SUMMARY
Severe congenital neutropenia (SCN) is characterized by a deficiency of mature neutrophils, leading to recurrent bacterial and fungal infections. Although mutations in Elastase-2, neutrophil (ELA2) predominate in human SCN, mutation of Ela2 in mice does not recapitulate SCN. The growth factor independent-1 (GFI1) transcription factor regulates ELA2. Mutations in GFI1 are associated with human SCN, and genetic deletion of Gfi1 results in murine neutropenia. We examined whether human SCN-associated GFI1N382S mutant proteins are causal in SCN and found that GFI1 functions as a rate-limiting granulopoietic molecular switch. The N382S mutation inhibited GFI1 DNA binding and resulted in a dominant-negative block to murine granulopoiesis. Moreover, Gfi1N382S selectively derepressed the monopoietic cytokine CSF1 and its receptor. Gfi1N382S–expressing Csf1−/− cells formed neutrophils. These results reveal a common transcriptional program that underlies both human and murine myelopoiesis, and that is central to the pathogenesis of SCN associated with mutations in GFI1. This shared transcriptional pathway may provide new avenues for understanding SCN caused by mutations in other genes and for clinical intervention into human neutropenias.
INTRODUCTION
Severe congenital neutropenia (SCN) is characterized by maturation arrest at the promyelocyte stage in the bone marrow. SCN patients demonstrate deficient numbers of mature neutrophils and are subsequently prone to recurrent bacterial and fungal infections (Horwitz et al., 2007). Current treatment with recombinant granulocyte-colony stimulating factor (G-CSF) increases the rate of granulopoiesis for most—but not all—patients yet fails to correct an underlying defect in the expression of granule proteins (Donini et al., 2007), resulting in increased numbers of, albeit somewhat functionally defective, neutrophils in the peripheral circulation. SCNpatients also frequently develop myelodysplasia or acute myelogenous leukemia (Rosenberg et al., 2006), and there is concern that G-CSF therapy might accelerate leukemogenesis. Autosomal-dominant mutation of Elastase 2, neutrophil (ELA2) is the most common cause of SCN (Horwitz et al., 2007), but autosomal-dominant mutation of the gene involved in Wiskott-Aldrich syndrome (WAS) (Ancliff et al., 2006) and Growth factor independent-1 (GFI1) (Person et al., 2003) and, recently, recessive mutations in HCLS1-associated protein X-1 (HAX1) (Klein et al., 2007) have also been described. Unfortunately, modeling SCN in mice is difficult because neither the targeting of SCN-associated mutations into murine Ela2 (Grenda et al., 2002) nor complete deletion of Ela2 (Belaaouaj et al., 1998) has an effect on murine hematopoiesis.
Gfi1 is a zinc-finger transcription factor with a SNAG (Snail + Gfi1) amino-terminal transcriptional-repressor domain (Grimes et al., 1996a; Zweidler-Mckay et al., 1996). Gfi1 is normally expressed in hematopoietic stem cells (HSCs) and progenitors and controls cell growth and differentiation (Hock et al., 2003; Karsunky et al., 2002b; Zeng et al., 2004). We previously described a family of SCN patients with GFI1N382S mutations and hypothesized that GFI1N382S functions as a dominant-negative-acting protein (Person et al., 2003). Similar to GFI1N382S humans, Gfi1−/− mice lack mature neutrophils (Hock et al., 2003; Karsunky et al., 2002b). Thus, we reasoned that Gfi1 might offer a unique opportunity to validate a human SCN-associated mutant protein as causal in SCN and to determine the molecular cause of GFI1N382S–induced human disease. Here, we show that wild-type Gfi1 functions as a rate-limiting molecular switch to induce granulopoiesis. In contrast, the expression of Gfi1N382S blocks granulopoiesis in primary murine lineage negative (Lin−) bone-marrow cells through the selective derepression of Gfi1 target genes. A computational model of the Gfi1 zinc fingers involved in DNA binding and biological validation of the model allowed us to predict new Gfi1 amino acid substitutions capable of inducing Gfi1N382S–like phenotypes. Notably, these results demonstrate the limitation of the Gfi1−/− mouse as a potential human SCN model and show that Gfi1N382S selectively functions through the critical deregulation of monopoietic inducers Csf1 and Csf1r.
RESULTS
Murine Gfi1N382S Mutant Blocks Granulopoiesis In Vitro
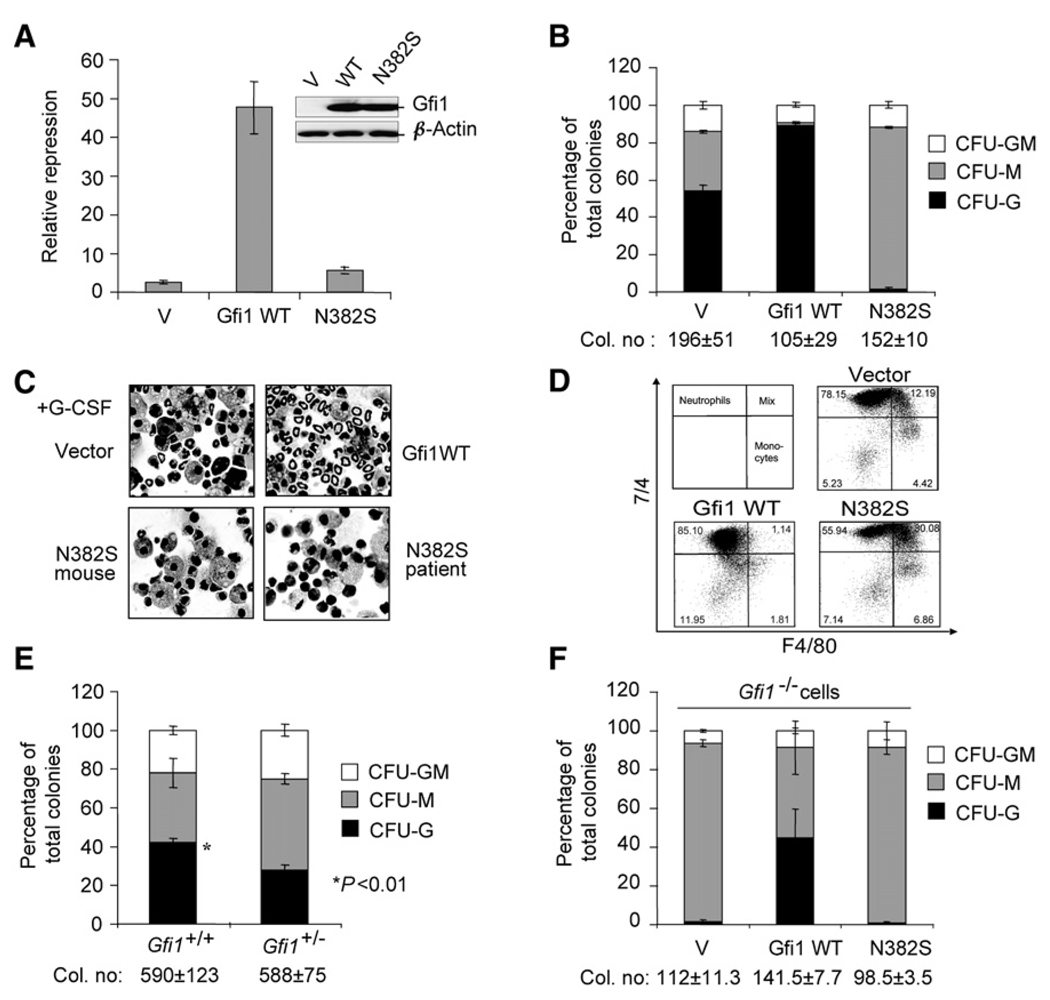
First, the Gfi1N382S mutation was introduced into a murine Gfi1 cDNA, and its ability to repress transcription was tested. A Gfi1-responsive reporter construct (Zweidler-Mckay et al., 1996) was transfected into HEK293 cells along with Gfi1 expression constructs. Both wild-type and Gfi1N382S proteins accumulated similarly in transfected cells (Figure 1A, inset). Similar to our results with human Gfi1N382S (Person et al., 2003), whereas wild-type Gfi1 strongly repressed the reporter, Gfi1N382S was unable to repress the reporter (Figure 1A), Thus, mutation of N382 in both human and murine Gfi1 cDNAs inhibits transcriptional repression on this reporter construct.
Figure 1. Wild-type Gfi1 Induces, whereas N382S Mutant Blocks, Granulopoiesis In Vitro.
(A) Transient transcription assay with reporters, empty MSCV control (V), and MSCV vectors encoding Gfi1 (WT) and Gfi1N382S transfected into 293 cells. Results ± SD from three independent experiments depicted as transcriptional repression (Gfi1-responsive reporter versus control reporter) are shown. The inset shows immunoblot analysis with a Gfi1-peptide-specific rabbit polyclonal anti-sera for detecting expression of MSCV vector-encoded proteins in transfected 293 cells.
