Abstract
The phytohormone abscisic acid (ABA) acts in seed dormancy, plant development, drought tolerance and adaptive responses to environmental stresses. Structural mechanisms mediating ABA receptor recognition and signaling remain unknown, but are essential for understanding and manipulating abiotic stress resistance. Here we report structures of PYR1, a prototypical PYR/PYL/RCAR protein that functions in early ABA signaling. The crystallographic structure reveals an α/β helix-grip fold and homodimeric assembly, verified in vivo by co-immunoprecipitation. ABA binding within a large internal cavity switches structural motifs distinguishing ABA-free “open-lid” from ABA-bound “closed-lid” conformations. Small angle X-ray scattering suggests that ABA signals by converting PYR1 to a more compact, symmetric closed-lid dimer. Site-directed PYR1 mutants designed to disrupt hormone binding lose ABA-triggered interactions with type 2C protein phosphatase partners in planta.
The phytohormone abscisic acid (ABA) plays key regulatory roles in physiological pathways for plant growth and development, and enables adaptation to abiotic stresses. In the half-century since ABA's discovery (1, 2), much has been learned about its downstream signaling network (3, 4), yet protein recognition mechanisms for this hormone have remained enigmatic. Recently, a cluster of homologous genes that activate ABA signaling was identified in Arabidopsis thaliana by groups using different methods: yeast two hybrid screening, chemical genetics and co-immunoprecipitation analyses (5, 6). In the presence of ABA, the gene products, designated PYR1 (Pyrabactin Resistance 1) and PYL (PYR1-Like) or RCAR (Regulatory Components of ABA Receptor), down regulate their binding partners, cluster A type 2C protein phosphatase (PP2C) family members (7, 8). These phosphatases, including ABI1, ABI2, PP2CA, HAB1 and HAB2, are negative regulators of early ABA signaling (9–18). Similarly, OST1/SnRK2.6/SnRK2E SNF1-related protein kinases 2 (SnRK2s) are important mediators of ABA signal transduction (19–22). PYR/PYL/RCAR family members exhibit functional redundancy in ABA perception, and variations in ABA regulation of their binding to PP2C family members (5, 6). Contrasting hypotheses suggest that ABA either binds to PYR/PYL/RCAR proteins directly or forms a molecular “glue” between these proteins and PP2Cs (5, 6, 23), similar to auxin's role in joining TIR1-related and AUX/IAA signaling proteins (24). Distinct but overlapping functions of different PYR/PYL/RCAR proteins complicate genetic analysis and mechanistic testing in plants, but may enhance flexible regulation of ABA signaling to maximize environmental adaptability of plants.
Plant hormone receptors have evolved from diverse protein families (25) and operate by distinct protein-hormone and protein-protein binding interactions. Understanding the mechanism of action of ABA receptor(s) has been controversial and challenging, due partly to the absence of a structure for an ABA-bound protein complex. Recent structural studies of hormone-bound auxin and gibberellin receptors identified binding sites, characterized protein assemblies, and enabled major advances in understanding hormone signaling in plants (24, 26, 27). Here, we report the ABA recognition mechanism by the PYR1 dimer assembly, as revealed by ABA-bound and unbound crystallographic structures and small angle X-ray scattering (SAXS) in solution, coupled with analysis of structure-based site-directed mutants by coimmunoprecipitation analyses in vivo. Our results show that ABA binds directly to PYR1 within a large internal water-filled cavity, rather than acting as a molecular glue at an interface with PP2Cs. The PYR1 structure reveals how both (+)-ABA and (−)-ABA enantiomers can show biological activity. We define the unusual asymmetric homodimeric assembly of PYR1 that allows hormone access and sequestration; discover structural motifs (Pro-Cap, Leu-Lock and Recoil) that mediate open-lid and closed-lid conformations; and deduce a probable mechanism for ABA signal transduction via hormone-induced conformational changes that promote binding of PP2C partners.
PYR1 architecture and dimeric assembly
To understand ABA binding and signaling, we crystallized and determined structures (28) of Arabidopsis thaliana PYR1 with the phytohormone (+)-cis,trans abscisic acid (S-ABA, fig. S1). Extensive screening produced crystals in space group P2, but only with ABA. Crystallization trials without hormone were unsuccessful, suggesting conformational flexibility. Initial crystals were obtained with enantiomorphic (+/−)-ABA at pH 5.8, the approximate isoelectric point for both protein and hormone. Diffraction quality crystals were reproducible with (+)-ABA alone, at pH values ranging from 5.4 to 6.8, but were always ABA dependent. Molecular replacement was accomplished with a probe structure from the pathogenesis-related protein Bet vI (Pfam: PF00407) (29) family.
