Abstract
The promoter regions of genes that are differentially regulated in the synovial membrane during the course of rheumatoid arthritis (RA) represent attractive candidates for application in transcriptionally targeted gene therapy. In this study, we applied an unbiased computational approach to define proximal-promoters from a gene expression profiling study of murine experimental arthritis. Synovium expression profiles from progressing stages of collagen-induced arthritis (CIA) were classified into six distinct groups using k-means clustering. Using an algorithm based on local over-representation and comparative genomics, we identified putatively functional transcription factor–binding sites (TFBS) in TATA-dependent proximal-promoters. Applying a filter based on spacing between TATA box and transcription start site (TSS) combined with the presence of over-represented nuclear factor κB (NFκB), AP-1, or CCAAT/enhancer-binding protein β (C/EBPβ) sites, 382 candidate murine and human promoters were reduced to 66, corresponding to 45 genes. In vitro, 9 out of 10 computationally defined promoter regions conferred cytokine-inducible expression in murine cells and human synovial fibroblasts. Under these conditions, the serum amyloid A3 (Saa3) promoter showed the strongest transcriptional induction and strength. We applied this promoter for driving therapeutically efficacious levels of the interleukin-1 receptor antagonist (Il1rn) in a disease-regulated fashion. These results demonstrate the value of bioinformatics for guiding the selection of endogenous promoters for transcriptionally targeted gene therapy.
Introduction
Selective spatial and temporal expression of a therapeutic transgene has a major impact on the safety and efficacy of gene therapeutic treatments for human disease. This can be accomplished by transcriptional targeting using endogenous or synthetic cis-regulatory DNA regions, which facilitate tissue/cell-specific or physiologically regulated expression. Predominantly in cancer gene therapy, numerous tumor/tissue-specific or drug/radiation-inducible promoters have been developed and applied.1
With emphasis on the discontinuous clinical course of rheumatoid arthritis (RA), which is characterized by spontaneous remissions and exacerbations of joint inflammation, transcriptionally targeted gene therapy appears ideally suited. The few endogenous promoters that have been tested in RA thus far were empirically derived. Efficacious inflammation-responsive expression in experimental arthritis has been accomplished using the promoter elements of the murine acute-phase response gene complement factor 3 (C3) (refs. 2,3) and the human cytokine gene interleukin-6 (IL6) (ref. 4). Although IL6 and C3 promoters showed high specificity toward inflammation, they only conferred weak transcriptional activity that had to be enhanced artificially using an interleukin-1 enhancer region5 or a recombinant human immunodeficiency virus-1-tat transcriptional activator,6 respectively. However, the application of the latter approach is hampered due to the immunogenic properties of human immunodeficiency virus-1 tat.
The promoter of a gene is defined as the cis-regulatory DNA region that drives transcription in response to environmental signals. Roughly, a promoter can be divided into a core-, proximal-, and distal-promoter region. The core-promoter encompasses the 80–100 base pair (bp) region surrounding the transcription start site (TSS) and is required for assembly of the preinitiation complex.7 Bioinformatic analyses of mammalian core promoters have revealed that the spacing between the TATA box and TSS affects the transcriptional specificity of the downstream transcript, and the highest specificity has been observed for TATA-TSS distances between −32 and −29 (ref. 8). In mouse, the most common TATA-TSS spacings are 30 and 31 bp (ref. 9), and most functional TATA boxes reside in a window ranging from positions −34 to −27 relative to the TSS.8 The proximal-promoter (−500/+200 relative to the TSS) contains modules of transcription factor–binding sites (TFBS) that confer tissue- and context-specific expression.10 Several bioinformatics tools have been developed for computational identification of these TFBS in eukaryotic promoters (reviewed in ref. 11). The accuracy of prediction of TFBS that are functional in vivo has been significantly enhanced using the combination of phylogenetic footprinting with sets of co-regulated genes.12 These methods have been used to successfully identify cartilage-13 or muscle-specific14 cis-regulatory elements in mammals.
In this study, we computationally designed and verified endogenous proximal promoters from a gene expression profiling study of murine collagen-induced arthritis (CIA) for transcriptionally targeted gene therapy for RA. The 201 genes that were significantly regulated during advancing CIA severity stages were divided in clusters of distinct expression profiles using k-means clustering. Next, over-represented TFBS per cluster were calculated in proximal-promoter sequences of the genes and their human orthologs that contained a putative TATA box within the −44 to −17 bp region relative to the TSS. We investigated the transcriptional properties of 10 computationally identified proximal-promoter regions in vitro using lentiviral luciferase reporters. Nine out of ten promoters drove gene expression in a cytokine-inducible fashion. The serum amyloid A3 (Saa3) proximal-promoter conferred high transcriptional specificity and activity toward inflammation, and we demonstrated that this promoter improved the efficacy of transcriptionally targeted adenoviral gene therapy using interleukin-1 receptor antagonist (Il1rn) as a transgene.
