Abstract
Drosophila melanogaster telomeres are composed of two retrotransposons, HeT-A and TART. Drosophila virilis has recently been shown to have telomere-specific TART elements with many of the characteristics of their D. melanogaster homologues. We now report identification of the second telomere-specific retrotransposon, HeT-A, from D. virilis. These results show that HeT-A and TART have been maintaining telomeres in Drosophila for more than the 60 million years that separate D. melanogaster and D. virilis. All Drosophila species and stocks studied have both of these telomeric elements, suggesting that the elements collaborate, an assumption supported by evidence from D. melanogaster that their Gag proteins interact. Although the HeT-A sequence evolves at a high rate, the element retains the unusual structural features that characterize all HeT-A homologues. These features may be involved in the role of HeT-A at the telomere. The Gag protein from HeT-Avir is as much like TART Gag from other species as it is like HeT-A Gag, suggesting that these Gags are evolving under similar constraints, probably to maintain appropriate interactions with host telomeres and possibly to allow collaborative interactions like those seen in D. melanogaster. In addition, we have identified a chimeric element, Uvir, carrying a pol coding sequence only distantly related to sequences thus far found in any telomere arrays.
Telomeres of eukaryotic chromosomes are composed of long arrays of DNA repeats. In most animals, plants, and unicellular eukaryotes, these repeats are short species-specific sequences (5–10 bp) reverse transcribed onto chromosome ends by the enzyme telomerase. Telomeres in Drosophila melanogaster are a surprising exception to the general type. D. melanogaster telomeres are arrays of repeated sequences but these arrays are produced by successive transpositions of two non-LTR retrotransposons. Rather than successive additions of a telomerase repeat (1), each repeat in these telomeres is a copy of one of the telomeric elements, HeT-A and TART.
HeT-A and TART group into the Jockey clade based on the sequences of their coding regions. The Jockey clade contains several of the more abundant retrotransposable elements in the D. melanogaster genome, including Doc, jockey, and X, but HeT-A and TART have several characteristics that set them apart from the other elements. The most obvious difference is the targeting of transposition: HeT-A and TART transpose only onto chromosome ends, whereas the other members of the clade (generally considered parasitic elements) transpose into many parts of the genome but are never found in these telomere regions. This targeting appears to involve Gag proteins because Gags from both HeT-A and TART move efficiently to specific intranuclear sites and associate with chromosome ends, whereas Gags from other members of the clade remain almost entirely in the cytoplasm (2). Another difference is the large amount of noncoding sequence (5′ and 3′ UTR) in HeT-A and TART in D. melanogaster and Drosophila yakuba; the other elements, like almost all other retroelements, have very little sequence that does not code for proteins involved in their own transposition. We have suggested that the noncoding sequence of the telomeric elements is involved in the chromatin structure of the telomere (3).
Although they appear to have similar roles at the telomere, HeT-A and TART have notable differences. HeT-A has a novel promoter that appears to be an evolutionary intermediate between the typical promoter of non-LTR elements and that of LTR retrotransposons and retroviruses. The HeT-A promoter is at the 3′ end of the element and promotes transcription of the adjacent downstream element, thus taking on the structure and function of a LTR. In contrast, TART has a strong promoter in its 3′ end, but this promoter directs transcription back into the element that contains it, yielding antisense transcripts. We assume that sense-strand TART RNA is transcribed from a promoter in the 5′ UTR, like promoters of other non-LTR elements (1).
TART contains a pol gene; HeT-A does not. Pol proteins provide enzymatic activities needed for retrotransposition, including reverse transcriptase (RT). The nearly ubiquitous presence of this coding region in retroelements suggests that Pol proteins are much more effective in transposing the element that encodes them (acting in cis) than acting on other elements (acting in trans). This suggestion is supported by evidence from mammalian LINEs (long interspersed nuclear elements) (4). Interestingly, there is also evidence that SINEs (short interspersed nuclear elements) have evolved to use enzymatic activity of LINEs (5). The possibility that TART provides RT for HeT-A raises the question of why HeT-A is both more abundant than TART in the genome and is involved in almost all of the experimentally detected healing events on broken chromosomes. Is HeT-A also contributing to a cooperative interaction? A possible answer has come from evidence in D. melanogaster that HeT-A Gag is needed to efficiently target TART Gag to the telomeres (6). Thus, HeT-A may provide telomere targeting while TART provides RT activity.
Telomere targeting and the similarity of their Gag proteins originally suggested that HeT-A and TART were diverging from a common ancestor. As additional differences are detected, it seems more likely that the two elements evolved from different ancestors and converged on their telomeric roles by acquiring similar gag genes. This scenario would require less drastic sequence changes than those needed to derive the two elements from a common ancestor. Once they converged on these roles, the two elements have maintained their separate identities, despite extensive sequence change during the evolution of the Drosophila genus.
