Abstract
We study the folding mechanism of a three-helix bundle protein at atomic resolution, including effects of explicit water. Using replica exchange molecular dynamics we perform enough sampling over a wide range of temperatures to obtain the free energy, entropy, and enthalpy surfaces as a function of structural reaction coordinates. Simulations were started from different configurations covering the folded and unfolded states. Because many transitions between all minima at the free energy surface are observed, a quantitative determination of the free energy barriers and the ensemble of configurations associated with them is now possible. The kinetic bottlenecks for folding can be determined from the thermal ensembles of structures on the free energy barriers, provided the kinetically determined transition-state ensembles are similar to those determined from free energy barriers. A mechanism incorporating the interplay among backbone ordering, sidechain packing, and desolvation arises from these calculations. Large Φ values arise not only from native contacts, which mostly form at the transition state, but also from contacts already present in the unfolded state that are partially destroyed at the transition.
The energy landscape theory and the funnel concept, along with a new generation of experiments, have created a “new view” of protein folding (1–4). Folding is best described as an ensemble of converging pathways toward the native structure. For good folding sequences, these paths are all energetically very similar with barriers between them on the kBT energy scale. Minor fluctuations due to environmental or mutational changes may vary the path probabilities, but the overall global landscape flow will be only weakly altered. This leads to a key result of the energy landscape theory: protein folding dynamics can be properly understood as the diffusion of an ensemble of protein configurations over a low-dimensional free energy surface, which may be constructed by using many different order parameters.
In this article, we describe the free energy landscape for protein folding simulated with full atomic details of both protein and solvent, over a broad range of temperatures. For our studies, a natural choice is the 10–55 helical fragment B of protein A from Staphylococcus aureus, using the replica exchange molecular dynamics (REMD) algorithm described by Sugita and Okamoto (5). Protein A folds into a simple three-helix bundle (6) whose folding has been widely studied by using minimalist and all-atom simulations (7–17) as well as experiments (18, 19). All of these different studies provided some information about the folding mechanism and the nature of the transition-state ensemble (TSE). The study presented here aims to establish a quantitative picture by integrating the earlier qualitative findings with detailed simulations. We simulated 82 replicas of the water–protein system, with temperature, T, ranging from 277 to 548 K, and each replica was started from a different configuration spanning the space of folded and unfolded states. Multiple transitions from the unfolded ensemble to the native minimum were observed in our computer simulations, thus ensuring the validity both of the sampling and the potential energy functions. Until now, evidence of folding/unfolding simulations has only been shown in the context of peptides by using a simpler implicit solvent representation (20, 21).
The events leading to folding span many time scales. Recent ultrafast spectroscopy experiments show that the formation of interresidue contacts occurs on the 10-ns time scale (22); formation of α-helices occurs in 200 ns (23); formation of β-hairpins occurs in 1–10 μs (24); and folding of single domain proteins occurs in 10–1,000 μs (25). The longest single-trajectory simulation of protein folding in explicit solvent previously published was not sufficiently long to show complete folding of the protein (26). Simple molecular dynamics cannot break the time scales needed to describe protein folding in explicit solvent models. By using new simulation tools, we are able to perform numerical investigations that were practically unfeasible even 1 year ago.
Methods
REMD simulations were carried out by using an explicit solvent model, under periodic boundary conditions. The 46-aa system is contained in a cubic box containing 5,107 water molecules and 16,055 atoms. The protein chain is the amino-acetylated and carboxy-amidated 46-aa fragment of fragment B of staphylococcal protein A. In our analysis, amino acids are numbered sequentially, starting with the acetyl group, such that amino acid Gln-10 in the protein fragment corresponds to amino acid Gln-2 in our numbering. The folded protein in aqueous solution was equilibrated for 1.1 ns at constant temperature (300 K) and pressure (1 atm; 1 atm = 101.3 kPa). We used Protein Data Bank file 1BDB as the folded structure (6). We used the force field of Cornell et al. (27), with modified backbone dihedral angle potentials (28), and the suite of programs in amber (University of California, San Francisco), with the Generalized Reaction Field (GRF) (29) treatment of electrostatic interactions (with a cut-off of 8.0 Å) and the REMD algorithm (5). Nonbonded pair lists were updated every 10 integration steps. The integration step in all simulations was 0.0015 ps. The system was coupled to an external heat bath with a relaxation time of 0.1 ps (30). All bonds involving hydrogen atoms were constrained in length. The solvated systems were subjected to 500 steps of steepest-descent energy minimization and a 1.1-ns molecular dynamics simulation at constant pressure (P) and temperature (T), with P = 1 atm and T = 300 K. The equilibrated system was contained in a cubic box of side dimension 54.39 Å. All replica calculations were done at constant volume.