(B) Methylcellulose colony formation of Lin− bone-marrow cells transduced with MSCV empty vector (V) or MSCV expressing either Gfi1 WT or Gfi1N382S. Results of CFU assays are displayed as the percentage of CFU-G-, -M, and -GM from three independent wells ± SD with total number of colonies ± SD.
(C) Cytospin from 4 day G-CSF-stimulated liquid cultures of Lin− bone-marrow cells transduced with MSCV empty vector (V) or MSCV expressing either Gfi1 WT or Gfi1N382S. Human GFI1N382S patient sample cultured with human recombinant G-CSF from (Person et al., 2003).
(D) Flow-cytometric analysis of liquid cultures in (C) with 7/4 and F4/80 for denoting neutrophil, mixed, and monocytic phenotypes.
(E) Methylcellulose colony formation of Lin− bone-marrow cells from wild-type and Gfi1+/− litter-mates.
(F) Methylcellulose colony formation of Lin− Gfi1−/− bone-marrow cells transduced with MSCV empty vector (V) or MSCV expressing either Gfi1 WT or Gfi1N382S.
To test the effect of murine Gfi1N382S on granulopoiesis, we transduced murine bone-marrow lineage negative (Lin−) cells with MSCV-puro retroviral vectors (Hawley, 1994) expressing either wild-type Gfi1 or the Gfi1N382S mutant. Transduced cells were assayed for colony formation in methylcellulose with inter-leukin-3 (IL3), interleukin-6 (IL6), stem cell factor (SCF), and puromycin selection. In comparison to cells transduced with the empty retrovirus vector, overexpression of wild-type or mutant Gfi1 sometimes decreased total colony formation; however, this was variable and did not reach statistical significance. Wild-type Gfi1 expressing cells almost exclusively formed granulocytic colonies (Figure 1B). In striking contrast, cells overexpressing Gfi1N382S gave rise to mostly monocytic colonies (Figure 1B). The assay was repeated with sorted common myeloid progenitors (CMPs) and granulocyte monoctye progenitors (GMPs) with basically the same result (Figure S1 available online). To eliminate potential variables introduced by our transduction and selection scheme, we repeated the assay with the CMMP-iresGFP Moloney-based vector (Klein et al., 2000). Murine Lin− cells were transduced with CMMP-iresGFP vectors encoding the wild-type Gfi1 or the Gfi1N382S mutant and expanded for 36 hr; then, GFP+ cells were sorted. Equal numbers of GFP+ cells were assayed for colony formation, and the results were essentially the same as those for MSCV-puro vector transduced cells selected in puromycin (data not shown). Thus, it is unlikely that differences in colony formation or number are due to unique properties of the MSCV vector or individual viral stocks. We conclude that Gfi1 appears to induce, whereas Gfi1N382S inhibits, granulocytic-colony formation.
The forced expression of Gfi1 or the Gfi1N382S mutant dramatically alters colony formation. However, it is also possible that the combination of IL3, IL6, and SCF are insufficient to facilitate granulocytic differentiation of Gfi1N382S–expressing cells. To distinguish between these possibilities, we assembled liquid cultures with an instructive cytokine signal (G-CSF) and analyzed their morphology and cell-surface markers. Murine bone-marrow Lin− cells overexpressing wild-type Gfi1 or Gfi1N382S were plated in liquid cultures and stimulated with G-CSF for 4 days. As shown in Figure 1C, vector-transduced cells differentiated mainly to the stage of myelocytes, metamyelocytes, and a few band neutrophils. In contrast, wild-type Gfi1 overexpressing cells mostly displayed morphological features of band neutrophils, whereas Gfi1N382S–expressing cells were blocked in differentiation at an unknown stage. Interestingly, these G-CSF-stimulated murine cells overexpressing a murine Gfi1N382S were morphologically similar to human GFI1N382S mutant patient stem cells similarly cultured (compare lower- right and left panels, Figure 1C). In sum, G-CSF signaling was insufficient to override Gfi1N382S–blocked granulopoiesis.
Gfi1 Resolves the Cell Fate of Early Myeloid Precursors
Flow-cytometric analysis of cells from G-CSF-stimulated liquid cultures stained with neutrophilic (7/4) (Hirsch and Gordon, 1983) and monocytic (F4/80) (Hume and Gordon, 1983) markers delineates granulocytes, monocytes, and cells with a mixed myeloid phenotype (Figure 1D), which we confirmed with sorted and stained cytospins (data not shown). In comparison to vector-transduced cells, cells overexpressing wild-type Gfi1 demonstrated a shift toward a granulocytic phenotype (compare vector 78% to Gfi1 WT 85%) and a dramatic reduction in undifferentiated cells (compare vector 12% to Gfi1 WT 1%) (Figure 1D). Surprisingly, with these markers, Gfi1N382S–overexpressing cells showed phenotypic granulocytes (Figure 1D), even though such cells did not have the cellular morphology of neutrophils (Figure 1C). However, the number of such phenotypic granulocytes was reduced (compare vector 78% to N382S 56%), whereas the number of mixed-lineage phenotype cells considerably increased (compare vector 12% to N382S 30%) (Figure 1D). Taken together, these assays suggest that overexpression of wild-type Gfi1 induces granulopoiesis and resolves a mixed lineage fate, whereas overexpression of the Gfi1N382S mutant blocks granulopoiesis at an intermediate stage and/or induces a phenotype in which differentiation-associated surface markers and nuclear morphology are disconnected.
Granulopoiesis Is Gfi1-Dose Dependent
Retroviral-vector-mediated forced expression of Gfi1 induced more granulocytes ex vivo than vector-control-transduced cells (Figure 1B–1D). To determine whether manipulation of endogenous Gfi1 gene dosage also affects cell differentiation, we compared Gfi1+/+ and Gfi1+/− littermate Lin− marrow stem and progenitors in colony-formation assays. Strikingly, the number of granulocytic colonies generated by Gfi1+/− cells was significantly less than that from Gfi1+/+ cells (p < 0.01) (Figure 1E). Thus, Gfi1 is rate limiting for granulopoiesis.
Gfi1-Induced Granulopoiesis Requires Sequence-Specific DNA Binding
Germline deletion of Gfi1 blocks the formation of mature neutrophils (Hock et al., 2003; Karsunky et al., 2002b). Gfi1−/− Lin− cells formed mainly monocytic colonies in vitro (Figure 1F). We transduced Gfi1−/− Lin− cells with wild-type Gfi1-expressing retroviruses and analyzed them for colony formation. As previously reported for sorted granulocyte-monoctye progenitors (Hock et al., 2003), forced expression of Gfi1 in Gfi1−/− Lin− cells restores granulopoiesis (Figure 1F). Interestingly, forced Gfi1 expression in Gfi1−/− cells did not occlude monopoiesis as it did in Gfi1+/+ cells (Figure 1B). It is formally possible that not all Gfi1−/− Lin− cells are capable of responding to Gfi1. However, given the dose-dependent granulopoiesis demonstrated for Gfi1+/− cells, it is likely that MSCV retroviral-vector-mediated expression of Gfi1 is consistent with a dose-dependent effect.
The Gfi1N382S mutant fails to bind a consensus Gfi1 DNA-binding site (see Figure 3B) (Person et al., 2003). We hypothesize that Gfi1N382S acts as a dominant negative by sequestering limiting cofactors that are required for wild-type Gfi1 to function when bound to DNA. However, it is alternatively possible that the Gfi1N382S mutant is a gain-of-function mutant related to physical interaction between Gfi1 and the Protein inhibitor of activated STAT 3 (Pias3) SUMO-E3 ligase or the U2AF26 splicing-factor proteins (Heyd et al., 2006; Rodel et al., 2000). To determine whether Gfi1N382S retains any wild-type Gfi1 activities relevant to myelopoiesis, we assayed Gfi1N382S activity in the absence of endogenous Gfi1. In contrast to forced expression of Gfi1, colony formation of Gfi1N382S–expressing cells was virtually indistinguishable from that of vector-transduced cells (Figure 1F), in that Gfi1N382S was similarly unable to rescue Gfi1−/− granulopoiesis. Importantly, the ability of Gfi1 to modulate granulopoiesis appears to be critically dependent upon sequence-specific DNA binding.
Figure 3. Gfi1 DNA Binding Is Required for G-CSF Induced Granulopoiesis.
(A) Amino acid sequence depicting Gfi1 zinc finger 3, 4, and 5. Amino acids predicted (by the computational model in Figure 2) to interact with DNA are in bold and large font.
(B) Alanine-substitution mutants or a luciferase control (Luc.) were in vitro transcribed and translated (IVT, bottom), then incubated with a radiolabeled “R21” oligonucleotide. EMSA (middle panel) revealed shifted oligonucleotides (arrow). Panels are derived from an autoradiogram of a single gel that has been divided for clarity of presentation.
