Abstract
Chromatin assembly and remodeling complexes alter histone–DNA interactions by using the energy of ATP hydrolysis catalyzed by nucleosome-dependent ATPase subunits. Several classes of ATP-dependent chromatin remodeling complexes exist, including the ISWI family. ISWI complexes disrupt histone–DNA interactions in vitro by facilitating nucleosome sliding. Snf2h is a widely expressed ISWI ATPase. We investigated the role of the Snf2h gene in mammalian development by generating a null mutation in mice. Snf2h heterozygous mutant mice are born at the expected frequency and appear normal. Snf2h-/- embryos die during the periimplantation stage. Blastocyst outgrowth experiments indicate that loss of Snf2h results in growth arrest and cell death of both the trophectoderm and inner cell mass. To investigate the effect of decreased Snf2h levels in adult cells, we performed antisense inhibition of Snf2h in human hematopoietic progenitors. Reducing Snf2h levels inhibited CD34+ progenitors from undergoing cytokine-induced erythropoiesis in vitro. Our results indicate that Snf2h is required for proliferation of early blastocyst-derived stem cells and adult human hematopoietic progenitors. Cells lacking Snf2h are thus prevented from further embryonic development and differentiation.
The enormous sequence content of eukaryotic chromosomes in relation to the size of the eukaryotic nucleus necessitates organization of the DNA into a highly compact nucleoprotein complex referred to as chromatin. The fundamental unit of organization of eukaryotic chromatin is the nucleosome core particle, consisting of an octamer of four core histones (H2A, H2B, H3, and H4) around which 147 bp of DNA is wrapped (1). The packaging of DNA into chromatin limits its accessibility to DNA-binding factors. Therefore, regulation of chromatin structure is expected to play an important role in most and perhaps all transactions occurring on DNA, including transcription, replication, recombination, repair, etc. (2). To cope with the barrier created by chromatin compaction, eukaryotic cells contain two major classes of chromatin-modifying activities: histone-modifying complexes that add or remove chemical residues from the histones and thereby alter their interactions with each other and with DNA and other proteins, and ATP-dependent chromatin remodeling complexes that use the energy of ATP hydrolysis to change the position or structure of the DNA with respect to individual nucleosome core particles (3).
Currently, three known families of ATP-dependent chromatin remodeling complexes exist, the SWI/SNF-family, the ISWI family, and the Mi-2 family, distinguished by their catalytic ATPase subunit (3). These ATPase subunits exhibit homology only in the ATPase domain. They each interact with different proteins, forming multiprotein complexes that in some cases have many subunits. The two most well studied families are the SWI/SNF and ISWI families, with representatives described in a wide array of organisms. Mammalian SWI/SNF complexes contain either brahma (BRM) or brahma-related gene 1 (BRG1) ATPases. SNF2h is the ATPase subunit of the major mammalian ISWI complex. BRG1 and SNF2h appear to differ in their mechanism of action on the DNA–nucleosome core complex. SNF2h mediates DNA accessibility by sliding the histone octamer, whereas BRG1 may facilitate access to DNA primarily by generating DNA loops on the surface of the nucleosome core particle (4).
Studies in several organisms strongly support a role for ATP-dependent chromatin remodeling complexes in regulation of gene expression (5, 6). SWI/SNF complexes have been shown to interact with numerous sequence-specific DNA-binding proteins (7, 8), which are thought to recruit the complexes to specific promoters (9). SWI/SNF complexes probably participate in regulating transcription of large numbers of genes (10). They also can interact with the retinoblastoma protein and participate in cell-cycle regulation (11, 12). ISWI ATPases also interact with a variety of DNA-binding factors. In several instances these interactions have been shown to result in transcriptional repression. For example, the NORC complex, containing the ISWI ATPase Snf2h and nucleolar protein Tip5, has been shown to repress ribosomal gene transcription by recruiting DNA methyltransferase and histone deacetylase activities to rDNA promoters (13, 14). Snf2h also localizes in heterochromatin through its interactions with Acf1 (15) and the Williams Syndrome transcription factor (16). These complexes accumulate at sites in heterochromatin coincident with their replication, suggesting a role for ISWI chromatin remodeling functions in replication of DNA in highly condensed chromatin (15). ISWI complexes also may have a role in facilitating repair and recombination of DNA in chromatin (17).