The temperatures of the replicas were chosen to maintain an exchange rate among replicas between 8% and 20%. Exchanges were attempted every 250 integration steps (0.375 ps). We simulated 82 replicas of the water–protein system, with T = 277–548 K. To generate a set of initial conditions that broadly covers the configuration space of the protein, we performed independent simulations, extending from 0.5 to 2 ns, at T = 275–1,000 K. We chose configurations at random from this sampling and kept structures with a radius of gyration, Rg, <15 Å and a total number of contacts, Z, >50 (i.e., we chose partially collapsed configurations) as initial structures for the replicas. The resulting configurations were assigned at random to one of 82 temperatures. Of the 82 initial configurations, 25 had an rms distance (rmsd) <2.0 Å, 27 had 2.0 < rmsd < 4.0 Å, and 30 had rmsd > 4.0 Å from the folded state. All replicas were equilibrated for 200 ps without exchanging temperatures, at the beginning of the simulations. After the 200 ps without exchanges, 44 replicas had rmsd < 4 Å, 38 had rmsd > 4 Å, 11 had rmsd > 8 Å, and 3 had rmsd > 10 Å. The largest rmsd is 13 Å, and the largest Rg is 16.3 Å. The largest Rg and rmsd adopted during the simulation were 19 and 15 Å, respectively. The REMD simulation was carried out for 13.085 ns per replica (1.07-μs total simulation time). The last 11.775 ns (31,400 configurations) per replica were used to calculate all of the averages reported here. Protein hydration was analyzed during the last 750 ps per replica of the simulation. We analyzed the configurations generated by the REMD simulations in terms of the α-helical content, θ, the rmsd from the Protein Data Bank structure, and the fraction of native contacts, Q. A helical segment was defined whenever three or more consecutive residues occupy the α-helical region of the φψ map (φ = –60 ± 35° and ψ = –47 ± 30°). Contacts were defined as occurring whenever two amino acid side chains, separated by five or more amino acids along the chain, have any two atoms within 6.4 Å of each other. We defined native contacts as those contacts with a 1/e probability of being present in the lowest T contact map. The folded state has 52 native contacts, and the maximum number of contacts is 104.
Results and Discussion
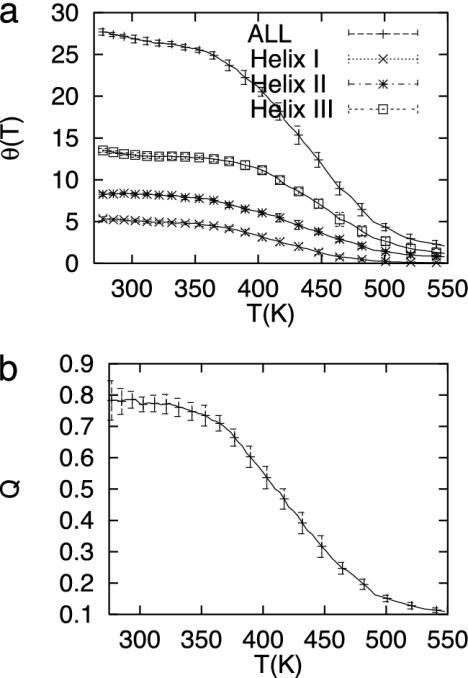
Thermodynamic Description of the Folding Energy Landscape. The improved sampling provided by the REMD technique allows us to explore the configuration space of protein A. Fig. 1 shows the average number of amino acids in α-helical configurations (θ) and the average Q as a function of temperature. The curves are typical of a broad two-state transition, although examination of the transition with multiple order parameters shows three major basins. The half-maximum helical content is near 440 K. At this temperature the average Q is 0.35. Rapid changes in the number of α-helical amino acids facilitate the fast convergence of the helical content as a function of temperature. Slow changes in the number of native contacts and rmsd make the convergence slow. However, the inclusion of multiple replicas enhances the sampling and reduces the correlation in the ensemble averages. The helical content and fraction of native contact profiles are well equilibrated within 1- to 3-ns block averages.
Fig. 1.
(a) Average number of amino acids in the α-helices, Θh, as a function of temperature for all amino acids (ALL) and for amino acids in α-helices I, II, and III. The temperature stability of the helices are helix III ∼ helix II > helix I. The temperature at which the total helical content is half the maximum is ≈440 K. (b) Average fraction of native contacts as a function of temperature. The error bars are calculated by block averages and represent three standard deviations (45).
