Abstract
Lentiviral vectors are very efficient at transducing dividing and quiescent cells, which makes them highly useful tools for genetic analysis and gene therapy. Traditionally this efficiency was considered dependent on provirus integration in the host cell genome; however, recent results have challenged this view. So called integration-deficient lentiviral vectors (IDLVs) can be produced through the use of integrase mutations that specifically prevent proviral integration, resulting in the generation of increased levels of circular vector episomes in transduced cells. These lentiviral episomes lack replication signals and are gradually lost by dilution in dividing cells, but are stable in quiescent cells. Compared to integrating lentivectors, IDLVs have a greatly reduced risk of causing insertional mutagenesis and a lower risk of generating replication-competent recombinants (RCRs). IDLVs can mediate transient gene expression in proliferating cells, stable expression in nondividing cells in vitro and in vivo, specific immune responses, RNA interference, homologous recombination (gene repair, knock-in, and knock-out), site-specific recombination, and transposition. IDLVs can be converted into replicating episomes, suggesting that if a clinically applicable system can be developed they would also become highly appropriate for stable transduction of proliferating tissues in therapeutic applications.
Introduction
The development of the prototypical lentivirus (human immunodeficiency virus, HIV) from disease-causing agent to efficient tool for therapeutic gene delivery was an accomplishment pioneered by a number of groups (for example, refs. 1,2). Lentiviral vectors have several highly advantageous properties for this purpose: (i) comparatively large coding capacity (~8-kb transgene cassettes in addition to all the required cis-acting signals);3 (ii) reduced immunogenicity upon in vivo administration4,5 compared to other gene transfer vehicles like adenovirus6 or adeno-associated virus vectors;7,8,9,10 (iii) similar transduction efficiency to adeno-associated virus vectors, at titers 3- to 4-log lower (R.J. Yáñez-Muñoz, K.S. Balaggan, A.J. Thrasher and R.R. Ali, unpublished results); (iv) transduction of quiescent cells, a crucial issue for gene transfer to differentiated cells like neurons,11 retinal cells,12 and hepatocytes;13 and (v) stable gene expression through their integration into the host genome. In the case of retroviral vectors, the latter property turned into a serious drawback when the well-known risk of provirus-mediated insertional mutagenesis (leading to disruption of host gene expression and malignant transformation of cells) was observed in a clinical trial.14
Proficient lentiviral transduction of quiescent cells was first shown by transgene expression in growth-arrested fibroblasts and terminally differentiated neurons,11 and relies on the ability of the lentiviral preintegration complex (PIC) to cross the intact nuclear membrane. HIV-1 PIC consists of the linear viral DNA, integrase (IN), reverse transcriptase (RT), matrix protein, the accessory protein vpr, and cellular proteins (see ref. 15 for a review). Several components of the PIC may facilitate nuclear import, including IN, the central DNA flap, vpr, and matrix, but controversy about the validity of various claims, the contributions of individual PIC components, and their mechanisms remains.16,17
Genomic insertion of retroviruses is mediated by the virus-encoded IN. This viral enzyme is encoded in the pol gene and translated as part of the gag–pol polyprotein, from which it is cleaved by the viral protease.18 Provirus insertion is a multistep process consisting of vector 3′-end processing (removal of a dinucleotide from both 3′-ends), strand transfer (staggered cutting of host cell genome and insertion of the viral DNA), gap repair, and ligation (likely to be accomplished by cellular proteins).19 Proviral integration is a relatively inefficient process20 and with both retroviruses and retroviral vectors a significant amount of full-length linear double-stranded vector DNA is converted into circular forms by nuclear proteins.21,22,23,24 The role of these episomal circles in the viral life cycle is not fully understood, and for a long time viral episomes were believed to be dead-end products24 with very low transcriptional activity.25 However, several reports have shown that two long terminal repeat (LTR)–containing circles can be cut by retroviral and HIV-1 integrases.26,27 Recent results have challenged the view that episomal circles are poor substrates for transcription. Using integration-deficient lentiviral vectors (IDLVs) several authors have reported efficient gene expression in vitro and in vivo. IDLVs are also proficient vehicles for homologous recombination, site-specific integration and transposition, and can be converted into stable, replicating episomes. These properties, combined with their highly reduced risk of causing insertional mutagenesis, have led to a renewed interest on IDLVs for genetic analysis and gene therapy, which will be reviewed here. Other recent relevant reviews describe gene expression from unintegrated HIV,28 replication of unintegrated HIV,20 features of retroviral episomes related to pathogenesis and potential for therapy,29 and toxicity of lentiviral integration, IN mutations, and other properties of IDLVs.30,31
Discovery of Retroviral Episomes
During the natural life cycle of retroviruses the target cell is infected and the viral RNA is reverse transcribed into linear double-stranded DNA. A fraction of this viral DNA is then integrated into the host cell genome and represents the major source of viral gene expression (reviewed in refs. 20,32). Circular forms of the retroviral double-stranded DNA, classically known as form X, have also been found during the normal replication cycle and exist intracellularly in parallel to the free linear form and the integrated provirus.23 These circular forms are hypothesized to support viral function for they show some gene expression activity, albeit lower than that of the integrated virus forms.25,33 HIV-1 circles are considerably stable after infection, with progressive vector episome dilution due to cell division. Thus, the apparent decrease in circularized HIV-1 DNA after infection of CD4+ MT-2 or SupT1 T-cells is the result of ongoing cell division causing the dilution of nonreplicating viral episomes in the cell population.34,35 Episomes are stable in macrophages for at least 21 days (ref. 36), while a turnover of episomes has been observed in vivo in human peripheral blood mononuclear cells over the course of several weeks, suggesting that they can be slowly degraded.37
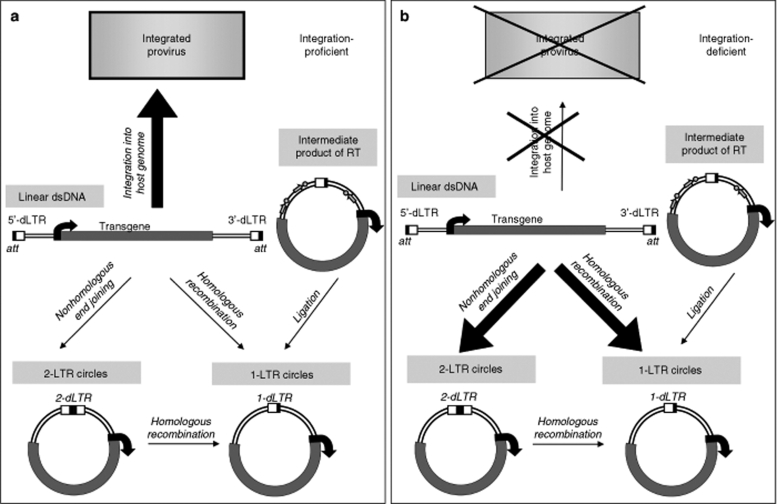
Circularization of linear viral DNA resulting from reverse transcription can occur by at least five ways: nonhomologous end-joining (NHEJ); homologous recombination via strand-invasion or single-strand annealing; closure of intermediate products of reverse transcription (Figure 1); and autointegration.38 The first four mechanisms result in the two major circular variants, the 1-LTR and 2-LTR circles, both containing the whole coding viral genome but bearing either one or two tandem LTR motifs. Autointegration results from a viral genome end inserting within the same linear viral DNA molecule, producing episomes of different length and structure, the so-called “mutant” circles,38 which will not be discussed further. NHEJ and homologous recombination are the main DNA double-strand break (DSB) repair pathways in mammalian cells. NHEJ brings together and ligates the 5′- and 3′-ends of the linear vector DNA, resulting in the formation of 2-LTR circles.24 1-LTR episomes can be produced by homologous recombination between the viral 5′- and 3′-LTRs through either the strand-invasion pathway or single-strand annealing, a minor DSB repair pathway which mediates recombination between repeated sequences (and extrachromosomal homologous recombination of transfected DNAs in mammalian cells).39,40 1-LTR circles could also potentially result from ligation of nicks present in intermediate products formed during reverse transcription.41 The variety of possible pathways rendering 1-LTR circles may explain the puzzling observation of these circles being several fold more abundant than 2-LTR episomes,42 when the latter are produced by the most common DSB repair pathway in mammalian cells. NHEJ and homologous recombination are mainly active in different cell cycle stages, G1 and early S phase for the former, late S and G2 for the latter, suggesting that the proportion of 1-LTR to 2-LTR circles potentially generated in an infected cell may also depend on cell cycle stage. It should be noted that DNA episomes are highly prone to intermolecular homologous recombination in mammalian cells, resulting in episomal and integrated concatemers (for example ref. 43), which may be reminiscent of concatemers observed in retrovirus-infected cells.44
Figure 1.
Generation of episomes from lentiviral vectors. The product of vector reverse transcription is a linear double-stranded DNA (dsDNA) with LTRs at both ends (deleted in the U3 region in self-inactivating vectors, here denoted as dLTR). This DNA is imported into the nucleus as part of the viral preintegration complex. (a) Conventional lentiviral vectors, harboring a functional IN, can be integrated into the host genome as proviruses. However, the linear DNA can also be circularized in several possible ways: nonhomologous end-joining produces 2-LTR circles, while intramolecular homologous recombination between the LTRs in linear DNA or 2-LTR episomes, or ligation of nicks (shown as arrow-bead structures) in intermediate products of reverse transcription, lead to 1-LTR circles. (b) When proviral integration is blocked through class I IN mutations, increased amounts of vector episomes are produced. att, IN attachment sites at the ends of viral DNA. LTR, long terminal repeat.
In summary, with both wild-type retro- and lentiviruses some of the linear double-stranded DNA resulting from reverse transcription is converted into circular episomes by cellular proteins. These episomes contain one or two copies of the viral LTR and are metabolically stable. As the viral episomes lack replication signals, they are stable in quiescent cells but progressively diluted out in proliferating cells.
Making Lentiviral Episomes: the Advent of IDLVS
The formation of retroviral circles negatively correlates with the ability of the virus to integrate into the host genome. Given that viral episomes can be adequate for various applications (e.g., transgene expression, recombination, transposition; see below), preventing integration in order to favor the formation of circles can be a desirable goal. In a way, retroviral episomes are similar to DNA minicircles (plasmids from which sequences required for bacterial amplification are removed prior to mammalian cell transfection to prevent transcriptional silencing45) with the added advantages of the efficient transduction provided by the viral machinery and the natural lack of bacterial sequences.
There are several possible ways to impair the integration ability of retroviruses. Currently, the most efficient way is to create mutated virus strains. This is easily done by replacing the standard gag/pol packaging plasmid with an IN-mutant version. Many studies have been reported examining the effects of mutating specific amino acids of IN to produce mutant strains with impaired integration in retroviruses46,47 and lentiviruses.33,48,49,50 Because IN is pleiotropic, being involved in virion morphogenesis, reverse transcription, PIC nuclear translocation, and integration, many of these mutations have effects on several viral functions not only related to integration and can render reduced amounts of viral DNA (class II mutations). In contrast, class I mutations specifically affect IN function regarding DNA cleavage and integration but result in normal levels of viral DNA.49,50,51
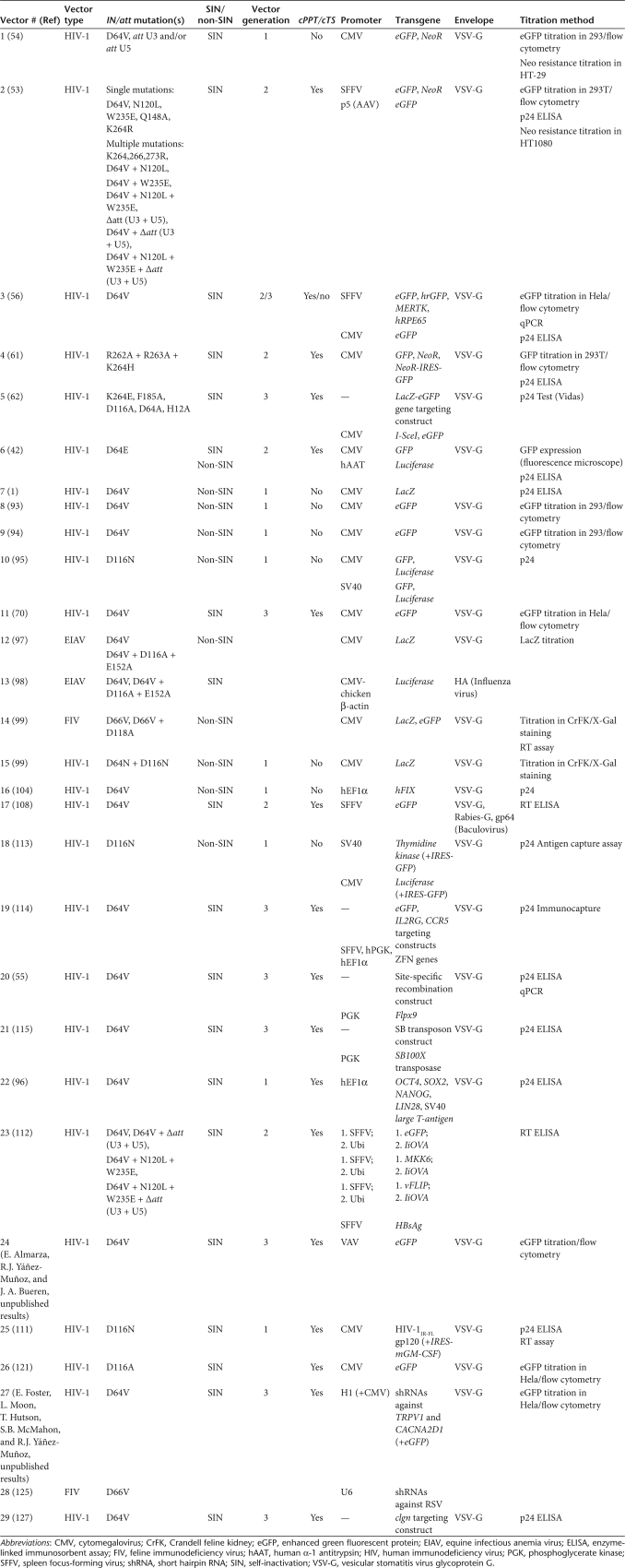
Class I mutants are normally created by substituting any one of the three amino acids of the catalytic triad (for HIV-1 IN these are D64, D116, and E152). Mutating or deleting the attachment (att) sites at the ends of the viral genome (Figure 1), involved in recognition by IN and cut by it, has also been shown to hamper provirus formation if performed on both U3 and U5 ends, but less efficiently than IN mutations.52 Furthermore, att site mutations do not lead to further reduction of integration frequencies when used in conjunction with IN mutations in IDLVs (vectors #1–2 on Table 1).53,54 For a comprehensive review on IN mutations of relevance to lentiviral vectors see Philpott and Thrasher.31 A number of studies have demonstrated increased frequencies of vector episomes relative to total vector DNA when IDLVs were compared with integrating vectors.42,53,55,56
Table 1.
