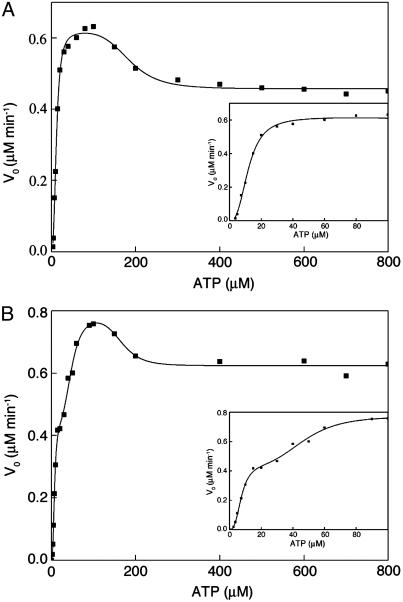
ATPase Activity. The mutation Asp-155 → Ala was introduced into a variant of GroEL containing the mutation Phe-44 → Trp so that its effects on ATP-induced conformational changes could be determined by monitoring time-resolved changes in fluorescence (wild-type GroEL has no tryptophan residues). The allosteric properties of the Phe-44 → Trp mutant are similar to those of wild-type GroEL (22). Plots of initial rates of ATP hydrolysis by wild-type GroEL and the Phe-44 → Trp mutant as a function of ATP concentration were previously found to be bisigmoidal (15) (Fig. 2A). The two sigmoidal phases reflect the ATP-induced allosteric transitions of the two rings. In contrast, the kinetic profile of the Phe-44 → Trp, Asp-155 → Ala mutant is found to be triphasic (Fig. 2B), thereby suggesting that it undergoes at least three ATP-induced allosteric transitions. The data in Fig. 2 for the Phe-44 → Trp variant and the Phe-44 → Trp, Asp-155 → Ala double mutant were fitted to Hill-type equations that describe two (24) or three (Eq. 1) allosteric transitions, respectively. The three allosteric transitions of the Phe-44 → Trp, Asp-155 → Ala mutant are found to have midpoints of 8 (± 1), 47 (± 4), and 165 (± 18) μM. The corresponding Hill coefficients of these three allosteric transitions have values of 2.9 (± 0.6), 4.0 (± 1.4), and 7 (± 4), respectively. In the case of the Phe-44 → Trp mutant, the two allosteric transitions were found to have midpoints of 12.0 (± 0.3) and 185 (± 12) μM and Hill coefficients with values of 2.7 (± 0.2) and 5.3 (± 1.8), respectively.
Fig. 2.
Initial velocities of ATP hydrolysis by the Phe-44 → Trp single mutant (A) and the Phe-44 → Trp, Asp-155 → Ala double mutant (B) as a function of ATP concentration. The reactions were carried out as described in Materials and Methods. The data for the Phe-44 → Trp single mutant (taken from ref. 22) and the Phe-44 → Trp, Asp-155 → Ala double mutant were fitted to a Hill-type equation for two allosteric transitions (24) or Eq. 1 for three allosteric transitions, respectively. (Insets) The data for low ATP concentrations.