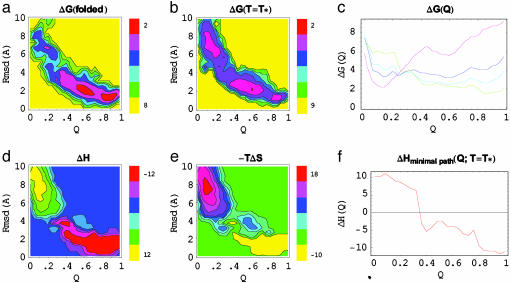
Free energy surfaces were calculated from histograms of the occurrence of selected order parameters (Q and the rmsd from the folded structure) in the configurational ensembles generated for each temperature (Fig. 2). The overall features of this surface are similar to that calculated previously with a different force field (9). At low T the free energy surface, ΔG(Q,rmsd), shows a large folding basin containing two minima weakly separated; one corresponding to the native state (folded unsolvated with Q > 0.8) and the other a nearly folded state with a hydrated core (folded solvated with 0.30 < Q < 0.8). The populations of these two “folded” regions are equal at 387 K. At very high T there is only one minimum corresponding to the unfolded basin. The populations of the two minima switch as T changes. At intermediate T we see two large basins separated by a free energy barrier. The transition temperature, T*, at which the two states are equally populated is 421 K. T* is significantly higher (≈30%) than expected. At the folding temperature, T*, ΔG(Q,rmsd|T*), exhibits two large energy basins corresponding to the folded (Q ≥ 0.30, rmsd ≤ 4 Å) and unfolded (Q < 0.30, rmsd > 4 Å) states, with the folded basin containing the unsolvated and folded solvated states. The barrier separating the two basins is ≈4 kcal/mol. The free energy profiles as a function of Q, for various temperatures, are shown in Fig. 2c. Any of the order parameters used, Q or rmsd, is sufficient to distinguish the two basins, suggesting that a collective folding transition occurs with concurrent secondary and tertiary structure formation and side chain packing, as expected from a funnel-like landscape. Also, the unfolded basin still shows some resemblance to the native state, indicating that this ensemble is not fully disordered. The free energy surface shows that Q and rmsd can be used to identify transitions between folded and unfolded states, with the transition region defined by Q* ∼ 0.30 and rmsd* ∼ 4.0 Å.
Fig. 2.
Contour maps of the free energy in the folded state (a), ΔG(folded T = 387 K), and at the transition temperature (b), ΔG(T = T* = 421 K), as a function of the rmsd from the experimental folded structure and Q. At low T the “folded” state basin has two minima separated by a small barrier. These two minima correspond to the native state (Q > 0.8, rmsd < 2 Å) and a nearly folded state with a hydrated core (0.30 < Q < 0.8). The population in these two folded basins is equal at 387 K. At T = T*, there are two large free energy basins that are equally populated. The folded state is defined by rmsd < 4 Å and Q > 0.30, and the unfolded state is defined by rmsd > 4.0 Å and Q < 0.30. At T » T*, the unfolded basin is the most populated. (c) Free energy profile as a function of Q, at temperatures given as a fraction T/T* (green, 0.75; cyan, 0.91; purple, 1.0; magenta, 1.17). The free energy as a function of T is decomposed into the enthalpy (d), ΔH, and entropy (e), TΔS. The folded basin is dominated by low enthalpy and low entropy. The unfolded basin is dominated by high entropy and high enthalpy. The deep minimum in ΔH covers the whole folded state basin (rmsd < 4 Å, Q > 0.30). The entropy has a deep minimum in the region corresponding to the desolvated folded state (Q > 0.8, rmsd < 4 Å). (f) Enthalpy along a minimal free energy path as a function of Q, at T = T*. The minimal path is defined by the pair of (Q,rmsd) coordinates for which ΔG(T = T*) is minimal for each value of Q. The enthalpy along this path shows a decrease characteristic of a funneled landscape, although larger cooperativity is observed in the 0.25–0.4 range of Q. This curve shows a similar profile to the complement of the helical fraction for helix I (1 – ΘI), and the total number of coordinated waters to the protein backbone and the helical content of helix I, as a function of Q, shown in Figs. 4 and 5, respectively. All energies are given in kcal/mol. The transition-state ensemble is characterized by Q* ∼ 0.25–0.4.
We decompose the free energy landscape into its enthalpic and entropic components by fitting all free energy surfaces at all sampled T. We fit all free energy surfaces at all sampled T to a function ΔG(Q,rmsd|T) =–(ΔH(Q,rmsd) – TΔS(Q,rmsd)), where ΔH(Q,rmsd) and ΔS(Q,rmsd) are the enthalpy and entropy of the system, respectively, as a function of the Q and rmsd order parameters. ΔH(Q,rmsd) and ΔS(Q,rmsd) are assumed to be T-independent. These quantities are changes in the total enthalpy and entropy of the system with contributions from both protein and solvent. ΔH(Q,rmsd) spans a range of values from –12 to 12 kcal/mol, where the folded basin has low enthalpy and the unfolded state has high enthalpy. The entropy, –TΔS(Q,rmsd), shows the opposite behavior, with low entropy for the folded state and high entropy for the unfolded state. The entropy near Q = 1 and rmsd ∼ 1.5 Å is the lowest. The enthalpy and entropy are down hill, without barriers. The imbalance between entropy and enthalpy results in the free energy barrier separating the folded and unfolded basins, shown in ΔG(Q,rmsd|T = T*).