Results
K-means clustering of CIA gene expression profiles
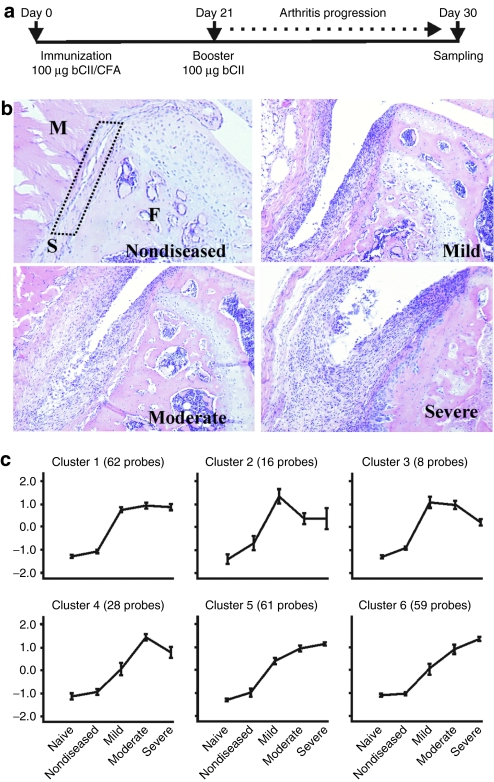
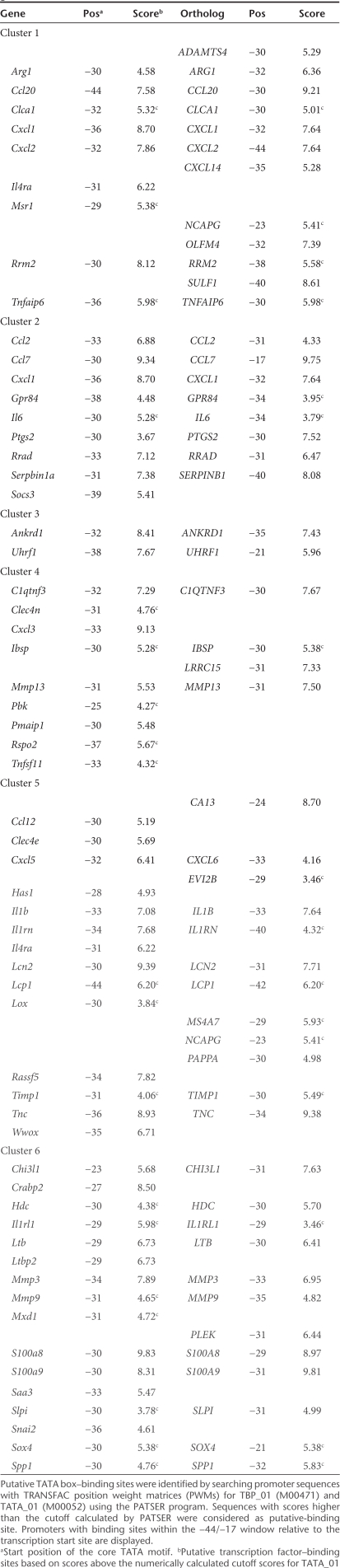
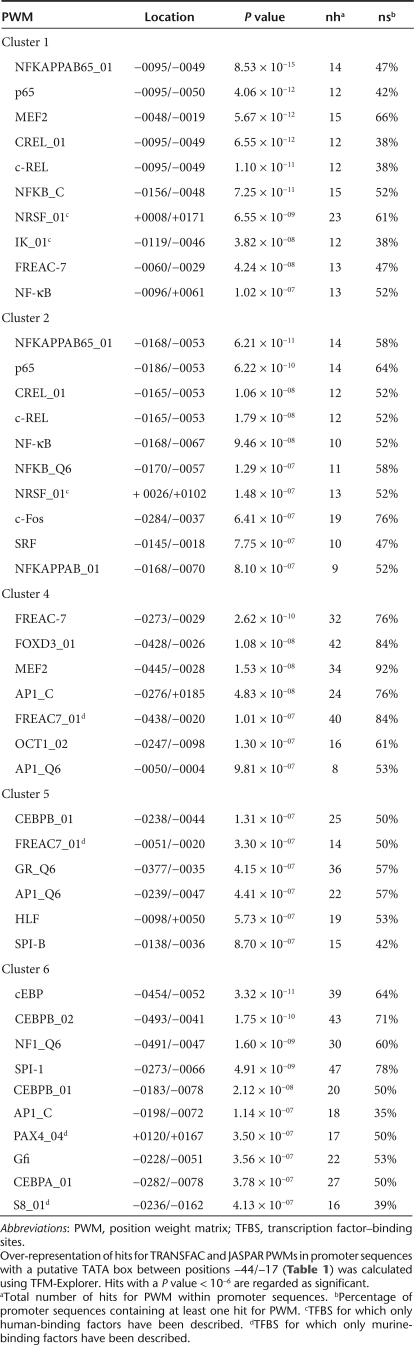
In local gene therapy for RA, the synovial tissue (which is) a thin layer of connective tissue consisting of fibroblast- and macrophage-like cells is transduced by viral vectors.15 To elucidate candidate genes whose proximal-promoter might confer inflammation-inducible expression, we analyzed gene expression profiles in inflamed synovium of mice with CIA. Whole genome expression profiles were obtained from synovial tissue biopsies of knee joints from both naive and DBA/1J mice at day 30 of CIA using the Mouse Genome 430 2.0 array (Figure 1a). Prior to isolation, we semiquantitatively scored the degree of joint inflammation and subdivided samples in four advancing stages of disease severity (Figure 1b). For each stage, hybridization experiments were performed in triplicate and data normalized using MAS 5.0 software. Using EXPANDER 4.1 software,16 we obtained 234 Affymetrix probes that correspond to 201 unique Ensembl genes whose expression was at least tenfold upregulated in at least three severity stages compared to naive conditions. Next, the standardized (mean 0 and variance 1) expression profiles were partitioned into distinct clusters using the k-means algorithm. Partitioning into six clusters resulted in the most optimal average separation (0.913) and homogeneity (0.938) of clusters. The average standardized expression levels of the clusters are plotted in Figure 1c. The annotation of the probe identities per cluster is documented in Supplementary Table S1.
Figure 1.
Cluster analysis of gene expression profiles from collagen-induced arthritis. (a) Schematic overview of the collagen-induced arthritis model. DBA/1J mice were immunized at day 0 by intradermal injection of bovine collagen type II (bCII). Immunized mice received an intraperitoneal booster injection of bCII at day 21 that triggers arthritis onset. After 10 days of arthritis progression, knee joints were macroscopically scored and divided in the four indicated severity stages. (b) Representative hematoxylin and eosin–stained tissue sections of knee joints from the four advancing severity stages are shown. M, muscle; S, synovium (dotted rectangle); F, femur. (c) Mean expression profiles of the eight clusters obtained with k-means clustering in EXPANDER 4.1. The y axis represents standardized expression levels. Error bars represent the ± 1 SD of the members of each cluster about the mean of the particular severity stage.
AP-1, C/EBPβ, and NF-κB-binding sites are over-represented in TATA proximal-promoters
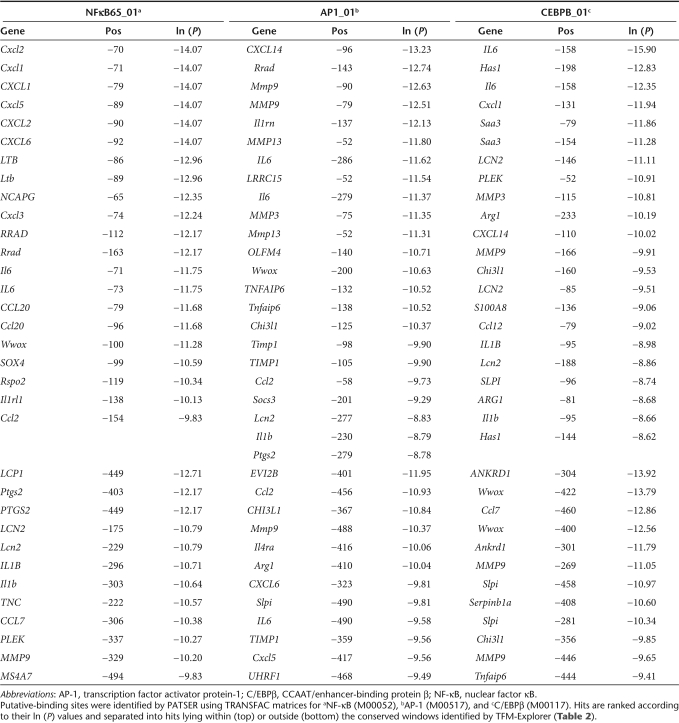
To filter for promoters with high transcriptional specificity toward inflammation, we analyzed the murine and human orthologous proximal-promoters (−500/+200 region) for the presence of a putative TATA box within a −44 to −17 bp window relative to the TSS. The promoter sequences of 191 murine and human orthologs (Supplementary Table S2) were retrieved from the Cold Spring Harbor Promoter database,17 and putative TATA box–binding sites were searched with the program PATSER18 using TATA_01 (M00252) and TBP_01 (M00471) position weight matrices (PWMs) in the TRANSFAC 7.0 (ref. 19) database. For 37 genes, a putative TATA box was predicted in both murine and human promoter sequences. Additionally, a single TATA TFBS was predicted in promoter sequences of 21 murine and 12 human genes (Table 1). The TATA proximal-promoters were used for subsequent identification of over-represented cis-regulatory elements. For this, we used Transcription Factor Matrix-Explorer software because it combines motif over-representation with comparative genomics and takes spatial conservation of cis-regulatory elements into account. Per cluster, over-represented TFBS were searched using PWMs corresponding to human- or mouse-binding factors in the public TRANSFAC and JASPAR20 databases. Proximal-promoters from clusters 1 and 2 were significantly (P < 10−6) enriched for PWMs corresponding to the transcription factor Rel/nuclear factor κB (NF-κB). In clusters 2, 4, 5, and 6, several PWMs for the transcription factor activator protein-1 (AP-1) were over-represented. In contrast, promoters of cluster 5/6 genes, whose transcriptional activity correlates most closely with disease severity, were enriched for CCAAT/enhancer-binding protein β (C/EBPβ) binding sites. The majority of over-represented cis-regulatory elements were spatially conserved in a narrow 200–100 bp window upstream (NF-κB −170/−50, AP-1 −290/−30, and C/EBPβ –240/−45) of the putative TATA box–binding site (Table 2).
Table 1.
Putative TATA-dependent murine and human orthologous genes
Table 2.
Over-represented transcription factor–binding sites in k means clusters
Target genes of these transcription factors were identified using PATSER with high quality TRANSFAC PWMs, compiled from >20 experimentally verified binding sites, NFκB65_01 (M00052), AP1_01 (M00517), and CEBPB_01 (M00109). Bona fide hits (P < 10−8) were ranked according to the log ratio of their p values (ln p) (Table 3). From the 97 TATA-dependent promoters, 66 contained a spatially conserved NF-κB, AP-1, or C/EBPβ TFBS. Without taking spatial conservation into account, 81 promoters contained a putative binding site. This strategy has narrowed down the number of candidate proximal-promoters for transcriptional targeting in arthritis and provides useful information for rational selection of the promoter region.