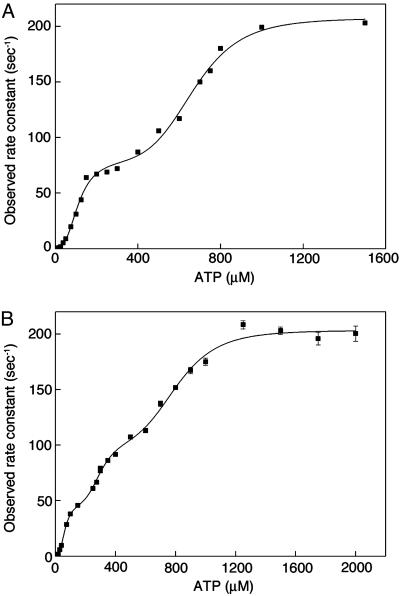
Transient Kinetic Analysis. Next, we wanted to determine whether the above-described triphasic steady-state kinetic profile of the Phe-44 → Trp, Asp-155 → Ala mutant is due to an effect of the mutation on its allosteric mechanism or to an effect on its catalytic activity (a V-system effect). Equal volumes of different concentrations of ATP and GroEL were rapidly mixed, and the time-resolved change in fluorescence emission at wavelengths >320 nm, on excitation at 295 nm, was followed. The data were fitted to a triple-exponential equation with a floating end point, yielding estimates for amplitudes and rate constants. Here, we consider only the observed rate constant corresponding to the fast phase with the largest amplitude that reflects the ATP binding-induced T → R conformational change (22). In the case of the Phe-44 → Trp mutant, the value of this observed rate constant was previously found to display a bisigmoidal dependence on ATP concentration (22) (Fig. 3A) that mirrors the steady-state ATPase data for this mutant (Fig. 2 A). These transient kinetic data were fitted to a Hill-type equation, yielding estimates for the values of the two Hill coefficients of 2.9 (± 0.5) and 5.9 (± 0.7) (ref. 22). In the case of the Phe-44 → Trp, Asp-155 → Ala mutant, the plot of the observed rate constant of the ATP binding-induced T to R conformational change as a function of ATP concentration is found to be triphasic (Fig. 3B). A fit of these data to a Hill-type equation for three allosteric transitions (Eq. 2) yielded estimates for the values of the Hill coefficients of 2.9 (± 1.6), 5.0 (± 3.0), and 6.0 (± 1.4). The steady-state ATPase data (Fig. 2B) and transient kinetic data (Fig. 3B) of also this mutant are, therefore, found to mirror each other. Taken together, the steady-state ATPase data and transient kinetic data, thus, indicate that the mutation Asp-155 → Ala caused a change in the mechanism of allosteric switching of the GroEL rings that is reflected in a triphasic kinetic profile.
Fig. 3.
Observed rate constants of the ATP binding-induced T to R conformational change of the Phe-44 → Trp single mutant and the Phe-44 → Trp, Asp-155 → Ala double mutant as a function of ATP concentration. The reactions were carried out as described in Materials and Methods. The data for the Phe-44 → Trp single mutant (A) and Phe-44 → Trp, Asp-155 → Ala double mutant (B) were fitted to a Hill-type equation for two allosteric transitions (22) or Eq. 2 for three allosteric transitions, respectively.
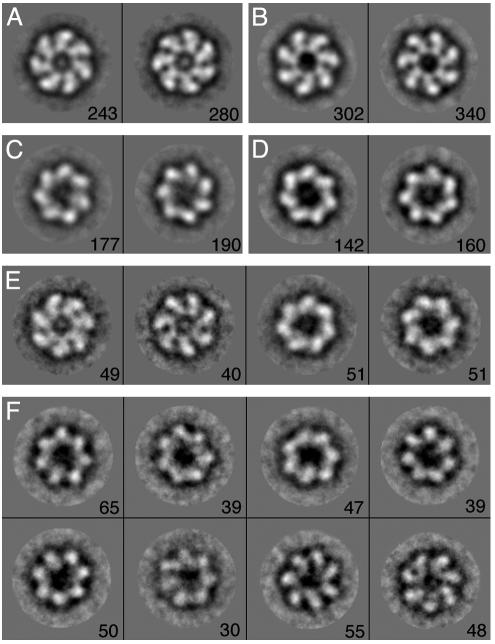
Electron Microscopy Analysis. The kinetic data suggested that the mutation Asp-155 → Ala introduces an ATP-induced intra-ring break in symmetry. This break in symmetry implies that the mutation Asp-155 → Ala converts the concerted t7 → r7 allosteric transition of a GroEL ring (t and r stand for the respective conformations of a subunit in the T and R states and tnr7–n stands for a ring with n adjacent subunits in the t state and 7 – n adjacent subunits in the r state) into a sequential allosteric transition such as t7 → t5r2 → r7 or t7 → t4r3 → r7 (other single sequential pathways or combinations of pathways are also possible). This interpretation suggested that a break in intra-ring symmetry might be visualized at nonsaturating ATP concentrations. We decided to test it by imaging negatively stained single-ring (23) versions of Phe-44 → Trp GroEL, either with or without the Asp-155 → Ala mutation, in the presence of different concentrations of ATP (Fig. 4). The ATP-induced break in intra-ring symmetry is also expected in the case of the single-ring version of Phe-44 → Trp, Asp-155 → Ala GroEL because it undergoes two allosteric transitions (see Fig. 6, which is published as supporting information on the PNAS web site) instead of the one allosteric transition that wild-type single-ring GroEL undergoes (30). Importantly, the symmetry analysis of single-ring particles is more straightforward than that of double-ring particles because projections through only one ring are observed.