We have recently reported the characterization of TART elements from Drosophila virilis, showing that retrotransposon telomeres have existed since before the divergence of the extant Drosophila species (7). One of the clones we sequenced had a small fragment of an unidentified element adjacent to the TART array but truncated by the phage arm. We have now used this fragment to clone and characterize this component of the D. virilis telomere. It is a homologue of HeT-A. We have also found a chimeric HeT-A-related element; however, this chimera does not appear to be a significant component of the D. virilis telomere.
Materials and Methods
Fly Stocks. D. virilis stock no. S170 was obtained from the European Drosophila Stock Center (Umea, Sweden).
Library Screening. A genomic library of D. virilis DNA in lambda phage was screened with the HeT-Avir probe (see below). Hybridization was performed overnight at 65° in 4× SET (1× SET = 0.15 M NaCl/0.03 M Tris, pH 7.4/2 mM EDTA), 5× Denhardt's solution, 0.5% SDS, and 50 μg/ml salmon sperm DNA. Washes were 2 × 20 min at 65° with 2× SSC/0.5% SDS, 1 × 20 min with 1× SSC/0.5% SDS, and 1 × 20 min with 0.5× SSC/0.5% SDS.
Sequence Analyses. Sequences were analyzed by blast searches. Alignments were made by clustalw (8). Nucleotide alignments from coding regions were corrected with gendoc (9) to agree with protein alignments. Phylogenetic analyses were made by mega, Version 2.1 (10), with both neighbor-joining and UPGMA algorithms. dotplot (11) analyses were made with a window of 25 and a stringency of 15.
In Situ Hybridization. Hybridization to polytene chromosomes was performed as described in ref. 12.
Probes. HeT-Avir probe: SalI fragment of 4,719 nt used for Southern hybridizations (Fig. 1). TARTvir probe: nucleotides 2679–5792 of GenBank accession no. AY219708. D. virilis histone H1 probe: nucleotides 252-1004 of accession no. L76558.
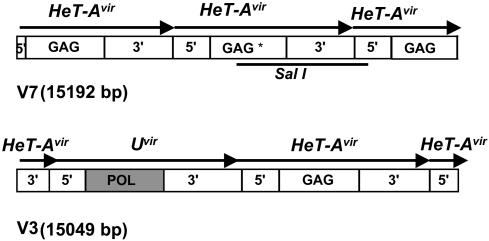
Fig. 1.
Diagrams of the D. virilis phage clones V3 and V7. Diagrams are approximately to scale. The arrows above the diagrams identify each element and indicate the 5′ → 3′ orientation of sense strand. Gag and Pol, coding regions; Gag*, gag gene 300 nt shorter than in other copies of HeT-Avir. The solid bar under Upper indicates the probe used.
Underreplication Studies. Genomic DNA was extracted from brains and salivary glands from D. virilis larvae. Equal amounts of DNA were digested with SalI and HindIII for 2 h, and Southern blots were made as in ref. 12. Hybridization with HeT-Avir or TARTvir probes was done after the protocol of library hybridization (see above). After exposure, the blots were stripped by placing them in 1% SDS at 100°C and allowing the solution to cool to room temperature. The procedure was repeated twice. After exposure, filters were hybridized with H1vir probe as described above.
Results
Isolation of HeT-Avir Arrays. As is typical of retroelements, the sequence of the telomeric transposons changes rapidly, making cross-species studies difficult. We obtained TARTvir clones by low-stringency hybridization with a probe from the TARTmel RT coding region, the most conserved region in retroelements (13). The two lambda phage clones we sequenced contained tandem arrays of TARTvir. Adjoining one TART element was the 5′ end of an element that had been truncated by cloning, leaving only the 5′ UTR and a fragment of N-terminal coding sequence in the clone (7). The sequence clearly was not TART but did not have sufficient similarity to any other known element to allow identification.
To obtain more sequence of this second telomeric element, we used the fragment from the TART array to screen a D. virilis genomic library. We obtained eight positive phages. Six were clearly different and nonoverlapping; two, V3 and V7, were sequenced and are presented here (Fig. 1).
Each phage contains slightly more than 15 kbp of D. virilis DNA, consisting of head-to-tail arrays of a non-LTR retrotransposon with 5′ UTRs and gag sequences that match the uncharacterized fragment found in the TART array. The newly found element has a complete gag gene followed by a long 3′ UTR. No other ORF was found in any copy of this element. The lack of a pol gene and the long 3′ UTR, two of the unusual characteristics of HeT-A, strongly suggest that this is a D. virilis homologue of HeT-A (Fig. 2).
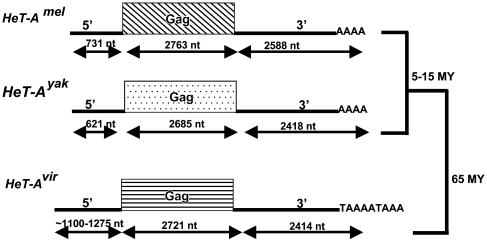
Fig. 2.