The role of chromatin remodeling ATPases in cell division and development has been studied by gene inactivation in several organisms. SWI2/SNF2, the core ATPase of the yeast SWI/SNF complex, is not an essential gene, although it participates in regulating expression of many genes in yeast (18–20). Neither of the two yeast ISWI proteins is an essential gene, but when null mutations in these genes are combined, they produce a lethal phenotype under certain stress conditions (21). Both ISWI (22) and BRM (23) ATPases are essential for proper Drosophila development. Surprisingly, however, in mice Brg1 is an essential gene (24) but Brm is not (25). Thus, in mice two very similar SWI/SNF complexes have different functions in development. Therefore, it is of great interest to investigate the role of the other major type of chromatin remodeling complex in mammals, ISWI. We report here studies in which we have inactivated the mouse gene for Snf2h, the core ATPase of ISWI complexes. Our results show that, like Brg1, Snf2h is an essential gene for early embryonic mammalian development. Studies of Snf2h-null blastocysts and hematopoietic progenitors inhibited for Snf2h expression suggest that ISWI complexes may be required for cell proliferation.
Materials and Methods
Isolation of Snf2h Genomic Clones and Preparation of the Snf2h Targeting Construct. A 1-kb human SNF2h cDNA fragment (26) corresponding to the 3′ region of SNF2h coding sequence was used to screen a 129Sv mouse genomic bacterial artificial chromosome (BAC) library (Genome Systems, Palo Alto, CA). Two overlapping BAC clones were identified. The BAC clone spanning all 24 exons was used to generate an EcoRI library of 90 subclones containing 8- to 12-kb inserts in the pZero vector (Invitrogen). Two plasmid subclones spanning exons 2–10 were identified by Southern blot-based screening with radioactively labeled cDNA (GenBank accession no. AF375046), sequence-verified, and used for the targeting construct. A 4.5-kb EcoRV15299–EcoRI19895 fragment (GenBank accession no. NT_039467) containing exons 9b and 10 was cloned into pBlueScript SK(+). A 2-kb HindIII–HpaI fragment from the plNTK plasmid containing the PGK-Neo cassette surrounded by LoxP1 sites (27) was introduced into a HindIII–EcoRV site upstream of the EcoRV–EcoRI fragment. A 4.5-kb (HindIII)EcoRI4931–HindIII9455 fragment (GenBank accession no. NT_039467) containing exons 3 and 4 was introduced into a HindIII site upstream of the PGK-Neo cassette. A herpes simplex virus thymidine kinase selection cassette was inserted at a NotI site downstream of the EcoRV–EcoRI Snf2h fragment.
Disruption of the Snf2h Gene in Embryonic Stem (ES) Cells. Twenty micrograms of the 16-kb targeting construct was linearized at a XhoI site upstream of the Snf2h HindIII fragment and electroporated into WW6 ES cells as described (28). Electroporated cells (108) were transferred to eight gelatinized plates containing irradiated 40% confluent, G418-resistant feeder cells (leukemia inhibitory factor-expressing STO fibroblasts, American Type Culture Collection). After 24 h, ganciclovir and G418 (both from Sigma) were added to the media at final concentrations of 2 μM and 250 μg/ml, respectively. After an additional 10 days of culture, a total of 44 individual proliferating colonies were observed. Colonies were removed from the plates by using a manual pipette, individual cell suspensions were prepared by trypsinization at 37°C, and the cells were expanded by growth in feeder-containing cultures. Genomic DNA was isolated from all 44 G418-ganciclovir-resistant clones and analyzed by long-template genomic PCR (AccuTaq, Sigma). Primer sequences are indicated in Fig. 1A in respect to their position (GenBank accession no. NT_039467). The annealing temperature was 55°C, and the polymerization reaction was carried out at 68°C for 12 min. The PCR bands were excised from the gel, purified, and analyzed by digestion with restriction enzymes to confirm the specificity of the PCR. The gels containing the PCR products were transferred onto nylon membranes and probed with radioactively labeled Snf2h DNA probes.