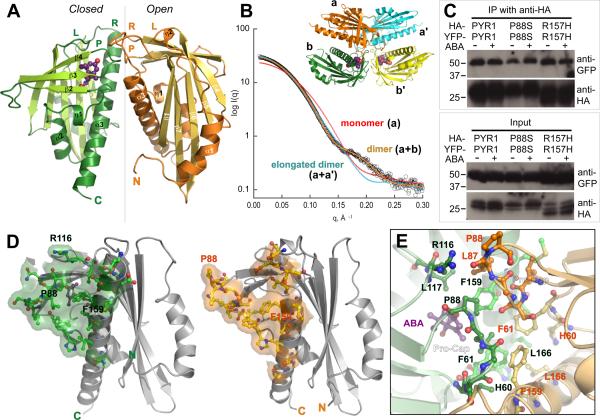
The PYR1 structure, determined to 1.7 Å resolution (Fig. 1A; table S1; movie S1) is a 7-stranded anti-parallel β-sheet wrapped around a long C-terminal α-helix. This α/β helix-grip fold is shared by plant pollen allergen Bet vI and mammalian steroidogenic acute regulatory lipid transfer (START) proteins (30–32). The variable N-terminus (fig. S2) forms PYR1 helix α1 arching back over the β-sheet (Fig. 1A). β1 is hydrogen-bonded with β7, but covalently linked by helices across the β-sheet to β2. In contrast, intervening β-strands exhibit nearest-neighbor (+1) connectivity. β7 is connected via a projecting loop to C-terminal α3.
Fig. 1.
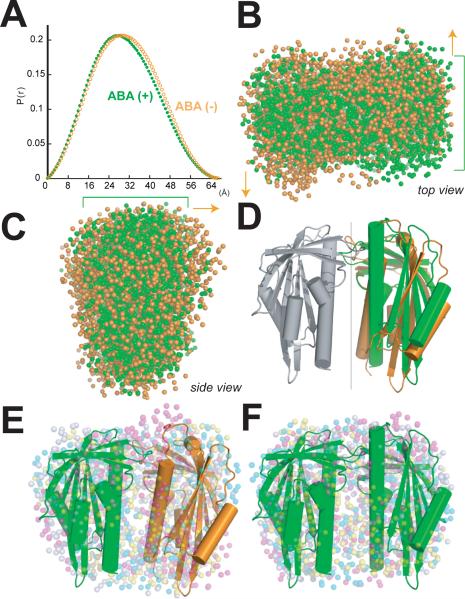
Dimeric structure of ABA sensor PYR1. (A) Crystallographic asymmetric dimer shown as ribbons (labeled β-strands and helices) with ABA (purple ball-and-stick model with red oxygen atoms) bound beneath the closed lid of one subunit (left), in a large cavity between the β-sheet and long C-terminal α-helix. Labeled Pro-Cap (P), Leu-Lock (L) and Recoil (R) structural motifs undergo ABA-induced conformational changes. Vertical line (center) indicates pseudo 2-fold axis relating the subunits. (B) Theoretical SAXS curve (orange line) for asymmetric crystallographic dimer (a+b in inset) matches experimental SAXS data for PYR1 without ABA (open circles), whereas curves calculated assuming a monomer (red) or elongated a+a' dimer (blue) from crystal lattice (see inset) do not. (C) Co-immunoprecipitation from extracts of plant leaves expressing YFP-tagged PYR1 and HA-tagged PYR1 without (−) and with (+) exogenously applied ABA confirm dimeric PYR1 assembly. After co-immunoprecipitation using an anti-HA matrix, immunoprecipitated (above) and input (below) samples were detected with anti-GFP and anti-HA antibodies. PYR1 wild-type and mutants P88S and R157H are homodimeric in planta, as shown by anti-GFP antibody labeling of YFP-tagged PYR1 co-immunoprecipitated with HA-tagged PYR1. (D) Residues and buried surface area contributed to dimer interface by closed-lid (left) and open-lid (right) subunit conformations in asymmetric dimer. (E) PYR1 dimer interface viewed looking down from top in (A) at the interacting lids: open (orange) and closed (green) over ABA (purple). Dimer contacts include the interacting Pro-Cap structural motifs (foreground), plus a side-chain-to-main-chain hydrogen bond from Arg116 in the ABA-bound subunit (top left) to Leu87 in the ABA-free subunit.
Interestingly, in the crystallographic asymmetric unit, PYR1 assembles into a homodimer of one ABA-bound (Fig. 1A, left) and one ABA-free subunit (Fig. 1A, right), related by an ~170° rotation around a pseudo 2-fold axis. Roughly perpendicular interactions of α3 helices align the β-sheets in parallel (Fig. 1A). To determine whether PYR1 (monomer Mr = 20 kDa) forms this dimer in solution, we used multi-angle laser light scattering (MALS) and SAXS (33, 34). MALS allowed simultaneous measurements of absolute molecular weight and hydrodynamic radius. In the absence of ABA, PYR1 forms a homodimer in solution (Mr = 40.8±0.4 to 40.9±0.3 kDa) under varying pH (5.4 to 7.6) and salt conditions (20 mM NaCl and 20–100 mM KCl), including those approximating physiological intracellular levels (100 mM K+, pH 7.6). Thus, ABA is not required for PYR1-PYR1 homodimer formation. SAXS measurements showed that the PYR1 dimer in solution matched the dimer assembly in the crystallographic asymmetric unit, rather than alternative monomeric, dimeric, or tetrameric packing assemblies within the crystal lattice (Fig. 1B; fig. S3). The maximum intramolecular distance (Dmax=68Å) determined from the SAXS-derived pair-distance distribution function also matched that measured from the crystallographic structure (69.8Å).