Diagrams of HeT-A homologues in D. melanogaster, D. yakuba, and D. virilis, drawn approximately to scale. 5′,5′ UTR; 3′,3′ UTR; Gag, gag gene; AAA or TAAAAA, 3′ oligo(A) that characterizes non-LTR elements; MY, million years. The solid bars on the right side indicate the phylogenic relationships.
Each cloned sequence contains several junctions between elements. These junctions have no interruptions or additional sequence between the 3′ end of one element and the 5′ end of its neighbor (see Fig. 7, which is published as supporting information on the PNAS web site). The high level of conservation at the 3′–5′ junctions suggests that these elements are complete transposition products, without the 5′ truncation frequently seen with non-LTR elements.
All HeT-Avir coding regions in the two phages are open. Their deduced Gag proteins are nearly identical, except for the second copy in the array in phage V7. This copy encodes a Gag protein that ends 100 aa before the predicted proteins in the other copies. In addition to this deletion in the coding region, the element contains small deletions within the 5′ and 3′ UTRs, whereas other copies do not.
blast comparisons of the nucleotide sequence of the full-length element yielded only one significant match, the expected match to the sequence from the TART array already in the database. The complete conceptual translation of the ORF showed the expected similarities with Gag proteins from the Jockey clade. The protein contains zinc knuckle (CCHC) and MHR (major homology region) motifs, which characterize Gags from the Jockey clade (6), as well as those from mammalian retroviruses. blastp comparisons with the amino acid sequence of this newly found Gag protein showed the highest similarity to the small fragment already in the database, followed by the Gag protein of the X element (E = 2e–22), the Gag protein of TARTvir (E = 2e–20), TARTBmel (E = 8e–19), and several entries for the Gag protein of HeT-Amel (E = 2e–15 to 4e–15). These sequence similarities are consistent with the assumption, based on sequence organization, that this element is HeT-Avir.
Phage V3 also contains one copy of what appears to be a second, very unusual non-LTR element (Fig. 1). The 5′ UTR of this element is very similar to that of HeT-Avir, but the element has no gag gene. In place of the gag gene, this element has an ORF encoding an apparently complete Pol protein. The element also has a long 3′ UTR with >90% identity to HeT-Avir 3′ UTR in the last 1 kb of sequence but no significant similarity in the rest of this region (see below).
Chromosome Location of HeT-Avir Elements. In the Drosophila species previously studied, D. melanogaster, D. yakuba, and Drosophila simulans, the most striking difference between the telomeric elements and other members of the Jockey clade is the difference in targets of transposition. HeT-A and TART are found only at telomeres with some related sequences at a few internal heterochromatic sites on the Y chromosome and in the pericentric region. None of these sequences are ever found in the euchromatic gene-rich regions. In contrast, the other elements are found at many sites in the euchromatic regions but have never been found in the telomeric arrays. (This difference in localization is surprising because none of these elements are thought to be targeted by specific DNA sequences.) The magnification provided by polytene chromosomes makes it possible to map sequences in the euchromatic regions very precisely, although underreplication and amorphous morphology make mapping to heterochromatin less reliable. For D. melanogaster, the conclusions from in situ hybridization are now supported by Release 3 of the DNA sequence of euchromatin (14).
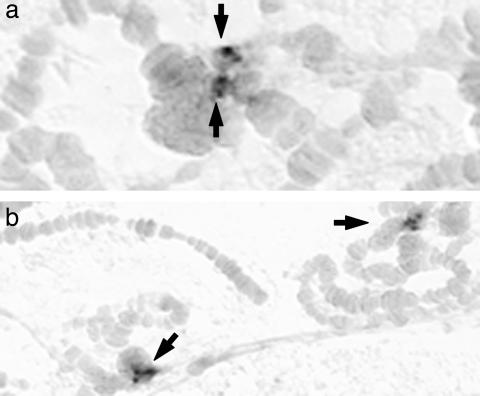
In situ hybridization of HeT-Avir showed that, like its homologues in other species, the element is not found in euchromatic regions. HeT-Avir was detected in four clusters within the chromocenter that is formed in each nucleus by fusion of the centromeric regions (Fig. 3). All of the D. virilis chromosomes are acrocentric, with one telomere very close to the centromere. These telomeres are present in the chromocenter, but because the region is very amorphous, it is not possible to follow specific chromosomes here. The reproducibility of the clusters of HeT-A hybridization from one nucleus to another indicates that the DNA sequences in the chromocenter have a more orderly organization than seen by cytological staining. It is attractive to think that the four clusters represent the telomeres of the short arms of the four pairs of large autosomes. We note that in the other species of Drosophila we have studied, these four pairs of autosomes found in D. virilis are fused at the centromeres to yield two pairs of metacentric chromosomes. Therefore, in these other species the chromocenter would not contain telomeres of these short arms.
Fig. 3.