(C) Methylcellulose colony formation of Lin− bone-marrow cells transduced with MSCV empty vector (V) or MSCV expressing Gfi1 WT or alanine substitution mutants. Results of CFU assays are displayed as the percentage of CFU-G-, -M, and -GM from three independent wells ± SD with total number of colonies ± SD.
(D) Cytospin and 7/4 versus F4/80 flow-cytometric analyses of 4 day G-CSF-stimulated liquid cultures of Lin− bone-marrow cells transduced with MSCV empty vector (V), Gfi1 WT, and Gfi1 alanine-substitution mutants.
A Computational Model of DNA-Bound Gfi1 Zinc Fingers 3, 4, and 5
The Gfi1N382S mutation in zinc finger 5 ablates sequence-specific DNA binding and has been identified in three related patients with SCN (Person et al., 2003). Zinc fingers 3, 4, and 5 are absolutely required for sequence-specific DNA binding (Zweidler-Mckay et al., 1996); however, other than N382S, neutropenic individuals with mutations in these zinc fingers have not been identified. We were interested in determining whether single amino acids (other than N382) in zinc fingers 3, 4, and 5 are critically required for DNA binding and their subsequent functional role in regulating granulopoiesis. To this end, we utilized the established crystal structure of a synthetic Cys-His zinc finger protein (1Mey) (Kim and Berg, 1996) and the Modeler, ClustalW, and molecular dynamics (Amber) computer programs to generate an optimized computational homology model of zinc fingers 3, 4, and 5 bound to a synthetic “R21” high-affinity Gfi1 binding site (Figure 2A). From the crystal structure of Cys-His zinc fingers, it is evident that amino acids at −1,2,3, and 6 along the alpha helix of the zinc-finger structure were frequently found to interact with DNA (Wolfe et al., 2001) (Figure 2B, amino acids depicted in bold). Indeed, the homology model predicted interaction between DNA and amino acids in the alpha helix at these positions, in addition to others (Figure 2B). The homology model predicts that amino acids in zinc finger 3 interact with bases outside the Gfi1-consensus core binding motif “AATC,” whereas those in zinc fingers 4 and 5 interact with both the AATC core and nucleotides in close proximity (Figure 2B, arrows). Interestingly, N382 in zinc finger 5 specifically interacted with two neighboring alanines in and outside the binding core (Figure 2C); however, when DNA docking of the SCN-associated Gfi1N382S mutant was forced in the model, only a nonspecific interaction with the backbone was predicted (Figure 2D). Thus, the homology model predicts how the N382S mutation affects DNA binding.
Figure 2. Predicted Gfi1-DNA Interactions.
(A) Computational model of Gfi1 zinc fingers3 (contact residues in red), 4 (contact residues in blue), and 5 (contact residues in orange) interacting with the “R21” synthetic high-affinity DNA-binding site (violet).
(B) Graphic representation of interactions between amino acids and nucleotides predicted by the model in (A). The line over DNA sequence indicates the AATC core of the Gfi1 binding site. Predicted contact nucleotides are shown in blue. Zinc finger amino acids that normally interact with DNA are in bold.
(C) Interaction of Gfi1N382 with DNA. Numbers indicate distances in angstroms.
(D) Interaction of Gfi1N382S with DNA. Numbers indicate distances in angstroms.
Single Amino Acids in Gfi1 Zinc Fingers 4 and 5 Are Critical for DNA Binding
It is possible that N382 interactions with DNA are more important than other amino acids for sequence-specific DNA binding. To investigate this possibility and to validate the virtual model, we first biochemically tested the importance of predicted DNA binding amino acids (Figure 3A, bold). First, each amino acid predicted to interact with DNA was mutated to alanine, and then the mutated Gfi1 cDNAs were in vitro transcribed and translated (Figure 3B, bottom panel, “IVT”). The resulting proteins were analyzed by electrophoretic mobility-shift assay (EMSA) with radiolabeled oligonucleotides encoding a concensus Gfi1 binding site (“R21”). Proteins with zinc finger 3 mutations, zinc finger 4 Gfi1K356A, and zinc finger 5 Gfi1S381A reproducibly demonstrated slightly reduced binding compared to wild-type Gfi1 (Figure 3B). On the other hand, both zinc finger 4 Gfi1D354A and Gfi1K357A mutants and zinc finger 5 Gfi1Q379A and Gfi1S380A mutants reproducibly demonstrated severely reduced binding (Figure 3B, middle panel). As expected, the Gfi1N382A and Gfi1N382S mutants failed to bind DNA. We conclude that Gfi1 amino acids D354, K357, Q379, S380, and N382 are critically required for sequence-specific DNA binding to the R21 consensus oligonucleotide.
We next determined whether N382 is uniquely required for granulopoiesis. The alanine-substitution mutants were retrovirally expressed in Lin− bone-marrow cells and subjected to colony assay in methylcellulose, G-CSF-stimulated liquid culture, and flow-cytometric analysis. As before, expression of wild-type Gfi1 excluded monocytic-colony development (Figure 3C). The zinc finger 3 mutants that demonstrated consistent but modest reduction in DNA binding capacity in EMSA (Figure 3B) were equivalently impaired in occluding monocytic colonies compared to wild-type Gfi1 (Figure 3C). Independent of measurable DNA binding to the R21 oligonucleotide, expression of zinc finger 4 and 5 mutants dramatically lowered CFU-G formation (Figure 3C). However, in G-CSF-stimulated liquid cultures, zinc finger 4 and 5 mutants that lack DNA binding directly correlated with defective neutrophilic differentiation. Specifically, G-CSF-stimulated cells expressing Gfi1D354A, Gfi1K357A, Gfi1Q379A, Gfi1S380A, and Gfi1N382A showed a dramatic decrease in band neutrophils (Figure 3D, cytospins). Gfi1N382S–expressing cells display more 7/4hi F4/80hi mixed neutrophilic-monocytic characteristics (Figure 1D). Similarly, Gfi1Q379A–expressing cells display an increase in this 7/4hi F4/80hi mixed population (Figure 3D). Thus, whereas Gfi1 D354A, K357A, Q379A, and S380A mutations substantially interfered with DNA binding to the R21 oligonucleotide (Figure 3B) and altered CFU-G formation, mutation of N382 seems to play a unique role in Gfi1 DNA binding and control of myelopoiesis, with the Gfi1Q379A mutant most similarly to N382A. Interestingly, N382 and Q379 specify neighboring core nucleotides in the Gfi1 DNA binding site (Figure 2B). Clearly, both Gfi1N382A- and Gfi1N382S–expressing cells display the most dramatic expansion of an atypical mixed 7/4hi F4/80hi population combined with a profound lack of band neutrophils in G-CSF-stimulated cultures.
Gfi1N382S Impairs Granulopoiesis by Restricting SNAG-Dependent Cofactors
Mutation of the first proline in the Gfi1 SNAG transcriptional repression domain (P2A) disrupts Gfi1-repression activity without altering subcellular localization, protein accumulation, or DNA binding (Grimes et al., 1996a). We reasoned that if the Gfi1N382S mutant functions by sequestering corepressors necessary for wild-type Gfi1 function, then a repressor-defective mutant such as Gfi1P2A might reproduce Gfi1N382S effects by excluding wild-type Gfi1 from DNA binding sites. To test this hypothesis, we introduced the P2A mutation in a murine Gfi1 cDNA and tested its ability to regulate granulopoiesis. First, we transduced Gfi1−/− Lin− cells with wild-type Gfi1- and Gfi1P2A–expressing retroviruses and analyzed them for colony formation. Whereas forced expression of Gfi1 in Gfi1−/− Lin− cells restored granulopoiesis (Figure 1F), colony formation of Gfi1P2A–expressing cells was virtually indistinguishable from that of vector-transduced cells (Figure 4A). Thus, it is unlikely that Gfi1P2A retains any ability to rescue Gfi1−/− granulopoiesis. The ability of Gfi1 to modulate granulopoiesis appears to critically depend upon an intact SNAG transcriptional-repressor domain.
Figure 4. N382S Functions as a SNAG-Domain-Dependent Dominant Negative.
(A) Methylcellulose colony formation of Gfi1−/− Lin− bone-marrow cells transduced with MSCV empty vector (V) or MSCV encoding Gfi1P2A (P2A). Results of CFU assays are displayed as the percentage of CFU-G-, -M, and -GM from three independent wells ± SD with total number of colonies ± SD.
(B) Methylcellulose colony formation of wild-type Lin− bone-marrow cells transduced with MSCV empty vector (V) or MSCV encoding Gfi1P2A or Gfi1P2A+N382S; *p < 0.01.
(C and D) Cytospin (C) and 7/4 (D) versus F4/80 flow-cytometric analysis of 4 day G-CSF-stimulated liquid cultures of Lin− bone-marrow cells transduced with MSCV empty vector (V) or MSCV encoding Gfi1P2A or Gfi1P2A+N382S.