To determine whether PYR1 is homodimeric in planta, we performed Agrobacterium-mediated infiltration of Nicotiana benthamiana leaves with both YFP- and HA-tagged PYR1, followed by co-immunoprecipitation analyses. PYR1 constitutively formed a dimer in vivo, both in the presence and absence of exogenously applied ABA (Fig. 1C). Thus, the dimer observed in the crystallographic asymmetric unit is evidently a biological unit for PYR1 in vivo.
In this asymmetric dimer, the interface lies between the crossed α3 helices (Fig. 1A). The roughly triangular surfaces shielded in each subunit are similar, but not identical (Fig. 1D). The core of the dimer interface is centered on the asymmetric hydrophobic packing interactions of Phe61 and Phe159 from both subunits (Fig. 1D and E), but the dimer packing specificity may depend on interactions (Fig. 1E) of clustered structural motifs that undergo ABA-induced conformational changes (Fig. 1A).
Abscisic acid binding site inside PYR1
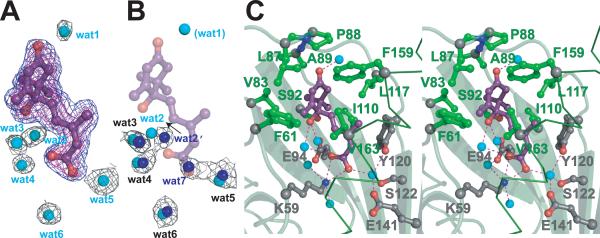
Naturally occurring phytohormone (+)-cis,trans-ABA, (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid (fig. S1), binds within a large interior cavity of PYR1 between the twisted β-sheet and long α3 helix (Fig. 1A). The electron density shows (+)-ABA bound in only one subunit of each PYR1 dimer (Fig. 2, A and B). Thus, the crystal structure resolves both ABA-bound and unbound forms of PYR1. ABA is tethered at both ends by hydrogen bonds to the protein (Fig. 2C). The planar, conjugated, 3-methylpenta-2-cis,4-trans-dienoic acid tail of ABA extends ~10Å into a large protein cavity, where the terminal carboxylate is anchored by the inward-pointing Lys59 side chain. At the cavity entrance, the ABA carbonyl group links two protein loops via hydrogen bonds with main-chain nitrogen atoms of Ala89 and, through a water molecule, Arg116. The adjacent Pro88 ring caps the ABA carbonyl to form a lid of the ABA-binding cavity (Fig. 2C); Leu87 from the ABA-free subunit reaches across the PYR1 dimer to block the remaining cavity access (Fig. 1E). Thus PYR1 dimer formation contributes to ABA sequestration.
Fig. 2.
Water-filled ABA-binding cavity. (A) ABA (purple ball and stick model, with red oxygen atoms) and adjacent, ordered, water molecules (cyan spheres) inside the PYR1 cavity, shown with electron density (mesh). Omit Fo-Fc density for ABA contoured at 3σ (blue) and 4σ (magenta); 2Fo-Fc electron density for water molecules contoured at 1σ (black). All maps were calculated after “shaking” coordinates to reduce phase bias. (B) Ordered water molecules (blue spheres) within the ABA-free subunit cavity, shown with associated 2Fo-Fc electron density, as in (A). ABA (purple) and water molecules (cyan) from ABA-bound PYR1 subunit (shown in A) are superimposed showing conserved, water positions. ABA displaces one water molecule (wat7) with the carboxylate, shifts a second (wat2), and introduces or stabilizes a third (wat1), which interacts with the ABA carbonyl to stabilize lid closure. (C) Stereo view of PYR1 residues contributing to the ABA binding site. Hydrophobic side chains (green ball-and-stick) surround the ABA ring, whereas hydrogen-bonded (red dashed lines) internal water molecules (cyan spheres) link ABA oxygen atoms (red) to PYR1 hydrophilic side chains (gray ball-and-stick with red oxygen and blue nitrogen atoms) projecting into the binding cavity. Larger gray spheres show Cα atoms. Lys59, Phe61, Arg79, Val83, Leu87, Pro88, Ala89, Ser92, Glu94, Ile110, Leu117, Tyr120, Ser122, Glu141, Phe159, Val163 and Asn167 contribute to forming this large internal cavity.