Characteristics of reported integration-deficient lentiviral vectors
It is also possible to impair IN activity with specific inhibitors. Two inhibitors of HIV-1 IN, raltegravir,57 and elvitegravir,58 both strand-transfer inhibitors, have been recently developed, with the former being now clinically available. In addition, there are promising attempts to interfere with the interaction of IN and cellular cofactors, which may be critically involved in viral replication and are becoming putative therapeutic targets.59 Among them, LEDGF/p75, integrase interactor 1, gemin2, emerin, barrier to autointegration formation, and others have been recently identified and are being explored as potential targets for pharmacological intervention (reviewed in ref. 59; see also ref. 60). Pharmacological treatment against IN or its cofactors alone is unlikely to prevent viral integration more efficiently than IN mutations, but it might contribute to reduce the frequency of (rare) integration events observed with IDLVs.53,54,61,62
In summary, IDLVs can be easily produced by using class I IN mutations in the catalytic triad (most authors using mutant D64V in the case of HIV vectors). Increased levels of circular vector episomes are produced upon IDLV transduction. Further mutations in the att sites do not lead to additional reductions in integration frequencies, but may be advantageous to reduce risks in a clinical setting (see below). The effect on residual integration frequencies of combining IN mutations and specific inhibitors remains to be tested.
Safety Issues for In Vivo Applications
General safety issues (replication-competent recombinants, mobilization, RT fidelity)
Integration of the retroviral provirus in the host genome can lead to stable transgene expression, one of the outstanding features of retroviral vectors. Unfortunately, it is also their Achilles' heel, because integration can cause insertional mutagenesis, whose undesired effects may lead to cancer in a worst-case scenario. This theoretical risk has unfortunately materialized during the clinical application of γ-retroviral vectors, resulting in uncontrolled T-cell proliferation in an X-linked severe combined immunodeficiency clinical trial.14 Such risk could be greatly increased by the potential generation of replication-competent recombinants (RCRs) or vector mobilization upon wild-type virus superinfection. Either of these possibilities would cause uncontrolled vector spread and an increase in the number of insertional events and hence magnify the risk of cancer. Therefore, lentivector production methods aim to minimize the chance of RCR generation and vector mobilization, and the vectors themselves can be modified to reduce the effects of the integrated provirus on adjacent genomic sequences. As a further development toward increased biosafety, the use of IDLVs can greatly reduce (albeit not completely eliminate, see below) the risk of genomic provirus insertion and thus insertional mutagenesis, a most desirable goal provided that the biological results expected from the vector can be obtained from an episomal configuration.
Lentiviral vectors are most commonly produced through transient cotransfection of three (packaging, envelope, and transfer) or more plasmids into producer cells. Increased biosafety has been achieved by modifications involving all plasmid components (reviewed in refs. 63,64). We will discuss some of these modifications because of their relevance to this review. Vector generations refer to modifications of the packaging plasmid, including a progressive reduction in the number of encoded lentiviral genes and the split of the viral genome into more than one plasmid. Thus, a first-generation packaging plasmid encodes all lentiviral genes except for env, which is interrupted by mutations. A second-generation plasmid additionally lacks the accessory genes vif, vpr, vpu, and nef (in the case of HIV-1, relevant genes in other lentiviruses), which are not essential for vector production and dispensable for viral gene expression in vivo, at least in some cell types.65,66 A third-generation packaging plasmid system is made of two or more plasmids (one of them normally encoding gag/pol and the other one rev) and lacks tat, which is required for high-level, lentiviral LTR–mediated transcription but dispensable for chimeric LTRs with a heterologous promoter.67,68,69 Splitting the viral genome adds further protection against generation of RCRs in producer cells. As we will describe in more detail below, IDLVs have been efficiently produced with first-, second-, and third-generation systems, simply replacing the relevant gag/pol packaging plasmid with an IN-mutated version.53,54,56,61,62,70
Modifications in the transfer plasmid include self-inactivation (SIN). SIN vectors are produced through the targeted elimination of the promoter sequences from the 3′-LTR U3 region, making transgene expression necessarily reliant on an internal promoter. SIN vectors have reduced risks of host gene transactivation, RCR generation, and mobilization upon superinfection with wild-type virus.3,68,71 However, it should be noted that the 3′-polyadenylation signal of the HIV-1 SIN LTR is leaky, potentially affecting downstream genes.72 For added safety SIN vectors can be endowed with polyadenylation enhancers,73 insulators,74 internal promoters without enhancer capacity,75 or regulatable transgene expression.76,77 Standard and SIN IDLVs have been produced, but the SIN deletion in the vector genome has been shown to lead to higher expression levels (see below).42
Integrating lentivectors and IDLVs are not equally susceptible to RCR generation and vector mobilization. With rare exceptions, active IN is a requirement for HIV replication,78 so IDLV episomes should not support HIV-1-based replication if an unlikely recombination event generated an RCR molecule. As a consequence, IDLVs are significantly safer than integrated proviruses in this respect. Concerning the risk of vector mobilization, integrated SIN vectors have been shown to produce full-length transcripts from promoters either upstream or internal to the vector genome, competent for encapsidation and integration.79 Unfortunately, as discussed by Cockrell and Kafri, IDLVs are not exempt from the risk of mobilization, because they can generate full-length, encapsidation-competent transcripts.63 Transcription has been shown to occur across the 2-LTR junction in episomal HIV-1 circles,80 and 1-LTR SIN HIV-1 episomes are competent for vector production in transiently transfected human embryonic kidney 293T (HEK293T) cells due to an alternative transcription start site within the deleted U3 region81 coupled with the leakiness of the 3′-polyadenylation signal in the SIN LTR.72 Although the combination of att and IN mutations does not reduce integration frequencies below the levels achieved by IN mutation alone, att mutations would reduce integration frequencies of vector mobilized by wild-type virus; therefore, it would be advisable to include both IN and att mutations in IDLVs for clinical application.
Regardless of integration proficiency, another concern with retroviral—and particularly with lentiviral—vectors is the low fidelity of RT, estimated in vitro for HIV-1 at 5.9 × 10−4 (ref. 82), i.e., five point mutations per reverse transcription cycle for an average 8-kb vector genome. Viral genes are only expressed from plasmids in producer cells, so they will be unaffected by the low fidelity of reverse transcription; this implies that reversion of IN mutations in producer cells resulting in the generation of integration-competent vectors is very unlikely. However, because of reverse transcription in the target cells, lentiviral transduction will result in a quasispecies of viral DNA molecules in the transduced cells rather than identical products in all cells, with possible mutations in the transgene as well as in the vector backbone, as has been shown for native HIV-1 (reviewed in ref. 83). The biological implications of this fact from the point of view of lentiviral vector–mediated therapies remain to be explored.
In conclusion, IDLVs are much less likely to generate RCRs than integrating vectors, but both vector types are potentially liable to mobilization by superinfection with replicating HIV-1. Of note, this has not been demonstrated in T-cells.63 IDLVs for clinical application should carry both IN and att mutations to minimize integration in the event of vector mobilization. The low fidelity of reverse transcription in target cells should affect integrating vectors and IDLVs in a similar manner, but its relevance for clinical applications remains to be determined.
Residual integration of IDLVs
IDLVs are by definition deficient in genomic integration, but this is a relative rather than absolute property. Few studies on integration of IDLVs have been published. Available data include analyses of residual proviruses and host DNA–vector junctions in a very limited data set of integration events, and quantitation of integration frequencies using selectable transgenes or enhanced green fluorescent protein (eGFP) expression.54,56,61,62,84
The product of HIV reverse transcription is a linear double-stranded DNA molecule (with a discontinuous plus strand41), which could undergo genomic integration by IN-dependent and IN-independent mechanisms. In either case it would be reasonable to find host DNA–vector junctions involving the ends of the vector LTRs, and assays for the analysis of integration junctions are normally biased to detect such LTR-mediated events. However, circular IDLV episomes may be subjected to DNA damage and metabolism like any other episomal DNA molecule existing in the nucleus of a cell, which could lead to breakage anywhere on the episome and hence non-LTR-mediated genomic integration. Very few attempts have been made to detect such host DNA–vector junctions not involving the LTR ends, but the stable detection of 2-LTR HIV-1 junctions in dividing cells is supporting evidence for non-LTR-mediated integration of viral circles.85
The viral locus responsible for retroviral IN function was originally described in 1984 landmark papers.86,87 One of these papers already showed that residually integrated IN mutant MoMLV proviruses had abnormal structures.86 Only in very rare cases had IN mutant proviruses the structures expected from wild-type virus, i.e., two nucleotides lost from each end of viral DNA and identical 4-bp (or 5-bp) flanking sequences of host origin.88 In the case of HIV-1, integrated proviruses from replication-defective, catalytic triad IN mutant variants encoding a hygromycin resistance gene appeared intact by Southern blot. However, sequencing of integration junctions isolated by inverse PCR showed that the integrated proviruses from IN mutants did not conform with the canonical characteristics of integrated HIV-1: (i) the 5-bp direct repeat of host DNA was often not maintained; (ii) integration often occurred with truncations of vector DNA ends, perhaps due to aberrant 3′ processing; (iii) in some cases integration caused a deletion of host DNA at the insertion site; and (iv) target site selection of mutant IN viruses showed sequence preferences not observed with wild-type proviruses.89
Regarding the structure of the integrated proviruses from IDLVs, a study in human colon adenocarcinoma HT-29 cells transduced with HIV-1 vector (vector #1 on Table 1) showed gross reorganizations by Southern blot in about 50% of G418-resistant colonies. Inverse PCR-based strategies on genomic DNA of nongrossly reorganized proviruses from IN mutant vectors, followed by DNA sequencing, demonstrated noncanonical patterns including deletions, duplications or reorganizations of vector sequence and lack of the characteristic 5-bp duplication of flanking host DNA.54 Our ongoing large-scale linear amplification-mediated (LAM)-PCR analysis of integration events from HIV-1 IDLVs (vectors #3 on Table 1) in cervical cancer Hela and feral mouse embryo SC-1 cells has revealed both canonical and noncanonical LTR-end sequences (C. Bartholomae, R.J. Yáñez-Muñoz, S.J. Howe, M. Schmidt, C. von Kalle, and A.J. Thrasher, unpublished results). However, this LAM-PCR approach renders DNA sequences from single ends of the integration event and no conclusions can be made regarding the structure of the other proviral end.
The peculiarities showed by proviruses from IN mutant HIV-1 and IDLVs, together with the lack of effect on residual integration frequencies from adding att mutations to IN mutants, have led to the conclusion that integration events from IN mutants are unlikely to be IN-mediated.54,89 In support of this, integration of an IDLV has been observed in a pre-existing DSB in human osteosarcoma U-2 OS cells,62 consistent with the known frequent capture of extrachromosomal DNA fragments and vector genomes by pre-existing genomic DSBs.90 However, the lack of IN involvement in some of the residual integration events has not been formally demonstrated, and it could be argued that residual IN activity could have aberrant characteristics and hence the products would not necessarily display all the features of wild-type proviruses.