(E) Wild-type Lin− bone-marrow cells transduced with CMMP empty vector (V) or CMMP encoding Gfi1 WT or Gfi1N382S were stimulated with G-CSF and then analyzed by flow cytometry for GFP and then for Annexin V versus 7AAD. The average percentage of GFP+ early apoptotic (Annexin V+ 7AAD−) cells from three independent experiments ± SD is shown.
Next, we assayed colony formation in wild-type Lin− cells. Similar to Gfi1N382S, Gfi1P2A–expressing cells gave rise to mainly monocytic colonies (Figure 4B), and G-CSF-stimulated liquid culture of Gfi1P2A–overexpressing cells resulted in a predominance of monocytic cells and a few immature neutrophilic cells (Figure 4C). Moveover, Gfi1P2A–expressing G-CSF-stimulated cultures showed a dramatic increase in mixed 7/4hi F4/80hi cells (Figure 4D). In a manner strikingly similar to Gfi1N382S–expressing cells, Gfi1P2A–overexpressing Lin− bone-marrow cells are blocked in granulopoiesis.
It is possible that both Gfi1N382S and Gfi1P2A act as dominant-negative mutants that interfere with endogenous Gfi1-directed granulopoiesis. We reasoned that if the dominant-negative activity of Gfi1P2A relies on DNA binding, whereas that of Gfi1N382S depends upon an intact repression domain, then a double mutant should lack dominant-negative activity. To test this hypothesis, we constructed a Gfi1P2A+N382S mutant and tested its ability to regulate granulopoiesis. In agreement with our hypothesis, Gfi1P2A+N382S–expressing cells gave rise to both monocytic and granulocytic colonies similarly to vector-transduced cells (Figure 4B). Notably, the number of purely granulocytic colonies was lower in the Gfi1P2A+N382S–expressing colonies than the empty-vector-transduced cells (p < 0.01), and the number of Gfi1P2A+N382S–expressing 7/4hi F4/80hi cells was variable. It is unlikely that the double mutant is completely inactive; however, from both colony data and morphology, Gfi1-P2A+N382S neutrophils were clearly formed (Figure 4C). Thus, Gfi1P2A and Gfi1N382S act through a dominant-negative mechanism to block granulopoiesis, and granulocytic-differentiation defects in Gfi1N382S–overexpressing cells can be rescued by simultaneous mutation of the SNAG domain. We conclude that Gfi1N382S most probably restricts granulopoiesis by interfering with wild-type Gfi1 function.
Gfi1 Instructs Granulopoiesis
The data in sum do not delineate whether Gfi1 functions as an instructive or a selective agent. Specifically, Gfi1 may function as a molecular switch to induce granulopoiesis, or Gfi1-overexpressing progenitors might only survive if they adopt a granulocytic fate. To determine whether Gfi1 switches cellular fate of progenitors or induces death of monocytic-determined progenitors, we monitored cell death daily in G-CSF-stimulated cultures by Annexin V staining. In comparison to empty-vector-transduced cells, Gfi1-expressing cultures demonstrated less apoptosis, whereas Gfi1N382S–expressing cells showed a modest increase in cell death in the third and fourth days of culture (Figure 4E). Thus, cell death of monocytic lineage cells is unlikely to explain Gfi1 effects on granulopoiesis. In agreement with this concept, the forced coexpression of the antiapoptotic proteins Bcl2 or Mcl1 did not reveal substantial differences in Gfi1 or Gfi1N382S colony formation (Figure S2). We conclude that Gfi1 is instructive for granulocytic differentiation.
Gfi1N382S Blocks Granulopoiesis through Csf1
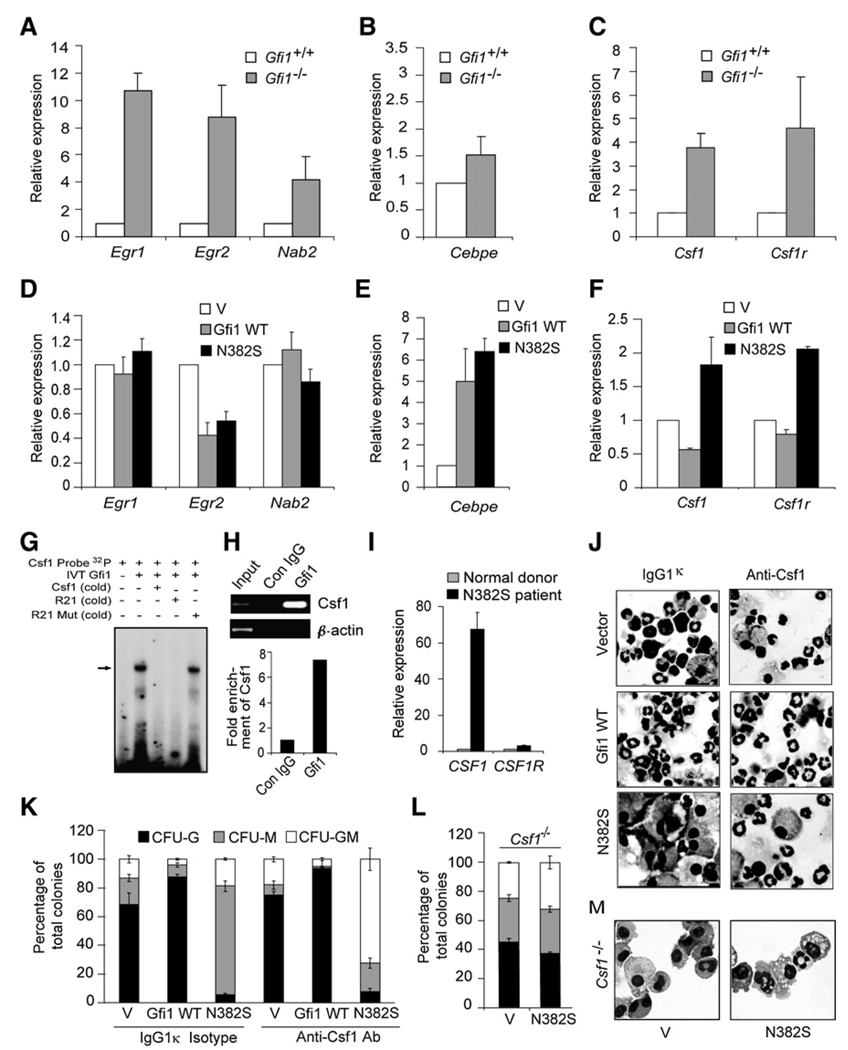
Because Gfi1-instructed granulopoiesis appears to require DNA binding and transcriptional repression, we wished to delineate relevant Gfi1 target gene(s). To this end, we analyzed RNA from lineage-negative bone-marrow cells for transcripts whose steady-state expression is increased in Gfi1−/− versus wild-type littermates. Recently, Laslo et al. demonstrated that Gfi1 may regulate the Egr1, Egr2, and Nab2 genes to resolve myeloid lineage-fate decisions (Laslo et al., 2006). In addition, Zhuang et al. have shown that in the 32D cell line, Gfi1N382S deregulates CCAAT/enhancer binding protein, epsilon (Cebpe), expression and induces apoptosis in response to G-CSF stimulation, subsequently blocking granulocyte formation (Zhuang et al., 2006). Previous reports noted deregulated Csf1r expression in Gfi1−/− myeloid cells (Hock et al., 2003) and suggested that this may be due to direct interaction between Gfi1 and Pu.1 on the Csf1r promoter (Dahl et al., 2007). Thus, we interrogated the gene expression profiles of Egr1, Egr2, Nab2, Cebpe, and Csf1r. Because deregulated expression of Csf1r would not be functionally relevant to in vitro cultures without its ligand, we also examined Csf1 expression. In comparison to wild-type lit-termate Lin− bone-marrow cells, we found that Egr1, Egr2, Nab2 (Figure 5A), Csf1, and Csf1r (Figure 5C) transcripts were indeed higher in Gfi1−/− Lin− cells. However, the expression of Cebpe was only mildly elevated in Lin− cells from Gfi1−/− bone marrow (Figure 5B). Thus, it is possible that Gfi1 regulation of Egr1, Egr2, Nab2, Csf1, and Csf1r might explain the Gfi1N382S phenotype.
Figure 5. Gfi1N382S Suppresses Granulopoiesis by Deregulating Csf1 and Csf1r.
(A–C) Taqman analyses of steady-state gene expression in RNA from wild-type (WT) or Gfi1−/− (KO) littermate Lin− bone-marrow cells (which were not cultured ex vivo). Taqman values were normalized to Gapdh, and the average fold expression from at least three independent samples + SD is shown.
(D–F) Taqman analyses of steady-state gene expression in Lin− cytokine-expanded bone-marrow cells transduced with MSCV empty vector (V) or MSCV encoding Gfi1 or Gfi1N382S.