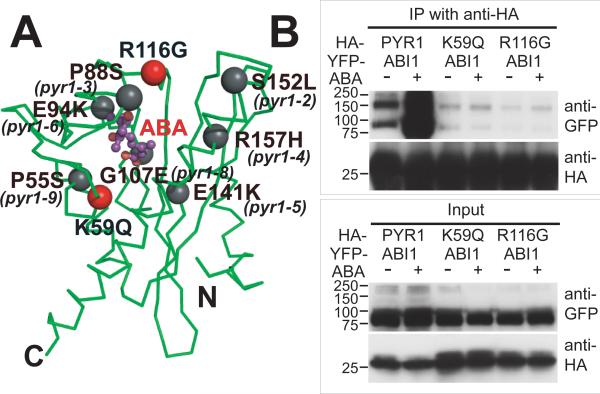
To biologically test the observed protein-hormone interactions, we made site-directed mutants of PYR1 (fig. S4) designed to weaken ABA binding. To examine their consequences on ABA signaling, these PYR1 mutants were transiently expressed in tobacco leaves, and analyzed for function by co-immunoprecipitation assays that detect ABA-triggered interactions between PYR1 and the ABI1 protein phosphatase (Fig. 3). The K59Q PYR1 mutation, designed to neutralize the counter-ion to the ABA carboxylate (Figs. 2C and 3A), disrupted exogenous ABA-induced PYR1 binding to ABI1 (Fig. 3B). Thirteen of 14 ABA sensor family members conserve Lys59, whereas PYL13 has Gln (fig. S2). Invariant Arg116, which contributes to both ABA binding and the dimer interface (Fig. 1E), was mutated to Gly. The R116G PYR1 mutation also abolished ABA-induced PYR1 binding to ABI1 (Fig. 3B). These mutational results support the biological relevance of the ABA-binding cavity and dimer interface characterized by our crystal structures.
Fig. 3.
Disruption of PYR1-ABI1 interactions by single-site PYR1 mutations. (A) PYR1 mutants designed from the structure (red) and identified after chemical mutagenesis and screening (gray) (6) are mapped to the PYR1 subunit structure (green Cα trace). (B) Co-immunoprecipitation from extracts of plant leaves expressing YFP-tagged ABI1 phosphatase with HA-tagged wild-type and mutant PYR1 proteins in planta, both in the absence (−) and presence (+) of exogenously applied ABA. After co-immunoprecipitation by an anti-HA matrix, immunoprecipitated (top) and input (bottom) samples were detected with anti GFP and anti-HA antibodies (labeled at right). Structure-based PYR1 mutants designed to disrupt ABA binding (K59Q and R116G) folded properly (fig. S5), but lost ABA-induced interactions with ABI1 phosphatase.
In PYR1, the ABA ring is surrounded by hydrophobic side chains and sequestered from solution (Fig. 2C). This hydrophobic enclosure can accommodate (+)- or (−)-ABA (fig. S1), as shown by our initial 1.8Å resolution structure determined with mixed enantiomorphic (+/−)-ABA (fig. S5). Within the PYR1 cavity, (−)-ABA maintains the tethering hydrogen bonds and tail position of (+)-ABA. To accommodate the changed chirality, the (−)-ABA ring is flipped ~180° from the (+)-ABA ring (fig. S5), swapping the ring pucker and axial methyl substituent to the opposite side of the cavity. The ring-flipped binding of (−)-ABA provides a structural basis for its varying bioactivity (35) in different PYR/PYL/RCAR proteins (6). The ABA hydroxyl group, located on the central chiral carbon (figs. S1 and S3), has no protein hydrogen-bonding partner. Instead, two water molecules bridge the ABA hydroxyl and proximal carboxylate with hydrogen bonds (Fig. 2). Another water molecule links the distal ABA carboxylate oxygen to invariant Tyr120, Ser122 and Glu141 (Fig. 2C). This complex water-bridged hydrogen-bonding network also interconnects ABA through Glu94 to Ser92 and Arg79, and through Glu141 to Asn167. Chemically induced Arabidopsis mutants identified by ABA signaling deficiencies (6) include PYR1 mutants with inward-facing Glu94 and Glu141 mutated to Lys (Fig. 3A). The ABA tail is sandwiched between Ile110 and Val163 (Fig. 2C), and the large internal PYR1 cavity extends beyond this tail. Thus, most structural elements of PYR1 (fig. S2) contribute to ABA-binding cavity formation: helix α3, strands β3–β7, and loops preceding β2 and joining β3–β4 and β5–β6 (Fig. 1A). This architectural design creates the ABA-binding cavity, hydrogen-bonding network, and conserved ordered waters in the absence of ABA, as seen in the ABA-free subunit of the PYR1 dimer (Fig. 2B).