Fig. 4.
Representative class averages of single-ring GroEL particles containing the Phe-44 → Trp single mutation or the Phe-44 → Trp, Asp-155 → Ala double mutation in the presence of different concentrations of ATP. The number of particles corresponding to each class average is indicated. The respective class averages for the Phe-44 → Trp single mutant and the Phe-44 → Trp, Asp-155 → Ala double mutant, in the absence of ATP, are shown in A and B. The respective class averages for the Phe-44 → Trp single mutant and the Phe-44 → Trp, Asp-155 → Ala double mutant, in the presence of 100 μM ATP, are shown in C and D. Class averages for the Phe-44 → Trp mutant, in the presence of 5 μM ATP, are shown in E. The class averages for the Phe-44 → Trp, Asp-155 → Ala mutant in the presence of 5 μM ATP are shown in F. Image processing was carried out as described in Materials and Methods.
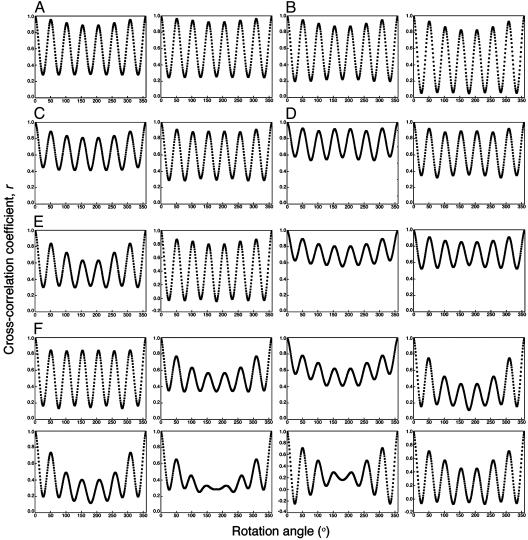
Single-ring particle images recognized as top views were manually selected and then aligned and classified by using eman (26) (Fig. 4). In the absence of ATP, two class averages with opposite handedness are observed for both the Phe-44 → Trp single mutant (Fig. 4A) and the Phe-44 → Trp, Asp-155 → Ala double mutant (Fig. 4B). The number of particles in each of the classes with opposite handedness is found to be about the same in the case of both mutants, thus reflecting equal affinities for the grid of the equatorial and apical domains when ATP is absent. The extent of 7-fold symmetry of each class average was evaluated by rotating it by N (an integer that runs from 1 to 360) × 1° and calculating the cross-correlation coefficient, r, between each of the rotated class averages and the nonrotated one by using spider (27). Plots of r as a function of the rotation angle are found to have maxima at integer multiples of 51.4° (360°/7) in agreement with the real or pseudo 7-fold symmetry of the particles. The values of r at the maxima are found to be high and relatively constant when ATP is absent (Fig. 5 A and B), thus reflecting the presence of the 7-fold symmetry in the apo structures of both mutants.
Fig. 5.
Extent of 7-fold symmetry of class averages of single-ring GroEL particles containing the Phe-44 → Trp single mutation or the Phe-44 → Trp, Asp-155 → Ala double mutation in the presence of different concentrations of ATP. Each class average was rotated by N (an integer that runs from 1 to 360) × 1°. The extent of the 7-fold symmetry of the different class averages was determined by calculating a cross-correlation coefficient, r, between the nonrotated class average and the rotated ones by using procedures written with spider (27). The respective rotational correlation plots for the Phe-44 → Trp single mutant and the Phe-44 → Trp, Asp-155 → Ala double mutant, in the absence of ATP, are shown in A and B. The respective rotational correlation plots for Phe-44 → Trp single mutant and the Phe-44 → Trp, Asp-155 → Ala double mutant, in the presence of 100 μM ATP, are shown in C and D. The respective rotational correlation plots for the Phe-44 → Trp single mutant and the Phe-44 → Trp, Asp-155 → Ala double mutant, in the presence of 5 μM ATP, are shown in E and F.