(G) EMSA analyses of in vitro-transcribed and -translated Gfi1 binding to Csf1 promotor sequences.
(H) Fold enrichment of Csf1 promoter sequences in chromatin-immunoprecipitation templates with a Gfi1-specific monoclonal antibody versus control mouse IgG from quantitative PCR amplification. The inset shows the ethidium-bromide-stained products of the amplification reactions for specific (Csf1) versus nonspecific β-actin) targets.
(I) Taqman analyses of human CD34+ bone-marrow cells from three normal donors (average normalized to one) versus a Gfi1N382S mutant patient.
(J and K) Methylcellulose colony formation with cytospin from colonies of Lin− bone-marrow cells transduced with MSCV empty vector (V) or MSCV encoding Gfi1 or Gfi1N382S and then incubated with a Csf1-inactivating monoclonal antibody or an isotope-matched control IgG1κ. Results of CFU assays are displayed as the percentage of CFU-G-, -M, and -GM from three independent wells ± SD.
(L and M) Methylcellulose colony formation with cytospin from colonies as in (J) and (K) with Csf1−/−(op/op) bone-marrow cells.
Given the hypothesis that Gfi1N382S acts as a dominant negative, putative target genes responsible for the Gfi1N382S phenotype would be anticipated to be bound and repressed by Gfi1 overexpression but activated by Gfi1N382S expression. However, neither Egr1 nor Nab2 were repressed by Gfi1, and although Egr2 transcripts were decreased in Gfi1-overexpressing cells, they were also decreased in Gfi1N382S–expressing cells (Figure 5D). Moreover, Cebpe was consistently upregulated by expression of either Gfi1 or Gfi1N382S (Figure 5E). In contrast, the accumulation of both Csf1 and Csf1r transcripts was decreased in bone-marrow cells overexpressing Gfi1 and elevated in those expressing Gfi1N382S (Figure 5F). Thus, although Gfi1 may interact with Egr1, Egr2, Nab2, and Cebpe to regulate aspects of myelopoiesis, only the expression patterns of Csf1 and Csf1r are in agreement with the hypothesized dominant-negative function of Gfi1N382S. To determine whether Gfi1 binds to Csf1, we utilized a computer program (Jegga et al., 2007) to reveal a conserved Gfi1 binding site (5′- GCCAGGGTGATTTCC-3′) at −57 to −72 from the transcription start site of the Csf1 promoter. EMSA revealed that in vitro-transcribed and -translated Gfi1 bound specifically to an oligonucleotide encoding the putative Gfi1 binding site in the Csf1 promoter (Figure 5G). Chromatin-immunoprecipitation assays in U937 human myelomonocytic cells revealed that endogenous Gfi1 was consistently and specifically associated with the Csf1 promoter sequences, but not a β-actin control (Figure 5H). Finally, quantitative real-time PCR analyses of RNA from CD34+ cells obtained from normal or Gfi1N382S patients revealed a dramatic CSF1 -transcript induction, which dwarfed a modest induction of CSF1R (Figure 5I). Thus, Gfi1 directly targets the Csf1 promoter for transcriptional repression in that Csf1 was repressed by Gfi1 overexpression, deregulated in Gfi1−/− and Gfi1N382S–expressing human and murine cells, and directly bound by Gfi1 in EMSA and ChIP.
Next, we directly tested the relevance of Csf1 and Csf1r deregulation to the Gfi1N382S phenotype. We transduced Lin− bone-marrow cells with retroviral vectors expressing Gfi1 or Gfi1N382S and performed colony assays with either a monoclonal antibody that binds and inactivates Csf1 or an IgG1κ isotype-matched control. The Csf1-inactivating antibody dramatically increased the number of Gfi1N382S mixed colonies at the expense of solely monocytic colonies (Figure 5K). Importantly, the morphology of many of the cells within these mixed colonies was clearly that of neutrophils (Figure 5J). Interestingly, the gene-expression profile of Egr1, Egr2, and Nab2 was similar, whereas the expression of MPO and NE were modestly increased, in Csf1-antibody-treated Gfi1N382S–expressing- versus vector-control-transduced cells (data not shown). Likewise, genetic ablation of Csf1 blocked the effects of Gfi1N382S expression in that colony formation was not different from wild-type littermate bone marrow (Figure 5L) and that neutrophil morphology was visible (Figure 5M). As expected from published reports (Wiktor-Jedrzejczak et al., 1990; Yoshida et al., 1990), abnormal vesicles are seen in Csf1−/− cells independent of Gfi1N382S expression (Figure 5M). With the same conditions for Lin− cell culture, the addition of recombinant murine Csf1 induced the dose-dependent conversion of colonies to a monocytic phenotype (Figure S3). Thus, restricting the biological activity of the Csf1 generated by Gfi1N382S–expressing cells permitted these cells to form granulocytes.
DISCUSSION
Patients with severe congenital neutropenia (SCN) display mutations in ELA2, GFI1,WAS, and HAX1 (Devriendt et al., 2001; Klein et al., 2007; Person et al., 2003), but mutations in ELA2 predominate (Horwitz et al., 2007). Modeling mutant Ela2 in primary murine cells has failed to mimic SCN (Grenda et al., 2002), and Ela2−/− mice have mature neutrophils (Belaaouaj et al., 1998). In contrast, our data clearly show that expression of murine Gfi1N382S in otherwise normal murine Lin− stem and progenitor cells blocks granulopoiesis in a manner strikingly similar to Gfi1N382S patient samples processed in a comparable manner. Although Gfi1−/− mice and Gfi1N382S patients have a similar block to granulopoiesis (Hock et al., 2003; Karsunky et al., 2002b; Person et al., 2003), our data reveal selective deregulation of putative Gfi1 targets by Gfi1N382S. Thus, deletion of murine Gfi1 engenders severe transcriptional changes that are apparently not relevant to Gfi1N382S–mediated human disease. Similarly, expression of Gfi1N382S in the 32D cell line results in profound G-CSF-induced cell death instead of differentiation (Zhuang et al., 2006), which did not occur in primary Lin− cells. Neither the 32D cell line nor Gfi1−/− mice represent a self-sufficient model of Gfi1 mutant SCN.
Of the putative Gfi1 target genes we examined, only Csf1 and Csf1r were deregulated in Gfi1−/− bone-marrow cells, repressed by Gfi1, and derepressed by Gfi1N382S. It is currently unclear why Gfi1N382S regulates Csf1 differently than Cebpe, Egr1, Egr2, and Nab2. However, we have shown that Gfi1 binds to the Csf1 promoter in living U937 cells. In SCN patients and murine Lin− cells, Gfi1N382S may require an intact SNAG repressor domain to compete for corepressors utilized by Csf1 -promoter-bound wild-type Gfi1. Regulation of Csf1 and its receptor are obviously critical for Gfi1N382S action because antibody absorption of Csf1 in culture or genetic ablation of Csf1 released the Gfi1N382S block to granulopoiesis. Although inactivation of Csf1 did not perfectly restore myelopoiesis, the granulocytes formed from Gfi1N382S–expressing cells without Csf1 support the hypothesized dominant-negative activity of Gfi1N382S. Clearly, Gfi1 regulates multiple targets and the expression of Gfi1N382S may be used to selectively identify target genes that are critical to granulopoiesis and to the pathogenesis of SCN.
Previously, mutations found in humans with SCN, introduced into the corresponding murine cDNA, and expressed in primary murine cells failed to block the production of mature murine granulocytes. Given our results, it is perhaps not surprising that Gfi1N382S, which engenders the most severe mutant phenotype we observed, was identified in the patient population. Other zinc-finger amino acid substitutions, although not as potent as Gfi1N382S, are clearly capable of interfering with granulopoiesis. Although neutrophil elastase mutants do not successfully model SCN in mice, and thus highlight differences in myelopoiesis between rodents and humans, expression of GFI1N382S (and other Gfi1 mutants) in primary Lin− bone-marrow cells illustrates a central transcriptional program that controls neutrophil development and that is relevant to the pathogenesis of SCN.
Although Gfi1 may have an array of transcriptional targets in different cell types, the subset of these genes relevant to Gfi1N382S–induced SCN may be restricted to monopoietic genes in progenitors. That Gfi1 influences granulopoiesis by restricting the expression of monopoietic genes in neutrophils is currently a matter of debate. Several studies (Dahl et al., 2007; Hock et al., 2003; Laslo et al., 2006) have suggested that monocytic genes are repressed by Gfi1, but recent work on Gfi1-protein degradation suggested that Gfi1 only accumulates in monocytic cells. Gfi1 may thus function to repress granulocytic genes in monocytes (Marteijn et al., 2006). Although this possibility has not been excluded, neutrophils from Gfi1+/− animals have deregulated transcription of secondary granule-protein-encoding genes (Khanna-Gupta et al., 2007), suggesting that there is sufficient Gfi1 protein in neutrophils to regulate granulocytic genes. Our own data show that Gfi1 instructs granulopoiesis and that Gfi1N382S critically deregulates the expression of Csf1 to induce monopoiesis.