ABA-induced subunit conformational changes
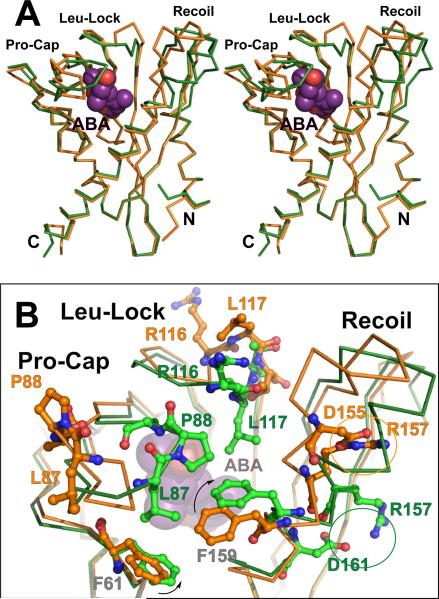
Superposition of ABA-bound and free subunits of the PYR1 dimer (Fig. 4A) revealed significant conformational differences in three loop motifs. Upon ABA binding, Proline Cap (“Pro-Cap”,Val83-Asn90) and Leucine Lock (“Leu-Lock”, Glu114-Thr118) motifs fold over ABA to close the lid on the cavity (Fig. 4B), and the “Recoil” motif (Met147–Phe159) coils into helix α3, allowing lid closure (Fig. 4). Between open-lid and closed-lid conformations, Pro88 cis-to-trans isomerization switches the direction in which flanking Leu87 and Ala89 side chains project (Fig. 4B). A hinge motion of the entire Pro-Cap, pivoting at Ile84 and Asn90, directs Leu87, Pro88 and Ala89 to close over ABA (Fig. 4B). The closed and open Pro-Caps of the two PYR1 subunits directly interact, contributing substantially to the asymmetric dimer interface (Fig. 1, D and E). This interaction suggests why substitution of Pro88 with smaller Ser in the P88S mutant (Fig. 3A) reduces ABA-induced PYR1 interactions with PP2Cs (6).
Fig. 4.
ABA-induced subunit conformational changes. (A) Stereo image showing superposition of ABA-free (orange) and ABA-bound (green) PYR1 Cα traces. ABA-induced helix coiling by the Recoil motif (upper right) is coupled to lid closure over ABA (purple with red oxygen atoms) by the Pro-Cap and Leu-Lock structural motifs (upper left). (B) Enlarged view of ABA-triggered conformational changes in these three structural motifs that close the lid over bound ABA, colored as in (A). ABA (beneath center) triggers rotation of Phe159 (arrow) to coil the Recoil motif into helix α3 (diagonal at right), switching the Arg157 charge-charge interaction (circled) to a new partner within (rather than outside) this helix. Pro88 isomerization from trans (orange, far left) to cis (green, center) converts the open-lid Pro-Cap to the closed-lid conformation, clamping Leu87, Pro88 and Ala89 over ABA. Leu117 (orange, top center) locks down (green, center) against ABA, closing the Leu-Lock, and flipping the preceding Arg116 side chain (orange, top center) toward the opposing subunit (forward and slightly to the right in this view) across the dimer interface (see also Fig. 1E).
Similar to the Pro-Cap, the Leu-Lock motif between Glu114 and Thr118 also undergoes a hinge motion, allowing Leu117 to swing inward to lock against the ABA ring (Fig. 4B). When Leu117 is locked in, the Arg116 side chain projects outward across the dimer interface (Fig. 1, D and E), and the His115 ring flips to block solvent access to the methyl substituents of ABA ring carbon C6′ (fig. S1). In the Recoil motif, Arg157 interacts with Asp155, capping the N-terminal end of helix α3 in the ABA-free conformation; whereas this helix is N-terminally extended in the ABA-binding form, and Arg157 interacts with Glu153 or Asp161 (Fig. 4B). The S152L and R157H mutants of PYR1 (Fig. 3B), like the P88S mutant, reduce ABA-induced PYR1 interactions with PP2C (6). Some PYR/PYL homologs have Cys at the Arg157 position of PYR1 (fig. S2), presumably contributing to structural modulation differently, perhaps by disulfide bond formation with nearby Cys30. Conformational changes in the Pro-Cap, Leu-Lock and Recoil motifs appear concerted (Fig. 4B): pivoting of invariant Phe159 of α3 to pack against ABA likely triggers the Recoil motif to coil back into helix α3, thus allowing lid closure by the Pro-Cap and Leu-Lock motifs (movie S2). Thus direct interactions of ABA with Phe159, Leu117 and Leu87 (Fig. 4B) coordinate interactions of the Recoil, Leu-Lock and Pro-Cap motifs to complete ABA enclosure.
ABA-induced conformational changes in dimer assembly
ABA-induced conformational changes, monitored by SAXS, render the PYR1 dimer more compact, flatter, and less irregular (Fig. 5). The crystallographic asymmetric dimer, containing one ABA-bound and one ABA-free subunit, produces an excellent fit to PYR1 SAXS curves (Fig. 1B), particularly for data collected in the absence of ABA (χ2 = 1.2). When excess ABA was added (~4 ABA molecules per protein subunit) to the PYR1 dimer in solution, the Guinier radius of gyration decreased (23.71±0.04 to 22.72±0.07 Å) and the pair-distance distribution function describing intramolecular distances became narrower and shifted to shorter distances (Fig. 5A), although the maximum intramolecular distance (Dmax = 68Å) remained constant. SAXS data collected in the presence of ABA were not fit as well (χ2 = 3.3 to 3.4) by the crystallographic dimer or a dimer modeled with two ABA-bound subunits.