Table 3.
Significant transcription factor–binding sites for NF-κB, AP-1, and C/EBPβ in TATA-dependent genes
Construction and validation of transcriptionally targeted SIN lentiviral vectors
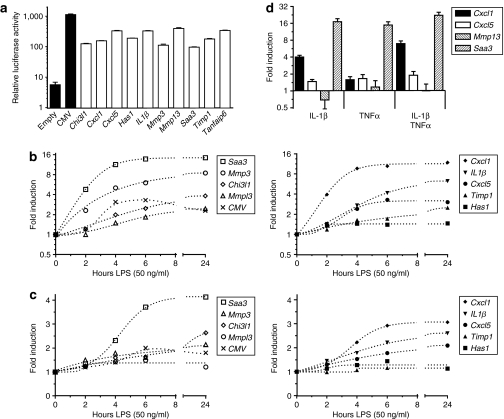
We developed 10 transcriptionally targeted lentiviral vectors, in which the firefly (Photinus Pyralis) luciferase complementary DNA (cDNA) expression is under control of computationally identified proximal-promoters. All promoters comprised the upstream region containing the identified over-represented TFBS, the TSS, and a part of the 5′ UTR-exon (Figure 2). To evaluate the basal transcriptional activity of the cloned promoter regions, murine NIH-3T3 fibroblasts were co-transfected with promoter constructs and as internal control reporter pRL-TK encoding constitutively active Renilla Reniformis luciferase. Cells were transfected with a promoterless or constitutively active cytomegalovirus immediate-early (CMV) promoter construct as negative and positive control, respectively. Assessment of the relative luciferase activity at 2 days after transfection revealed that the basal transcriptional activity of all generated constructs was approximately three- to tenfold lower than obtained with the CMV promoter (1,141 ± 48). The strongest and lowest basal activity in murine fibroblasts was observed for the promoter regions of Cxcl5 (332 ± 12), Il1b (331 ± 10), Mmp13 (398 ± 26), Tnfaip6 (342 ± 8), Saa3 (96 ± 3), and Mmp3 (112 ± 10), respectively (Figure 3a). Next, we determined the responsiveness and kinetics of promoter activities to a pro-inflammatory stimulus. Murine RAW 264.7 macrophages (Figure 3b) or NIH-3T3 fibroblasts (Figure 3c) were transduced with lentiviral promoter-luciferase vectors and after 2 days, challenged with lipopolysaccharide (50 ng/ml). Luciferase activity was consecutively measured at 2-hour intervals (0–6 hours) and 24 hours. With the exception of the Tnfaip6 (−499/+63) promoter, all constructs showed induction of promoter activity in response to toll-like receptor 4 triggering by lipopolysaccharide in macrophages. In contrast, a clear promoter response in fibroblasts was only observed for Saa3 (−314/+50), Cxcl1 (−455/+43), Cxcl5 (−319/+31), and Il1b (−350/−53) promoters. Because our computational analyses included the promoter regions of human orthologs (Table 1) and the majority of over-represented TFBS were conserved between species (Table 3), we investigated whether the murine promoters would be responsive in synovial fibroblasts isolated from RA patients (RASF). Primary synovial fibroblasts (n = 4) were transduced with lentiviral vectors containing the Saa3, Cxcl1, Cxcl5, or Mmp13 promoter. Two days after transduction, RASF were stimulated for 24 hours with either recombinant human IL-1β (0.25 ng/ml) or hTNFα (1 ng/ml) alone, or the combination of the two cytokines (Figure 3d). The Saa3 promoter activity was most strongly induced by stimulation with hIL-1β (17 ± 2) or hTNFα (15 ± 2), and the induction was less than additive for the combination of stimuli (22 ± 3). As in murine fibroblasts, the Cxcl5 and Mmp13 were modestly and nonresponsive, respectively. Strikingly, the Cxcl1 promoter activity was induced by hIL-1β (4.0 ± 0.3) but only marginally by hTNFα (1.6 ± 0.2), whereas the combination of cytokines synergistically activated the Cxcl1 promoter (7.0 ± 0.8).
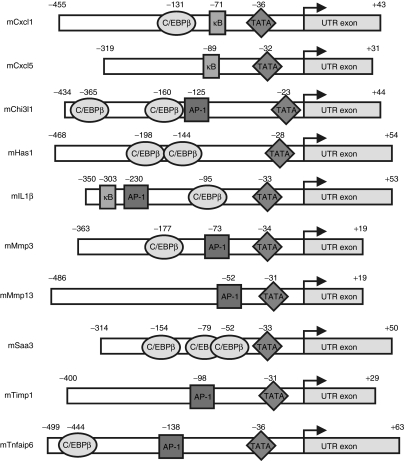
Figure 2.
Schematic map of promoter regions with putative cis-regulatory elements. Indicated promoter regions were inserted upstream of the firefly luciferase complementary DNA in a self-inactivating lentiviral backbone. The position of putative binding sites for TATA, NF-κB, AP-1, and C/EBPβ identified by PATSER (Tables 1 and 3) are indicated. Promoter regions were derived from the murine genes CXC ligand 1/5 (Cxcl1/5), chitinase 3–like 1 (Chi3l1), hyaluronan synthase 1 (Has1), interleukin-1 β (IL1β), matrix metalloproteinase 3/13 (Mmp3/13), serum amyloid A3 (Saa3), tissue inhibitor of metalloproteinase 1 (Timp1), and TNF α–induced protein 6 (Tnfaip6). C/EBPβ, CCAAT/enhancer-binding protein β.
Figure 3.
Experimental verification of transcriptionally targeted vectors. (a) Dual luciferase assay of basal promoter activity in NIH-3T3 fibroblasts. Cells were co-transfected with 500 ng of transcriptional targeted vector and 50 ng of pRL-TK. Promoterless (empty) and cytomegalovirus-promoter vectors (black bars) served as negative and positive controls, respectively. Promoter activity is expressed as relative (firefly/Renilla) luciferase activity ± SEM (n = 3). Kinetics of promoter induction by toll-like receptor 4 triggering in (b) murine RAW 264.7 macrophages and (c) NIH-3T3 fibroblasts. Two days after transduction with lentiviral vectors, cells were stimulated for indicated time points with lipopolysaccharide (50 ng/ml). Luciferase activities are represented as fold induction over unstimulated conditions. (d) Induction of promoter activity in rheumatoid arthritis synovial fibroblasts (n = 4 donors). Fibroblasts were transduced with lentiviral vectors and stimulated for 24 hours with either recombinant human IL-1β (0.25 ng/ml) or hTNFα (1 ng/ml) alone, or the combination. Luciferase activities are expressed as fold induction ± SEM. LPS, lipopolysaccharide.