An analysis of relative integration frequencies of replication-defective, integrating and IN mutant HIV-1 variants encoding a hygromycin resistance gene showed that mutations in the catalytic triad led to reductions of 3–4 logs in the number of drug-resistant colonies with the latter.50 Similar results have been obtained in the context of SIN IDLV HIV-1 vectors. Relative integration frequencies in HT-29 cells have been estimated as about 4-log lower than wild-type for the D64V mutant, regardless of the presence of additional att site mutations (vectors #1 on Table 1).54 Studies in human fibrosarcoma HT1080 cells and HeLa cells with various IN mutant vectors including D64V (Table 1, vectors #2 and 4) showed at most 3-log reductions in relative integration frequency compared to wild-type, even with vectors carrying multiple IN mutations and lacking att sites.53,61
Absolute integration frequencies in human Jurkat T-cells and CD34+ cells were determined with vectors carrying IN D64V (and/or att site mutations) and expressing eGFP (Table 1, vectors #1). Using a low vector dose (multiplicity of infection 3, based on titering by flow cytometry for GFP expression) on Jurkat cells and a high dose (multiplicity of infection 40) on CD34+ cells, frequencies of stable eGFP expression ranged between 0.1 and 0.5% for the most disabled vectors.54 The absolute integration frequency of similar eGFP vectors (Table 1, vectors #4–6) carrying a variety of IN mutations in HEK293T cells ranged from <0.1 to 2.3%, depending on mutation and dose.42,61,62 Our own analysis of integration events from HIV-1 D64V IDLVs (vectors #3 on Table 1) in Hela cells using a high vector dose (multiplicity of infection 25, based on eGFP titer) detected absolute integration frequencies of 1.1–1.3%, judged by stable eGFP expression (C. Bartholomae, R.J. Yáñez-Muñoz, S.J. Howe, M. Schmidt, C. von Kalle, and A.J. Thrasher, unpublished results).
The only published analysis of genomic integration of IDLVs in vivo was performed on genomic DNA from mouse and rat eyecups subretinally injected with D64V vectors (#3 on Table 1). We used LAM-PCR strategies that could detect LTR- and non-LTR-mediated integration, which we estimated allowed scanning of 30–50% of the vector genome for host DNA–vector junctions. LAM-PCR on genomic DNA of these nonclonal samples only analyses one end of the integration event, so no conclusions are possible regarding the presence or absence of the flanking 5-bp host DNA duplication. An extensive analysis showed a single integration event in a rat eyecup 2.5 months postinjection, LTR-mediated and with canonical vector sequence. We also used a quantitative PCR method for detection of 2-LTR junction-containing molecules and observed that eyecups injected with IDLVs had levels of 2-LTR junctions 8 times higher than those observed in samples injected with integrating lentivectors. As our LAM-PCR search for integration events in these samples only revealed a single case, we concluded that the high levels of IDLV 2-LTRs corresponded to episomal molecules, present at much higher levels than with integrating vectors.56
Summarizing, optimum IDLVs have residual integration frequencies 3–4 log below those of their wild-type counterparts, at least in cell culture. Initial data sets from in vivo experiments are equally encouraging. Estimates of stable integration frequency of IDLVs in cultured cells (0.1–2.3%) are within the range described for plasmid transfection.91,92 Integrated proviruses from IDLVs are frequently noncanonical regarding gross or minor vector rearrangements, terminal nucleotides, deletions in host cell DNA, and abnormal or missing DNA flanking repeats. As discussed by Nightingale et al. even this minor level of residual integration may be unacceptable in some clinical applications, like those involving mitosis-promoting genes, which may require non-DNA-based methods to prevent any genomic modification.54 However, the risk of insertional mutagenesis from IDLVs is highly reduced compared to that from integrating lentivectors, and from a biosafety standpoint IDLVs should be considered highly suitable for applications involving quiescent tissues.
In Vitro Gene Expression from IDLVS
Studies on gene expression from IDLVs have been carried out mostly using vesicular stomatitis virus glycoprotein G pseudotyped vectors derived from human as well as nonhuman lentiviruses. Similar to studies with HIV-1 episomes,25,33 early reports described relatively low gene expression from non-SIN IDLVs in cell culture. The first report was from Naldini et al. in 1996, describing that the transduction efficiency of IDLVs (vector #7 on Table 1) was markedly reduced in dividing (17-fold) and nondividing (sevenfold) HeLa cells compared to integrating vectors.11 Similar non-SIN IDLVs (vectors #8–10 on Table 1) were tested in cultured human CD34+ cells and the HEK293 and HEK293T cell lines. Transient gene expression from IDLVs was confirmed (for 10 days in CD34+ cells, around 14 days in HEK293 and HEK293T cells), albeit at lower levels than from integrating vectors.93,94,95 First-, second-, and third-generation SIN HIV-1 vectors (#1–6 and 11 on Table 1) with various internal expression cassettes and class I IN mutations (alone or in combination with att site mutations) have more recently shown efficient but transient transduction of mouse myoblast C2C12 cells, HeLa and HEK293T cells, Jurkat T-cells, and human cord blood CD34+ cells.42,53,54,56,61,62,70 As described below, IDLVs have also been used on human fetal IMR90 and adult skin 1087sk fibroblasts, with the aim of producing induced pluripotent stem (iPS) cells.96
The effects of IN mutations on a nonhuman lentivirus vector were first investigated in equine infectious anemia virus (vectors #12 on Table 1). A class I IN mutation (D64V) was reported to reduce 450-fold the transduction titer on the D17 canine sarcoma cell line. A triple mutation of the entire DDE motif (D64V, D116A, and E152A) reportedly led to a 3,000-fold reduction.97 However, recently produced SIN equine infectious anemia virus IDLVs (vectors #13 on Table 1) with the single or triple mutations express eGFP transiently in HEK293T cells, and their titers are only reduced about sixfold compared with integrating vectors.98
Transient gene expression has similarly been observed from non-SIN feline immunodeficiency virus (#14 on Table 1) IDLVs in dividing cells in culture, with 3–5 log lower transduction efficiencies in several cell lines being observed with D66V or double D66V/D118A mutants compared with their integrating counterpart (these residues being analogous to HIV-1 D64 and D116). However, these authors made an important observation: very similar 48-hour transduction frequencies were obtained when nondividing rat retinal ganglion cells, growth-arrested Crandell feline kidney or HT1080 cells were treated with wild-type and IN mutant vectors. Release from growth arrest in Crandell feline kidney or HT1080 cells led to steady decline of gene expression from IN mutant vectors. A similar cell cycle dependence was observed with non-SIN D64N HIV-1 vectors (vector #15 on Table 1).99 Confirming these initial observations, efficient, stable transduction of growth-arrested (differentiating) C2C12 cells, postmitotic primary rat dorsal root ganglia neurons, cortical neurons, and astrocytes with second- and third-generation SIN HIV-1 IDLVs (vectors #2, 4, and 11 on Table 1) has recently been described.53,61,70
In summary, early work reported poor transduction with non-SIN IDLVs in cultured cells. More recently, efficient transient transduction has been described with SIN IDLVs in proliferating cells, albeit at lower expression levels than with integrating vectors. A direct comparison of SIN and non-SIN vectors (vectors #6 on Table 1) has recently shown that lentiviral episome gene expression is significantly improved by the extended SIN deletion in the 3′-LTR,42 consistent with a negative regulatory effect arising from the U3 region of the full-length LTR.100 Importantly, transduction of quiescent cells in culture is efficient and stable with IDLVs. An additional advantage of IDLVs may be a reduced risk of transgene silencing in episomes101 compared to integrated proviruses, which are the subject of epigenetic regulation and silencing.102,103
In Vivo Gene Expression From IDLVS
Early in vivo studies with IDLVs described virtually no gene expression from non-SIN vectors. Stereotactic delivery into rat striatum or hippocampus of HIV-1 IDLV (vector #7 on Table 1) showed transduction efficiencies <2% compared to the integration-proficient virus, measured 2 or 6 weeks after injection.1,4 Subsequent work appeared to confirm this initial failure. Portal vein injection of a human coagulation factor IX-encoding HIV-1 IDLV (vector #16 on Table 1) after partial hepatectomy did not lead to detectable levels of human factor IX in mouse serum over a 16-week period.104 Equine infectious anemia virus injected into rat brain gave no significant levels of transduction.105 Lack of transgene expression was also reported with feline immunodeficiency virus IDLV (vector #14 on Table 1). This vector was injected subretinally in 7-day-old rats, resulting in very poor gene expression in the retinal pigment epithelium (RPE) 2–7 months after administration.99,106
A number of factors may have contributed to explaining why recent in vivo studies with IDLVs have given completely different results (see below), including vector optimization, experimental factors and target tissue. First, it should be noted that early lentiviral vectors like the ones used in some of the experiments described above had infectivities (measured as transducing units/ng p24) at least tenfold lower than those currently achieved. Thus, early vectors may have reached infectivities of 103 for integrating configurations and lower for IDLVs,11,97 compared with 104 or higher for current vectors regardless of integration proficiency.53,56 Second, lentiviral episome gene expression is significantly improved both in vitro and in vivo by an extended SIN deletion in the 3′-LTR.42 Early IDLVs had full-length 3′-LTRs and hence should have led to lower expression levels. The compounded effect of lower infectivities, lower expression levels from full-length 3′-LTR vectors, and in some cases experiment or tissue-specific factors (hepatectomy-induced hepatocyte proliferation or RPE cell proliferation in immature rat eyes, both possibly causing IDLV episome dilution) may all have contributed to the very poor expression levels initially reported in vivo from IDLVs. The presence of central polypurine tract/central termination sequence (cPPT/cTS) and/or Woodchuck hepatitis virus post-transcriptional regulatory element motifs in modern vectors may have favored gene expression, but is certainly not strictly required for efficient in vivo gene expression from IDLVs.56,107
In 2006, we published the first study showing efficient transgene expression in vivo with IDLVs. Using second-generation SIN HIV-1 vectors (vectors #3 on Table 1) we demonstrated long-term eGFP expression in the adult RPE after subretinal injection (followed for 9 months in mice, 3 months in rats). In the mouse experiments, we also tested vectors lacking cPPT/cTS or Woodchuck hepatitis virus post-transcriptional regulatory element, which showed efficient RPE eGFP expression. Therapeutic efficiency of the IDLVs was shown in two rodent models of retinal degeneration, the Rpe65rd12/rd12 mouse and the Royal College of Surgeons (RCS) Mertk-deficient rat. Stereotactic injections of vector in mouse (striatum or hippocampus) or rat brain (red nucleus) demonstrated efficient eGFP expression in adult neurons and their axon tracts, which was followed for up to 1 month.56,107 In all cases the proficiency of titer-matched integrating and nonintegrating vectors was indistinguishable in in vivo experiments in these quiescent tissues.56 Further work with these vectors showed efficient eGFP expression for at least 1 month in mouse skeletal muscle after direct intramuscular injection in neonates.70 Additionally, with third-generation SIN HIV-1 IDLVs (vector #11 on Table 1) we have further demonstrated highly efficient gene expression in: (i) rat spinal cord after direct intraparenchymal injection; and (ii) rat brain subventricular zone and olfactory bulb after stereotactic injection at the beginning of the rostral migratory stream.70
Shortly after our initial report, brain eGFP expression from SIN HIV-1 IDLVs (vector #4 on Table 1) was independently confirmed. These authors demonstrated efficient expression for at least 4 weeks poststereotactic injection in adult mice, reporting similar efficiencies for integrating vector and IDLV. They also referred to their unpublished results of stable eGFP expression for 6 months after subretinal injection of IDLV in the dog eye.61 Further confirmation of IDLV proficiency in the central nervous system has been obtained by intracraneal injection in adult rats or mice in utero, using SIN HIV-1 vectors pseudotyped with various envelope proteins (vectors #17 on Table 1).108
Muscle gene expression from IDLVs has also been independently demonstrated. Apolonia et al. used HIV-1 SIN IDLVs (vectors #2 on Table 1) with IN mutations alone or in combination with att site mutations, and showed efficient in vivo eGFP expression. This was followed for up to 3 months after intramuscular injection of mouse neonates. eGFP levels at 3 months were similar to those obtained with wild-type vector, and levels of 2-LTR circles were elevated with mutant vectors.53 Later work by these authors has shown eGFP expression for at least 8 months in injected muscle.84 Efficient, long-term gene expression from IDLVs in muscle after neonatal injection is remarkable because of the considerable increase in muscle size between injection and sample analysis. This size increase is brought about by muscle fiber growth, involving the fusion of satellite cells to pre-existing fibers. Given the relatively inefficient transduction of adult muscle by lentiviral vectors,109 it is possible that neonatal muscle transduction may involve satellite cells, which subsequently fuse with the muscle fiber.
Until recently, efficient liver transduction from IDLVs seemed an elusive goal. We have already mentioned that Park et al. failed to detect factor IX expression in mouse experiments involving a hepatectomy (vector #16 on Table 1).104 In contrast, it has been very recently shown that stable mouse liver expression can be obtained from IDLVs. Using SIN HIV-1 IDLV (vector #6 on Table 1) and intraperitoneal injection, expression of luciferase was observed for at least 6 months.42 Additionally, systemic administration of IDLV expressing canine coagulation factor IX from a potent hepatocyte-specific promoter in hemophilia B mice resulted in sustained therapeutic factor IX levels for at least 3 months (T. VandenDriessche, personal communication). However, unlike in the previous reports of efficient central nervous system gene expression from IDLVs,56,61 liver luciferase and blood factor IX levels were considerably lower with IDLVs than with their integrating counterparts (ref. 42 and T. VandenDriessche, personal communication). Importantly, the relatively low transgene expression level from IDLVs in hepatocytes can be used to induce antigen-specific regulatory T-cells and immune tolerance if the vector also encodes a micro-RNA target to mediate hematopoietic cell de-targeting.110
Specific immune responses can also be obtained with IDLVs (see Vaccination section below), as shown in mice with vectors encoding an HIV-1 gp120 variant,111 ovalbumin, or hepatitis B virus surface antigen.112 Finally, it has also been very recently demonstrated that non-HIV-1 IDLVs are capable of efficient in vivo expression. For this, SIN equine infectious anemia virus–based IDLVs (vector #13 on Table 1) were inoculated intranasally into 6-week-old mice. Whole-body imaging demonstrated strong luciferase expression in the nasal epithelium for 120 days, and much lower expression levels in the lung (ref. 98 and J. Olsen, personal communication).