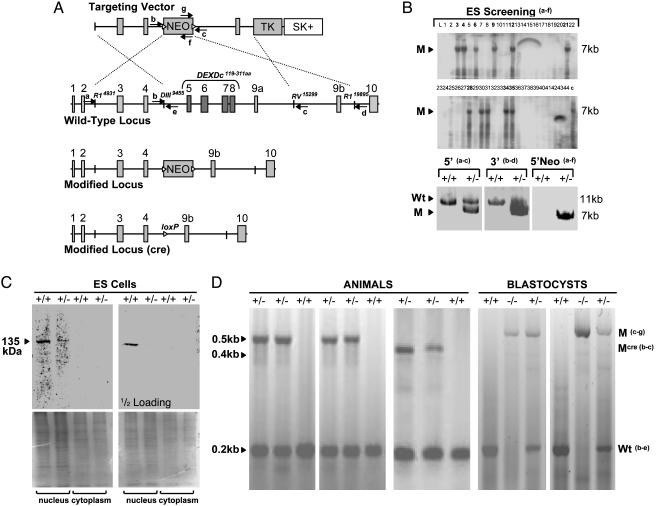
Fig. 1.
Targeted disruption of the mouse SNF2h gene. (A) Strategies for modifying the Snf2h locus in ES cells and mice. Shown is a schematic diagram of the targeting vector, wild-type locus, and modified Snf2h locus either before or after excision of the PGK-Neo cassette with Cre recombinase. Small solid boxes represent Snf2h exons (the DEXDc helicase domain is localized within the deletion), a line between them depicts noncoding regions, large solid boxes represent resistance markers PGK-Neo (flanked by loxP sites, triangles) and herpes simplex virus thymidine kinase, and the clear box represents pBlueScript SK(+) plasmid sequences. Arrows accompanied by letters indicate the position and orientation of primers (a–g) used for genotype analysis. The restriction enzyme recognition sites (EcoRI, HindIII, and EcoRV), delineating the ends of the homology regions in the targeting vector, are accompanied by numbers indicating their distances from the Snf2h gene start site (NT_039467). (B) Identification of ES cell clones containing the modified Snf2h allele. DNA from Snf2h ES clones was analyzed by using long-template genomic PCR with primers a4830–4855–fneo (shown in A) that are expected to produce an ≈7-kb DNA fragment from the modified Snf2h locus. PCR products were analyzed by agarose gel electrophoresis and ethidium bromide staining and further confirmed by Southern blot hybridization (Upper) with a DNA probe derived from a portion of intron 4 that is expected to be present at the modified Snf2h locus. The arrowhead with M indicates the position of the expected ≈7-kb PCR product from the modified locus. The clones (SNF2h KO nos. 72 and 83) analyzed in lanes 28 and 34 gave germ-line transmission. DNA from correctly targeted ES cell clones was subsequently further analyzed by long-template genomic PCR (Lower) by using primer pairs a4830–4855 and c15462–15489 and b9277–9297 and d20229–20260. DNA from parental ES cells (+/+) was used as a control. Each primer is expected to produce an ≈11-kb DNA fragment from the wild-type locus (Wt with arrowhead) and an ≈7-kb fragment from the modified locus. (C) Snf2h expression is decreased in heterozygous ES cells. Shown is Western blot analysis of Snf2h levels in separated nuclear and cytoplasmic cell lysates of the wild-type and Snf2h-targeted (+/-) ES cells by using a polyclonal antibody directed against the C-terminal portion of Snf2h. The antibody specifically recognizes the ≈135-kDa full-length Snf2h protein. No smaller protein products were detected in targeted cell lines. Two concentrations of the lysates (50 μg per lane, Upper Left, and 25 μg per lane, Upper Right) were used to demonstrate the difference of Snf2h band intensities in wild-type and mutant ES cells. The lower gels show Coomassie staining after transfer. (D) Genotype analysis of DNA from tails of offspring animals or from E3.5 dpc blastocysts produced from parents heterozygous for the modified Snf2h allele. Duplex PCR reactions were carried out with primer pairs specific for the wild-type Snf2h locus (b9437–9458–e9605–9624) and either the neo-containing mutant locus (gneo–c) or the mutant locus (b–c) from which the neo cassette has been excised by breeding with ZP3 Cre mice (see text and Materials and Methods for details). The sizes of the expected PCR products are indicated on the left side and labeled mutant (M) or wild type (WT) on the right side, and the deduced genotypes are indicated above each lane.