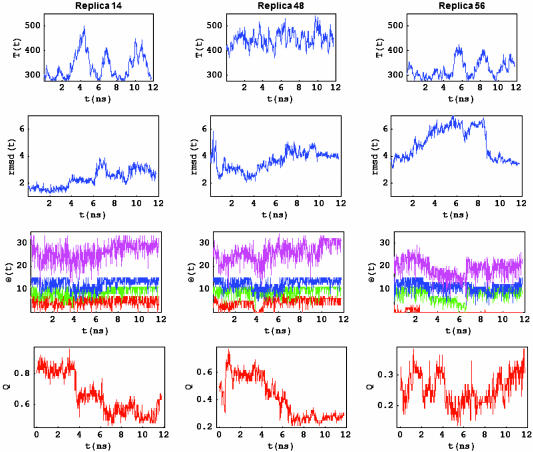
Now that we have characterized the folded and unfolded states in terms of Q and rmsd order parameters, we can explore the completeness of the sampling performed by the REMD. To illustrate the ample and detailed sampling of the folding/unfolding energy landscape obtained with the REMD, we show the time history T, rmsd, the number of α-helical content in each one of the three helices (Θh), and the fraction of native contacts for three representative replicas (replicas 14, 48, and 56) (Fig. 3). Replica 14 covers mostly the folded state basin and shows a transition between the folded solvated to unsolvated states. Many other replicas exhibit similar transitions and the reverse transition. Replica 48 shows multiple transitions between the folded solvated and unfolded basins, without getting too far from the transition region between the unfolded and folded basins. Replica 56 shows the most drastic change in rmsd while undergoing a transition from a folded solvated state (with rmsd ∼ 2.7 Å) to an unfolded state (with rmsd ∼ 7.3 Å) and back to the folded solvated state (with rmsd ∼ 3.2 Å). These trajectories cover a large region of the configuration space, because they vary their temperature during their time evolution. Because of the variations in temperature used in the REMD algorithm, the time history of the replicas is not directly related to the folding/unfolding pathways at constant temperature, but it provides a reasonable description of the order of the events during folding.
Fig. 3.
Time series (at 50-ps intervals) of the temperature, the rmsd from the folded structure, the helical content for helix I (red), helix II (green), and helix III (blue), the total helix content (magenta), and Q, sampled by replicas 14, 48, and 56. All three replicas span a broad region of the allowed temperatures. Replica 14 remains folded and T remains mostly below T*; however, all three helices fold and unfold, although for short periods of time. This replica shows a transition from the unsolvated folded state (Q > 0.8, rmsd < 2 Å) to the solvated folded state (0.30 < Q < 0.8, rmsd > 2 Å). Replica 48 unfolds and refolds along the trajectory. The system switches back and forth between folded (Q > 0.30, rmsd < 4.0 Å) and unfolded (Q < 0.30, rmsd > 4.0 Å) during the last 6 ns. All three helices break and form at some point along the trajectory. Replica 56 shows variations in the rmsd from 4 to 7 Å within the first 4 ns, remains unfolded for 5 ns, and then folds back to the solvated native state, with a rmsd of 3.5 Å. Helix II and III unfold and refold, with helical content spanning 0–11 and 0–15 aa, respectively. The chain segment corresponding to helix I in the folded state forms a transient β-hairpin for a few nanoseconds. In this trajectory, helix I remains mostly disordered. Helix III remains largely formed during the trajectory, although the helical content spans a range of helical content from 0 to 25 aa. The fraction of native contacts switches back and forth between unfolded (Q < 0.25) and folded (Q > 0.35).