In the presence of a saturating ATP concentration of 100 μM, class averages with opposite handedness are again observed for both mutants (Fig. 4 C and D), thus indicating that the two orientations of the ATP-bound state also have similar affinities for the grid. It may be seen that the 7-fold symmetry is preserved also in the ATP-bound R state of both mutants (Fig. 5 C and D). In the presence of a nonsaturating ATP concentration of 5 μM, the class averages of the Phe-44 → Trp mutant (Fig. 4E) are found to correspond to those of the unbound T state (Fig. 4A) or the ATP-bound R state (Fig. 4C). The 7-fold symmetry is preserved also in all of these class averages (Fig. 5E), and class averages with opposite handedness are again observed.
In contrast, a break in the 7-fold symmetry is observed in most of the class averages corresponding to the Phe-44 → Trp, Asp-155 → Ala mutant when a nonsaturating ATP concentration of 5 μM is present (Figs. 4F and 5F). This observation is in agreement with the interpretation of the kinetic results for this mutant described above. In addition, a larger number of particle classes are observed in the case of this mutant (Fig. 4F), suggesting that it may undergo several different sequential allosteric transitions. Insight into the nature of the allosteric intermediates of this mutant can be obtained by considering how the value of r at the maxima changes as a function of the rotation angle (see Fig. 7, which is published as supporting information on the PNAS web site). In the case of a particle in the t6r1 state, for example, the value of r at the maxima will decrease after the first rotation by 51.4° and then stay constant on further rotations by 51.4° because the number of superimposed subunits that are in different conformations does not change. In the case of a particle in the t5r2 state, the value of r at the maxima will decrease after both the first and second rotations by 51.4° whereas, in the case of a particle in the t4r3 state, it will decrease after each of the first three rotations by 51.4°. Inspection of the plots of r as a function of the rotation angle for the different class averages of the Phe-44 → Trp, Asp-155 → Ala mutant at 5 μM ATP, therefore, indicates that a major allosteric intermediate is t4r3 although other allosteric intermediates are likely to also exist.
Discussion
Plots of initial rates of ATP hydrolysis and observed rates of the T → R conformational change for the Phe-44 → Trp, Asp-155 → Ala GroEL mutant as a function of ATP concentration were found to be triphasic. These kinetic data suggest that there are two types of break in symmetry in the Phe-44 → Trp, Asp-155 → Ala GroEL mutant: one within rings and the second between rings. The break in symmetry between rings is reflected in the uneven number of observed allosteric transitions (i.e., the two rings undergo a different number of allosteric transitions) whereas the break in symmetry within rings is reflected in the fact that more than two allosteric transitions are observed. The rotational correlation plots (Figs. 5 and 7) suggest that the ATP-induced intra-ring break in symmetry is due to the stabilization of allosteric intermediates such as t4r3. Class average images with four distinct t-like subunits and three distinct r-like subunits are not observed probably because of conformational changes in the unbound subunits as might be expected in the case of a sequential mechanism. In other words, the mutation Asp-155 → Ala converts the concerted allosteric switch of GroEL into a sequential one such as t7 → t4r3 → r7, in agreement with the observed Hill coefficients for the two allosteric transitions of the first ring. A break in intra-ring symmetry in the allosteric transition of the second ring is not observed, probably owing to the much stronger positive cooperativity of that transition (15).