In summary, early in vivo uses of non-SIN lentiviral vectors led to negligible levels of transduction. More recent studies have demonstrated effective transduction with SIN IDLVs in the eye, brain, spinal cord, muscle, and to a lesser extent in the liver. The improvement in efficiency is probably due to several reasons, including better vector preparations, higher expression levels from SIN backbones and, even though not strictly required, from the inclusion of cPPT/cTS and Woodchuck hepatitis virus post-transcriptional regulatory element in many current vectors.
Practical Considerations for the Use of IDLVS
Published examples of efficient production of IDLVs and effective gene expression from them are accumulating steadily,53,54,56,61,99,107,108,111,112 but anecdotal unpublished examples exist of problems with IDLV production or poor transgene expression levels. This section summarizes information gained from successful studies with the aim of helping future attempts to develop IDLVs.
Vector choice
First-, second-, and third-generation IN-deficient packaging plasmids have been successfully used to produce IDLVs. Biosafety reasons should bias the choice toward later generations, at a cost of slight losses in vector yield resulting from the need of more plasmids for cotransfection into producer cells. The choice of IN mutation is a crucial issue because affecting IN functions other than integration could have an effect on vector yield and infectivity. Catalytic triad mutations do not affect vector titers or infectivity more than a few fold (vectors #1–3 on Table 1),53,54,56 demonstrate minimal integration frequencies (vectors #2 and 5),53,62 and may perhaps offer better transgene expression levels (vectors #5).62
As discussed in the previous sections, one of the likely main reasons for improved expression levels from IDLVs in recent studies is the use of SIN transfer plasmid backbones. These also offer reduced risks of RCR generation, vector mobilization and, if integrated, host gene transactivation. att site mutations in the transfer plasmid are less efficient at preventing integration than IN mutations, and do not provide further reductions in residual integration frequencies when combined with IN mutations. However, att site mutations would minimize the risk of RCR generation and vector mobilization, so for future clinical vectors both IN and att mutations should be combined.
Gene expression levels
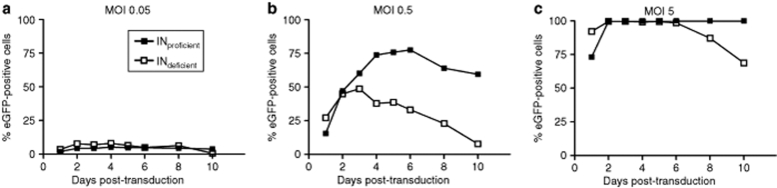
At least two parameters should be considered when analyzing the efficiency of transduction experiments: percentage of transduced cells and transgene expression level per cell. A typical example of transduction with integrating vectors and IDLVs is shown in Figure 2, in which proliferating HeLa cells were treated with vector at multiplicity of infection 0.05, 0.5, or 5 (based on titering by flow cytometry for eGFP expression) and percentages of eGFP-positive cells recorded over time. Regardless of integration proficiency, the percentages of transduced cells were very similar at early time points, but this was followed by loss of expression from IDLVs at late time points due to episome dilution as the cells proliferated (Figure 2b). The reduction in the percentage of eGFP-expressing cells with IDLVs was delayed at high vector dose (Figure 2c) because cells had high levels of episomes after transduction. Conversely, at very low dose (Figure 2a) eGFP expressors disappeared faster.
Figure 2.
Stability of eGFP expression in cells transduced with integrating vectors and IDLVs. Proliferating HeLa cells were transduced at MOI (a) 0.05, (b) 0.5, or (c) 5 (based on titering by flow cytometry for GFP expression) with integrating vector or IDLV (vector #11 on Table 1). The percentage of eGFP-expressing cells was estimated at the indicated times by flow cytometry. eGFP, enhanced green fluorescent protein; IDLV, integration-deficient lentiviral vector; MOI, multiplicity of infection.
The expression levels per cell and the effect of promoter choice are exemplified in Figure 3, where eGFP vectors carrying either immediate early cytomegalovirus (CMV) or spleen focus-forming virus internal promoters are compared in integrating or IDLV configurations at low vector doses to avoid saturation in proliferating HeLa cells. IDLV and integrating configurations clearly differed in their pattern of expression. IDLVs with the CMV promoter displayed less variation in expression levels than integrating counterparts, specifically lacking high-level expressors (Figure 3a). Judging by mean fluorescence intensity of eGFP-expressing cells, Cornu and Cathomen have estimated that a tenfold higher dose of IDLV (vector #5 on Table 1) is necessary to reach the CMV-mediated expression level of an equivalent integrating vector.62 The spleen focus-forming virus promoter clearly rendered lower expression levels in IDLVs than in integrating vectors in these cells (Figure 3b). However, both CMV and spleen focus-forming virus IDLVs led to efficient gene expression in quiescent cells in vitro and in vivo (vectors #1–4, 15, and others on Table 1).53,54,56,61,99 Other strong viral promoters successfully used in IDLVs are those from simian virus 40 (SV40; vectors #10 and 18),95,113 adeno-associated virus p5 (vector #2)53 and a chimeric CMV-chicken β-actin promoter (vector #13).98 Regarding mammalian promoters, human α-1 antitrypsin (vector #6),42 phosphoglycerate kinase (vectors #19, 20, and 21),55,114,115 elongation factor-1α (vectors #19 and 22),96,114 ubiquitin (vectors #23),112 and VAV (vector #24) (E. Almarza, R.J. Yáñez-Muñoz, and J.A. Bueren, unpublished results) have been used successfully.
Figure 3.
Transgene expression levels and effect of internal promoter with IDLVs. Proliferating HeLa cells were transduced at low vector doses (<0.1 eGFP vector unit/cell) with integration-proficient vectors or IDLVs encoding eGFP, and eGFP fluorescence analyzed by flow cytometry 3 days post-transduction. (a) eGFP expression levels in mock-transduced cells or cells transduced with integrating or integration-deficient lentivector carrying a CMV-eGFP-WPRE expression cassette, shown schematically above the dot-plots (vector #11 on Table 1). (b) eGFP expression levels in mock-transduced cells or cells transduced with integrating or integration-deficient lentivector carrying an SFFV-eGFP-WPRE expression cassette, shown schematically above the dot-plots (vector #3 on Table 1). CMV, cytomegalovirus; eGFP, enhanced green fluorescent protein; IDLV, integration-deficient lentiviral vector; SFFV, spleen focus-forming virus; WPRE, Woodchuck hepatitis virus post-transcriptional regulatory element.
Titration method
The lower expression levels from IDLVs and their progressive dilution in dividing cells highlight the need for a careful selection of titration method for objective comparisons of IDLVs and integrating vectors. Suitable methods (see Table 1) include detection of components of the vector particle (i.e., p24 ELISA or RT amount/activity, with the caveat that the particles detected are not necessarily complete and functional) and quantitative PCR measurement of vector reverse transcribed genomes (i.e., late reverse transcript assay,116 provided that it is performed on samples harvested shortly after transduction; we use a 24-hour time point). Titration by quantification of protein products like eGFP in transduced cells can be more problematic, given the relatively slow expression kinetics from lentiviral vectors (see Figure 2b) and expected lower expression levels from IDLVs (Figure 3). Nonetheless, provided that the cells used for titration grow slowly (i.e., HeLa cells, further slowed by the use of polybrene used to enhance transduction efficiency), eGFP titration at 2 or 3 days post-transduction can also be a suitable method. For example, when we transduced cells with vectors normalized by eGFP expression and analyzed total vector DNA at an early time point by Southern blot, the results were consistent regardless of integration proficiency (Supplementary Table S1 and Supplementary Figure S1c in ref. 56).
In summary, catalytic active site IN mutations are the most efficient for generation of IDLVs. Vectors of any generation can be used but later generations should be preferred on safety grounds. SIN configurations provide increased expression and biosafety levels and can be designed with a variety of internal promotors of viral and mammalian origin. The titration method of IDLVs should be considered with care; we favor detection of vector particle components or quantitative PCR titration of vector DNA in samples harvested 24 hours post-transduction.
Applications and Ongoing Development of IDLVS
Transient gene expression
As discussed in the previous sections, IDLVs can provide transient gene expression in proliferating cells. A major application would be the production of human iPS cells and their differentiation into lineages of interest. iPS cell production requires overexpression of 2–4 transgenes and so far has been successful with a number of delivery systems including integrating retroviral and lentiviral vectors.117 A study has reported that a cocktail of four IDLVs with elongation factor-1α–driven transgenes (vectors #22 on Table 1) can lead to the generation of iPS cell colonies from human fetal IMR90 fibroblasts, albeit with a 1-week delay and 50-fold less efficiently than with equivalent integrating lentivectors. These authors included the SV40 T-antigen as one of the four transgenes and observed that regardless of the integration proficiency of the vector, T-antigen was always integrated in the resulting iPS cell colonies. It was not reported whether the other three transgenes were also integrated or what expression levels were achieved. When they used a single IDLV (encoding T-antigen) on adult skin 1087sk fibroblasts the efficiency of iPS colony generation was fivefold lower than with an all-integrating cocktail, and they again observed genomic integration of T-antigen in the resulting iPS cell colonies. The authors interpreted that the selective pressure under these conditions is such that sustained presence of T-antigen is required for iPS colony generation.96
Another important application of IDLV-mediated transient expression would be to increase the efficiency of engraftment of hematopoietic stem cells (HSCs), particularly in clinical settings where their number is limited (ex vivo gene therapy of diseases like Fanconi anemia where HSC numbers are much reduced; or in cord blood transplants). The transcription factor HoxB4 and the chemokine receptor CXCR4 are prime candidates for this approach, but their stable overexpression is not a clinical option.118,119 Retroviral vector–mediated overexpression of the chemokine receptor CXCR4 by HSCs enhances their bone marrow homing in NOD/SCID mice. The effect is maximal with a truncated form of CXCR4 that leads to persistent signaling, but stable expression of this variant is detrimental to marrow function.119 These authors have now overexpressed wild-type and truncated CXCR4 by IDLV transduction in mobilized, healthy human peripheral blood stem cells. Their results showed high expression at day 4, which persisted for several days. Transgene expression was undetectable by culture day 14, and vector copy by 22. Maximal effects were observed with truncated CXCR4, which led to enhanced in vitro peripheral blood stem cell migration in response to its ligand SDF1, and three- to sixfold improvement in engraftment in the NOD/SCID mouse model.120
Vaccination
IDLVs are also able to induce specific immune responses. A HIV-1 IDLV encoding a codon-optimized HIV-1JR-FL gp120 and mGM-CSF (murine granulocyte monocyte–colony stimulatory factor, to improve antigen-specific responses; vector #25 on Table 1) was injected once intramuscularly into 6- to 8-week-old mice. Thirty days after vector administration, cellular immune responses (frequency of IFN-γ-expressing splenocytes) were similar regardless of integration proficiency. After 90 days the frequency of effector memory and cytotoxic T lymphocytes was lower with IDLV but still significant. Frequencies of antigen-specific memory T-cells in bone marrow were similar for integrating vector and IDLV 90 days postimmunization. Humoral responses (anti-gp120 immunoglobulin G titer) were higher with integrating vector at both 30- and 90-day time-points. No attempt was made in this study to determine the nature of the cells targeted by the immunizing lentivectors.111 Further confirmation of efficient immunization has recently been provided using ovalbumin and hepatitis B virus surface antigen–expressing IDLVs (vectors #23 on Table 1).112 It may be possible to boost these immune responses by co-injecting the immunizing IDLV with simian immunodeficiency virus vpx virus-like particles, an approach that improves the transduction of dendritic cells in culture with both integrating and nonintegrating HIV-1 vectors (vector #26 on Table 1).121 Simian immunodeficiency virus vectors may be particularly suitable for transduction of monocytes,122 and simian immunodeficiency virus IDLVs have been developed and are currently undergoing in vivo testing (J.-L. Darlix, personal communication).