Saa3 promoter drives therapeutically efficacious Il1rn in a disease-regulated fashion
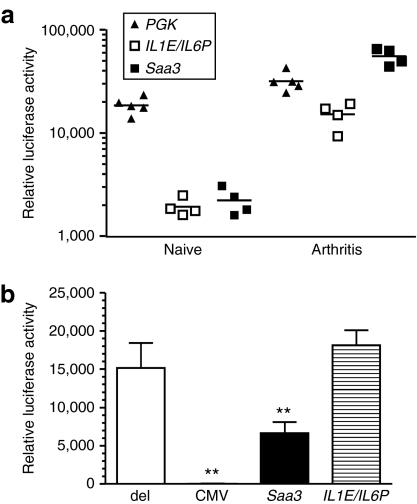
Previously, we have demonstrated therapeutic efficacy of transcriptionally targeted IL-4 gene therapy in CIA using a hybrid promoter consisting of the enhancer region of IL1B (−3690/−2720) fused to the proximal promoter of IL6 (−163/+12) (IL1E/IL6P).4 However, when the IL4 cDNA was replaced for the Il1rn cDNA, we failed to prevent the development and progression of CIA (data not shown) as described for constitutive overexpression of this transgene.2,21 We sought to address this efficacy issue by substituting the IL1E/IL6P promoter for a computationally identified promoter. Based on in vitro validations of promoter strength and responsiveness (Figure 3), the Saa3 promoter was selected as the most promising candidate for such a substitution. First, we compared the promoter strength under arthritic conditions in vivo. Knee joints of C57Bl/6 mice were transduced by lentiviral vectors, and after 7 days, an acute arthritis was induced by intra-articular injection of zymosan A (180 µg). Twenty-four hours after challenge, we assessed luciferase activity ex vivo (Figure 4a). To control for amplification of transgene expression by inflammation-induced proliferation of lentivirally transduced synovium,22 knee joints were transduced with a vector encoding phosphoglycerate kinase-1 (PGK) promoter-driven, constitutively expressed luciferase. Under naive conditions, transcriptionally targeted vectors exhibited equal and low luciferase activities, approximately a log-fold lower compared to PGK-driven expression. One day after induction of arthritis, we measured a twofold upregulation of PGK-driven luciferase expression. In contrast, under arthritic conditions, Saa3 and IL1E/IL6P promoter activities were 25- and eightfold upregulated, respectively, and Saa3 promoter strength was approximately sixfold higher compared to IL1E/IL6P. Next, we generated adenoviral vectors encoding Saa3 or IL1E/IL6P-driven Il1rn, and measured the Il1rn protein levels under basal and stimulated conditions. HeLa cells were transduced [multiplicity of infection (MOI) 10] at day 1, and the day thereafter, left untreated or stimulated for 24 hours with hTNFα (10 ng/ml). Basal and induced levels were 152.9 ± 34.2 versus 325.0 ± 50.8 ng/ml (CMV) and 2.8 ± 0.2 versus 49.6 ± 8.1 ng/ml (Saa3) (n = 4). Protein levels for IL1E/IL6P-driven Il1rn were around the detection limit of the enzyme-linked immunosorbent assay (ELISA) (1 ng/ml). Next, we tested therapeutic efficacy of these vectors in an in vitro assay system (Figure 4b). NIH-3T3 fibroblasts stably transfected with the luciferase gene downstream of five tandem repeats of NF-κB-binding sites were transduced (MOI 10) at day 1 with adenovirus encoding transcriptionally targeted Il1rn and at day 2 with constitutively expressed Il1b. As a positive and negative control, cells were transduced at day 1 with adenovirus encoding constitutively expressed Il1rn (CMV) or nonencoding adenovirus (del). At day 3, we assessed Il1rn protein levels by ELISA and IL-1β-induced NF-κB activation by luciferase assay at day 3. As expected, CMV-driven Il1rn expression (56.8 ± 11.8 ng/ml) completely suppressed IL-1β-induced NF-κB activation (P < 0.01). Empty control virus and IL1E/IL6P-driven Il1rn expression (undetectable) failed to significantly inhibit NF-κB activation. In contrast, Saa3-driven Il1rn expression (11.1 ± 1.7 ng/ml) led to a significant (P < 0.01) reduction (~56%) of IL-1β-induced NF-κB activation. These results indicate that the promoter strength and responsiveness of Saa3 might be sufficient to provide therapeutically efficacious protein levels in a transcriptionally targeted Il1rn gene therapy approach for experimental arthritis.
Figure 4.
Comparison of therapeutic efficacy using Saa3 versus IL1E/IL6P promoter for disease-regulated Il1rn expression. (a) Promoter activity in transduced synovium of naive and arthritic C57Bl/6 mice. Knee joints were injected with 300 ng p24gag equivalent lentivirus encoding PGK, Saa3, or IL1E/IL6P-luciferase. Seven days after transduction, arthritis was induced by intra-articular injection of 180 µg zymosan A. After 24 hours, luciferase activity was assessed ex vivo. Data are represented as individual relative luciferase activities; horizontal bars indicate the means per group. (b) Efficacy of transcriptionally targeted adenoviral vectors expressing Il1rn. NIH-3T3-5xNF-κB-Luc were transduced at a multiplicity of infection (MOI) of 10 with control vector (del) or adenovirus encoding CMV, Saa3, and IL1E/IL6P-driven Il1rn. After 24 hours, cells were transduced at an MOI of 10 with control vector (del) or Ad5.CMV-Il1b. The day thereafter, IL-1β-induced NF-κB activation was assessed by luciferase assay. Data are represented as relative luciferase activities ± SEM (n = 4), and differences were determined using analysis of variance with Dunnett's post-test. **P < 0.01.
Discussion
Endogenous proximal-promoters that confer a range of transcriptional activities in an inflammation-specific fashion are of great value toward tailor-made transcriptionally targeted gene therapy for RA. To this end, we used a gene expression profiling study of CIA for elucidating disease-regulated genes and performed computational analyses on the proximal-promoter regions to define DNA regulatory elements that can be applied for transcriptional targeting. Using this approach, we narrowed down the number of candidate murine and human promoters from 382 to 66, corresponding to 45 unique genes.
In our approach, we took the spacing between TATA box and TSS as a filtering parameter for transcriptional specificity. The rationale for this was derived from recent studies8,9 showing the association of TATA-dependent transcription with tissue/context-specific gene expression. Moreover, these studies demonstrate that the TATA-TSS spacing affects the transcriptional specificity of the downstream transcript. We predicted putative bona fide TATA boxes in using matching of PWM models. Generally, the score of a predictive PWM model is highly correlated with the strength of the protein–DNA interaction.23 However, a genome-wide characterization of the interaction between the TATA box and the preinitiation complex has revealed the absence of a global correlation between PWM score and tissue specificity.8 These findings are reflected in our computations in which we could not establish any correlation between PWM score and spacing. Accordingly, in the experimental verifications using promoter-luciferase constructs, we did neither find an apparent correlation between TATA box scores and basal promoter activity in a murine fibroblast cell line.
Apart from the TATA-TSS spacing, the identification of putative TFs that govern a particular expression profile was the second key determinant for selection of promoter regions. As demonstrated in similar approaches,13,14 the combination of co-regulated genes with phylogenetic footprinting proved fruitful for identification of over-represented TFBS. Supporting evidence for the functional relevance of the predicted over-represented TFBSs in RA is substantial. The over-representation of NF-κB- and AP-1-binding sites correspond to the pivotal role that has been implied for these factors in human RA, CIA, and immunity.24,25,26,27 Enhanced expression and DNA-binding activity of C/EBPβ in synovial tissue of RA patients has been implicated in the pathology28 and chronicity29 of disease. The latter coincides with the enrichment of C/EBPβ-binding sites in the CIA clusters in which the gene expression profile was closely correlated with disease severity. The additional enrichment of Spi-1 (Pu.1) and C/EBPα binding sites in cluster 6 is expected to arise from the increasing infiltration of the inflamed joint in CIA by myeloid (monocytes, dendritic cells, and neutrophils) and lymphoid (T and B cells) cells. Pu.1 and C/EBPα are key factors in development of myeloid and B cells.30 Interestingly, promoters of myeloid and B-cell-specific genes often contain a regulatory module consisting of a C/EBPα motif in close proximity (<60 bp) of the Pu.1 motif.31 Indeed, the conserved locations of Pu.1 (−273/−66) and C/EBPα (−282/−78) in our analyses indicate the presence of such myeloid-specific modules. Because synovial fibroblasts represent the target cells for local gene therapy,15,32 the identification of myeloid-specific modules might be exploited to further refine the number of candidate promoters. Besides aforementioned TFs, we expect a contribution of the TF signal transducers and activators of transcription-1/3 (STAT-1/3) (ref. 33), and interferon regulatory factor-1 (ref. 34) in synovial inflammation. Indeed, scanning TATA-dependent promoters with corresponding PWMs (STAT_01 and IRF1_01), we found 15 and 36 promoters with a putative binding site, respectively (data not shown). However, these hits were evenly dispersed over the clusters on a wide range (−500/−100) of positions, which explains the absence of over-representation for these TFBS in our analyses. The accuracy of the approach for inferring functional cis-regulatory elements is not only demonstrated by our experimental verifications but also by literature confirming the functionality of computationally identified top-ranking TFBS in several promoters such as Il1b,35 Cxcl1(ref. 36), Saa3 (ref. 37), Timp1, and Mmp3/13 (ref. 38).