Expression of SNF2h in Targeted ES Cells. For reverse transcription PCR analysis, DNase I-treated total RNA was isolated from ES cells and reverse transcribed into cDNA (SuperScript, Invitrogen); in a parallel experiment, the reverse transcriptase was not added. PCR (AccuTaq, Sigma) was carried out with Snf2h-specific primers spanning from the ATG to the TGA codons: forward (aacatgtcgtccgcggtggagcctccgccg) and reverse (ttcatagtttcagcttcttttttcttccac). Nuclear and cytoplasmic fractions of ES cell lysates were prepared by using described procedures (29). Lysates were electrophoresed on 6.5% denaturing polyacrylamide gels, and immunoblotting was performed as described (29) by using a goat anti-Snf2h antiserum (C-16, Santa Cruz Biotechnology), followed by incubation with horseradish peroxidase-linked secondary antibody (sc-2314, Santa Cruz Biotechnology). In a parallel experiment, a Snf2h-blocking peptide (sc-8760P), preincubated with the primary antibody, completely abolished staining of the 135-kDa Snf2h band.
Animal Procedures and Genotyping. Eight independent Snf2h heterozygous ES clones were injected into C57BL/6 mouse blastocysts. The chimeric blastocysts were then injected into pseudopregnant CD1 female recipients. Fifteen chimeric mice were obtained and bred with C57BL/6 mice. Snf2h heterozygotes were backcrossed with C57BL/6 and 129Sv mice. Snf2h heterozygous mice were also bred with a strain containing the ZP3 Cre transgene (30) to produce strains in which the PGK-Neo cassette was excised from the modified Snf2h locus. For mouse genotype determinations, tail DNA was prepared as described (28) and analyzed by duplex PCR by using primers (b9437–9458–e9605–9624, 200 bp) specific for the wild-type locus (NT_039467) and (gneo–c15462–15489, 520 bp) specific for the modified locus, or (b9437–9458–c15462–15489, 450 bp) specific for the modified locus from which the PGK-Neo cassette has been excised (Fig. 1 A).
Blastocyst Outgrowth Study and Histological Analysis of Implanted Embryos. Snf2h heterozygotes were mated and coitus was monitored by daily inspection for plugs. Embryos at 3.5 days postcoitum (dpc) were isolated by flushing the uterine tract with serum-containing medium. The embryos were transferred to gelatinized wells containing 100 μl of ES cell medium supplemented with lymphocyte-inhibitory factor. The cultures were incubated at 37°C in 5% CO2, humidified atmosphere, monitored each day and photographed at selected intervals. After 6 days, an aliquot of cells from each culture was taken for DNA isolation and genotyping by PCR. Embryos at 7.5 dpc and later were recovered by microdissection under the microscope and genotyped.