Gfi1 is rate limiting for granulopoiesis. Our data demonstrate that Lin− bone-marrow cells from Gfi1+/− mice generated substantially fewer granulocytic colonies than Gfi1+/+ littermates. Thus, physiological manipulation of Gfi1 dosage affects lineage fate. In addition, retroviral-vector-mediated expression of Gfi1 rescued a normal ratio of granulopoietic and monopoietic colonies from Gfi1−/− Lin− bone-marrow cells, whereas it ablated monocytic-colony formation in Gfi1+/+ Lin− bone-marrow cells. The amount of Gfi1 achieved in our experiments (and not deregulated expression) appears to influence lineage choice.
Both DNA-binding and transcription-repression functions appear critical for Gfi1 instruction of granulopoiesis. Gfi1 can apparently function as a molecular sink for the Pias3 SUMO E3 ubiquitin ligase and the U2AF26 splicing factor (Heyd et al., 2006; Rodel et al., 2000). Notably, these functions do not require DNA binding or an intact SNAG domain. If the biologically available amount of Pias3 or U2AF26 were the critical regulator of lineage fate, then either Gfi1N382S or Gfi1P2A should have bound these proteins and rescued Gfi1−/− granulopoiesis, but they did not. In agreement with these data, Fiolka recently showed that mice with homozygous Gfi1P2A knockin alleles have Gfi1−/− phenotypes (Fiolka et al., 2006). On the other hand, we note that although granulopoiesis was restored in Gfi1P2A+N382S–expressing cells, the number of granulocytic colonies is not completely normal. Because neither Gfi1P2A nor Gfi1N382S mutations would be expected to disrupt Pias3 or U2AF26 interactions, the residual activity of the Gfi1P2A+N382S mutant might be explained by Gfi1 binding to these proteins. In contrast, Gfi1 antagonism of Pu.1 required the SNAG domain, but not DNA binding (Dahl et al., 2007). We observed a lack of granulopoiesis in both Gfi1N382S- and Gfi1P2A–expressing Gfi1−/− Lin− bone-marrow cells. If the major defect in Gfi1−/− Lin− cells were overactive Pu.1, then the Gfi1N382S but not the Gfi1P2A mutant should have been capable of some rescue, but neither mutant rescued Gfi1−/− granulopoiesis. These data emphasize the requirement for both Gfi1 DNA binding and transcription-repression functions to instruct granulopoiesis.
Our data support the hypothesis that a greater amount of Gfi1 resolves the fate of mixed-lineage cells (Laslo et al., 2006). It is alternatively possible that Gfi1 functions through cell-cycle or cell-death pathways to limit monocytic-colony formation. However, cell-cycle analysis of Gfi1- or Gfi1N382S–expressing cytokine-stimulated Lin− bone-marrow cells did not reveal substantial differences (data not shown). Moreover, Gfi1 was antiapoptotic in cytokine-stimulated bone-marrow progenitors; as already established in T cells (Grimes et al., 1996b; Karsunky et al., 2002a). Although Gfi1 was recently shown to block Pu.1-induced macrophage development in a cell line (Dahl et al., 2007), our work demonstrates that forced expression of Gfi1 instructs granulopoiesis in a DNA-binding and SNAG-domain-dependent manner not explained by Pu.1 interactions. Instead, primary wild-type Lin− cells yielded almost exclusively granulocytic colonies with dramatic diminution of monocytic colonies. When the same cells were stimulated with G-CSF in liquid culture, Gfi1-expressing cells had a uniform neutrophilic surface phenotype (7/4+ F4/80−) and were morphologically more mature than vector-transduced cells. In addition, a striking reduction of not only monocytic (7/4− F4/80+) but also mixed lineage (7/4+ F4/80+) phenotype cells occurred. In sum, Gfi1 is a rate-limiting granulocytic switch that represses critical monopoietic target genes (such as Csf1 and Csf1r) to resolve cell-fate decisions in progenitor cells.
EXPERIMENTAL PROCEDURES
Plasmids and Virus Production
For the construction of Gfi1 mutants, the Bci6-HincII DNA fragment of the murine Gfi1 cDNA (gift from T. Möröy) was first cloned into pCR-BluntII-TOPO vector (Invitrogen); then, the SNAG domain P2A and zinc finger N382S mutations were introduced with Quickchange II site-directed mutagenesis kit (Stratagene) and confirmed by sequencing. The BamHI-NotI DNA fragments were cloned into pcDNA3.1(+) for IVT. For virus production, the EcoRI DNA fragments were cloned into either the MSCV-puro (Hawley, 1994) or MMP-ires-GFP (Klein et al., 2000) retroviral vectors. Phoenix-Eco packaging cells were transfected with the viral vectors for viral supernatant generation.
Transient Transcription Assay and Immunoblotting
A chloramphenicol acetyltransferase (CAT) assay was performed as described elsewhere (Zweidler-Mckay et al., 1996). In brief, 400 ng of either B30TKCAT or TKCAT were cotransfected with 10 ng of MSCV vectors encoding Gfi1 or Gfi1N382S into HEK293 cells with Lipofectamine2000 (Invitrogen). Thirty-six hours later, cells were lysed and CAT activity was determined by the scintillation method, and proteins were detected through immunoblot analyses with a rabbit polyclonal antisera directed against a peptide encoding Gfi1 amino acids 12–26.
Bone-Marrow Lin− Cells Isolation and Retroviral Infection
Mice were bred and housed by CCHMC Veterinary Services, and mouse manipulations were reviewed and approved by the Children’s Hospital Research Foundation IACUC. Bone-marrow Lin− cells were isolated from both 6- to 8-week-old C57Bl/6 WT mice and Gfi1 mutant littermates or Csf1−/−(op/op)-deficient mice (Yoshida et al., 1990). Bone marrow was flushed from both femurs and tibias with PBS containing 0.5% FBS, then labeled with the mouse Lineage Cell Depletion kit (Miltenyi Biotec); this was followed by separation on AutoMacs magnetic sorter (Miltenyi Biotec). Cells were maintained in serum-free StemSpan medium (StemCell Technologies) supplemented with IL-3 (10 ng/ml), IL-6 (20 ng/ml), SCF (25 ng/ml), and TPO (25 ng/ml) and various concentrations of Csf1 (all from PeproTech). After 48 hr of cytokine expansion, the cells were subjected to retroviral transduction. In brief, six-well non-tissue-culture-treated plates were coated with Retronectin (TaKaRa) according to the manufacturer’s protocol and preloaded with viral particles. A total of 2 × 106 cells were plated per well and spinfected at 1000 g for 1.5 hr.
Methylcellulose Assay, Liquid Cultures, and Flow Cytometry
One day after MSCV-puro retroviral vector transduction, 6 × 103 cells/ml were plated in MethoCult GF M3534 containing IL-3, IL-6, and SCF (StemCell Technologies) with 2.5 mg/ml of puromycin (Sigma). Alternatively, 36 hr after CMMP retroviral-vector transduction, GFP+ cells were sorted and plated in MethoCult. Hematopoietic colonies containing more then 50 cells were scored at day 7 or 8 and differentiated on the basis of their morphology. For experiments involving neutralization of Csf1 bioactivity, Lin− cells were transduced with retroviral vectors and plated in triplicate in Methocult in the presence of 350 ng/mL monoclonal antibody that neutralizes Csf1 (clone 5A1, BD Biosciences) or a rat IgG1, k isotype control (BD Biosciences). Results are displayed as the percentage of CFU-G, -M, and -GM from three independent wells ±SD with total number of colonies ± SD (“col. no:”). Assays were repeated at least three times with similar results. For liquid cultures, 1 day after retroviral-vector transduction, the cells were plated in Iscove’s DMEM with 10% FBS and 2.5 mg/ml of puromycin and stimulated for 4 days with 5 ng/ml of murine G-CSF (Pepro Tech). The cells were then subjected to either cytospin (and then Giemsa or Hematoxylin-Eosin staining) or flow cytometry. In brief, cells were stained with antibodies to 7/4 (clone 7/4, Serotec) and F4/80 (clone CI:A3-1, Serotec), or Annexin-V-APC and 7-AAD (BD-PharMingen), then analyzed on an LSRII flow cytometer (BD-Biosciences) with FloJo software (TreeStar). Representative flow plots are shown with statistical analyses from at least three independent experiments.
RNA Isolation and Real-Time PCR
We isolated total RNA with the RNeasy Mini Kit (QIAGEN) and digested it with DNase I (Ambion) to remove DNA contamination. A total of 1 µg of RNA was used for reverse transcription with random hexamers in the TaqMan Reverse Transcription System according to the manufacturer’s instructions (Applied Biosystems) and then analyzed with TaqMan probes targeting Egr1, Egr2, Nab2, Cebpe, Csf1, Csf1r, and β-actin transcripts (Applied Biosystems). The expression of the glyceraldehyde-3-phosphate dehydrogenase (Gapdh) gene was used for normalizing samples. The average fold expression from at least three independent samples ± SD is shown.