In situ hybridization of HeT-Avir probes to polytene chromosomes. Arrows point to the clusters of hybridization dots in different chromocenters. In each chromocenter, four clusters can be resolved, most easily seen at higher magnification in a.In b, two chromocenters are seen, each with four dots. No hybridization is seen in the banded chromosome arms. (Magnification: a, ×8,000; b, ×4,000.)
Surprisingly, we have not seen hybridization of HeT-A to the telomeres of the chromosome arms that are not in the chromocenter. These are the telomeres where TART sequence has been detected (7). This result suggests that there may be less intermingling of the two elements in D. virilis than in the other species studied, although our cloned sequence shows that at least one HeT-A element is adjacent to a TART element.
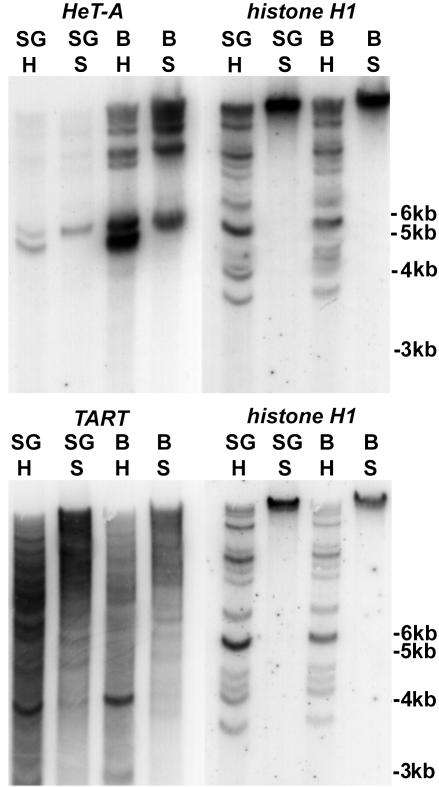
HeT-Avir Is Underreplicated in Polytene Nuclei. Satellite DNAs are underreplicated in D. virilis polytene nuclei (15), and it is possible that other sequences undergo the same fate. To test this possibility for HeT-Avir we compared the amount of HeT-Avir in polytene salivary glands with that in diploid larval brain tissue (Fig. 4). Southern blots of DNA were hybridized with HeT-Avir or TARTvir probes. As a measure of the number of genome equivalents in each lane of the gel, blots were stripped and reprobed with a probe from the D. virilis histone H1 gene, thought to be fully polytenized. Hybridization to the histone probe shows that the lanes contain approximately the same number of genome equivalents, whereas hybridization with the HeT-Avir probe shows that genomes in the polytene tissue have many fewer copies of HeT-Avir than do genomes in the diploid tissue. In contrast, the blots show that TARTvir is present at approximately the same level in the two tissues.
Fig. 4.
Southern hybridization showing underreplication of HeT-Avir in salivary gland DNA. SG, lanes with salivary gland DNA; B, lanes with larval brain DNA; H, DNA digested with HindIII; S, DNA digested with SalI. Probes are indicated at the top of each filter. See Materials and Methods for probe information.
HeT-A Gags Have Low Levels of Sequence Conservation. Retroviral Gags have rapidly changing sequences (13). HeT-A Gags show this same characteristic. Table 1 compares sequence identity of HeT-A homologues from D. melanogaster, D. yakuba, and D. virilis. We include three host genes: rough, a homeobox gene; histone H1, the most variable histone gene; and histone H3, a highly conserved gene. For comparison with a retroelement gag gene, we use sequence from R1, a non-LTR retrotransposon from Drosophila mercatorum, because no sequence from D. virilis is available.
Table 1. Comparisons of sequence identity and synonymous and nonsynonymous substitutions.
| nt, % identity | aa, % identity | Ks | Ka | |
|---|---|---|---|---|
| HeT-A gag | ||||
| mel—yak | 58 (62) [68] | 54 (57) [75.3] | 1.06 ± 0.07 | 0.31 ± 0.016 |
| mel—vir | 30 (30.3) [45] | 16 (18) [35.8] | 1.96 ± 0.49 | 1.27 ± 0.301 |
| yak—vir | 29 (29.5) [44] | 14.2 (17) [36] | 1.96 ± 0.71 | 1.27 ± 0.390 |
| rough | ||||
| mel—vir | 55 (65.6) | 55.4 (66) | 1.20 ± 0.18 | 0.27 ± 0.024 |
| histone H1 | ||||
| mel—vir | 61.4 (65.4) | 62 (66) | 1.41 ± 0.21 | 0.27 ± 0.026 |
| histone H3 | ||||
| mel—hyd | 76.7 | 97 | — | — |
| R1 gag | ||||
| mel—mer | 39 (42) | 29 (31) | 1.68 ± 0.26 | 0.89 ± 0.056 |
Values in parentheses were calculated by omitting the residues that fall in gaps. Values in brackets correspond to the MHR and zinc knuckle region only. Ks, synonymous substitutions; Ka, replacement substitutions (± SE). GenBank accession nos. were as follows: R1mel, P16424; R1mer, AAB94026; D. virilis histone H1, L76558; D. melanogaster histone H1, X04073; D. hydei histone H3, X52576; D. melanogaster histone H3, X14215; D. melanogaster rough, AAA56800; D. virilis rough, M35372; HeT-Amel Gag, AAC17188; HeT-Ayak gag, AAC01742. virilis, mercatorum, and hydei are all approximately equidistant from melanogaster.