Antisense Inhibition of SNF2h. Human erythroleukemia K562 cells (104 per well) and magnetically sorted CD34-positive, glycophorin A (GlyA)-negative (CD34+GlyA-) cells (104 per well) derived from normal bone marrow (obtained from Cord Blood Bank, Czech Republic) were cultured for 4 days in the presence of erythropoietin (Amgen) in 100 μl of serum-free medium (31). Magnetic separation of CD34+GlyA- cells was accomplished with the CD34 Isolation Kit, magnetic apparatus, and columns from Miltenyi Biotec (Auburn, CA). Gel-purified and desalted SNF2h sense (acggaacatcatgtcgtccgcggc) and antisense (gccgcggacgacctgatgttccgt) oligonucleotides (Invitrogen) were added directly to the medium at the indicated final concentrations. Cells from aliquots of the cultures were monitored daily by flow cytometry (FACSCalibur, Becton Dickinson) for CD34 antigen [anti-CD34PE, clone 8G12(HPCA-2) from Pharmingen] and GlyA (anti-TER119FITC, clone D 2.10 from Pharmingen). RT-PCR analysis was carried out by isolating RNA from aliquots containing 103 cells with RNAzol B (Tel-Test) and incubating with SuperScript (Invitrogen) and gene-specific primers (32). After 4 days of culture, cells were washed once with PBS and transferred (103 per ml) into semisolid media supplemented with erythropoietin (3 units/ml). Semisolid cultures also contained 30% FBS and 0.9% methylcellulose in Iscove's modified Dulbecco's medium (33). Erythroid and myeloid colonies were counted after 14 days' incubation by using an inverted microscope according to standard criteria (34).
Results
Targeted Disruption of the Snf2h Gene in Mouse ES Cells. To investigate the function of Snf2h in mammalian development, the gene was inactivated by homologous recombination in mouse ES cells. A gene-targeting vector was constructed in which two 4.5-kb genomic DNA segments lying 5′ and 3′ of Snf2h exons 5 and 9a were placed upstream and downstream, respectively, of a phosphoglycerate kinase (PGK) promoter-neomycin resistance cassette (Fig. 1A). The PGK-Neo cassette was flanked by loxP sites so that it could be deleted with Cre recombinase. Additionally, a PGK-thymidine kinase cassette was inserted downstream of the 3′ homology region for negative selection with ganciclovir. Homologous recombination between the targeting vector and the Snf2h locus results in production of a modified Snf2h allele in which exons 5–9a, encoding the catalytic ATPase domain (DEXDc) of Snf2h, are deleted. The targeting vector was linearized and electroporated into WW6 ES cells, and cells resistant to G418 and ganciclovir were selected (28). Genomic DNA from 44 doubly resistant clones was analyzed by long-template genomic PCR, and 11 correctly targeted ES cell clones were identified (Fig. 1B). Western blot analysis of nuclear and cytoplasmic protein extracts with a Snf2h antiserum showed that the targeted ES cells had reduced levels of a 135 kDa, nuclear localized protein representing full-length Snf2h (Fig. 1C). No smaller protein products capable of reacting with the Snf2h antiserum were detected in the targeted cells. Furthermore, longtemplate RT-PCR analysis of Snf2h mRNA expression in the targeted ES cells produced only the expected ≈3.2-kb cDNA fragment representing the Snf2h coding sequence. No shorter products that might result from transcription of the targeted Snf2h allele were detected (data not shown). These results indicate that the targeting produced a null allele.