RNA interference
Sequence-dependent gene silencing by small interfering RNAs has a myriad of applications in research and potentially the clinic.123 Lentiviral vectors are effective tools to deliver expression cassettes for short hairpin RNAs,124 but stable expression may not always be an adequate option. We have used HIV-1 IDLVs to express H1 promoter-driven short hairpin RNAs (vectors #27 on Table 1) against target genes expressed in cultured rat neurons (the ion channel-encoding TRPV1, which has a well-established role in pain, and the voltage gated N-type calcium channel subunit α2δ1-encoding CACNA2D1, which is thought to be the target for gabapentin-mediated analgesia in vivo). The efficiency of target gene down-regulation (three- to fourfold) was identical to that of equivalent integrating vectors (E. Foster, L. Moon, T. Hutson, S.B. McMahon, and R.J. Yáñez-Muñoz, unpublished results). Another ongoing study is making use of feline immunodeficiency virus IDLVs encoding short hairpin RNAs driven by the U6 promoter (vector #28), to inhibit respiratory syncytial virus infection in respiratory epithelial cell models.125
Homologous recombination (gene repair, knock-in, knock-out)
Gene repair by homologous recombination is the ultimate goal of gene therapy for monogenic diseases. Although it is not particularly difficult to produce isolated clones of cells modified by such gene targeting, clinically relevant frequencies of corrected cells have not been achieved with traditional gene targeting methods.91,126 IDLVs (vector #29 on Table 1) have been used to produce similarly low frequencies of gene targeting in mouse embryonic stem cells,127 but there is also unpublished anecdotal evidence of failures to achieve gene targeting with IDLVs. However, the frequency of gene targeting can be increased up to 10,000-fold if DNA DSBs are introduced near the desired genomic recombination site. Proof-of-principle for this was provided using engineered recognition sites for the yeast meganuclease I-SceI, whose 30-bp target site is unlikely to appear at random in a eukaryotic genome.128,129 HIV-1 IDLVs (vector #5 on Table 1) have recently been used to deliver an eGFP targeting cassette and the I-SceI gene to human osteosarcoma U-2 OS, HEK293T or HT1080 cells carrying an I-SceI/eGFP-based model system of gene repair, with targeting frequencies reaching 1.9% of cells. Frequencies were higher (up to 12%) if the I-SceI gene was delivered with an integrating vector, this difference correlating with the relative expression levels of I-SceI from the integrating vector and the IDLV. Integration of the I-SceI IDLV vector in the DSB induced by the nuclease was observed in U-2 OS cells.62
The recent availability of designer nucleases, able to cut at specific genomic sequences, allows the gene repair field to move from model systems to therapeutic genes. Designer nucleases belong to two major groups: meganucleases derived from natural enzymes130 and chimeric zinc-finger nucleases (ZFNs) in which sequence-specific zinc-finger DNA-binding motifs are coupled with the nuclease domain of the type II restriction endonuclease FokI to produce a dimeric site-specific nuclease.131 High-efficiency gene conversion has been demonstrated with both chimeric meganucleases and ZFNs at chromosomal target loci.132,133 HIV-1 IDLVs have been recently used to deliver ZFN genes and an IL2RG targeting construct (vectors #19 on Table 1) to K-562 myeloid leukemia cells and Epstein–Barr virus–transformed B-lymphocytes, with homologous recombination (gene conversion) frequencies approaching 30 and 8%, respectively.114
Homology-dependent, site-specific gene addition (knock-in) is also very efficient using IDLVs for delivery of ZFN genes and targeting construct. Such knock-ins would be useful to correct all the mutations downstream of the insertion site in a given gene using a single targeting construct, rather than having to employ mutation-specific targeting constructs; they could also be used to direct transgene integration to genomic regions where detrimental effects would be unlikely, a possible option to avoid insertional mutagenesis. IDLV-mediated knock-in frequencies at the IL2RG and CCR5 loci (vectors #19 on Table 1) ranged 3.4–40% in cell lines and 1.8–6% in transformed B-lymphocytes, these frequencies being 3- to 30-fold above random integration of the targeting construct in the absence of the nuclease. In human embryonic stem cell lines and CD34+ hematopoietic progenitors targeted knock-in at the CCR5 locus rendered frequencies of 3.5% and 0.06%, respectively, 10-fold higher than background without the ZFNs. The CD34+ result may reflect intrinsically low recombination frequencies in HSCs, because IDLV transduction is an efficient delivery method.114 Of note, in some cases these authors have observed integration of head-to-tail concatemers rather than single-copy knock-in.114
Gene disruption can also be obtained at high frequency in human embryonic stem cell lines (13%, ref. 114) and HSCs (4%, P. Gregory, personal communication) by NHEJ repair of a ZFN-induced DSB at the CCR5 locus (vector #19 on Table 1). This CCR5 knockout strategy is paradigmatic for the generation of CD4+ T-cells resistant to HIV-1 infection.134 Finally, a cautionary note should be added regarding the potential genotoxic effects caused by designer nucleases through off-target DNA cutting (reviewed in ref. 135).
Site-specific integration and transposition
Homology-dependent knock-ins have been discussed in the previous section as a possible way to target transgene integration to genomic regions potentially devoid of insertional mutagenesis risk. This could also be achieved through the use of site-specific recombinases136 and, to a lesser extent, transposases.137 IDLVs have recently been used to deliver both a substrate for site-specific recombination (an FRT-containing episome) and the gene encoding the FRT site-specific flp recombinase (vectors #20 on Table 1). These IDLVs were used to transduce HEK293 cell lines carrying several genomic integrants of an FRT-containing cassette. The results showed that flp could be expressed from an IDLV and that both 1-LTR and 2-LTR circles supported flp-mediated recombination with the chromosomal FRT target site, albeit at lower frequencies than those achieved by calcium phosphate-mediated plasmid transfection of recombinase gene and recombination substrate. However, quantitations of extrachromosomal DNA copy numbers indicated that IDLV episomes were more efficient substrates than transfected plasmids for flp-mediated recombination in a “per copy” basis.55 FRT sites are not naturally present in the human genome, but this work provides proof-of-principle for the development of more clinically applicable systems that could target endogenous chromosomal sites.
The Sleeping Beauty DNA transposon is also undergoing development as a potentially applicable gene delivery system, in the hope that its integration profile may prove safer than that of other integrating vectors.137 In this strategy separate IDLVs are used to deliver a Sleeping Beauty transposase gene and a drug resistance transgene cassette flanked by the transposon's terminal inverted repeats (vectors #21 on Table 1). Results indicate that transposase activity and transposon dose are important factors for success with IDLVs, with the hyperactive transposase version SB100X leading to a maximum 12-fold increase in the number of drug-resistant colonies above background integration. Importantly, integration of the transposon seems to follow a random pattern in a limited data set of integration events.115
The final frontier: replicating IDLV episomes
The evidence reviewed in the previous sections provides compelling proof of the efficiency of IDLVs for transient delivery to dividing cells and stable transduction of quiescent tissues. Stable episomal maintenance in dividing cells would extend the range of IDLV applications significantly, provided that genomic integration remained low. Other episomal replicating vectors have been described, including scaffold/matrix attachment region–, Epstein–Barr virus– or SV40-based systems, conditionally replicating viral vectors, and artificial chromosomes.138,139,140,141,142 Scaffold/matrix attachment region plasmids or minicircles can provide stable maintenance, but have shortcomings including inefficient delivery to some tissues and low frequency of episomal establishment.138 Systems based on viral proteins and origins of replication (Epstein–Barr virus EBNA-1/oriP, SV40's T-antigen/oriT) can be extremely efficient but their possible clinical application is limited by concerns regarding the oncogenic potential of EBNA-1 and large T-antigen.143,144 Adenoviral vectors have been made conditionally replicating in p53-deficient and other cancer cells,141 but no system is currently available to make them replicate and segregate in synchrony with the cellular genome in normal cells. Artificial chromosomes are a promising option145 but delivery and maintenance of large DNAs pose significant challenges. Thus, there is still an unmet need for an efficient episomal system for transgene delivery to proliferating cells. IDLVs can fulfill at least some of the theoretical requirements, including defined structure, efficient delivery, metabolic stability, and capacity to be converted into replicating episomes (see below).
Proof-of-principle exists that IDLVs can be converted into replicating episomes, but this relies on the use of the SV40 large T-antigen and its cognate sequence oriT. In the context of native HIV-1, replication of catalytically inactive IN mutants is only possible in certain permissive T-cell lines. However, incorporation of oriT in the viral genome in place of nef leads to stable replication in T-antigen-expressing Jurkat cells. Alternatively, T-antigen can also be encoded by the oriT-containing viral genome.146 Of note, a similar strategy with the Epstein–Barr virus EBNA-1/oriP system failed to generate replicating IN-defective HIV-1.146 Similarly, use of the SV40 large-T/oriT system leads to replicating IDLVs. In this case, the authors incorporated oriT into an HIV-1 IDLV (vector #10). The presence of oriT led to stable IDLV episome maintenance specifically in HEK293T cells, which express the large T-antigen. Lack of IDLV episome integration was established at the detection level of quantitative fluorescent in situ hybridization experiments.95 Therapeutic proof-of-principle for replicating IDLVs has been shown by incorporating the herpes simplex virus thymidine kinase gene in the stabilized IDLV episome (vector #18 on Table 1), which renders cells susceptible to suicide gene therapy with ganciclovir.113 The development of a clinically applicable replicating IDLV system would allow stable episomal delivery to proliferating cells, with major implications for the improvement of lentiviral vector biosafety. Such system is being hotly pursued but remains to be achieved, with scaffold/matrix attachment regions and chimeric proteins being actively explored. However, a cautionary word is of relevance: a replicating IDLV system could support expansion of an RCR, as shown with IN-deficient HIV-1.146
To summarize, IDLVs can be applied for transient transduction of dividing cells and stable transduction of quiescent cells. They support RNA polymerase II and III gene expression, can mediate RNA interference and vaccination, and are being developed as efficient vectors for homologous recombination (gene repair, knock-in, and knock-out), site-specific recombination, and transposition. There is also scope for the production of clinically applicable replicating IDLVs, which would allow efficient and stable transduction of proliferating cells while minimizing the risk of insertional mutagenesis.
Conclusions
We have reviewed work demonstrating that IDLVs generate high levels of nonreplicating episomal molecules in transduced cells and mediate efficient transduction. They have an outstanding range of research and therapeutic uses, and it is tempting to discuss the remaining hurdles before IDLVs can reach the clinic and speculate on the most likely initial clinical applications. As vectors particularly appropriate for transgene delivery to quiescent tissues, long-term, high-level transgene expression should be demonstrated with therapeutic transgenes in animal models of human diseases. Long-term evidence of lack of genomic integration beyond residual levels should also be provided. IDLVs are also very promising vectors for gene repair, but one of the major targets, human HSCs, remains refractory to high-efficiency homologous recombination. This failure is cause for concern because given the clinical experience with retroviral vector–mediated gene transfer and the selective advantage of corrected cells in several immunodeficiency models, gene repair in HSCs would be a system likely to progress quickly toward a clinical application. Finally, transduction of dividing cells with IDLVs would benefit from higher expression levels and particularly from episomal maintenance. IDLV-optimized internal promoters and clinically useful episomal replication/segregation systems would have wide applicability.
Regarding the most likely initial systems for clinical application, IDLV gene therapies for inherited retinal dystrophies would be prime candidates. Therapeutic effects have been demonstrated in rodent models, and long-term marker gene expression in both rodents and dogs has also been shown.56,61 The ongoing adeno-associated virus–mediated clinical trials for RPE65 deficiency have promising initial results,147,148,149 but if long-term or efficient correction were not achieved IDLVs would be an alternative worth exploring. IDLVs also show potential for vaccination and could be used to induce immunization against HIV and tumors. Finally, efficient targeted genomic modifications are a very attractive development for ex vivo gene therapy. Given the current inefficiency of gene repair in HSCs, patient-specific iPS cells could be prime targets for gene correction followed by expansion, differentiation into appropriate cell types and transplantation, particularly in diseases where corrected cells have a selective advantage. Proof-of-principle for this approach has been obtained in a mouse model of sickle cell anemia.150
IDLVs are efficient delivery vectors, have very low genomic integration frequencies (in the range described for plasmid transfection), and have much lower risk of insertional mutagenesis and RCR generation than integrating lentivectors. They provide a very significant enhancement of biosafety compared to integrating lentivectors and should be considered as strong candidates for transgenesis in quiescent tissues. If a clinically applicable system can be developed to convert IDLVs into replicating episomes they may provide stable transgenesis in proliferating tissues, thus displacing standard integrating lentivectors from their last stronghold. Based on all the preclinical work reviewed here we look forward to the development of clinical applications based on IDLVs.
Acknowledgments
We thank Andrew Porter, Jonathan Beauchamp, Martin Broadstock, Steven Howe, Takis Athanasopoulos, and Ian Graham for critical reading of the manuscript. We are indebted to Thierry VandenDriessche, Jean-Luc Darlix, John Olsen, Philip Gregory, and many of our collaborators for allowing us to cite ongoing work. Research in the Yáñez lab has been funded by Genoma España, the European Union, the Primary Immunodeficiency Association, the Friends of Guy's Hospital, the Medical Research Council, the SouthWest London Academic Network, Action Medical Research, Clinigene and Royal Holloway-University of London.