HeT-A gag is the most rapidly changing sequence in Table 1. Sequence conservation, both nucleotide and amino acid, for HeT-A gag in the melanogaster/yakuba comparison (divergence 5–15 million years) is already as low as or lower than conservation of host genes in the melanogaster/virilis comparisons (divergence 60 million years).
Two regions thought to be important for Gag protein–protein interactions are the zinc knuckles and the MHR (major homology region) (6). These motifs represent one sixth of the full-length HeT-Avir Gag protein. The two domains have significantly higher identity than the entire protein (Table 1, bracketed values). This result suggests that only these two domains are under selective pressure. Analyses of the gag gene from TART homologues gave a similar result (7).
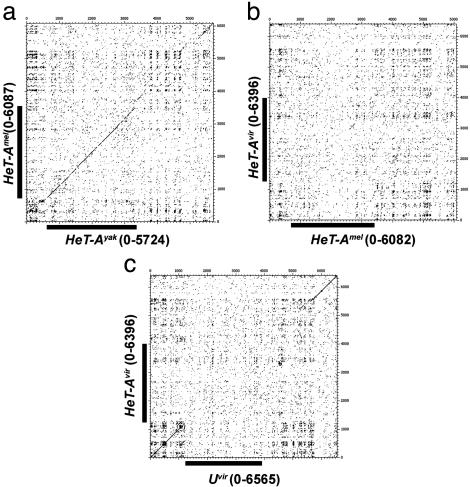
Comparisons of HeT-A homologues show that the level of nucleotide identity in the coding region correlates with evolutionary distance (Table 1), declining to 30% in the melanogaster/virilis comparison. In a surprising contrast, the identity of the UTRs (both 5′ and 3′) is essentially the same (55–62%) in every species comparison, reaching a plateau in 5–15 million years, suggesting that the sequence for the UTR has constraints that set a higher threshold of nucleotide conservation than seen for the gag region. Dot-matrix analyses (Fig. 5) show that sequences in the UTRs exhibit patterns of off-diagonal similarities that might result from such a constraint. These patterns correspond to adenine-rich regions that have a conserved composition bias and distribution but not identical sequence. As for all other HeT-A and TART elements, HeT-Avir has a strong A+C bias in the coding strand (59.39% A+C). It is interesting that this resembles the strand bias in the telomerase template. These sequence distributions might be important either in the ribonuclear protein particle that facilitates retrotransposition or in the chromatin structure of the telomere after the elements are added to the chromosome.
Fig. 5.
Dot-matrix comparisons of HeT-Amel–HeT-Ayak (a), HeT-Amel–HeT-Avir (b), and HeT-Avir–Uvir (c). The comparisons were made with a window of 25 and a stringency of 12. GenBank accession nos. were as follows: HeT-Amel, U06920; HeT-Ayak, AF043258. The coding region in the mel–yak comparison has sufficient similarity to produce a diagonal line, but the mel–vir does not (coding regions are indicated by black bars on the axes). The 3′-UTR sequences do not have sufficient similarity to give a diagonal line, but all have a pattern of sequence repeats that produces regular off-diagonal clusters.
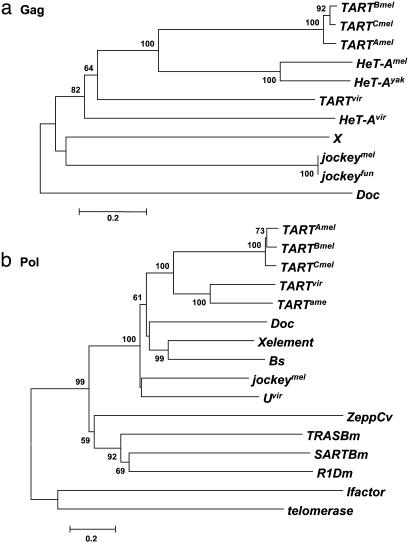
Phylogeny of HeT-Avir. In studying the phylogeny of HeT-A and TART, we have compared sequences from all known HeT-A and TART homologues with Gag proteins from four other non-LTR elements of the Jockey clade, jockeymel, jockeyfun, X, and Doc (Fig. 6a).
Fig. 6.