What is the molecular mechanism responsible for the conversion, promoted by the Asp-155 → Ala mutation, of the allosteric switch of GroEL from concerted to sequential? Two dominant contributions to the positive intra-ring cooperativity in GroEL come from (i) steric interference that would arise if one subunit underwent a t → r transition while its neighbors had not (20); and (ii) the inter-subunit salt-links between Glu-386 at the N terminus of helix M in the intermediate domain and Arg-197 in the apical domain that stabilize the T state (31) (Fig. 1) and are broken during the T → R transition (10, 17, 20). Breaking the Asp-155-Arg-395 salt-link on the mutation Asp-155 → Ala seems to also weaken the Arg-197-Glu-386 salt-link, thereby freeing the apical domains. Such energetic coupling between salt bridges has been demonstrated before in other systems (32). Evidence for the freeing of the apical domains has come from cryo-electron microscopy analysis of the apo state of the Phe-44 → Trp, Asp-155 → Ala GroEL mutant at 14 Å resolution that shows large disordering of the apical domains in one but not the other ring (not shown). The asymmetry between rings in the disordering of the apical domains is consistent with the kinetic data that indicate a break in intra-ring symmetry in one ring only. In the presence of MgATP, there is a downward movement of the intermediate domain that is stabilized by the interaction of Asp-398 in helix M with the coordination sphere of the ATP-bound Mg2+. Hence, the equilibrium of an ATP-bound subunit is shifted from t to r. Owing to the freeing of the apical domains in the Phe-44 → Trp, Asp-155 → Ala mutant, the transition t → r of an ATP-bound subunit is not blocked by a repulsive (steric) interaction with a neighboring subunit without bound ATP although the equilibrium of this unbound subunit is consequently shifted toward the r state. Our data suggest that the t → r transition of one ATP-bound subunit shifts the equilibrium of the two neighboring subunits (in the direction from Arg-197 to Glu-386 around the ring) from t toward r. It is striking that breaking an intra-subunit interaction between Asp-155 and Arg-395 results in conversion of the allosteric switch from concerted to sequential. This finding thus provides direct experimental evidence that the allosteric behavior of GroEL arises from large coupled tertiary conformational changes, as suggested by simulations (19).
Supplementary Material
Acknowledgments
We thank Drs. Debbie Fass, Tammy Horovitz, and Ofer Yifrach for critical reading of this paper and Dr. Miriam Eisenstein for discussion and help with preparing Fig. 1. This work was supported by the Israel Science Foundation administered by The Israel Academy of Sciences and Humanities. A.H. is an incumbent of the Carl and Dorothy Bennett Professorial Chair in Biochemistry.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Perutz, M. F. (1989) Q. Rev. Biophys. 22, 139–237. [DOI] [PubMed] [Google Scholar]
- 2.Monod, J., Wyman, J. & Changeux, J.–P. (1965) J. Mol. Biol. 12, 88–118. [DOI] [PubMed] [Google Scholar]
- 3.Koshland, D. E. Jr., Némethy, G. & Filmer, D. (1966) Biochemistry 5, 365–385. [DOI] [PubMed] [Google Scholar]
- 4.Eigen, M. (1967) Nobel Symp. 5, 333–369. [Google Scholar]
- 5.Thirumalai, D. & Lorimer, G. H. (2001) Annu. Rev. Biophys. Biomol. Struct. 30, 245–269. [DOI] [PubMed] [Google Scholar]
- 6.Saibil, H. R., Horwich, A. L. & Fenton, W. A. (2001) Adv. Protein Chem. 59, 45–72. [DOI] [PubMed] [Google Scholar]