Molecular Modeling
An alignment of the zinc fingers of 1MEYto zinc fingers 3,4, and 5 of Gfi1 was carried out with ClustalW (Eddy, 1995). On the basis of this alignment, a homology-modeled structure for Gfi1 zinc fingers 3,4, and 5 was generated by Modeler V8.0, employing the crystal structure of 1MEY (Kim and Berg, 1996) as the template. Zinc ions were included and crystal-structure-derived distance constraints were used for maintaining the zinc finger topology. The resulting structure was used as a starting point for molecular dynamics simulations with AMBER8.0 (Case et al., 2005). Specifically, we used Xleap to generate the water-solvated system comprising a homology model of Gfi1 with TIP3P solvent molecules and ions added for neutrality. Standard equilibrium and production-run protocols were used (Trent, 2001). Molecular-dynamics simulations were performed with the parm99 force field with AMBER 8.0 and the mpi Sander module in the isothermal isobaric ensemble (p = 1 atmosphere); isotropic pressure scaling (ntp = 1), periodic boundary conditions with PME, 1.5 fs time step, and frozen hydrogen atoms were calculated with SHAKE. The models were solvated in 10 Å box of water with xleap. Counter ions were added randomly for overall charge neutrality. The system was equilibrated for 125 ps, with restraints on the position of the zinc being kept constant with weights of 100 Kcal. Unrestrained production run of 2 ns was carried out. Analysis was carried out of the snapshots as well as on the averaged structure generated from the last 25 psof the production run. Calculations were performed on 16 R12000 processor Silicon Graphics Origin 2000 server.
Electrophoretic Mobility-Shift Assay
The pcDNA3.1(+) vector templates were in vitro transcribed-translated with the TnT Coupled Reticulocyte Lysate System and T7 polymerase (Promega) according to the manufacturer’s protocol. For confirming protein synthesis, the reaction was also performed in the presence of S35 Cysteine and Methionine, resolved on SDS-PAGE, dried, and exposed to BiomaxMR film (Kodak) with a Biomax Transcreen-LE intensifying screen (Kodak). To perform EMSA, we incubated 0.5 µl of in vitro transcribed-translated proteins with 50,000 cpm of P32 end-labeled R21 probe 5′-GGACCGGGGTGCAGTGATTT GGTGTGGCGATC-3′, R21 mutant 5′-GGACCGGGGTGCAGTGACCTGGTG TGGCGATC-3′, or Csf1 probe 5′-GGAGCCAGGGTGATTTCCCATAAACCA-3′ in a reaction mixture containing 10 mM HEPES (pH 7.6), 10% glycerol, 10% NP-40, 50 mM NaCl, 0.5 mM DTT, 50 ng/µl poly (dI-dC) (Sigma), and 2.5µg/µl nonfat milk for 30 min at RT. Complexes were resolved by electrophoresis on 6% acrylamide gels in 0.5× TBE buffer at 4°C. The gels were dried and exposed to BioMax MS films (Kodak) with a TranScreen-HE intensifying screens (Kodak) at −70°.
Statistical Analysis
All experiments were repeated at least three times and analyzed with ANOVA test and the StatView computer program.
Chromatin-Immunoprecipitation Analysis
The chromatin immunoprecipitation (ChIP) assay was essentially performed as described previously (Doan et al., 2004). U937 cells (2 × 108) were fixed in 1% formaldehyde for 10 min on ice and terminated with 0.125 M Glycine. Cells were resuspended in cell-lysis buffer (50 mM Tris-HCl [pH 8.1], 10 mM EDTA [pH 8.0], 10% glycerol, 1% SDS, and 13 complete protease inhibitor) and incubated on ice for 10 min.Cells were then sonicated with a Sonicator 3000 cup horn (Misonix) on ice for generation of soluble chromatin complex with DNA fragments of <1 kb length. Approximately 1.5 ml of soluble chromatin was used per reaction for immunoprecipitation with antibody to Gfi1 (clone 2.5D.17) and control mouse IgG (NA931V, GE Health care). Quantitative PCR was performed with SYBR Green (Applied Biosystems) on an iCycler (Bi-oRad) with Csf1 (5′-GGGCCTCTGGGGTGTAGTAT-3′ and 5′-CCGAGGCAA ACTTTCACTTT-3′; product size 189 bp) or β-actin (5′-AGCGCGGCTACAGCT TCA-3′ and 5′-CGTAGCACAGCTTCTCCTTAATGTC-3′; product size 120 bp) primer pairs. β-actin was used as a control for nonspecific enrichment.
Supplementary Material
ACKNOWLEDGMENTS
We thank S. Orkin for the Gfi1 null mice, R. Hawley for the MSCV vectors, J. Walsh and R. Mulligan for the CMMP-iresGFP vectors, and C. Jay who constructed the murine Gfi1 mutant cDNA expressing vectors. We also thank H. Geiger, J. Cancelas, J. Mulloy, and M. Wills-Karp for helpful comments and manuscript review and CCHMC Biomedical Informatics for assistance. This work was partially supported by NIH grants R01 HL079574 and CA105152 and a Leukemia Lymphoma Society Scholar award (H.L.G.).
Footnotes
SUPPLEMENTAL DATA
Three figures are available at http://www.immunity.com/cgi/content/full/28/3/370/DC1/.
REFERENCES
- Ancliff PJ, Blundell MP, Cory GO, Calle Y, Worth A, Kempski H, Burns S, Jones GE, Sinclair J, Kinnon C, et al. Two novel activating mutations in the Wiskott-Aldrich syndrome protein result in congenital neutropenia. Blood. 2006;108:2182–2189. doi: 10.1182/blood-2006-01-010249. [DOI] [PubMed] [Google Scholar]
- Belaaouaj A, McCarthy R, Baumann M, Gao Z, Ley TJ, Abraham SN, Shapiro SD. Mice lacking neutrophil elastase reveal impaired host defense against gram negative bacterial sepsis. Nat. Med. 1998;4:615–618. doi: 10.1038/nm0598-615. [DOI] [PubMed] [Google Scholar]
- Case DA, Cheatham TE, 3rd, Darden T, Gohlke H, Luo R, Merz KM, Jr, Onufriev A, Simmerling C, Wang B, Woods RJ. The Amber biomolecular simulation programs. J. Comput. Chem. 2005;26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl R, Iyer SR, Owens KS, Cuylear DD, Simon MC. The transcriptional repressor GFI-1 antagonizes PU.1 activity through protein-protein interaction. J. Biol. Chem. 2007;282:6473–6483. doi: 10.1074/jbc.M607613200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devriendt K, Kim AS, Mathijs G, Frints SG, Schwartz M, Van Den Oord JJ, Verhoef GE, Boogaerts MA, Fryns JP, You D, et al. Constitutively activating mutation in WASP causes X-linked severe congenital neutropenia. Nat. Genet. 2001;27:313–317. doi: 10.1038/85886. [DOI] [PubMed] [Google Scholar]
- Doan LL, Porter SD, Duan Z, Flubacher MM, Montoya D, Tsichlis PN, Horwitz M, Gilks CB, Grimes HL. Targeted transcriptional repression of Gfi1 by GFI1 and GFI1B in lymphoid cells. Nucleic Acids Res. 2004;32:2508–2519. doi: 10.1093/nar/gkh570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donini M, Fontana S, Savoldi G, Vermi W, Tassone L, Gentili F, Zenaro E, Ferrari D, Notarangelo LD, Porta F, et al. G-CSF treatment of severe congenital neutropenia reverses neutropenia but does not correct the underlying functional deficiency of the neutrophil in defending against microorganisms. Blood. 2007;109:4716–4723. doi: 10.1182/blood-2006-09-045427. [DOI] [PubMed] [Google Scholar]
- Eddy SR. Multiple alignment using hidden Markov models. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1995;3:114–120. [PubMed] [Google Scholar]
- Fiolka K, Hertzano R, Vassen L, Zeng H, Hermesh O, Avraham KB, Duhrsen U, Moroy T. Gfi1 and Gfi1b act equivalently in haematopoiesis, but have distinct, non-overlapping functions in inner ear development. EMBO Rep. 2006;7:326–333. doi: 10.1038/sj.embor.7400618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grenda DS, Johnson SE, Mayer JR, McLemore ML, Benson KF, Horwitz M, Link DC. Mice expressing a neutrophil elastase mutation derived from patients with severe congenital neutropenia have normal granulopoiesis. Blood. 2002;100:3221–3228. doi: 10.1182/blood-2002-05-1372. [DOI] [PubMed] [Google Scholar]