REFERENCES
- Naldini L, Blömer U, Gage FH, Trono D., and , Verma IM. Efficient transfer, integration, and sustained long-term expression of the transgene in adult rat brains injected with a lentiviral vector. Proc Natl Acad Sci USA. 1996;93:11382–11388. doi: 10.1073/pnas.93.21.11382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada T, Fujii H, Mitsuya H., and , Nienhuis AW. Targeted and highly efficient gene transfer into CD4+ cells by a recombinant human immunodeficiency virus retroviral vector. J Clin Invest. 1991;88:1043–1047. doi: 10.1172/JCI115365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zufferey R, Dull T, Mandel RJ, Bukovsky A, Quiroz D, Naldini L, et al. Self-inactivating lentivirus vector for safe and efficient in vivo gene delivery. J Virol. 1998;72:9873–9880. doi: 10.1128/jvi.72.12.9873-9880.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blömer U, Naldini L, Kafri T, Trono D, Verma IM., and , Gage FH. Highly efficient and sustained gene transfer in adult neurons with a lentivirus vector. J Virol. 1997;71:6641–6649. doi: 10.1128/jvi.71.9.6641-6649.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abordo-Adesida E, Follenzi A, Barcia C, Sciascia S, Castro MG, Naldini L, et al. Stability of lentiviral vector-mediated transgene expression in the brain in the presence of systemic antivector immune responses. Hum Gene Ther. 2005;16:741–751. doi: 10.1089/hum.2005.16.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Nunes FA, Berencsi K, Furth EE, Gönczöl E., and , Wilson JM. Cellular immunity to viral antigens limits E1-deleted adenoviruses for gene therapy. Proc Natl Acad Sci USA. 1994;91:4407–4411. doi: 10.1073/pnas.91.10.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirmule N, Propert K, Magosin S, Qian Y, Qian R., and , Wilson J. Immune responses to adenovirus and adeno-associated virus in humans. Gene Ther. 1999;6:1574–1583. doi: 10.1038/sj.gt.3300994. [DOI] [PubMed] [Google Scholar]
- Mastakov MY, Baer K, Symes CW, Leichtlein CB, Kotin RM., and , During MJ. Immunological aspects of recombinant adeno-associated virus delivery to the mammalian brain. J Virol. 2002;76:8446–8454. doi: 10.1128/JVI.76.16.8446-8454.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mingozzi F, Maus MV, Hui DJ, Sabatino DE, Murphy SL, Rasko JE, et al. CD8(+) T-cell responses to adeno-associated virus capsid in humans. Nat Med. 2007;13:419–422. doi: 10.1038/nm1549. [DOI] [PubMed] [Google Scholar]
- Murphy SL, Li H, Mingozzi F, Sabatino DE, Hui DJ, Edmonson SA, et al. Diverse IgG subclass responses to adeno-associated virus infection and vector administration. J Med Virol. 2009;81:65–74. doi: 10.1002/jmv.21360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naldini L, Blömer U, Gallay P, Ory D, Mulligan R, Gage FH, et al. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science. 1996;272:263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- Miyoshi H, Takahashi M, Gage FH., and , Verma IM. Stable and efficient gene transfer into the retina using an HIV-based lentiviral vector. Proc Natl Acad Sci USA. 1997;94:10319–10323. doi: 10.1073/pnas.94.19.10319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kafri T, Blömer U, Peterson DA, Gage FH., and , Verma IM. Sustained expression of genes delivered directly into liver and muscle by lentiviral vectors. Nat Genet. 1997;17:314–317. doi: 10.1038/ng1197-314. [DOI] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S, von Kalle C, Schmidt M, Le Deist F, Wulffraat N, McIntyre E, et al. A serious adverse event after successful gene therapy for X-linked severe combined immunodeficiency. N Engl J Med. 2003;348:255–256. doi: 10.1056/NEJM200301163480314. [DOI] [PubMed] [Google Scholar]
- Suzuki Y., and , Craigie R. The road to chromatin – nuclear entry of retroviruses. Nat Rev Microbiol. 2007;5:187–196. doi: 10.1038/nrmicro1579. [DOI] [PubMed] [Google Scholar]
- Follenzi A, Ailles LE, Bakovic S, Geuna M., and , Naldini L. Gene transfer by lentiviral vectors is limited by nuclear translocation and rescued by HIV-1 pol sequences. Nat Genet. 2000;25:217–222. doi: 10.1038/76095. [DOI] [PubMed] [Google Scholar]
- Zennou V, Petit C, Guetard D, Nerhbass U, Montagnier L., and , Charneau P. HIV-1 genome nuclear import is mediated by a central DNA flap. Cell. 2000;101:173–185. doi: 10.1016/S0092-8674(00)80828-4. [DOI] [PubMed] [Google Scholar]
- Hindmarsh P., and , Leis J. Retroviral DNA integration. Microbiol Mol Biol Rev. 1999;63:836–843. doi: 10.1128/mmbr.63.4.836-843.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craigie R. HIV integrase, a brief overview from chemistry to therapeutics. J Biol Chem. 2001;276:23213–23216. doi: 10.1074/jbc.R100027200. [DOI] [PubMed] [Google Scholar]
- Wu Y. The second chance story of HIV-1 DNA: Unintegrated? Not a problem! Retrovirology. 2008;5:61. doi: 10.1186/1742-4690-5-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu TW, Sabran JL, Mark GE, Guntaka RV., and , Taylor JM. Analysis of unintegrated avian RNA tumor virus double-stranded DNA intermediates. J Virol. 1978;28:810–818. doi: 10.1128/jvi.28.3.810-818.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shank PR, Hughes SH, Kung HJ, Majors JE, Quintrell N, Guntaka RV, et al. Mapping unintegrated avian sarcoma virus DNA: termini of linear DNA bear 300 nucleotides present once or twice in two species of circular DNA. Cell. 1978;15:1383–1395. doi: 10.1016/0092-8674(78)90063-6. [DOI] [PubMed] [Google Scholar]
- Gianni AM, Smotkin D., and , Weinberg RA. Murine leukemia virus: detection of unintegrated double-stranded DNA forms of the provirus. Proc Natl Acad Sci USA. 1975;72:447–451. doi: 10.1073/pnas.72.2.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Olvera JM, Yoder KE, Mitchell RS, Butler SL, Lieber M, et al. Role of the non-homologous DNA end joining pathway in the early steps of retroviral infection. EMBO J. 2001;20:3272–3281. doi: 10.1093/emboj/20.12.3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cara A, Cereseto A, Lori F., and , Reitz MS. HIV-1 protein expression from synthetic circles of DNA mimicking the extrachromosomal forms of viral DNA. J Biol Chem. 1996;271:5393–5397. doi: 10.1074/jbc.271.10.5393. [DOI] [PubMed] [Google Scholar]
- Delelis O, Petit C, Leh H, Mbemba G, Mouscadet JF., and , Sonigo P. A novel function for spumaretrovirus integrase: an early requirement for integrase-mediated cleavage of 2 LTR circles. Retrovirology. 2005;2:31. doi: 10.1186/1742-4690-2-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandgenett DP., and , Vora AC. Site-specific nicking at the avian retrovirus LTR circle junction by the viral pp32 DNA endonuclease. Nucleic Acids Res. 1985;13:6205–6221. doi: 10.1093/nar/13.17.6205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y. HIV-1 gene expression: lessons from provirus and non-integrated DNA. Retrovirology. 2004;1:13. doi: 10.1186/1742-4690-1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cara A., and , Klotman ME. Retroviral E-DNA: persistence and gene expression in nondividing immune cells. J Leukoc Biol. 2006;80:1013–1017. doi: 10.1189/jlb.0306151. [DOI] [PubMed] [Google Scholar]
- Sarkis C, Philippe S, Mallet J., and , Serguera C. Non-integrating lentiviral vectors. Curr Gene Ther. 2008;8:430–437. doi: 10.2174/156652308786848012. [DOI] [PubMed] [Google Scholar]
- Philpott NJ., and , Thrasher AJ. Use of nonintegrating lentiviral vectors for gene therapy. Hum Gene Ther. 2007;18:483–489. doi: 10.1089/hum.2007.013. [DOI] [PubMed] [Google Scholar]
- Frankel AD., and , Young JA. HIV-1: fifteen proteins and an RNA. Annu Rev Biochem. 1998;67:1–25. doi: 10.1146/annurev.biochem.67.1.1. [DOI] [PubMed] [Google Scholar]
- Wiskerchen M., and , Muesing MA. Human immunodeficiency virus type 1 integrase: effects of mutations on viral ability to integrate, direct viral gene expression from unintegrated viral DNA templates, and sustain viral propagation in primary cells. J Virol. 1995;69:376–386. doi: 10.1128/jvi.69.1.376-386.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler SL, Johnson EP., and , Bushman FD. Human immunodeficiency virus cDNA metabolism: notable stability of two-long terminal repeat circles. J Virol. 2002;76:3739–3747. doi: 10.1128/JVI.76.8.3739-3747.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierson TC, Kieffer TL, Ruff CT, Buck C, Gange SJ., and , Siliciano RF. Intrinsic stability of episomal circles formed during human immunodeficiency virus type 1 replication. J Virol. 2002;76:4138–4144. doi: 10.1128/JVI.76.8.4138-4144.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillim-Ross L, Cara A., and , Klotman ME. HIV-1 extrachromosomal 2-LTR circular DNA is long-lived in human macrophages. Viral Immunol. 2005;18:190–196. doi: 10.1089/vim.2005.18.190. [DOI] [PubMed] [Google Scholar]
- Sharkey M, Triques K, Kuritzkes DR., and , Stevenson M. In vivo evidence for instability of episomal human immunodeficiency virus type 1 cDNA. J Virol. 2005;79:5203–5210. doi: 10.1128/JVI.79.8.5203-5210.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker C, Goff S, Gilboa E, Paskind M, Mitra SW., and , Baltimore D. Structure of a cloned circular Moloney murine leukemia virus DNA molecule containing an inverted segment: implications for retrovirus integration. Proc Natl Acad Sci USA. 1980;77:3932–3936. doi: 10.1073/pnas.77.7.3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin FL, Sperle K., and , Sternberg N. Model for homologous recombination during transfer of DNA into mouse L cells: role for DNA ends in the recombination process. Mol Cell Biol. 1984;4:1020–1034. doi: 10.1128/mcb.4.6.1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidman MM. Intermolecular homologous recombination between transfected sequences in mammalian cells is primarily nonconservative. Mol Cell Biol. 1987;7:3561–3565. doi: 10.1128/mcb.7.10.3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MD, Wang B., and , Bushman FD. Human immunodeficiency virus type 1 preintegration complexes containing discontinuous plus strands are competent to integrate in vitro. J Virol. 1995;69:3938–3944. doi: 10.1128/jvi.69.6.3938-3944.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer M, Kantor B, Cockrell A, Ma H, Zeithaml B, Li X, et al. A large U3 deletion causes increased in vivo expression from a nonintegrating lentiviral vector. Mol Ther. 2008;16:1968–1976. doi: 10.1038/mt.2008.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folger KR, Wong EA, Wahl G., and , Capecchi MR. Patterns of integration of DNA microinjected into cultured mammalian cells: evidence for homologous recombination between injected plasmid DNA molecules. Mol Cell Biol. 1982;2:1372–1387. doi: 10.1128/mcb.2.11.1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goubin G., and , Hill M. Monomer and multimer covalently closed circular forms of Rous sarcoma virus DNA. J Virol. 1979;29:799–804. doi: 10.1128/jvi.29.2.799-804.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartikka J, Sawdey M, Cornefert-Jensen F, Margalith M, Barnhart K, Nolasco M, et al. An improved plasmid DNA expression vector for direct injection into skeletal muscle. Hum Gene Ther. 1996;7:1205–1217. doi: 10.1089/hum.1996.7.10-1205. [DOI] [PubMed] [Google Scholar]
- Jonsson CB, Donzella GA, Gaucan E, Smith CM., and , Roth MJ. Functional domains of Moloney murine leukemia virus integrase defined by mutation and complementation analysis. J Virol. 1996;70:4585–4597. doi: 10.1128/jvi.70.7.4585-4597.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz RA, Mack JP, Merkel G, Kulkosky J, Ge Z, Leis J, et al. Requirement for a conserved serine in both processing and joining activities of retroviral integrase. Proc Natl Acad Sci USA. 1992;89:6741–6745. doi: 10.1073/pnas.89.15.6741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon PM, Wilson W, Byles E, Kingsman SM., and , Kingsman AJ. Human immunodeficiency virus type 1 integrase: effect on viral replication of mutations at highly conserved residues. J Virol. 1994;68:4768–4775. doi: 10.1128/jvi.68.8.4768-4775.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman A, Englund G, Orenstein JM, Martin MA., and , Craigie R. Multiple effects of mutations in human immunodeficiency virus type 1 integrase on viral replication. J Virol. 1995;69:2729–2736. doi: 10.1128/jvi.69.5.2729-2736.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leavitt AD, Robles G, Alesandro N., and , Varmus HE. Human immunodeficiency virus type 1 integrase mutants retain in vitro integrase activity yet fail to integrate viral DNA efficiently during infection. J Virol. 1996;70:721–728. doi: 10.1128/jvi.70.2.721-728.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman A. In vivo analysis of retroviral integrase structure and function. Adv Virus Res. 1999;52:411–426. doi: 10.1016/s0065-3527(08)60309-7. [DOI] [PubMed] [Google Scholar]