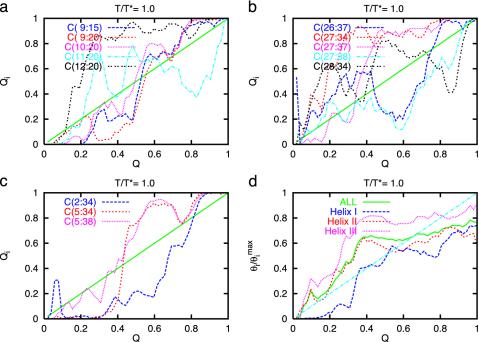
Folding Routes on the Energy Landscape. The broad sampling of configurations covered by the simulation data allows us to examine the equilibrium distribution of configurations covering a large region of the configuration space, as delineated by Q and rmsd. We describe this distribution at the transition temperature, T*. Figs. 4 shows the probability of forming contacts between helices I and II (thus forming turn I–II), contacts between helices II and III (forming turn II–III), long range contacts between helices I and III, and the helical content, as a function of Q. Notice that the configurations at the unfolded region (Q < 0.30) already include native features, particularly contacts in the loop regions and the formation of helix III. More interesting are the features of the TSE. Some of the early formed native contacts at the loop regions need to be rearranged (broken and reformed) when the system is near the transition region (0.25 < Q < 0.4). The possibility of nonmonotonic formation of native contacts as the folding reaction progresses has been ignored in most of the previous transition-state analysis, and they can have a substantial effect on the experimentally measured Φ values that have been used to explore this transition region (8, 31–35). An additional feature at the TSE is a substantial formation of helix I with the concurrent (or followed by the) formation of tertiary contacts between helices I and II and helices II and III, and of some long range contacts between helices I and III. Helix I is completely unstable unless tertiary contacts with other helices are formed. Therefore, the TSE is characterized by many simultaneous structural changes, i.e., the loops are partially unformed to allow for the packing of helices (specially between helices I and II) and the formation of helix I. Although helix III is formed very early in the folding event, only a fraction of its contacts with the other helices are formed in the transition region. The full packing with the remaining of the protein occurs only at the late stages of folding.
Fig. 4.
Probability of forming selected native contacts as a function of the average Q at T*. Specifically, we show the contacts involved in the formation of the turns I–II (a) and II–III (b), the long-range contacts between helices I and III (c), and the helical content (d), as a function of Q. The diagonal line is drawn as a reference for contacts that perfectly follow the average fraction of contacts. In turn I–II, the contacts 9–15, 9–20, and 10–20 form near Q = Q* ∼ 0.25–0.4, and after the helices are formed. Contacts 11–20 and 12–20 form at Q ∼ 0.2, suggesting that the I–II turn is formed early and is, on average, formed before Q reach Q*(Q* ∼ 0.25–0.4). In the II–III turn, contact 27–34 forms early (Q ∼ 0.2), but longer range contacts (26–37, 27–37, 27–38, and 28–34), involving the packing of helices II and III, occur late, indicating that turn II–III forms after Q*. The longest-range native contact is 2–34 (contact involving helices I–III). This contact forms late in the folding, catching up with Q for Q > 0.80. Contacts 5–34 and 5–38 form near Q*, after helix I forms. From d we see that helix III forms first and leads the total helix formation, whereas helix II follows the total helix content and the average Q. However, helix I lags behind the total helix formation and forms near Q*. Overall, this picture suggests a mechanism in which turns form early, without the formation of helix I. The TSE is characterized by many simultaneous structural changes where the turns are partially unformed to allow for the packing of helices and concurrent formation of helix I. Helix I is unstable unless contacts with helix II are formed.
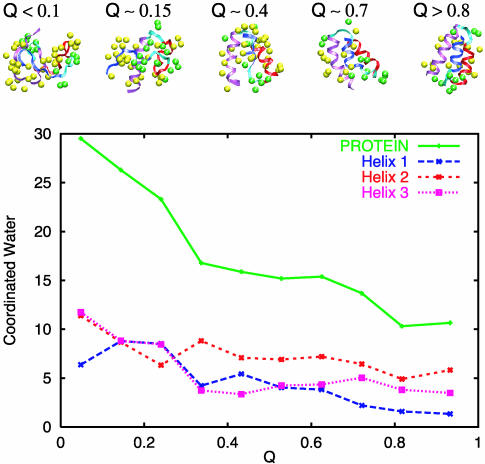
Finally, we can investigate the interplay between protein ordering and dehydration of the protein core; this question has been addressed in several recent papers (36–40), and it is now quantified so that a unified view for the folding mechanism becomes possible. The average number of water molecules coordinated to the backbone carbonyl oxygen atoms provides a measure of the degree of protein hydration. We monitor the protein hydration by examining along the folding reaction Q the number of water molecules coordinated to the backbone carbonyl oxygen atoms along the folding progress variable. From previous simulations of α-helical systems we know that an isolated α-helix in solution has, on average, one coordinated water per carbonyl oxygen atom near the center of the helix (28). In an unfolded helix, and at the C-terminal end of helices, the backbone carbonyl oxygen atoms have, on average, two coordinated water molecules. Regions of an α-helix that are buried in the protein core do not have coordinated water. Fig. 5 shows a plot of the average number of water molecules coordinated to various regions of the protein. Helix III has sufficiently large side chains that it gets its backbone carbonyl oxygen atoms protected as soon as the helix is formed. The number of coordinated water molecules to helix III is large (10) in the unfolded state but decreases to <5 as soon as the helix forms (Q < 0.30). Helix II, because it is more exposed and has smaller side chains, has a reduced level of protection during the entire folding event. The carbonyl groups of helix I, however, are the best probe for the dehydration of the protein core. A sharp transition in the number of coordinated waters (from eight to less than four) occurs concurrent with the packing of helix III against helices I and II (Fig. 4). The plot of the water coordination number for all carbonyl oxygen atoms in the protein shows two regions with large changes occurring as a function of Q. The first occurs at the folding transition region (Q < Q* ∼ 0.30) and the second at Q ∼ 0.8, where the helices pack closely and the protein changes from a solvated folded state to a desolvated folded structure. The coordination number is constant for the folded solvated state (0.30 < Q < 0.8). The TSE of the protein is characterized by structures where the loops are partially unformed to allow for the packing of helices I and II and the formation of helix I. This TSE transition is followed by a later dehydration transition that is concurrent with the final packing of helix III with helices I and II.