Phylogenetic relationships of Gag and Pol sequences. Neighbor-joining trees are shown (UPGMA trees gave the same result). Bootstrap values (at corresponding nodes) were calculated with 500 replications and a cutoff value of 50%. The scale bar indicates the number of differences per residue. (a) Gag proteins. (b) Pol proteins. GenBank accession nos. or sources were as follows: jockeymel, M22874; jockeyfun, PIR B38418; Doc, CAA35587; X, AF237761; TARTBmel, U14101; TARTAmel, F. Sheen and R. Levis; TARTCmel, L. Tolar, J. Stolk, and R. Levis; TARTvir, AAO67564; TARTame, AAO67565; jockeymel, AAA28675; SARTB.m, T18196; TRASB.m, T18199; ZeppC.v, T00078; I factor, AAA70222; R1D.m, P16425; Caenorhabditis elegans telomerase, NP_492374; Bs, S55543; X element, AAF81411.
The Gag protein of HeT-Avir groups with the Gag proteins of the HeT-A homologues and those of the TART homologues. This group is clearly separated from the rest of the Jockey clade, suggesting that the telomeric retrotransposons constitute a subclade in the Jockey clade, based on Gag phylogeny.
The tree also indicates that the Gag protein from HeT-Avir is as closely related to the TART homologues in other species as to the HeT-A homologues, suggesting that the Gag proteins from HeT-A and TART are evolving together. One characteristic that is found in the Gag proteins of HeT-Avir and TARTvir, but not in any of their homologues, is a high level of the amino acid glutamine. Gags from HeT-Amel and HeT-Ayak contain 3.1% and 3.8% glutamine, respectively, and HeT-Avir Gag contains 11.8%. As in TARTvir (7) the glutamine is concentrated at the C-terminal end of the HeT-A Gag protein, a region implicated in homologous and heterologous interactions (6); however, we have no evidence about the function of the glutamine repeats.
Uvir: A Possible New Non-LTR Element with Partial Resemblance to HeT-Avir. The HeT-A array in phage V3 contains one copy of a newly found sequence. It appears to be a non-LTR element with a single coding sequence and a long 3′ UTR. The sequence of the 5′ UTR and the last ≈1 kb of the 3′ UTR is highly similar to HeT-Avir, but the rest of the 3′ UTR is a newly found sequence. Surprisingly, in this element the gag gene has been replaced by an open and apparently complete pol gene (Fig. 1). In blastp analysis, this Pol protein showed similarity to Pol proteins from several non-LTR elements. The highest scores correspond to different entries for jockeymel (E = 5e–81 and 7e–81), TARTame (E = 6e–67), the X element (E = 1e–65), and TARTBmel (E = 8e–65).
It was not possible to clearly relate this newly found element to any of the non-LTR retrotransposons in the database. We performed phylogenetic analyses on Pol proteins from the Jockey, I factor, and R1 clades of Drosophila non-LTR retrotransposons. We also included Pol proteins from three elements that have a special affinity for telomeres in organisms that also have telomerase repeats: SART and TRAS from Bombyx mori, Zepp from Chlorella vulgaris, and the catalytic subunit of telomerase from C. elegans.We have named this non-LTR element “U,” for previously unknown, highly unexpected, and apparently unique.
The analysis shows Uvir grouping with the rest of the non-LTR elements from the Jockey clade (Fig. 6b). Note that the Jockey clade in this tree has the highest bootstrap value possible, 100, suggesting that Uvir is a new member of this clade. The tree also shows that the Pol proteins from SART, TRAS, and R1Dm are clearly related to each other and also more closely related to the Jockey clade than to the I factor or to the telomerase subunit of C. elegans.
Dot-matrix comparisons of full-length HeT-Avir with Uvir suggest that this newly found pol gene and an unidentified 3′ sequence have been inserted into the 5′ and 3′ UTRs of HeT-Avir (Fig. 5c). Note the perfect diagonal for the entire 5′-UTR sequence and the last 1 kb of the 3′ UTR. The region of the 3′ UTR where sequence identity is not detected is also less conserved between different HeT-Avir elements. The typical pattern of conserved adenine-rich regions, identified by the off-diagonal clusters, seen in this dot plot is also seen for HeT-A in other species.
This newly found element appears to be present in only one copy in the D. virilis genome. Southern blots of D. virilis DNA probed with Uvir sequence detected only elements with the same flanking restriction fragments as the original clone (data not shown). Had Uvir been inserted in other sites, the different flanking sequences would be detected as new restriction fragments. We conclude that Uvir is not inserted in other sites. Thus, there appears to be only one copy and it is not possible to determine whether Uvir is an element or a “pseudoelement,” incapable of autonomous transposition. Its structure, open coding region, and localization in the HeT-A array make Uvir extremely interesting.
We have probed DNA from Drosophila americana and Drosophila lummei with sequence from the Uvir coding region in high-stringency hybridization (data not shown). Both species are closely related to D. virilis, and both genomes contain one or a few copies of this sequence. This shows that the RT sequence is conserved, but we do not know whether this RT sequence is flanked by HeT-A UTR sequence in the other species.