- Masuda T, Kuroda MJ., and , Harada S. Specific and independent recognition of U3 and U5 att sites by human immunodeficiency virus type 1 integrase in vivo. J Virol. 1998;72:8396–8402. doi: 10.1128/jvi.72.10.8396-8402.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apolonia L, Waddington SN, Fernandes C, Ward NJ, Bouma G, Blundell MP, et al. Stable gene transfer to muscle using non-integrating lentiviral vectors. Mol Ther. 2007;15:1947–1954. doi: 10.1038/sj.mt.6300281. [DOI] [PubMed] [Google Scholar]
- Nightingale SJ, Hollis RP, Pepper KA, Petersen D, Yu XJ, Yang C, et al. Transient gene expression by nonintegrating lentiviral vectors. Mol Ther. 2006;13:1121–1132. doi: 10.1016/j.ymthe.2006.01.008. [DOI] [PubMed] [Google Scholar]
- Moldt B, Staunstrup NH, Jakobsen M, Yáñez-Muñoz RJ., and , Mikkelsen JG. Genomic insertion of lentiviral DNA circles directed by the yeast Flp recombinase. BMC Biotechnol. 2008;8:60. doi: 10.1186/1472-6750-8-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yáñez-Muñoz RJ, Balaggan KS, MacNeil A, Howe SJ, Schmidt M, Smith AJ, et al. Effective gene therapy with nonintegrating lentiviral vectors. Nat Med. 2006;12:348–353. doi: 10.1038/nm1365. [DOI] [PubMed] [Google Scholar]
- Markowitz M, Nguyen BY, Gotuzzo E, Mendo F, Ratanasuwan W, Kovacs C, et al. Rapid and durable antiretroviral effect of the HIV-1 Integrase inhibitor raltegravir as part of combination therapy in treatment-naive patients with HIV-1 infection: results of a 48-week controlled study. J Acquir Immune Defic Syndr. 2007;46:125–133. doi: 10.1097/QAI.0b013e318157131c. [DOI] [PubMed] [Google Scholar]
- Shimura K, Kodama E, Sakagami Y, Matsuzaki Y, Watanabe W, Yamataka K, et al. Broad antiretroviral activity and resistance profile of the novel human immunodeficiency virus integrase inhibitor elvitegravir (JTK-303/GS-9137) J Virol. 2008;82:764–774. doi: 10.1128/JVI.01534-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Mawsawi LQ., and , Neamati N. Blocking interactions between HIV-1 integrase and cellular cofactors: an emerging anti-retroviral strategy. Trends Pharmacol Sci. 2007;28:526–535. doi: 10.1016/j.tips.2007.09.005. [DOI] [PubMed] [Google Scholar]
- Jacque JM., and , Stevenson M. The inner-nuclear-envelope protein emerin regulates HIV-1 infectivity. Nature. 2006;441:641–645. doi: 10.1038/nature04682. [DOI] [PubMed] [Google Scholar]
- Philippe S, Sarkis C, Barkats M, Mammeri H, Ladroue C, Petit C, et al. Lentiviral vectors with a defective integrase allow efficient and sustained transgene expression in vitro and in vivo. Proc Natl Acad Sci USA. 2006;103:17684–17689. doi: 10.1073/pnas.0606197103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornu TI., and , Cathomen T. Targeted genome modifications using integrase-deficient lentiviral vectors. Mol Ther. 2007;15:2107–2113. doi: 10.1038/sj.mt.6300345. [DOI] [PubMed] [Google Scholar]
- Cockrell AS., and , Kafri T. Gene delivery by lentivirus vectors. Mol Biotechnol. 2007;36:184–204. doi: 10.1007/s12033-007-0010-8. [DOI] [PubMed] [Google Scholar]
- Vigna E., and , Naldini L. Lentiviral vectors: excellent tools for experimental gene transfer and promising candidates for gene therapy. J Gene Med. 2000;2:308–316. doi: 10.1002/1521-2254(200009/10)2:5<308::AID-JGM131>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Reiser J, Harmison G, Kluepfel-Stahl S, Brady RO, Karlsson S., and , Schubert M. Transduction of nondividing cells using pseudotyped defective high-titer HIV type 1 particles. Proc Natl Acad Sci USA. 1996;93:15266–15271. doi: 10.1073/pnas.93.26.15266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zufferey R, Nagy D, Mandel RJ, Naldini L., and , Trono D. Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nat Biotechnol. 1997;15:871–875. doi: 10.1038/nbt0997-871. [DOI] [PubMed] [Google Scholar]
- Kim VN, Mitrophanous K, Kingsman SM., and , Kingsman AJ. Minimal requirement for a lentivirus vector based on human immunodeficiency virus type 1. J Virol. 1998;72:811–816. doi: 10.1128/jvi.72.1.811-816.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi H, Blömer U, Takahashi M, Gage FH., and , Verma IM. Development of a self-inactivating lentivirus vector. J Virol. 1998;72:8150–8157. doi: 10.1128/jvi.72.10.8150-8157.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dull T, Zufferey R, Kelly M, Mandel RJ, Nguyen M, Trono D, et al. A third-generation lentivirus vector with a conditional packaging system. J Virol. 1998;72:8463–8471. doi: 10.1128/jvi.72.11.8463-8471.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yáñez-Muñoz RJ, Foster E, Yip P, McMahon SB, Waddington S., and , Antoniou M. Integration-deficient lentiviral vectors mediate efficient gene transfer to nervous system and muscle. Mol Ther. 2007;15:s378. [Google Scholar]
- Yu SF, von Rüden T, Kantoff PW, Garber C, Seiberg M, Rüther U, et al. Self-inactivating retroviral vectors designed for transfer of whole genes into mammalian cells. Proc Natl Acad Sci USA. 1986;83:3194–3198. doi: 10.1073/pnas.83.10.3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaiss AK, Son S., and , Chang LJ. RNA 3′ readthrough of oncoretrovirus and lentivirus: implications for vector safety and efficacy. J Virol. 2002;76:7209–7219. doi: 10.1128/JVI.76.14.7209-7219.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schambach A, Galla M, Maetzig T, Loew R., and , Baum C. Improving transcriptional termination of self-inactivating γ-retroviral and lentiviral vectors. Mol Ther. 2007;15:1167–1173. doi: 10.1038/sj.mt.6300152. [DOI] [PubMed] [Google Scholar]
- Arumugam PI, Scholes J, Perelman N, Xia P, Yee JK., and , Malik P. Improved human β-globin expression from self-inactivating lentiviral vectors carrying the chicken hypersensitive site-4 (cHS4) insulator element. Mol Ther. 2007;15:1863–1871. doi: 10.1038/sj.mt.6300259. [DOI] [PubMed] [Google Scholar]
- Zychlinski D, Schambach A, Modlich U, Maetzig T, Meyer J, Grassman E, et al. Physiological promoters reduce the genotoxic risk of integrating gene vectors. Mol Ther. 2008;16:718–725. doi: 10.1038/mt.2008.5. [DOI] [PubMed] [Google Scholar]
- Kafri T, van Praag H, Gage FH., and , Verma IM. Lentiviral vectors: regulated gene expression. Mol Ther. 2000;1:516–521. doi: 10.1006/mthe.2000.0083. [DOI] [PubMed] [Google Scholar]
- Reiser J, Lai Z, Zhang XY., and , Brady RO. Development of multigene and regulated lentivirus vectors. J Virol. 2000;74:10589–10599. doi: 10.1128/jvi.74.22.10589-10599.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaFemina RL, Schneider CL, Robbins HL, Callahan PL, LeGrow K, Roth E, et al. Requirement of active human immunodeficiency virus type 1 integrase enzyme for productive infection of human T-lymphoid cells. J Virol. 1992;66:7414–7419. doi: 10.1128/jvi.66.12.7414-7419.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan AC, Haas DL, Kafri T., and , Kohn DB. Integrated self-inactivating lentiviral vectors produce full-length genomic transcripts competent for encapsidation and integration. J Virol. 2004;78:8421–8436. doi: 10.1128/JVI.78.16.8421-8436.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brussel A., and , Sonigo P. Evidence for gene expression by unintegrated human immunodeficiency virus type 1 DNA species. J Virol. 2004;78:11263–11271. doi: 10.1128/JVI.78.20.11263-11271.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H., and , Kafri T. A single-LTR HIV-1 vector optimized for functional genomics applications. Mol Ther. 2004;10:139–149. doi: 10.1016/j.ymthe.2004.04.012. [DOI] [PubMed] [Google Scholar]
- Roberts JD, Bebenek K., and , Kunkel TA. The accuracy of reverse transcriptase from HIV-1. Science. 1988;242:1171–1173. doi: 10.1126/science.2460925. [DOI] [PubMed] [Google Scholar]
- Wain-Hobson S. Human immunodeficiency virus type 1 quasispecies in vivo and ex vivo. Curr Top Microbiol Immunol. 1992;176:181–193. doi: 10.1007/978-3-642-77011-1_12. [DOI] [PubMed] [Google Scholar]
- Apolonia L, Waddington SN, Ward NJ, Philpott NJ, Collins MK., and , Thrasher AJ. Factor IX expression from non-integrating lentiviral vectors in muscle. Mol Ther. 2008;16:s328. doi: 10.1038/sj.mt.6300281. [DOI] [PubMed] [Google Scholar]
- Brussel A., and , Sonigo P. Analysis of early human immunodeficiency virus type 1 DNA synthesis by use of a new sensitive assay for quantifying integrated provirus. J Virol. 2003;77:10119–10124. doi: 10.1128/JVI.77.18.10119-10124.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donehower LA., and , Varmus HE. A mutant murine leukemia virus with a single missense codon in pol is defective in a function affecting integration. Proc Natl Acad Sci USA. 1984;81:6461–6465. doi: 10.1073/pnas.81.20.6461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panganiban AT., and , Temin HM. The retrovirus pol gene encodes a product required for DNA integration: identification of a retrovirus int locus. Proc Natl Acad Sci USA. 1984;81:7885–7889. doi: 10.1073/pnas.81.24.7885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagino-Yamagishi K, Donehower LA., and , Varmus HE. Retroviral DNA integrated during infection by an integration-deficient mutant of murine leukemia virus is oligomeric. J Virol. 1987;61:1964–1971. doi: 10.1128/jvi.61.6.1964-1971.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaur M., and , Leavitt AD. Mutations in the human immunodeficiency virus type 1 integrase D,D(35)E motif do not eliminate provirus formation. J Virol. 1998;72:4678–4685. doi: 10.1128/jvi.72.6.4678-4685.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y., and , Waldman AS. Capture of DNA sequences at double-strand breaks in mammalian chromosomes. Genetics. 2001;158:1665–1674. doi: 10.1093/genetics/158.4.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasquez KM, Marburger K, Intody Z., and , Wilson JH. Manipulating the mammalian genome by homologous recombination. Proc Natl Acad Sci USA. 2001;98:8403–8410. doi: 10.1073/pnas.111009698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yáñez RJ., and , Porter ACG. Influence of DNA delivery method on gene targeting frequencies in human cells. Somat Cell Mol Genet. 1999;25:27–31. doi: 10.1023/b:scam.0000007137.28557.73. [DOI] [PubMed] [Google Scholar]
- Case SS, Price MA, Jordan CT, Yu XJ, Wang L, Bauer G, et al. Stable transduction of quiescent CD34(+)CD38(-) human hematopoietic cells by HIV-1-based lentiviral vectors. Proc Natl Acad Sci USA. 1999;96:2988–2993. doi: 10.1073/pnas.96.6.2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas DL, Case SS, Crooks GM., and , Kohn DB. Critical factors influencing stable transduction of human CD34(+) cells with HIV-1-derived lentiviral vectors. Mol Ther. 2000;2:71–80. doi: 10.1006/mthe.2000.0094. [DOI] [PubMed] [Google Scholar]
- Vargas J, Gusella GL, Najfeld V, Klotman ME., and , Cara A. Novel integrase-defective lentiviral episomal vectors for gene transfer. Hum Gene Ther. 2004;15:361–372. doi: 10.1089/104303404322959515. [DOI] [PubMed] [Google Scholar]
- Mali P, Ye Z, Hommond HH, Yu X, Lin J, Chen G, et al. Improved efficiency and pace of generating induced pluripotent stem cells from human adult and fetal fibroblasts. Stem Cells. 2008;26:1998–2005. doi: 10.1634/stemcells.2008-0346. [DOI] [PubMed] [Google Scholar]
- Mitrophanous K, Yoon S, Rohll J, Patil D, Wilkes F, Kim V, et al. Stable gene transfer to the nervous system using a non-primate lentiviral vector. Gene Ther. 1999;6:1808–1818. doi: 10.1038/sj.gt.3301023. [DOI] [PubMed] [Google Scholar]
- Patel M, Giddings AM., and , Olsen JC. Integration defective EIAV-based lentiviral vector gene transfer to the nasal epithelium of mice. Mol Ther. 2008;16:s338. [Google Scholar]
- Saenz DT, Loewen N, Peretz M, Whitwam T, Barraza R, Howell KG, et al. Unintegrated lentivirus DNA persistence and accessibility to expression in nondividing cells: analysis with class I integrase mutants. J Virol. 2004;78:2906–2920. doi: 10.1128/JVI.78.6.2906-2920.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Stenzel M, Sodroski JG., and , Haseltine WA. Effects of long terminal repeat mutations on human immunodeficiency virus type 1 replication. J Virol. 1989;63:4115–4119. doi: 10.1128/jvi.63.9.4115-4119.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenke AC, Scinteie MF, Stehle IM., and , Lipps HJ. Expression of a transgene encoded on a non-viral episomal vector is not subject to epigenetic silencing by cytosine methylation. Mol Biol Rep. 2004;31:85–90. doi: 10.1023/b:mole.0000031363.35839.46. [DOI] [PubMed] [Google Scholar]