- 7.Horovitz, A., Fridmann, Y., Kafri, G. & Yifrach, O. (2001) J. Struct. Biol. 135, 104–114. [DOI] [PubMed] [Google Scholar]
- 8.Braig, K., Otwinowski, Z., Hegde, R., Boisvert, D. C., Joachimiak, A., Horwich, A. L. & Sigler, P. B. (1994) Nature 371, 578–586. [DOI] [PubMed] [Google Scholar]
- 9.Roseman, A. M., Chen, S., White, H., Braig, K. & Saibil, H. R. (1996) Cell 87, 241–251. [DOI] [PubMed] [Google Scholar]
- 10.Ranson, N. A., Farr, G. W., Roseman, A. M., Gowen, B., Fenton, W. A., Horwich, A. L. & Saibil, H. R. (2001) Cell 107, 869–879. [DOI] [PubMed] [Google Scholar]
- 11.Staniforth, R. A., Burston, S. G., Atkinson, T. & Clarke, A. R. (1994) Biochem. J. 300, 651–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yifrach, O. & Horovitz, A. (1996) J. Mol. Biol. 255, 356–361. [DOI] [PubMed] [Google Scholar]
- 13.Yifrach, O. & Horovitz, A. (2000) Proc. Natl. Acad. Sci. USA 97, 1521–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shtilerman, M., Lorimer, G. H. & Englander, S. W. (1999) Science 284, 822–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yifrach, O. & Horovitz, A. (1995) Biochemistry 34, 5303–5308. [DOI] [PubMed] [Google Scholar]
- 16.Shiseki, K., Murai, N., Motojima, F., Hisabori, T., Yoshida, M. & Taguchi, H. (2001) J. Biol. Chem. 276, 11335–11338. [DOI] [PubMed] [Google Scholar]
- 17.Yifrach, O. & Horovitz, A. (1998) J. Am. Chem. Soc. 120, 13262–13263. [Google Scholar]
- 18.Horovitz, A. & Yifrach, O. (2000) Bull. Math. Biol. 62, 241–246. [DOI] [PubMed] [Google Scholar]
- 19.Ma, J. & Karplus, M. (1998) Proc. Natl. Acad. Sci. USA 95, 8502–8507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma, J., Sigler, P. B., Xu, Z. & Karplus, M. (2000) J. Mol. Biol. 302, 303–313. [DOI] [PubMed] [Google Scholar]
- 21.Horovitz, A, Bochkareva, E. S., Kovalenko, O. & Girshovich, A. S. (1993) J. Mol. Biol. 231, 58–64. [DOI] [PubMed] [Google Scholar]
- 22.Yifrach, O. & Horovitz, A. (1998) Biochemistry 37, 7083–7088. [DOI] [PubMed] [Google Scholar]
- 23.Weissman, J. S., Hohl, C. M., Kovalenko, O., Kashi, Y., Chen, S., Braig, K., Saibil, H. R., Fenton, W. A. & Horwich, A. L. (1995) Cell 83, 577–587. [DOI] [PubMed] [Google Scholar]
- 24.Kafri, G., Willison, K. R. & Horovitz, A. (2001) Protein Sci. 10, 445–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kafri, G. & Horovitz, A. (2003) J. Mol. Biol. 326, 981–987. [DOI] [PubMed] [Google Scholar]
- 26.Ludtke, S. J., Jakana, J., Song, J.-L., Chuang, D. T. & Chiu W. (2001) J. Mol. Biol. 314, 253–262. [DOI] [PubMed] [Google Scholar]
- 27.Frank, J., Shimkin, B. & Dowse, H. (1981) Ultramicroscopy 6, 343–357. [Google Scholar]
- 28.Xu, Z., Horwich, A. L. & Sigler, P. B. (1997) Nature 388, 741–750. [DOI] [PubMed] [Google Scholar]
- 29.Rye, H. S., Burston, S. G., Fenton, W. A., Beechem, J. M., Xu, Z., Sigler, P. B. & Horwich. A. L. (1997) Nature 388, 792–798. [DOI] [PubMed] [Google Scholar]
- 30.Inobe, T., Makio, T., Takasu-Ishikawa, E., Terada, T. P. & Kuwajima, K. (2001) Biochim. Biophys. Acta 1545, 160–173. [DOI] [PubMed] [Google Scholar]
- 31.Yifrach, O. & Horovitz, A. (1994) J. Mol. Biol. 243, 397–401. [DOI] [PubMed] [Google Scholar]
- 32.Horovitz, A., Serrano, L., Avron, B., Bycroft, M. & Fersht, A. R. (1990) J. Mol. Biol. 216, 1031–1044. [DOI] [PubMed] [Google Scholar]
- 33.Braig, K., Adams, P. D. & Brunger, A. T. (1995) Nat. Struct. Biol. 2, 1083–1094. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.