Fig. 5.
Water coordination of the backbone carbonyl oxygen atoms in helices I–III and turns, as a function of Q. (Upper) Selected configurations of the protein and the water molecules within 3.5 Å of the carbonyl oxygen atoms as the folding progresses along the reaction coordinate Q. Water molecules coordinating to carbonyl oxygen atoms in the helical regions are colored yellow, all others are colored green; helix I is blue, helix II is red, and helix III is magenta. (Lower) Average coordination number of water molecules to the carbonyl oxygen atoms of helices and the whole protein as a function of Q. This plot shows that helix III, which forms early, desolvates as the helix is formed, and reaches its folded-state coordination number near Q*. Helix I forms and partially desolvates at Q ∼ Q* = 0.25 and loses two more water molecules at Q ∼ 0.8. Helix II desolvates slightly, for Q ∼ 0.25, but then is partially resolvated as helix I forms and desolvates uniformly along Q. The coordination number of all carbonyl oxygen atoms shows two regions where large changes occurs as a function of Q. The first occurs as the system folds (Q < Q* ∼ 0.30) and the second at Q ∼ 0.7, where the helices pack closely and the protein changes from a solvated folded structure to a desolvated folded structure. The coordination number is constant for the folded solvated structure (0.30 < Q < 0.8).
Description of the Folding Mechanism. Folding a protein in a computer is now possible without the help of any structural constraints, other than those imposed by the limitation of the simulation box size, which may increase stability of the folded state but does not affect the folding mechanism (21). Multiple folding and unfolding transitions have been observed for a protein by using explicit solvation along with currently available force fields. These transitions are defined by using the features of the calculated free energy surface showing three basins, folded unsolvated (Q > 0.8, rmsd < 4), folded solvated (0.8 < Q < 0.30, rmsd < 4.0 Å), and unfolded (Q < 0.30, rmsd > 4 Å). The all-atom simulation reveals the quantitative details of the protein folding event that involve the funnel-guided evolving ensembles of structures [with an overall agreement with the picture presented by others (12, 15, 17)]. The unfolded state of the protein is compact, having Rg within 2 Å of the folded state and Q* ∼ 0.25–0.40, consistent with previous calculations (7). These evolving ensembles exhibit a folding mechanism where secondary structure formation is coupled with solvent exclusion and core formation. We find that helix III forms early, consistent with experiments (19) and most calculations (12, 15, 17). The results of Bockzo and Brooks (7) differ from ours on this specific detail. This is surprising because most other features of the folding mechanism that we calculated agree with theirs. However, we are using different force fields and simulation methods. Analysis of the helix content as a function of T shows that, as T increases, helix I unfolds first, followed by helix II and helix III. We find that the unfolded ensemble exhibits a stable β-hairpin in the helix I segment. This β-hairpin persists for a few nanoseconds in one replica and converts quickly to a persistent PPII-type of structure, which then converts to an α-helix (data not shown). The folding transition temperature of 421 K is higher than expected. Assuming a two-state folding transition, we calculated ΔGf/u (T = 300 K) = –4 kcal/mol, which is larger in magnitude than the –1 kcal/mol value estimated by Shea and Brooks (41). The truncated fragment of protein A is partially unfolded at room temperature (18) (suggesting ΔGf/u ∼ 0). Simulations on the isolated helix III fragment show that helix III unfolds at ≈420 K. This large stability comes from strong side chain contacts in helix III that are consistent with a helix conformation. The use of a constant volume in the REMD simulations overstabilizes hydrophobic contact formation at high T and may be partially responsible for the high transition T (42–44). The high transition T should be of no major concern because the folding temperature is extremely sensitive to the detailed cancellation between entropy and enthalpy. The important fact is that force fields are already able to show folding transitions of a protein without any constraints.