The strength of our approach lies in the combination of gene expression profiling for identification of candidate genes with computational prediction of functional endogenous proximal promoters. These are expected to contain evolutionarily conserved optimal combinations of and spacings between TFBSs. This represents a major advantage over recent efforts that aim at tailoring gene expression using genetic engineering of synthetic promoters.25,39,40 Although these studies have demonstrated encouraging results for modulating transgene expression in prokaryotes, the extrapolation to eukaryotic transcriptional regulation by modules proves far more complex and the screening of hundreds of randomly assembled synthetic promoters is laborious and time-consuming. Still, the identified over-represented TFBS and their distance constraints might be useful for guiding the rational design of a synthetic promoter. For example, it was demonstrated for RA gene therapy that a synthetic promoter consisting of six tandem repeats of NF-κB consensus sites drives efficacious41 expression of anti-TNFα. The functionality of this promoter might be explained by the fact that the construct consists of a TFBS that is over-represented in inflammation-induced genes and positioned within the evolutionary conserved −170/−50 region upstream of the TSS.
The pitfall of using Il1rn as a transgene lies in the fact that it needs to be present in at least 100-fold molar excess in order to block the effects of IL-1β on synoviocytes and chondrocytes.42 However, using Saa3-driven Il1rn expression, we obtained >50% inhibition of an CMV-driven, exaggerated, and nonphysiological amount of IL-1β. The relatively high transcriptional strength of the Saa3 proximal promoter has been demonstrated by Varley and co-workers.43 In their study, Saa3-driven luciferase expression exceeded that of the CMV promoter in response to cytokine-rich conditioned medium prepared from the culture supernatant of lipopolysaccharide-stimulated human peripheral blood monocytes. In combination with our computational analyses and verifications, the Saa3 promoter is the most appropriate endogenous promoter for transcriptionally targeted Il1rn gene therapy for arthritis.
For the panel of inflammation-responsive promoters, we envisage two major applications. First, these promoters can be applied in noninvasive imaging approaches of joint inflammation or activation of particular transcription factors, e.g., AP-1 (Mmp13) versus NFκB (Cxcl1), in human and experimental arthritis. Whether these proximal promoters confer differential expression patterns during experimental arthritis will be investigated using an in vivo imaging approach. Second, differential transcriptional activities can be exploited for gene therapy tailored to transgene properties, e.g., a promoter with strong transcriptional activity for generating the required excess of Il1rn expression and a promoter with low basal activity for minimizing Il4-induced side effects.
Materials and Methods
Animals. Male 10–12-week-old DBA/1J mice were obtained from Janvier (Le Genest-Saint-Isle, France). C57Bl/6 mice were obtained from Charles River Laboratories (Sulzfeld, Germany). During viral experiments, mice were housed in low-pressure isolator cages. The animals were fed a standard diet with food and water ad libitum. All in vivo studies complied with national legislation and were approved by the local authorities of the Care and Use of Animals.
Induction of CIA. Bovine collagen type II (bCII) was dissolved in 0.05 mol/l acetic acid to a concentration of 2 mg/ml and was emulsified in equal volumes of Freund's complete adjuvant (2 mg/ml of Mycobacterium tuberculosis strain H37Ra; Becton Dickinson, Detroit, MI). DBA-1/J mice were immunized intradermally at the base of the tail with 100 µl of emulsion (100 µg of bCII). On day 21, the mice were given an intraperitoneal booster injection of 100 µg bCII dissolved in phosphate-buffered saline (PBS). Mice were sacrificed on day 30 by cervical dislocation. Prior to biopsy, knee joints were scored visually using four grades: nondiseased, mild (minor swelling and color change due to infiltration), moderate (marked swelling), and severe (severe swelling and patellar tendon structure not visible). Synovial tissue samples from the lateral and medial sites (n = 24, 12 knee joints per severity stage) were isolated in a standardized manner using a 3 mm biopsy punch (Stiefel, Wächtersbach, Germany), as described previously.44 Samples were pooled in a randomized fashion to generate triplicates consisting of eight biopsies.
RNA isolation. Total RNA from biopsy punches was prepared by TRIzol extraction (Invitrogen Life Technologies, Carlsbad, CA), and purified on an affinity resin (RNeasy Kit; Qiagen, Hilden, Germany) according to the manufacturer's instructions. Quantity and purity were assessed by the absorbance at Λ = 260 nm (A260nm), and the ratio A260nm/A280nm. Integrity of the RNA was confirmed by nondenaturing agarose gel electrophoresis. Total RNA was stored at −80 °C until further processing.
Oligonucleotide array. One microgram of total RNA was used as a starting material for cDNA preparation. Generation of biotinylated cRNA and subsequent hybridization, washing and staining of MOE 430_2 oligonucleotide arrays (Affymetrix, Santa Clara, CA) were performed according to the Affymetrix Expression Analysis Technical Manual for one-cycle amplification. The arrays were then scanned using a laser scanner GeneChip Scanner (Affymetrix), and data were analyzed and normalized using Affymetrix Microarray Suite (MAS) 5.0 software according to the manufacturer's instruction. The gene expression data were deposited in the Gene Expression Omnibus database under accession number GSE13071.
Cluster analysis. Filtering, standardization, and cluster analysis were performed using Expression Analyzer and Displayer (EXPANDER) software version 4.1 (ref. 16). The mean (n = 3) expression values calculated from the normalized microarray data using Excel served as input. After filtering for tenfold regulated probes, the expression values were standardized using the mean 0 and variance 1 algorithm. Cluster analyses were performed using k-means clustering with the number of clusters set to six.
Promoter sequence retrieval. Affymetrix probe identifiers were converted into Ensembl gene identifiers (National Center for Biotechnology Information m37 mouse assembly) and RefSeq DNA identifiers (mm8) using BioMart (http://www.biomart.org). Human orthologs were retrieved from the Mouse Genome Database.45 We extracted the proximal-promoter regions of these genes (−500/+200) from the Cold Spring Harbor Laboratory mammalian promoter database17 using RefSeq identifiers. When multiple promoters existed for a gene, we selected either a promoter from the Eukaryotic Promoter Database or Database of Transcription Start Sites. When the prior two were unavailable, we selected the promoter with the shortest distance to the gene.
Identification of TFBS. The PATSER program18 at the Regulatory Sequence Analysis Tools server (http://rsat.ulb.ac.be/rsat)46 was used to search position weight matrix models collected from the public TRANSFAC 7.0 database.19 Both strands of the promoter sequences were scanned in our analyses. Sequences with scores higher than the cutoff calculated by PATSER were considered as putative TFBS.47
Identification of over-represented TFBS. Promoter sequences with a putative TATA-binding site were scanned, per cluster, for 322 vertebrate matrices from the TRANSFAC 7.0 and JASPAR20 databases using TFM-Explorer (http://bioinfo.lifl.fr/TFME).12 The ratio parameter, indicating the minimal average density of hits in the cluster relative to the reference model, was set to 4.0. PWMs with a P value < 10−6 were regarded as significantly over-represented.
Cell culture. Mouse embryonic fibroblasts (NIH-3T3), macrophages (RAW 264.7), and human HeLa cells were cultivated in Dulbecco's modified Eagle's medium (DMEM) with 1 mmol/l pyruvate, 40 µg/ml gentamicin, and 5 or 10% fetal calf serum (FCS), respectively. Early-passage RA synovial fibroblasts (kind gift from R.W. Kinne, University Hospital Jena, Eisenberg, Germany) were maintained in DMEM supplemented with 1 mmol/l pyruvate, 80 µg/ml gentamicin, and 10% FCS. Cells were kept at 37 °C in a humid atmosphere containing 5% CO2.