Discussion
HeT-A Has a Homologue in D. virilis, HeT-Avir. All stocks of the three Drosophila species previously studied, D. melanogaster, D. yakuba, and D. simulans, have both HeT-A and TART elements in their telomeres. Our studies now show that both elements are present in the telomeres of D. virilis, separated from D. melanogaster by 60 million years. As expected from the evolutionary distance between D. virilis and the other Drosophila species (16), the sequence of the D. virilis element is significantly different from other HeT-A elements. However, it maintains so many of the unusual features that characterize HeT-A that we can confidently identify it as HeT-Avir. (i) All copies of HeT-Avir in the arrays are in the same orientation, consistent with the assumption that the array was produced by successive events of reverse transcription onto the end of the chromosome. (ii) This element is composed of a 5′ UTR, a gag gene, and a very long 3′ UTR. Both lack of a pol gene and possession of a long 3′ UTR are extremely rare among retroelements; thus, there is strong evidence that the element is HeT-A. (iii) Both 5′ and 3′ UTRs have a pattern of A-rich repeats on the coding strand, another unusual characteristic of HeT-A in other species. (iv) The element is found only in telomeres and associated heterochromatin.
HeT-A and TART Appear to Have Nonrandom Chromosome Distributions in D. virilis. HeT-Avir hybridizes to specific chromocentral regions that appear to represent the telomeres of chromosome short arms. Surprisingly, we did not detect HeT-Avir hybridization on telomeres of the ends that were not in the chromocenter, sites where TARTvir hybridized (7). Thus, the in situ hybridization suggests that HeT-Avir and TART vir tend to be localized at different telomeres, although we have cloned sequence with a HeT-A element linked to TART vir. This biased distribution of the elements in D. virilis contrasts with the apparent random mixing of the two elements in telomeres of other species.
The first evidence that specific sequences are underreplicated in polytene nuclei came from studies of the satellite DNA of D. virilis (15). Our experiments now show that HeT-Avir also undergoes underreplication in these chromosomes. This underreplication of HeT-A may be responsible for at least some of the apparent bias in the distribution of HeT-A and TART on chromosome ends because we cannot determine which regions of the chromosome are under-replicated from Southern blots. However, we cannot eliminate the possibility that the two D. virilis elements do not have the same relationship that they have in other Drosophila species and have preferential localization on different telomeres.
What HeT-Avir Tells Us About HeT-A Evolution. Retroelement sequences tend to change at higher rates than nuclear genes, as we have seen for the telomeric elements. Because Ka measures the rate of replacement of amino acids and therefore might be affected by selection for function, it is generally assumed that Ks (rate of synonymous substitution) will change more freely than Ka. Surprisingly, the difference in the rate of change for the gag genes is due more to changes in amino acid residues (HeT-A Ka is 4.7 times that of host genes) than to conservative substitutions (HeT-A Ks is 1.5 times that of host genes). The gag gene of the retroelement, R1, follows the same pattern, although the differences are lower than for HeT-A.
The unexpectedly high levels in Ka for these gag genes may be related to a second unusual characteristic of the genes. Nucleotide identity in the two gag genes is significantly higher than the amino acid identities, in contrast to the nuclear genes. This finding suggests that selective pressure on the Gag protein is low, although it is also possible that there are constraints on the nucleotide sequence that are stronger than those on the amino acid sequence.
The low value for Ks/Ka found here (1.5 for the mel–vir and yak–vir HeT-A comparisons and 1.8 for the mel–mer R1 comparison) appears to be characteristic of retrotransposon gag genes, but not their pol genes. Studies of the RT sequences of R1 (17) found this retroelement evolving at a rate comparable to that of the nuclear genes (average Ks/Ka 6.6). Studies of TART evolution showed a Ks/Ka for gag sequences of 1.6, whereas values for pol sequences ranged from 2.0 to 4.0. It has been shown that the average Ks/Ka is between 4 and 20 for nuclear genes (18). The Ks/Ka for the host genes in our study also falls within this range (4.4 for rough and 5.2 for histone H1).
The values in Table 1 suggest that the HeT-A Gag protein evolves faster than the R1 Gag protein. However, we note that the R1 sequences used in the analysis did not include the entire protein and that other parts could evolve at higher rates.
The phylogeny presented in Fig. 6a shows that the Gag protein from HeT-Avir is as related to the TART homologues in other species as it is to the HeT-A homologues, suggesting that the Gag proteins from the two telomeric retrotransposons are evolving under similar constraints. It has been shown that the HeT-Amel Gag protein is important for targeting of both HeT-Amel and TARTmel Gags to the telomeres (6). Such a functional interaction may constrain the evolution of these two proteins, although it is likely that the evolution of both proteins is also driven by interactions with other telomere components.
Are There More than Two Telomere Elements? It is remarkable that, although D. melanogaster has many active transposable elements, none other than HeT-A and TART has been detected in the HeT-A/TART arrays. In this study, we have found what is, to our knowledge, the first telomeric sequence not closely related to either HeT-A or TART. That sequence is a pol coding sequence that, although distantly related to TART, still belongs to the Jockey clade.