Fig. 5.
ABA-induced changes in dimer assembly analyzed by SAXS. (A) The pair distance distribution function describing intramolecular distances in the PYR1 dimer in the absence of ABA (orange) becomes narrower and shifts to shorter distances in the presence of saturating ABA (green). (B, C) Two sets of eight independent ab initio bead models for the PYR1 dimer, representing SAXS experimental data in the absence (orange) or presence (green) of saturating ABA. Green brackets mark flatter PYR1 disk in the presence of saturating ABA. Orange arrows indicate greater thickness and asymmetry of PYR1 dimer in absence of ABA. ABA-induced changes in subunit orientation make the PYR1 dimer disk flatter and more compact, as seen from top (B) and side (C) relative to orientation in D. (D) Cartoons (depicting α-helices as cylinders and β-strands as arrows) of the asymmetric crystallographic dimer and a symmetric closed-lid dimer model, aligned by superposition of their common subunit (gray). The ~10° difference in orientation between the second subunits of each dimer (right), highlights the differences between the pseudo 2-fold axis (~170°) relating subunits (gray and orange) of the asymmetric crystallographic dimer and the exact 2-fold axis (vertical line) relating subunits (gray and green) of the symmetric dimer model. (E, F) Independently determined bead models (four sets of colored dots) representing SAXS results in the absence (E) and presence (F) of ABA, each aligned with the corresponding PYR1 structural model (D). The PYR1 dimer assembly shapes determined by SAXS show excellent fits to the crystallographic asymmetric dimer (E) and symmetric dimer model (F). The biconcave, red blood cell shape of the PYR1 dimer is seen by decreased bead density in the center of the PYR1 disks, as well as in cross section (B), particularly with saturating ABA.
Ab initio bead models, derived from solution SAXS experiments for PYR1 in the absence of ABA, are asymmetric and match the crystallographically determined dimer (Fig. 5, B to E). In contrast, ab initio models from SAXS experiments for PYR1 with excess ABA are flatter and more compact (Fig. 5, B to C), consistent with a structural model consisting of two ABA-bound PYR1 subunits related by exact 2-fold symmetry (Fig. 5, D and F). These shapes depict a flattened biconcave disk resembling a red blood cell (Fig. 5, B and F). The 2-fold symmetry axis and perpendicular direction traversing both subunits of the dimer form ~60Å disk diameters, whereas the cross-strand width of the β-sheet roughly aligns with the ~30Å disk thickness. In contrast, the ab initio shapes and crystallographic model representing the PYR1 dimer in the absence of ABA, depict a more irregular biconcave disk (Fig. 5, B to E) resulting from asymmetric interactions between open-lid and closed-lid subunit conformations (Fig. 1E and movie S2) and their ~10° deviation from 2-fold (180°) symmetry (Fig. 5D).
Implications for ABA perception and signaling
The crystal structure, solution SAXS assemblies and co-immunoprecipitation in vivo provide key insights into molecular and structural mechanisms mediating hormone recognition and signaling by the ABA sensor PYR1. ABA binding inside an occluded protein cavity shows PYR1 to be a direct ABA receptor and signal transduction partner, like the gibberellin receptor GID1 (26, 27), rather than one of two hormone-linked coreceptors, like auxin-linked TIR1 and AUX/IAA (24).
The PYR1 dimer crystal structure is unexpectedly asymmetric, revealing structures of both unbound, open-lid and ABA-bound, closed-lid, subunit conformations (Fig. 1A). SAXS analyses of PYR1 without ABA confirm an asymmetric dimer in solution (Fig. 1B). Interestingly, at the crystallographic asymmetric dimer interface, the open and closed Pro-Caps pack tightly with each other (Fig. 1E). This arrangement provides open access for ABA and possible allosteric interchange of open-lid and closed-lid subunits upon hormone binding (movies S1 and S2). Co-immunoprecipitation assays confirm homodimeric PYR1-PYR1 assembly in vivo, both with and without exogenous ABA (Fig. 1C). In solution, SAXS analyses indicate that saturating ABA converts PYR1 into a flatter, more compact dimer, reflecting an orientation change between subunits (Fig. 5). Together, our data support ABA-induced conformational changes producing a 2-fold symmetric closed-lid dimer. Consistent with these results, 2-dimensional nuclear magnetic resonance spectra show a single PYR1 conformer with saturating ABA, but multiple conformers without ABA (6).