Plasmids. Renilla luciferase vector pRL-TK was obtained from Promega (Madison, WI). For generation of recombinant lentiviral vectors, we made use of the third-generation self-inactivating (SIN) transfer vectors pRLL cPPT-PGK-mcs-PRE-SIN containing the human phosphoglycerate kinase (PGK) promoter and the promoterless pRLL-cPPT-mcs-PRE-SIN (kind gift from J. Seppen, AMC Liver Center, Amsterdam, the Netherlands). For cloning, we used Pfu DNA polymerase (Stratagene, La Jolla, CA) and T4 DNA Ligase (New England Biolabs, Ipswich, MA). All generated constructs were verified by sequencing. The firefly luciferase cDNA from pGL3b (Promega) was transferred as an NheI/XbaI fragment into the multiple cloning site generating pRLL-cPPT-PGK-Luc-PRE-SIN and pRLL-cPPT-Luc-PRE-SIN. The plasmid pRLL-cPPT-CMV-Luc-Pre-SIN was generated by PCR cloning the CMV promoter from pShuttle-CMV (Stratagene) into SalI/HpaI sites using primers FW 5′ GTCGACTAGTAATCAATTACGGGG-3′ and RV 5′GTTAACGGATCTGACGGTTCAC-3′. The murine promoter sequences were PCR cloned from liver genomic DNA into SalI/NheI sites of pRLL-cPPT-mcs-Luc-PRE-SIN using primers in Supplementary Table S3. The IL-1E/IL-6P promoter was transferred as a SalI/NheI fragment from pGL3-IL-1E/IL-6P5 into pRLL-cPPT-mcs-Luc-PRE-SIN. The cDNA of the mouse Il1rn gene was transferred as a SalI/XbaI fragment from pShuttle-CMV-Il1rn48 into pRLL-cPPT-PGK-mcs-PRE-SIN. For constructing pRLL cPPT-Saa3-Il1rn-PRE-SIN, Il1rn was amplified from pShuttle-CMV-Il1rn using primers FW 5′-GCTAGCGCCACCATGGAAATCTGCTGGGGAC-3′ and RV 5′-TCTAGACTATTGGTCTTCCTGGAAG-3′ introducing a 5′ NheI and 3′ XbaI site, and blunt ligated into the SrfI site of pCR-Script Amp SK(+) (Stratagene). The luciferase cDNA was removed from pRLL-cPPT-Saa3-Luc-PRE-SIN by restriction with NheI/NsiI and religated with a NheI/PstI fragment from pPCR-Script-Il1rn (−). A SalI/XbaI fragment from pRLL-cPPT-Saa3-Il1rn-PRE-SIN containing the Saa3 promoter and Il1rn cDNA was transferred to pShuttle-polyA to give pShuttle-Saa3-Il1rn-polyA. The cDNA of mouse interleukin-1 β (Il1b) was PCR cloned from reverse transcribed C57Bl/6 synovial tissue RNA into the KpnI/XhoI sites of pShuttle-CMV using primers FW 5′-AAAGGTACCGCTATGGCAACTGTTCC-3′ and RV 5′-TTTCTCGAGTTAGGAAGACAC-3′.
Lentiviral vector production. Packaging of VSV-G pseudotyped recombinant lentiviruses was performed by transient transfection of 293T cells. The day prior to transfection, 293T cells were seeded in a T75 flask at 1 × 105 cells/cm2 in DMEM supplemented with 10% FCS, 1 mmol/l pyruvate, 40 µg/ml gentamicin, and 0.01 mmol/l water-soluble cholesterol (Sigma, St Louis, MO). Cells were co-transfected with 19 µg transfer vector, 14 µg gag/pol packaging plasmid (pMDL-g/p-RRE), 4.7 µg rev expression plasmid (RSV-REV), and 6.7 µg VSV-G expression plasmid (pHIT-G) by calcium phosphate precipitation. Transfections were performed in 6 ml DMEM without antibiotics and cholesterol, and proceeded for 16 hours. Thereafter, medium was replaced with fully supplemented DMEM and supernatant harvested after 24 and 48 hours. Cell debris was removed by centrifugation at 1,500 rpm for 5 minutes at 4 °C, followed by passage through a 0.45 µm pore polyvinylidene fluoride Durapore filter (Millipore, Bedford, MA). For concentration by ultracentrifugation, 28 ml supernatant was layered on 4 ml 20% sucrose solution and centrifuged at 25,000 rpm in a Surespin 630 rotor (Sorvall; Thermo Fisher Scientific, Waltham, MA). Pelleted viruses were resuspended in sterile PBS and stored at −80 °C. Viral titers were determined by assaying p24gag values with a commercial ELISA kit (Abbott Diagnostics, Hoofddorp, The Netherlands) and expressed as ng p24gag/µl.
Adenoviral vectors. Replication-deficient adenoviral vectors (E1/E3 deleted) Ad5.Saa3-Il1rn, Ad5.IL-1E/IL-6P-Il1rn, Ad5.CMV-Il1rn, Ad5.CMV-Il1b, and Ad5.del were prepared according to the AdEasy system (Stratagene), with the exception that replication-competent recombinant free viral particles were produced in E1 transformed N52E6 amniocyte cells.49 Viruses were purified by two consecutive CsCl2 gradient purifications and stored in small aliquots at −80 °C in buffer containing 25 mmol/l Tris, pH 8.0, 5 mmol/l KCl, 0.2 mmol/l MgCl2, 137 mmol/l NaCl, 730 µmol/l Na2HPO4, 0.1% (wt/vol) ovalbumin, and 10% (vol/vol) glycerol. The infectious particle titer (focus-forming units) was determined by titrating vector stocks on 911 indicator cells and measuring viral capsid protein immunohistochemically 20 hours after transduction.
Luciferase measurements. For in vitro reporter studies, cells were seeded at 5 × 104 cells per well in a Krystal 2000 96-well plate (Thermo Fisher Scientific). The day after, cells were transduced with 50 ng p24gag equivalent lentivirus in 50 µl medium supplemented with 8 µg/ml polybrene (Sigma) for 4 hours at 37 °C. Cells were serum-starved (1% FCS) for 2 days and subsequently stimulated with recombinant human IL-1β (R&D Systems Europe, Oxford, UK), TNFα (Abcam, Cambridge, UK), or E. Coli lipopolysaccharide (Sigma) for indicated hours and subsequently lysed in ice-cold lysis buffer (0.5% NP-40, 1 mmol/l DTT, 1 mmol/l EDTA, 5 mmol/l MgCl2, 100 mmol/l KCl, and 10 mmol/l Tris-HCl pH 7.5). Luciferase activity was quantified using the Bright-Glo luciferase assay system (Promega) by adding an equal volume of Bright-Glo to the cell lysate. Luminescence was quantified in a luminometer (LUMIstar; BMG, Offenburg, Germany), expressed as relative light units and normalized to total protein content of the cell/tissue extracts. For transient transfection experiments, cells were seeded at 70% confluency in a 24-well plate and co-transfected with 500 ng firefly luciferase reporter and 50 ng Renilla luciferase reporter (pRL-TK) using Arrest-In (Open Biosystems, Huntsville, AL) according to the manufacturer's instructions. Cells were serum-starved (1% FCS) for 2 days, and firefly and Renilla luciferase activities were quantified using the Dual-Luciferase Reporter Assay System (Promega). For in vivo studies, knee joints were injected with 300 ng p24gag equivalent lentivirus in a total volume of 6 µl. Seven days after transduction, knee joints were injected with 180 µg zymosan A/6 µl PBS (Sigma). After 1 day, patellae with surrounding tissue were dissected, put in 250 µl cell culture lysis buffer (Promega) and snap frozen in liquid nitrogen. Supernatant was centrifuged at 13,000 rpm for 5 minutes and luciferase activity assayed as described above.
In vitro Il1rn-efficacy assay system. NIH-3T3 fibroblasts stably transfected with a 5xNF-κB luciferase reporter were seeded at 5 × 104 cells per well in a Krystal 2000 96-well plate. The day thereafter, cells were transduced at an MOI of 10 with Il1rn-encoding or control non-encoding adenovirus (Ad5.del) in DMEM for 4 hours at 37 °C. After 24 hours, medium was aspirated, cells were rinsed and transduced at an MOI of 10 with Il1b-encoding or Ad5.del virus in DMEM for 4 hours at 37 °C. The day after the second transduction, IL-1β-induced NF-κB activation was measured by assessing the luciferase activity as described above.