- Ellis J. Silencing and variegation of γ-retrovirus and lentivirus vectors. Hum Gene Ther. 2005;16:1241–1246. doi: 10.1089/hum.2005.16.1241. [DOI] [PubMed] [Google Scholar]
- Pannell D, Osborne CS, Yao S, Sukonnik T, Pasceri P, Karaiskakis A, et al. Retrovirus vector silencing is de novo methylase independent and marked by a repressive histone code. EMBO J. 2000;19:5884–5894. doi: 10.1093/emboj/19.21.5884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park F, Ohashi K., and , Kay MA. Therapeutic levels of human factor VIII and IX using HIV-1-based lentiviral vectors in mouse liver. Blood. 2000;96:1173–1176. [PubMed] [Google Scholar]
- Mazarakis ND, Azzouz M, Rohll JB, Ellard FM, Wilkes FJ, Olsen AL, et al. Rabies virus glycoprotein pseudotyping of lentiviral vectors enables retrograde axonal transport and access to the nervous system after peripheral delivery. Hum Mol Genet. 2001;10:2109–2121. doi: 10.1093/hmg/10.19.2109. [DOI] [PubMed] [Google Scholar]
- Loewen N, Leske DA, Chen Y, Teo WL, Saenz DT, Peretz M, et al. Comparison of wild-type and class I integrase mutant-FIV vectors in retina demonstrates sustained expression of integrated transgenes in retinal pigment epithelium. J Gene Med. 2003;5:1009–1017. doi: 10.1002/jgm.447. [DOI] [PubMed] [Google Scholar]
- Fabes J, Anderson P, Yáñez-Muñoz RJ, Thrasher A, Brennan C., and , Bolsover S. Accumulation of the inhibitory receptor EphA4 may prevent regeneration of corticospinal tract axons following lesion. Eur J Neurosci. 2006;23:1721–1730. doi: 10.1111/j.1460-9568.2006.04704.x. [DOI] [PubMed] [Google Scholar]
- Rahim AA, Wong AM, Howe SJ, Buckley SM, Acosta-Saltos AD, Elston KE, et al. Efficient gene delivery to the adult and fetal CNS using pseudotyped non-integrating lentiviral vectors. Gene Ther. 2009;16:509–520. doi: 10.1038/gt.2008.186. [DOI] [PubMed] [Google Scholar]
- Li S, Kimura E, Fall BM, Reyes M, Angello JC, Welikson R, et al. Stable transduction of myogenic cells with lentiviral vectors expressing a minidystrophin. Gene Ther. 2005;12:1099–1108. doi: 10.1038/sj.gt.3302505. [DOI] [PubMed] [Google Scholar]
- Annoni A, Brown BD, Cantore A, Sergi Sergi L, Naldini L., and , Roncarolo MG. In vivo delivery of a micro-RNA regulated antigen-encoding transgene induces antigen-specific regulatory T cells and promotes immunological tolerance. Mol Ther. 2009;17:s294. doi: 10.1182/blood-2009-04-214569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negri DR, Michelini Z, Baroncelli S, Spada M, Vendetti S, Buffa V, et al. Successful immunization with a single injection of non-integrating lentiviral vector. Mol Ther. 2007;15:1716–1723. doi: 10.1038/sj.mt.6300241. [DOI] [PubMed] [Google Scholar]
- Karwacz K, Mukherjee S, Apolonia L, Blundell MP, Bouma G, Escors D, et al. Nonintegrating lentivector vaccines stimulate prolonged T-cell and antibody responses and are effective in tumor therapy. J Virol. 2009;83:3094–3103. doi: 10.1128/JVI.02519-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas J, Klotman ME., and , Cara A. Conditionally replicating lentiviral-hybrid episomal vectors for suicide gene therapy. Antiviral Res. 2008;80:288–294. doi: 10.1016/j.antiviral.2008.06.015. [DOI] [PubMed] [Google Scholar]
- Lombardo A, Genovese P, Beausejour CM, Colleoni S, Lee YL, Kim KA, et al. Gene editing in human stem cells using zinc finger nucleases and integrase-defective lentiviral vector delivery. Nat Biotechnol. 2007;25:1298–1306. doi: 10.1038/nbt1353. [DOI] [PubMed] [Google Scholar]
- Staunstrup NH, Moldt B, Mátés L, Villesen P, Jakobsen M, Ivics Z, et al. Hybrid lentivirus-transposon vectors with a random integration profile in human cells Mol Ther 2009. doi : 10.1038/mt.2009.10 . [DOI] [PMC free article] [PubMed]
- Butler SL, Hansen MS., and , Bushman FD. A quantitative assay for HIV DNA integration in vivo. Nat Med. 2001;7:631–634. doi: 10.1038/87979. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Zhang XB, Beard BC, Trobridge GD, Wood BL, Sale GE, Sud R, et al. High incidence of leukemia in large animals after stem cell gene therapy with a HOXB4-expressing retroviral vector. J Clin Invest. 2008;118:1502–1510. doi: 10.1172/JCI34371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai T, Choi U, Cardwell L, DeRavin SS, Naumann N, Whiting-Theobald NL, et al. WHIM syndrome myelokathexis reproduced in the NOD/SCID mouse xenotransplant model engrafted with healthy human stem cells transduced with C-terminus-truncated CXCR4. Blood. 2007;109:78–84. doi: 10.1182/blood-2006-05-025296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai T, Choi U, Changpriroa C, Berg K, Lantz LM., and , Malech HL. Human hematopoietic stem cells infected with integration defective lentiviral vector encoding C-terminus truncated hyperfunctional CXCR4 demonstrate enhanced engraftment in NOD/SCID mice. Mol Ther. 2008;16:s114. [Google Scholar]
- Berger G, Goujon C, Darlix JL., and , Cimarelli A. SIVMAC Vpx improves the transduction of dendritic cells with nonintegrative HIV-1-derived vectors. Gene Ther. 2009;16:159–163. doi: 10.1038/gt.2008.128. [DOI] [PubMed] [Google Scholar]
- Mühlebach MD, Wolfrum N, Schüle S, Tschulena U, Sanzenbacher R, Flory E, et al. Stable transduction of primary human monocytes by simian lentiviral vector PBj. Mol Ther. 2005;12:1206–1216. doi: 10.1016/j.ymthe.2005.06.483. [DOI] [PubMed] [Google Scholar]
- Kim D., and , Rossi J. RNAi mechanisms and applications. BioTechniques. 2008;44:613–616. doi: 10.2144/000112792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abbas-Terki T, Blanco-Bose W, Déglon N, Pralong W., and , Aebischer P. Lentiviral-mediated RNA interference. Hum Gene Ther. 2002;13:2197–2201. doi: 10.1089/104303402320987888. [DOI] [PubMed] [Google Scholar]
- Banasik MB, Fischer AJ., and , McCray PB., Jr Development of an integrase deficient lentiviral vector for transient shRNA expression in respiratory epithelia. Mol Ther. 2007;15:s385. [Google Scholar]
- Yáñez RJ., and , Porter ACG. Therapeutic gene targeting. Gene Ther. 1998;5:149–159. doi: 10.1038/sj.gt.3300601. [DOI] [PubMed] [Google Scholar]
- Okada Y, Ueshin Y, Hasuwa H, Takumi K, Okabe M., and , Ikawa M. Targeted gene modification in mouse ES cells using integrase-defective lentiviral vectors. Genesis. 2009;47:217–223. doi: 10.1002/dvg.20469. [DOI] [PubMed] [Google Scholar]
- Choulika A, Perrin A, Dujon B., and , Nicolas JF. Induction of homologous recombination in mammalian chromosomes by using the I-SceI system of Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:1968–1973. doi: 10.1128/mcb.15.4.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smih F, Rouet P, Romanienko PJ., and , Jasin M. Double-strand breaks at the target locus stimulate gene targeting in embryonic stem cells. Nucleic Acids Res. 1995;23:5012–5019. doi: 10.1093/nar/23.24.5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epinat JC, Arnould S, Chames P, Rochaix P, Desfontaines D, Puzin C, et al. A novel engineered meganuclease induces homologous recombination in yeast and mammalian cells. Nucleic Acids Res. 2003;31:2952–2962. doi: 10.1093/nar/gkg375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YG, Cha J., and , Chandrasegaran S. Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc Natl Acad Sci USA. 1996;93:1156–1160. doi: 10.1073/pnas.93.3.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urnov FD, Miller JC, Lee YL, Beausejour CM, Rock JM, Augustus S, et al. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435:646–651. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- Arnould S, Perez C, Cabaniols JP, Smith J, Gouble A, Grizot S, et al. Engineered I-CreI derivatives cleaving sequences from the human XPC gene can induce highly efficient gene correction in mammalian cells. J Mol Biol. 2007;371:49–65. doi: 10.1016/j.jmb.2007.04.079. [DOI] [PubMed] [Google Scholar]
- Perez EE, Wang J, Miller JC, Jouvenot Y, Kim KA, Liu O, et al. Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases. Nat Biotechnol. 2008;26:808–816. doi: 10.1038/nbt1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cathomen T., and , Joung JK. Zinc-finger nucleases: the next generation emerges. Mol Ther. 2008;16:1200–1207. doi: 10.1038/mt.2008.114. [DOI] [PubMed] [Google Scholar]
- Calos MP. The phiC31 integrase system for gene therapy. Curr Gene Ther. 2006;6:633–645. doi: 10.2174/156652306779010642. [DOI] [PubMed] [Google Scholar]
- Ivics Z., and , Izsvak Z. Transposons for gene therapy! Curr Gene Ther. 2006;6:593–607. doi: 10.2174/156652306778520647. [DOI] [PubMed] [Google Scholar]
- Piechaczek C, Fetzer C, Baiker A, Bode J., and , Lipps HJ. A vector based on the SV40 origin of replication and chromosomal S/MARs replicates episomally in CHO cells. Nucleic Acids Res. 1999;27:426–428. doi: 10.1093/nar/27.2.426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates JL, Warren N., and , Sugden B. Stable replication of plasmids derived from Epstein-Barr virus in various mammalian cells. Nature. 1985;313:812–815. doi: 10.1038/313812a0. [DOI] [PubMed] [Google Scholar]
- Gething MJ., and , Sambrook J. Cell-surface expression of influenza haemagglutinin from a cloned DNA copy of the RNA gene. Nature. 1981;293:620–625. doi: 10.1038/293620a0. [DOI] [PubMed] [Google Scholar]
- Bischoff JR, Kirn DH, Williams A, Heise C, Horn S, Muna M, et al. An adenovirus mutant that replicates selectively in p53-deficient human tumor cells. Science. 1996;274:373–376. doi: 10.1126/science.274.5286.373. [DOI] [PubMed] [Google Scholar]
- Farr CJ, Bayne RA, Kipling D, Mills W, Critcher R., and , Cooke HJ. Generation of a human X-derived minichromosome using telomere-associated chromosome fragmentation. EMBO J. 1995;14:5444–5454. doi: 10.1002/j.1460-2075.1995.tb00228.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humme S, Reisbach G, Feederle R, Delecluse HJ, Bousset K, Hammerschmidt W, et al. The EBV nuclear antigen 1 (EBNA1) enhances B cell immortalization several thousandfold. Proc Natl Acad Sci USA. 2003;100:10989–10994. doi: 10.1073/pnas.1832776100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moens U, Van Ghelue M., and , Johannessen M. Oncogenic potentials of the human polyomavirus regulatory proteins. Cell Mol Life Sci. 2007;64:1656–1678. doi: 10.1007/s00018-007-7020-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshiya H, Kazuki Y, Abe S, Takiguchi M, Kajitani N, Watanabe Y, et al. A highly stable and nonintegrated human artificial chromosome (HAC) containing the 2.4 Mb entire human dystrophin gene. Mol Ther. 2009;17:309–317. doi: 10.1038/mt.2008.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R, Nakajima N, Hofmann W, Benkirane M, Jeang KT, Sodroski J, et al. Simian virus 40-based replication of catalytically inactive human immunodeficiency virus type 1 integrase mutants in nonpermissive T cells and monocyte-derived macrophages. J Virol. 2004;78:658–668. doi: 10.1128/JVI.78.2.658-668.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bainbridge JW, Smith AJ, Barker SS, Robbie S, Henderson R, Balaggan K, et al. Effect of gene therapy on visual function in Leber's congenital amaurosis. N Engl J Med. 2008;358:2231–2239. doi: 10.1056/NEJMoa0802268. [DOI] [PubMed] [Google Scholar]
- Maguire AM, Simonelli F, Pierce EA, Pugh EN, Mingozzi F, Bennicelli J, et al. Safety and efficacy of gene transfer for Leber's congenital amaurosis. N Engl J Med. 2008;358:2240–2248. doi: 10.1056/NEJMoa0802315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauswirth WW, Aleman TS, Kaushal S, Cideciyan AV, Schwartz SB, Wang L, et al. Treatment of leber congenital amaurosis due to RPE65 mutations by ocular subretinal injection of adeno-associated virus gene vector: short-term results of a phase I trial. Hum Gene Ther. 2008;19:979–990. doi: 10.1089/hum.2008.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna J, Wernig M, Markoulaki S, Sun CW, Meissner A, Cassady JP, et al. Treatment of sickle cell anemia mouse model with iPS cells generated from autologous skin. Science. 2007;318:1920–1923. doi: 10.1126/science.1152092. [DOI] [PubMed] [Google Scholar]