- Grimes HL, Chan TO, Zweidler-McKay PA, Tong B, Tsichlis PN. The Gfi-1 proto-oncoprotein contains a novel transcriptional repressor domain, SNAG, and inhibits G1 arrest induced by interleukin-2 withdrawal. Mol. Cell. Biol. 1996a;16:6263–6272. doi: 10.1128/mcb.16.11.6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimes HL, Gilks CB, Chan TO, Porter S, Tsichlis PN. The Gfi-1 protooncoprotein represses Bax expression and inhibits T-cell death. Proc. Natl. Acad. Sci. USA. 1996b;93:14569–14573. doi: 10.1073/pnas.93.25.14569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley RG. High-titer retroviral vectors for efficient transduction of functional genes into murine hematopoietic stem cells. Ann. N Y Acad. Sci. 1994;716:327–330. doi: 10.1111/j.1749-6632.1994.tb21724.x. [DOI] [PubMed] [Google Scholar]
- Heyd F, ten Dam G, Moroy T. Auxiliary splice factor U2AF26 and transcription factor Gfi1 cooperate directly in regulating CD45 alternative splicing. Nat. Immunol. 2006;7:859–867. doi: 10.1038/ni1361. [DOI] [PubMed] [Google Scholar]
- Hirsch S, Gordon S. Polymorphic expression of a neutrophil differentiation antigen revealed by monoclonal antibody 7/4. Immunogenetics. 1983;18:229–239. doi: 10.1007/BF00952962. [DOI] [PubMed] [Google Scholar]
- Hock H, Hamblen MJ, Rooke HM, Traver D, Bronson RT, Cameron S, Orkin SH. Intrinsic requirement for zinc finger transcription factor Gfi-1 in neutrophil differentiation. Immunity. 2003;(18):109–120. doi: 10.1016/s1074-7613(02)00501-0. [DOI] [PubMed] [Google Scholar]
- Horwitz MS, Duan Z, Korkmaz B, Lee HH, Mealiffe ME, Salipante SJ. Neutrophil elastase in cyclic and severe congenital neutropenia. Blood. 2007;109:1817–1824. doi: 10.1182/blood-2006-08-019166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hume DA, Gordon S. Mononuclear phagocyte system of the mouse defined by immunohistochemical localization of antigen F4/80. Identification of resident macrophages in renal medullary and cortical interstitium and the juxtaglomerular complex. J. Exp. Med. 1983;157:1704–1709. doi: 10.1084/jem.157.5.1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jegga AG, Chen J, Gowrisankar S, Deshmukh MA, Gudivada R, Kong S, Kaimal V, Aronow BJ. GenomeTrafac: A whole genome resource for the detection of transcription factor binding site clusters associated with conventional and microRNA encoding genes conserved between mouse and human gene orthologs. Nucleic Acids Res. 2007;35:D116–D121. doi: 10.1093/nar/gkl1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karsunky H, Mende I, Schmidt T, Moroy T. High levels of the onco-protein Gfi-1 accelerate T-cell proliferation and inhibit activation induced T-cell death in Jurkat T-cells. Oncogene. 2002a;21:1571–1579. doi: 10.1038/sj.onc.1205216. [DOI] [PubMed] [Google Scholar]
- Karsunky H, Zeng H, Schmidt T, Zevnik B, Kluge R, Schmid KW, Duhrsen U, Moroy T. Inflammatory reactions and severe neutropenia in mice lacking the transcriptional repressor Gfi1. Nat. Genet. 2002b;30:295–300. doi: 10.1038/ng831. [DOI] [PubMed] [Google Scholar]
- Khanna-Gupta A, Sun H, Zibello T, Lee HM, Dahl R, Boxer LA, Berliner N. Growth factor independence 1 (Gfi-1) plays a role in mediating specific granule deficinecy (SGD) in a patient lacking a gene inactivating mutation in the C/EBP{epsilon} gene. Blood. 2007;109:4181–4190. doi: 10.1182/blood-2005-05-022004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim CA, Berg JM. A 2.2 A resolution crystal structure of a designed zinc finger protein bound to DNA. Nat. Struct. Biol. 1996;3:940–945. doi: 10.1038/nsb1196-940. [DOI] [PubMed] [Google Scholar]
- Klein C, Bueler H, Mulligan RC. Comparative analysis of genetically modified dendritic cells and tumor cells as therapeutic cancer vaccines. J. Exp. Med. 2000;191:1699–1708. doi: 10.1084/jem.191.10.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein C, Grudzien M, Appaswamy G, Germeshausen M, Sandrock I, Schaffer AA, Rathinam C, Boztug K, Schwinzer B, Rezaei N, et al. HAX1 deficiency causes autosomal recessive severe congenital neutropenia (Kostmann disease) Nat. Genet. 2007;39:86–92. doi: 10.1038/ng1940. [DOI] [PubMed] [Google Scholar]
- Laslo P, Spooner CJ, Warmflash A, Lancki DW, Lee HJ, Sciammas R, Gantner BN, Dinner AR, Singh H. Multilineage transcriptional priming and determination of alternate hematopoietic cell fates. Cell. 2006;126:755–766. doi: 10.1016/j.cell.2006.06.052. [DOI] [PubMed] [Google Scholar]
- Marteijn JA, van der Meer LT, van Emst L, de Witte T, Jansen JH, van der Reijden BA. Diminished proteasomal degradation results in accumulation of Gfi1 protein levels in monocytes. Blood. 2006;109:100–108. doi: 10.1182/blood-2006-02-003590. [DOI] [PubMed] [Google Scholar]
- Person RE, Li FQ, Duan Z, Benson KF, Wechsler J, Papadaki HA, Eliopoulos G, Kaufman C, Bertolone SJ, Nakamoto B, et al. Mutations in proto-oncogene GFI1 cause human neutropenia and target ELA2. Nat. Genet. 2003;34:308–312. doi: 10.1038/ng1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodel B, Tavassoli K, Karsunky H, Schmidt T, Bachmann M, Schaper F, Heinrich P, Shuai K, Elsasser HP, Moroy T. The zinc finger protein Gfi-1 can enhance STAT3 signaling by interacting with the STAT3 inhibitor PIAS3. EMBO J. 2000;19:5845–5855. doi: 10.1093/emboj/19.21.5845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg PS, Alter BP, Bolyard AA, Bonilla MA, Boxer LA, Cham B, Fier C, Freedman M, Kannourakis G, Kinsey S, et al. The incidence of leukemia and mortality from sepsis in patients with severe congenital neutropenia receiving long-term G-CSF therapy. Blood. 2006;107:4628–4635. doi: 10.1182/blood-2005-11-4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trent JO. Molecular modeling of drug-DNA complexes: An update. Methods Enzymol. 2001;340:290–326. doi: 10.1016/s0076-6879(01)40428-9. [DOI] [PubMed] [Google Scholar]
- Wiktor-Jedrzejczak W, Bartocci A, Ferrante AW, Jr, Ahmed-Ansari A, Sell KW, Pollard JW, Stanley ER. Total absence of colony-stimulating factor 1 in the macrophage-deficient osteopetrotic (op/op) mouse. Proc. Natl. Acad. Sci. USA. 1990;87:4828–4832. doi: 10.1073/pnas.87.12.4828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe SA, Grant RA, Elrod-Erickson M, Pabo CO. Beyond the “recognition code”: Structures of two Cys2His2 zinc finger/TATA box complexes. Structure. 2001;9:717–723. doi: 10.1016/s0969-2126(01)00632-3. [DOI] [PubMed] [Google Scholar]
- Yoshida H, Hayashi S, Kunisada T, Ogawa M, Nishikawa S, Okamura H, Sudo T, Shultz LD, Nishikawa S. The murine mutation osteopetrosis is in the coding region of the macrophage colony stimulating factor gene. Nature. 1990;345:442–444. doi: 10.1038/345442a0. [DOI] [PubMed] [Google Scholar]
- Zeng H, Yucel R, Kosan C, Klein-Hitpass L, Moroy T. Transcription factor Gfi1 regulates self-renewal and engraftment of hematopoietic stem cells. EMBO J. 2004;23:4116–4125. doi: 10.1038/sj.emboj.7600419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang D, Qiu Y, Kogan SC, Dong F. Increased CCAAT enhancer-binding protein epsilon (C/EBPepsilon) expression and premature apoptosis in myeloid cells expressing Gfi-1 N382S mutant associated with severe congenital neutropenia. J. Biol. Chem. 2006;281:10745–10751. doi: 10.1074/jbc.M510924200. [DOI] [PubMed] [Google Scholar]
- Zweidler-Mckay PA, Grimes HL, Flubacher MM, Tsichlis PN. Gfi-1 encodes a nuclear zinc finger protein that binds DNA and functions as a transcriptional repressor. Mol. Cell. Biol. 1996;16:4024–4034. doi: 10.1128/mcb.16.8.4024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.