This newly found pol sequence is embedded in 5′- and 3′-UTR sequences from HeT-Avir to form an element that appears to be a non-LTR retrotransposon. We refer to this putative element as Uvir. The level of identity with the HeT-A sequences is remarkable. Noncoding regions are expected to evolve much faster than coding regions. The high level of identity with the HeT-A 5′- and 3′-UTR sequences suggests that this element is a chimera between HeT-Avir and an unknown coding sequence.
The nature and origin of Uvir is unclear. The form of the element and its junctions in the HeT-Avir array suggest that it moved into the array by transposition. However, its UTR sequences might have allowed Uvir to use the HeT-A transposition machinery, in analogy to pseudogene transposition. Our finding that the element seems to be present only in one copy suggests that Uvir is not very successful in transposition. On the other hand, the pol coding region has not undergone the decay expected of an inactive element; perhaps this indicates that this chimera is recently derived from an active element or a nuclear pol gene. The sequences could have been combined by template switches during reverse transcription of RNA or by recombination or gene conversion of double-stranded DNA. In either case, two switches are required because the element has HeT-Avir sequence on both ends. Identification of the donor of the pol gene would help settle this question and perhaps give insight into the evolution of telomeric retrotransposons.
Conclusion
The evolution of HeT-A during the 60 million years that separate D. melanogaster and D. virilis gives us important insight into both telomere conservation and retroelement evolution. HeT-A and TART have been coevolving with the Drosophila genome for >60 million years, performing the essential cellular role of telomere maintenance without losing their personalities as non-LTR retrotransposons. Both elements are found in all of the Drosophila stocks and cell lines that have been studied, strongly suggesting that they must collaborate in formation and/or the function of the Drosophila telomere.
Supplementary Material
Acknowledgments
We thank Ron Blackmun and Thom Kaufman for the D. virilis library, and Josep M. Casacuberta, Ky Lowenhaupt, and the members of the Pardue laboratory for helpful discussions and comments on the manuscript. This work was supported by National Institutes of Health Grant GM50315.
Abbreviation: RT, reverse transcriptase.
Data deposition: The sequences reported in the paper have been deposited in the GenBank database [accession nos. AY369259 (Drosophila virilis telomeric clone V3) and AY369260 (D. virilis telomeric clone V7)].
References
- 1.DeBaryshe, P. G. & Pardue, M.-L. (2003) Annu. Rev. Genet., in press. [DOI] [PubMed]
- 2.Rashkova, S., Karam, S. E. & Pardue, M.-L. (2002) Proc. Natl. Acad. Sci. USA 99, 3621–3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Danilevskaya, O. N., Lowenhaupt, K. & Pardue, M.-L. (1998) Genetics 148, 233–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wei, W., Gilbert, N., Ooi, S. L., Lawler, J. F., Ostertag, E. M., Kazazian, H. H., Boeke, J. D. & Moran, J. V. (2001) Mol. Cell. Biol. 21, 1429–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kajikawa, M. & Okada, N. (2002) Cell 111, 433–444. [DOI] [PubMed] [Google Scholar]
- 6.Rashkova, S., Athanasiadis, A. & Pardue, M.-L. (2003) J. Virol. 77, 6376–6384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Casacuberta, E. & Pardue, M.-L. (2003) Proc. Natl. Acad. Sci. USA 100, 3363–3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thompson, J. D., Higgins, D. G. & Gibson, T. J. (1994) Nucleic Acids Res. 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nicholas, K. B., Nicholas, J. B., Jr., & Deerfield, D. W. (1999) EMBO NEWS 4, 14. [Google Scholar]
- 10.Kumar, S., Tamura, K., Jacobsen, I. B. & Nei, M. (2001) Bioinformatics 17, 1244–1245. [DOI] [PubMed] [Google Scholar]
- 11.Maizel, J. V., Jr., & Lenk, R. P. (1981) Proc. Natl. Acad. Sci. USA 78, 7665–7669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Casacuberta, E. & Pardue, M. L. (2002) Genetics 161, 1113–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McClure, M. A., Johnson, M. S., Feng, D.-F. & Doolittle, R. F. (1988) Proc. Natl. Acad. Sci. USA 85, 2469–2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaminker, J. S., Bergman, C. M., Kronmiller, B., Carslon, J., Svirskas, R., Patel, S., Frise, E., Wheeler, D. A., Lewis, S. E., Rubin, G. M., et al. (2002) Genome Biol. 3, 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gall, J. G., Cohen, E. H. & Polan, M. L. (1971) Chromosoma 33, 319–344. [DOI] [PubMed] [Google Scholar]
- 16.Beverly, S. M. & Wilson, A. C. (1984) J. Mol. Evol. 21, 1–13. [DOI] [PubMed] [Google Scholar]
- 17.Eickbush, D. G., Lathe, W. C., III, Francino, M. P. & Eickbush, T. H. (1995) Genetics 139, 685–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Akashi, H. (1994) Genetics 136, 927–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.