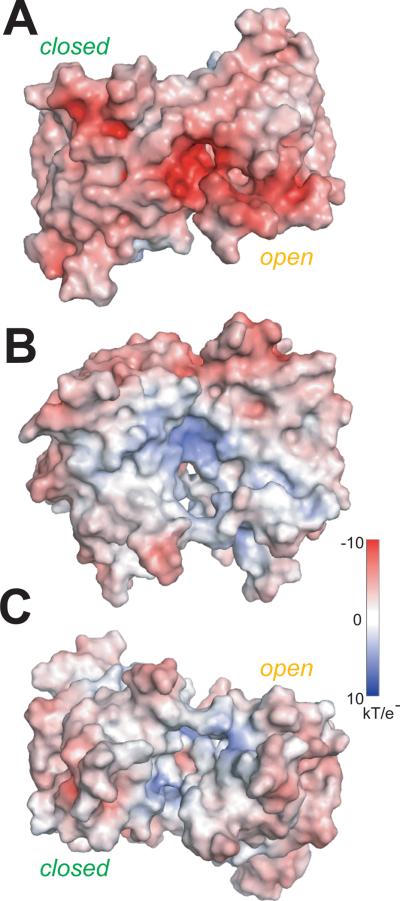
ABA-induced conformational differences in PYR1 subunit structure (Fig. 4) and dimer assembly (Fig. 5) point to a structural mechanism for PYR/PYL/RCAR protein-mediated ABA signal transduction to downstream proteins. The molecular surface of the PYR1 dimer exposes several likely interfaces for ABA-dependent assembly of signalosome complexes with PP2Cs or other potential partners (Fig. 6). Major ABA-induced subunit conformational changes cluster (Fig. 4) around the interacting lids at the “top” of the asymmetric PYR1 dimer (Fig. 1, A and E; Fig. 6A). Thus, ABA-induced binding of PP2Cs may occur at interfaces overlapping the closed lids of PYR1. PP2C binding would then favor PYR1 lid closure and decrease the ABA off rate, thus explaining observations that ABA binds more tightly to RCAR1 and PYL5 in the presence of PP2Cs (5, 23). Surfaces of the PYR1 dimer altered by ABA-induced subunit reorientation (Fig. 5D) also provide promising interfaces for signaling to protein partners (Fig. 6).
Fig. 6.
PYR1 molecular surface color-coded by electrostatic potential. ABA-induced changes in subunit orientation produce conformational changes in the disk-shaped PYR1 dimer at the (A) interacting lids (top), (B) concave sides, aligned as in Fig. 5 (D and E), and (C) the cleft (bottom) between C-terminal helices. Conformational changes of charged residues in the Leu-Lock (Glu114 and Arg116) and Recoil (Glu149, Glu153, Asp154, Asp155 and Arg157) motifs of each subunit reduce electrostatic surface potential upon lid closure (A).
The phytohormone ABA mediates resistance to abiotic stresses including drought, salinity and cold (3, 4). The characterized PYR1 motifs and dimer conformations are key to understanding and future chemical manipulation of phytohormone-induced abiotic stress resistance responses. The crystallographic structure shows that PYR1 is a direct ABA receptor and sensor, signaling hormone binding within an internal cavity through conformational changes impacting the dimer interface and assembly. Furthermore, the mechanistic basis of ABA binding reported here provides a framework for future design of alternate ligands for the large ABA-binding cavity to enable chemical activation of abiotic stress resistance in plants.
Supplementary Material
Acknowledgments
We thank G. Bhabha, J. Christie, M. Hammel, G.L. Hura , D.G. Mendoza-Cozatl, M.E. Pique, D.S. Shin and J.A. Tainer for discussions, plus the SIBYLS beamline staff and Arabidopsis Biological Resource Center. This research was supported by NIH grants GM060396, ES010337 (J.I.S.) and GM37684 (E.D.G.), NSF grants MCB-0918220 (J.I.S.) and IOS0820508 (S.R.C.), DOE grants DE-FG02-03ER15449 (J.I.S.) and DE-AC02-05CH11231 (R.P.R.), and The Skaggs Institute for Chemical Biology (K.H.). J.I.S. and E.D.G. conceived of the project. E.D.G., J.I.S., K.H., N.N., A.S.A. and R.P.R. designed research. N.N., K.H., A.S.A., R.P.R. and C.H. performed experiments. S.R.C. provided reagents and results prior to publication. K.H., N.N., A.S.A., R.P.R., J.I.S., and E.D.G. analyzed data. E.D.G., J.I.S., K.H and N.N. wrote the paper. Crystallographic coordinates and structure factors are deposited in the Protein Data Bank (code 3K3K).
Footnotes
Single Sentence Summary: Signaling by the plant hormone responsible for drought tolerance occurs through binding and conformational changes in its dimeric protein receptor.