Il1rn ELISA. White high-binding flat bottom 96-well plates (Greiner Bio-One, Alphen a/d Rijn, the Netherlands) were coated with the capture antibody rat anti-murine Il1rn (MAB480; R&D Systems, Minneapolis, MN) at 3 µg/ml in 0.1 mol/l carbonate buffer pH 9.6 and incubated overnight at 4 °C. Nonspecific binding sites were blocked with 1% bovine serum albumin in PBS for 1 hour at room temperature. Between subsequent incubations, wells were rinsed three times with 0.1% Tween-20 in PBS. Twenty-four hour culture supernatants (100 µl) from 3 × 105 HeLa cells transduced with adenoviral Il1rn-expressing vectors at an MOI of 10 were added to coated wells and incubated for 3 hours at room temperature. The plates were then incubated with the biotinylated goat anti-murine Il1rn (BAF 480; R&D Systems) at 0.2 µg/ml in 0.1% bovine serum albumin/PBS for 2 hours, followed by a 30-minute incubation with streptavidin-conjugated horseradish peroxidase (Dako, Glostrup, Denmark) at 0.25 µg/ml in PBS. Antibody complexes were detected by incubation with the SuperSignal ELISA Pico Chemiluminescent Substrate (Pierce, Rockford, IL) and luminescence quantified in a luminometer.
Statistics. Data are represented as means ± SEM, and significant differences were calculated using one-way analysis of variance (ANOVA) followed by Dunnett's multiple comparisons test (GraphPad Prism, San Diego, CA). P values < 0.05 were regarded significant.
SUPPLEMENTARY MATERIALTable S1. Annotation of the probe identities per k-means cluster.Table S2. Proximal (−500/+200) promoter sequences used in this study.Table S3. Supplementary Methods: Primers.
Supplementary Material
Annotation of the probe identities per k-means cluster.
Proximal (−500/+200) promoter sequences used in this study.
Supplementary Methods: Primers.
Acknowledgments
We are grateful to Martin Letzkus, Nicole Hartmann, and Frank Staedtler of the Genomics Factory, Novartis Institutes for BioMedical Research, Basel, Switzerland for performing the RNA isolation and oligonucleotide array procedures. This research is supported by a VIDI grant (917.46.373) from the Netherlands Organization for Scientific Research and an IOP Genomics grant (IGE02032). This research was conducted in Nijmegen, the Netherlands.
REFERENCES
- Robson T., and , Hirst DG. Transcriptional targeting in cancer gene therapy. J Biomed Biotechnol. 2003;2003:110–137. doi: 10.1155/S1110724303209074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker AC, van de Loo FA, Joosten LA, Arntz OJ, Varley AW, Munford RS, et al. C3-Tat/HIV-regulated intraarticular human interleukin-1 receptor antagonist gene therapy results in efficient inhibition of collagen-induced arthritis superior to cytomegalovirus-regulated expression of the same transgene. Arthritis Rheum. 2002;46:1661–1670. doi: 10.1002/art.10481. [DOI] [PubMed] [Google Scholar]
- Miagkov AV, Varley AW, Munford RS., and , Makarov SS. Endogenous regulation of a therapeutic transgene restores homeostasis in arthritic joints. J Clin Invest. 2002;109:1223–1229. doi: 10.1172/JCI14536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geurts J, Arntz OJ, Bennink MB, Joosten LA, van den Berg WB., and , van de Loo FA. Application of a disease-regulated promoter is a safer mode of local IL-4 gene therapy for arthritis. Gene Ther. 2007;14:1632–1638. doi: 10.1038/sj.gt.3303022. [DOI] [PubMed] [Google Scholar]
- van de Loo FA, de Hooge AS, Smeets RL, Bakker AC, Bennink MB, Arntz OJ, et al. An inflammation-inducible adenoviral expression system for local treatment of the arthritic joint. Gene Ther. 2004;11:581–590. doi: 10.1038/sj.gt.3302182. [DOI] [PubMed] [Google Scholar]
- Varley AW, Geiszler SM, Gaynor RB., and , Munford RS. A two-component expression system that responds to inflammatory stimuli in vivo. Nat Biotechnol. 1997;15:1002–1006. doi: 10.1038/nbt1097-1002. [DOI] [PubMed] [Google Scholar]
- Butler JE., and , Kadonaga JT. The RNA polymerase II core promoter: a key component in the regulation of gene expression. Genes Dev. 2002;16:2583–2592. doi: 10.1101/gad.1026202. [DOI] [PubMed] [Google Scholar]
- Ponjavic J, Lenhard B, Kai C, Kawai J, Carninci P, Hayashizaki Y, et al. Transcriptional and structural impact of TATA-initiation site spacing in mammalian core promoters. Genome Biol. 2006;7:R78. doi: 10.1186/gb-2006-7-8-r78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carninci P, Sandelin A, Lenhard B, Katayama S, Shimokawa K, Ponjavic J, et al. Genome-wide analysis of mammalian promoter architecture and evolution. Nat Genet. 2006;38:626–635. doi: 10.1038/ng1789. [DOI] [PubMed] [Google Scholar]
- Smith AD, Sumazin P, Xuan Z., and , Zhang MQ. DNA motifs in human and mouse proximal promoters predict tissue-specific expression. Proc Natl Acad Sci USA. 2006;103:6275–6280. doi: 10.1073/pnas.0508169103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang MQ.2007Computational analyses of eukaryotic promoters BMC Bioinformatics 8suppl. 6): S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Defrance M., and , Touzet H. Predicting transcription factor binding sites using local over-representation and comparative genomics. BMC Bioinformatics. 2006;7:396. doi: 10.1186/1471-2105-7-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies SR, Chang LW, Patra D, Xing X, Posey K, Hecht J, et al. Computational identification and functional validation of regulatory motifs in cartilage-expressed genes. Genome Res. 2007;17:1438–1447. doi: 10.1101/gr.6224007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao G, Schriefer LA., and , Stormo GD. Identification of muscle-specific regulatory modules in Caenorhabditis elegans. Genome Res. 2007;17:348–357. doi: 10.1101/gr.5989907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouze E, Gouze JN, Palmer GD, Pilapil C, Evans CH., and , Ghivizzani SC. Transgene persistence and cell turnover in the diarthrodial joint: implications for gene therapy of chronic joint diseases. Mol Ther. 2007;15:1114–1120. doi: 10.1038/sj.mt.6300151. [DOI] [PubMed] [Google Scholar]
- Sharan R, Maron-Katz A., and , Shamir R. CLICK and EXPANDER: a system for clustering and visualizing gene expression data. Bioinformatics. 2003;19:1787–1799. doi: 10.1093/bioinformatics/btg232. [DOI] [PubMed] [Google Scholar]
- Xuan Z, Zhao F, Wang J, Chen G., and , Zhang MQ. Genome-wide promoter extraction and analysis in human, mouse, and rat. Genome Biol. 2005;6:R72. doi: 10.1186/gb-2005-6-8-r72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stormo GD, Schneider TD, Gold L., and , Ehrenfeucht A. Use of the ‘Perceptron' algorithm to distinguish translational initiation sites in E. coli. Nucleic Acids Res. 1982;10:2997–3011. doi: 10.1093/nar/10.9.2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingender E, Chen X, Hehl R, Karas H, Liebich I, Matys V, et al. TRANSFAC: an integrated system for gene expression regulation. Nucleic Acids Res. 2000;28:316–319. doi: 10.1093/nar/28.1.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandelin A, Alkema W, Engström P, Wasserman WW., and , Lenhard B.2004JASPAR: an open-access database for eukaryotic transcription factor binding profiles Nucleic Acids Res 32Database issue): D91–D94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker AC, Joosten LA, Arntz OJ, Helsen MM, Bendele AM, van de Loo FA, et al. Prevention of murine collagen-induced arthritis in the knee and ipsilateral paw by local expression of human interleukin-1 receptor antagonist protein in the knee. Arthritis Rheum. 1997;40:893–900. doi: 10.1002/art.1780400517. [DOI] [PubMed] [Google Scholar]