The folding landscape that comes from these simulations is funnel-like but it still exhibits a reasonable amount of residual frustration. Several native contacts, especially around the loop regions, need to be broken and reformed during the folding transition. Nonnative structures, such as helix 1 in a strand conformation, are observed in the unfolded ensemble. Globally, the free energy profile is mostly flat for regions with Q ≥ 0.7–0.8. Although this computer-observed landscape gives the signature of a good folder, its residual frustration appears to be larger than that observed experimentally for small fast-folding proteins such as that studied here. Even with these minor differences, it is amazing how well the current force fields, with no further adjustments, are able to reproduce the experimentally observed folding event.
The analysis of the folding routes shows that the formation of α-helices and turns does not proceed monotonically along a reaction coordinate that measures the fraction of native contacts formed. We find that the formation of the turns occurs precipitously, and that they partially reverse their formation near the transition region to accommodate the packing tertiary structural elements. The picture we obtain from the simulations is that helix I can adopt multiple stable secondary structures and disordered conformations while helix II and III remain relatively fixed. Our simulation suggests that fluctuations occur in different time scales, where helix I forms and breaks multiple times while helix II and III move relative to each other.
This folding simulation shows that, because proteins have a sufficiently smooth folding landscape, folding is a very robust event for good folding sequences (true for most small or intermediate-size proteins) and, therefore, that current physically based force fields can already fold proteins. Although some refinement is still needed on existing force fields, and all-atom simulations still require very large amounts of computer time, our results fully support the energy landscape theory for protein folding and open a new era in folding simulations.
Acknowledgments
We thank C. L. Brooks III, G. Makhatadze, J. J. Portman, K. Y. Sanbonmatsu, and P. G. Wolynes for useful discussions. This work was supported by Department of Energy Contract W-740-ENG-36 and the Laboratory-Directed Research and Development program at Los Alamos National Laboratory. Work at the University of California was supported by National Science Foundation Grant MCB-0084797 with additional support from Grants PHY-0216576 and PHY-0225630.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: REMD, replica exchange molecular dynamics; rmsd, rms distance; TSE, transition-state ensemble.
References
- 1.Bryngelson, J. & Wolynes, P. (1987) Proc. Natl. Acad. Sci. USA 84, 7524–7528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leopold, P. E., Montal, M. & Onuchic, J. N. (1992) Proc. Natl. Acad. Sci. USA 89, 8721–8725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baldwin, R. L. (1995) J. Biomol. NMR 5, 103–109. [DOI] [PubMed] [Google Scholar]
- 4.Onuchic, J. N., Luthey-Schulten, Z. & Wolynes, P. G. (1997) Annu. Rev. Phys. Chem. 48, 545–600. [DOI] [PubMed] [Google Scholar]
- 5.Sugita, Y. & Okamoto, Y. (1999) Chem. Phys. Lett. 314, 141–151. [Google Scholar]
- 6.Gouda, H., Torigoe, H., Saito, A., Sato, M., Arata, Y. & Shimada, I. (1992) Biochemistry 31, 9665–9672. [DOI] [PubMed] [Google Scholar]
- 7.Boczko, E. M. & Brooks, C. L., III (1995) Science 269, 393–396. [DOI] [PubMed] [Google Scholar]
- 8.Shea, J. E., Onuchic, J. N. & Brooks, C. L., III (1999) Proc. Natl. Acad. Sci. USA 96, 12512–12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo, Z., Brooks, C. L., III, & Boczko, E. M. (1997) Proc. Natl. Acad. Sci. USA 94, 10161–10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takada, S., Luthey-Schulten, Z. & Wolynes, P. (1999) J. Chem. Phys. 110, 11616–11629. [Google Scholar]
- 11.Favrin, G., Irback, A. & Wallin, S. (2002) Proteins 47, 99–105. [DOI] [PubMed] [Google Scholar]
- 12.Ghosh, A., Elber, R. & Scheraga, H. A. (2002) Proc. Natl. Acad. Sci. USA 99, 10394–10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kolinski, A. & Skolnick, J. (1994) Proteins 18, 353–366. [DOI] [PubMed] [Google Scholar]