References
- 1.Ohkuma K, Lyon JL, Addicott FT, Smith OE. Science. 1963;142:1592. doi: 10.1126/science.142.3599.1592. [DOI] [PubMed] [Google Scholar]
- 2.Eagles CF, Wareing PF. Nature. 1963;199:874. [Google Scholar]
- 3.Finkelstein RR, Gampala SS, Rock CD. Plant Cell. 2002;14:S15. doi: 10.1105/tpc.010441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yamaguchi-Shinozaki K, Shinozaki K. Annu Rev Plant Biol. 2006;57:781. doi: 10.1146/annurev.arplant.57.032905.105444. [DOI] [PubMed] [Google Scholar]
- 5.Ma Y, et al. Science. 2009;324:1064. doi: 10.1126/science.1172408. [DOI] [PubMed] [Google Scholar]
- 6.Park SY, et al. Science. 2009;324:1068. doi: 10.1126/science.1173041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leung J, et al. Science. 1994;264:1448. doi: 10.1126/science.7910981. [DOI] [PubMed] [Google Scholar]
- 8.Meyer K, Leube MP, Grill E. Science. 1994;264:1452. doi: 10.1126/science.8197457. [DOI] [PubMed] [Google Scholar]
- 9.Gosti F, et al. Plant Cell. 1999;11:1897. doi: 10.1105/tpc.11.10.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Merlot S, Gosti F, Guerrier D, Vavasseur A, Giraudat J. Plant J. 2001;25:295. doi: 10.1046/j.1365-313x.2001.00965.x. [DOI] [PubMed] [Google Scholar]
- 11.Saez A, et al. Plant J. 2004;37:354. doi: 10.1046/j.1365-313x.2003.01966.x. [DOI] [PubMed] [Google Scholar]
- 12.Leonhardt N, et al. Plant Cell. 2004;16:596. doi: 10.1105/tpc.019000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Allen GJ, Kuchitsu K, Chu SP, Murata Y, Schroeder JI. Plant Cell. 1999;11:1785. doi: 10.1105/tpc.11.9.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murata Y, Pei ZM, Mori IC, Schroeder J. Plant Cell. 2001;13:2513. doi: 10.1105/tpc.010210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoshida T, et al. Plant Physiol. 2006;140:115. doi: 10.1104/pp.105.070128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuhn JM, Boisson-Dernier A, Dizon MB, Maktabi MH, Schroeder JI. Plant Physiol. 2006;140:127. doi: 10.1104/pp.105.070318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nishimura N, et al. Plant J. 2007;50:935. doi: 10.1111/j.1365-313X.2007.03107.x. [DOI] [PubMed] [Google Scholar]
- 18.Rubio S, et al. Plant Physiol. 2009;150:1345. doi: 10.1104/pp.109.137174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mustilli AC, Merlot S, Vavasseur A, Fenzi F, Giraudat J. Plant Cell. 2002;14:3089. doi: 10.1105/tpc.007906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yoshida R, et al. Plant Cell Physiol. 2002;43:1473. doi: 10.1093/pcp/pcf188. [DOI] [PubMed] [Google Scholar]
- 21.Fujii H, Verslues PE, Zhu JK. Plant Cell. 2007;19:485. doi: 10.1105/tpc.106.048538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fujii H, Zhu JK. Proc Natl Acad Sci U S A. 2009;106:8380. doi: 10.1073/pnas.0903144106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Santiago J, et al. Plant J. in press (available at http://www3.interscience.wiley.com/cgi-bin/fulltext/122514643/HTMLSTART). DOI: 10.1111/j.1365-313X.2009.03981.x.
- 24.Tan X, et al. Nature. 2007;446:640. doi: 10.1038/nature05731. [DOI] [PubMed] [Google Scholar]
- 25.Santner A, Estelle M. Nature. 2009;459:1071. doi: 10.1038/nature08122. [DOI] [PubMed] [Google Scholar]
- 26.Murase K, Hirano Y, Sun TP, Hakoshima T. Nature. 2008;456:459. doi: 10.1038/nature07519. [DOI] [PubMed] [Google Scholar]
- 27.Shimada A, et al. Nature. 2008;456:520. doi: 10.1038/nature07546. [DOI] [PubMed] [Google Scholar]
- 28.Materials and methods are available as supporting material on Science Online.
- 29.Finn RD, et al. Nucleic Acids Res. 2008;36:D281. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iyer LM, Koonin EV, Aravind L. Proteins. 2001;43:134. doi: 10.1002/1097-0134(20010501)43:2<134::aid-prot1025>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 31.Gajhede M, et al. Nat. Struct. Biol. 1996;3:1040. doi: 10.1038/nsb1296-1040. [DOI] [PubMed] [Google Scholar]
- 32.Tsujishita Y, Hurley JH. Nat. Struct. Biol. 2000;7:408. doi: 10.1038/75192. [DOI] [PubMed] [Google Scholar]
- 33.Putnam CD, Hammel M, Hura GL, Tainer JA. Q Rev Biophys. 2007;40:191. doi: 10.1017/S0033583507004635. [DOI] [PubMed] [Google Scholar]
- 34.Hura GL, et al. Nat Methods. 2009;6:606. doi: 10.1038/nmeth.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sondheimer E, Galson EC, Chang YP, Walton DC. Science. 1971;174:829. doi: 10.1126/science.174.4011.829. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.