- Gouze E, Pawliuk R, Gouze JN, Pilapil C, Fleet C, Palmer GD, et al. Lentiviral-mediated gene delivery to synovium: potent intra-articular expression with amplification by inflammation. Mol Ther. 2003;7:460–466. doi: 10.1016/s1525-0016(03)00024-8. [DOI] [PubMed] [Google Scholar]
- Stormo GD. DNA binding sites: representation and discovery. Bioinformatics. 2000;16:16–23. doi: 10.1093/bioinformatics/16.1.16. [DOI] [PubMed] [Google Scholar]
- Han Z, Boyle DL, Manning AM., and , Firestein GS. AP-1 and NF-kappaB regulation in rheumatoid arthritis and murine collagen-induced arthritis. Autoimmunity. 1998;28:197–208. doi: 10.3109/08916939808995367. [DOI] [PubMed] [Google Scholar]
- Li X, Eastman EM, Schwartz RJ., and , Draghia-Akli R. Synthetic muscle promoters: activities exceeding naturally occurring regulatory sequences. Nat Biotechnol. 1999;17:241–245. doi: 10.1038/6981. [DOI] [PubMed] [Google Scholar]
- Tak PP, Gerlag DM, Aupperle KR, van de Geest DA, Overbeek M, Bennett BL, et al. Inhibitor of nuclear factor kappaB kinase beta is a key regulator of synovial inflammation. Arthritis Rheum. 2001;44:1897–1907. doi: 10.1002/1529-0131(200108)44:8<1897::AID-ART328>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Aikawa Y, Morimoto K, Yamamoto T, Chaki H, Hashiramoto A, Narita H, et al. Treatment of arthritis with a selective inhibitor of c-Fos/activator protein-1. Nat Biotechnol. 2008;26:817–823. doi: 10.1038/nbt1412. [DOI] [PubMed] [Google Scholar]
- Nishioka K, Ohshima S, Umeshita-Sasai M, Yamaguchi N, Mima T, Nomura S, et al. Enhanced expression and DNA binding activity of two CCAAT/enhancer-binding protein isoforms, C/EBPbeta and C/EBPdelta, in rheumatoid synovium. Arthritis Rheum. 2000;43:1591–1596. doi: 10.1002/1529-0131(200007)43:7<1591::AID-ANR24>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Pope RM, Lovis R, Mungre S, Perlman H, Koch AE., and , Haines GK. C/EBP beta in rheumatoid arthritis: correlation with inflammation, not disease specificity. Clin Immunol. 1999;91:271–282. doi: 10.1006/clim.1999.4723. [DOI] [PubMed] [Google Scholar]
- Friedman AD. C/EBPalpha induces PU.1 and interacts with AP-1 and NF-kappaB to regulate myeloid development. Blood Cells Mol Dis. 2007;39:340–343. doi: 10.1016/j.bcmd.2007.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher RC., and , Scott EW. Role of PU.1 in hematopoiesis. Stem Cells. 1998;16:25–37. doi: 10.1002/stem.160025. [DOI] [PubMed] [Google Scholar]
- Evans CH, Robbins PD, Ghivizzani SC, Wasko MC, Tomaino MM, Kang R, et al. Gene transfer to human joints: progress toward a gene therapy of arthritis. Proc Natl Acad Sci USA. 2005;102:8698–8703. doi: 10.1073/pnas.0502854102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Hooge AS, van de Loo FA, Koenders MI, Bennink MB, Arntz OJ, Kolbe T, et al. Local activation of STAT-1 and STAT-3 in the inflamed synovium during zymosan-induced arthritis: exacerbation of joint inflammation in STAT-1 gene-knockout mice. Arthritis Rheum. 2004;50:2014–2023. doi: 10.1002/art.20302. [DOI] [PubMed] [Google Scholar]
- Shiraishi A, Dudler J., and , Lotz M. The role of IFN regulatory factor-1 in synovitis and nitric oxide production. J Immunol. 1997;159:3549–3554. [PubMed] [Google Scholar]
- Zhang Y, Saccani S, Shin H., and , Nikolajczyk BS. Dynamic protein associations define two phases of IL-1beta transcriptional activation. J Immunol. 2008;181:503–512. doi: 10.4049/jimmunol.181.1.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortés-Canteli M, Wagner M, Ansorge W., and , Pérez-Castillo A. Microarray analysis supports a role for ccaat/enhancer-binding protein-beta in brain injury. J Biol Chem. 2004;279:14409–14417. doi: 10.1074/jbc.M313253200. [DOI] [PubMed] [Google Scholar]
- Huang JH., and , Liao WS. Induction of the mouse serum amyloid A3 gene by cytokines requires both C/EBP family proteins and a novel constitutive nuclear factor. Mol Cell Biol. 1994;14:4475–4484. doi: 10.1128/mcb.14.7.4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborti S, Mandal M, Das S, Mandal A., and , Chakraborti T. Regulation of matrix metalloproteinases: an overview. Mol Cell Biochem. 2003;253:269–285. doi: 10.1023/a:1026028303196. [DOI] [PubMed] [Google Scholar]
- Cox RS, Surette MG., and , Elowitz MB. Programming gene expression with combinatorial promoters. Mol Syst Biol. 2007;3:145. doi: 10.1038/msb4100187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinkhabwala A., and , Guet CC. Uncovering cis regulatory codes using synthetic promoter shuffling. PLoS ONE. 2008;3:e2030. doi: 10.1371/journal.pone.0002030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adriaansen J, Khoury M, de Cortie CJ, Fallaux FJ, Bigey P, Scherman D, et al. Reduction of arthritis following intra-articular administration of an adeno-associated virus serotype 5 expressing a disease-inducible TNF-blocking agent. Ann Rheum Dis. 2007;66:1143–1150. doi: 10.1136/ard.2006.064519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arend WP, Welgus HG, Thompson RC., and , Eisenberg SP. Biological properties of recombinant human monocyte-derived interleukin 1 receptor antagonist. J Clin Invest. 1990;85:1694–1697. doi: 10.1172/JCI114622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varley AW, Coulthard MG, Meidell RS, Gerard RD., and , Munford RS. Inflammation-induced recombinant protein expression in vivo using promoters from acute-phase protein genes. Proc Natl Acad Sci USA. 1995;92:5346–5350. doi: 10.1073/pnas.92.12.5346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Meurs JB, Van Lent PL, Joosten LA, Van der Kraan PM., and , Van den Berg WB. Quantification of mRNA levels in joint capsule and articular cartilage of the murine knee joint by RT-PCR: kinetics of stromelysin and IL-1 mRNA levels during arthritis. Rheumatol Int. 1997;16:197–205. doi: 10.1007/BF01330296. [DOI] [PubMed] [Google Scholar]
- Bult CJ, Eppig JT, Kadin JA, Richardson JE., and , Blake JA.2008The Mouse Genome Database (MGD): mouse biology and model systems Nucleic Acids Res 36Database issue): D724–D728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Helden J. Regulatory sequence analysis tools. Nucleic Acids Res. 2003;31:3593–3596. doi: 10.1093/nar/gkg567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R. Methods for discovering novel motifs in nucleic acid sequences. Comput Appl Biosci. 1989;5:293–298. doi: 10.1093/bioinformatics/5.4.293. [DOI] [PubMed] [Google Scholar]
- Smeets RL, Joosten LA, Arntz OJ, Bennink MB, Takahashi N, Carlsen H, et al. Soluble interleukin-1 receptor accessory protein ameliorates collagen-induced arthritis by a different mode of action from that of interleukin-1 receptor antagonist. Arthritis Rheum. 2005;52:2202–2211. doi: 10.1002/art.21108. [DOI] [PubMed] [Google Scholar]
- Schiedner G, Hertel S., and , Kochanek S. Efficient transformation of primary human amniocytes by E1 functions of Ad5: generation of new cell lines for adenoviral vector production. Hum Gene Ther. 2000;11:2105–2116. doi: 10.1089/104303400750001417. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Annotation of the probe identities per k-means cluster.
Proximal (−500/+200) promoter sequences used in this study.
Supplementary Methods: Primers.