- 14.Zhou, Y. & Karplus, M. (1999) J. Mol. Biol. 293, 917–951. [DOI] [PubMed] [Google Scholar]
- 15.Alonso, D. O. & Daggett, V. (2000) Proc. Natl. Acad. Sci. USA 97, 133–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Onuchic, J. N., Wolynes, P. G., Luthey-Schulten, Z. & Socci, N. D. (1995) Proc. Natl. Acad. Sci. USA 92, 3626–3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berriz, G. & Shakhnovich, E. (2001) J. Mol. Biol. 310, 673–685. [DOI] [PubMed] [Google Scholar]
- 18.Witte, K., Skolnick, J. & Wong, C. (1998) J. Am. Chem. Soc. 120, 13042. [Google Scholar]
- 19.Bai, Y., Karimi, A., Dyson, H. J. & Wright, P. E. (1997) Protein Sci. 6, 1449–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Snow, C. D., Nguyen, H., Pande, V. S. & Gruebele, M. (2002) Nature 420, 102–106. [DOI] [PubMed] [Google Scholar]
- 21.Klimov, D. K., Newfield, D. & Thirumalai, D. (2002) Proc. Natl. Acad. Sci. USA 99, 8019–8024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lapidus, L. J., Eaton, W. A. & Hofrichter, J. (2000) Proc. Natl. Acad. Sci. USA 97, 7220–7225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Williams, S., Causgrove, T. P., Gilmanshin, R., Fang, K. S., Callender, R. H., Woodruff, W. H. & Dyer, R. B. (1996) Biochemistry 35, 691–697. [DOI] [PubMed] [Google Scholar]
- 24.Munoz, V., Thompson, P. A., Hofrichter, J. & Eaton, W. A. (1997) Nature 390, 196–199. [DOI] [PubMed] [Google Scholar]
- 25.Mayor, U., Johnson, C. M., Daggett, V. & Fersht, A. R. (2000) Proc. Natl. Acad. Sci. USA 97, 13518–13522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Duan, Y. & Kollman, P. A. (1998) Science 282, 740–744. [DOI] [PubMed] [Google Scholar]
- 27.Cornell, W. D., Cieplak, P., Bayley, C. I., Gould, R., Merz, K. M., Ferguson, D. M., Spellmeyer, D. C., Fox, T., Caldwell, J. & Kollman, P. A. (1995) J. Am. Chem. Soc. 117, 5179–5197. [Google Scholar]
- 28.García, A. E. & Sanbonmatsu, K. Y. (2002) Proc. Natl. Acad. Sci. USA 99, 2782–2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hummer, G., Soumpasis, D. & Neumann, M. (1994) J. Phys. Condens. Matter 6, A141–A144. [Google Scholar]
- 30.Berendsen, H. J. C., Postma, J. P., van Gunsteren, W. F., DiNola, A. & Haak, J. R. (1984) J. Chem. Phys. 81, 3684–3690. [Google Scholar]
- 31.Matouschek, A., Kellis, J. T., Jr., Serrano, L. & Fersht, A. R. (1989) Nature 340, 122–126. [DOI] [PubMed] [Google Scholar]
- 32.Mayor, U., Guydosh, N. R., Johnson, C. M., Grossmann, J. G., Sato, S., Jas, G. S., Freund, S. M. V., Alonso, D. O. V., Daggett, V. & Fersht, A. R. (2003) Nature 421, 863–867. [DOI] [PubMed] [Google Scholar]
- 33.Blanco, F. J., Serrano, L. & Forman-Kay, J. D. (1998) J. Mol. Biol. 284, 1153–1164. [DOI] [PubMed] [Google Scholar]
- 34.Kim, D. E., Fisher, C. & Baker, D. (2000) J. Mol. Biol. 298, 971–984. [DOI] [PubMed] [Google Scholar]
- 35.Oliveberg, M. (2001) Curr. Opin. Struct. Biol. 11, 94–100. [DOI] [PubMed] [Google Scholar]
- 36.Sheinerman, F. B. & Brooks, C. L., III (1998) Proc. Natl. Acad. Sci. USA 95, 1562–1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hillson, N., Onuchic, J. N. & García, A. E. (1999) Proc. Natl. Acad. Sci. USA 96, 14848–14853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cheung, M., García, A. & Onuchic, J. (2002) Proc. Natl. Acad. Sci. USA 99, 685–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.ten Wolde, P. R. & Chandler, D. (2002) Proc. Natl. Acad. Sci. USA 99, 6539–6543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shea, J., Onuchic, J. & Brooks, C. (2002) Proc. Natl. Acad. Sci. USA 99, 16064–16068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shea, J. E. & Brooks, C. L., III (2001) Annu. Rev. Phys. Chem. 52, 499–535. [DOI] [PubMed] [Google Scholar]
- 42.Hummer, G., Garde, S., García, A. E., Pohorille, A. & Pratt, L. R. (1996) Proc. Natl. Acad. Sci. USA 93, 8951–8955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Garde, S., Hummer, G., García, A. E., Paulaitis, M. E. & Pratt, L. R. (1996) Phys. Rev. Lett. 77, 4966–4968. [DOI] [PubMed] [Google Scholar]
- 44.Hummer, G., Garde, S., García, A. E. & Pratt, L. R. (2000) Chem. Phys. 258, 349–370. [Google Scholar]
- 45.Flyvbjerg, H. & Petersen, H. (1989) J. Chem. Phys. 91, 461–466. [Google Scholar]