Early Lethality of Snf2h-Null Embryos. Two lines of Snf2h+/- mice were established from chimeric mice derived from two independently targeted ES cell clones. These lines were backcrossed to both C57BL/6 and 129Sv strains. Snf2h+/- mice in both backgrounds were phenotypically normal up to 12 months of age. To determine whether Snf2h is required for normal mouse development, mice heterozygous for the mutant allele in both backgrounds were interbred. Of 91 F2 animals of both backgrounds, 35 were wild-type and 56 were heterozygotes, but no homozygous animals were born (Fig. 1D and Table 1). Genotyping of embryos showed that Snf2h-null embryos are not present at embryonic day (E) 7.5, but they are represented at the expected Mendelian frequency in litters at E3.5. Failure to detect Snf2h-null embryos at day 7.5 or later indicates that it is required for early embryonic development. Snf2h heterozygous mutants were also bred with animals containing a transgene encoding Cre recombinase under control of the ZP3 promoter (30). By further breedings, we produced a line in which the PGK-Neo cassette is deleted. F2 litters of this line also did not contain Snf2h-null pups, indicating that the presence of the PGK-Neo gene was not responsible for the early lethality of Snf2h-null embryos. Whereas the newborn litter sizes from heterozygous intercrosses were reduced because of the death of Snf2h-null embryos, we observed that the actual number of decidual swellings at E5.5 was normal. Furthermore, approximately one-fourth of decidual nodules at E7.5 lacked embryos. We attempted to detect blastocysts that had not implanted at E4.5 by flushing embryos from the uterus, but no embryos were recovered. Taken together these results suggest that Snf2h-null embryos implant into the uterus but that by E7.5 these embryos have died and been resorbed.
Table 1. Genotype distribution of progeny in SNF2h intercross litters.
|
Snf2h genotypes*
|
|||
|---|---|---|---|
| Stage of development | +/+ | +/- | -/- |
| Postnatal | 35 (38) | 56 (62) | 0 |
| Embryo,† 7.5 dpc | 16 (39) | 25 (61) | 0 |
| Blastocyst, 3.5 dpc | 21 (28) | 35 (46) | 20 (26) |
Values in parentheses are percentages.
Embryos also were recovered at E8.5-11.5 dpc, of which nine were +/+, 20 were +/-, and none were -/-.
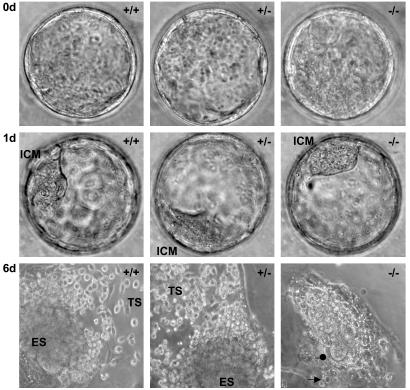
Snf2h Is Required for Survival of the Inner Cell Mass and Trophectoderm ex Vivo. To investigate the nature of the defect in early Snf2h-null embryos, we isolated blastocysts from Snf2h intercross matings and cultured them for several days, followed by genotyping of individual blastocyts by PCR. Snf2h-null blastocysts isolated at E3.5 appeared normal, consisting of viable inner cell mass (ICM) and trophoectoderm (Fig. 2). Beginning at 12 h of culture, blastocysts of all three genotypes were characterized by firm attachment of trophoblast cells to the cell-culture wells and rapid proliferation of trophoblast and ICM cells. However, after 2 days in culture Snf2h-null blastocysts began to degenerate and, subsequently, both trophoblast and ICM cells arrested and underwent massive apoptosis between 3 and 6 days in culture (Fig. 2). Furthermore, we consistently observed the formation of ES cell colonies from ICM cells of wild-type and heterozygous blastocysts, but such colonies were not observed in cultures of Snf2h-null blastocysts. ES cell lines were readily derived from either Snf2h wild-type or heterozygous blastocysts, but we were unable to obtain such lines from Snf2h-null blastocysts. These results suggest that Snf2h may be a general cell proliferation and survival factor. The blastocyst cell-culture experiments indicate that a defect first develops in Snf2h-null blastocysts at 48 h ex vivo. This time corresponds to ≈24 h after the expected time of implantation in vivo. These observations are also consistent with the foregoing suggestion that Snf2h-null embryos undergo implantation but then die in utero because of defective development occurring between E5.5 and E7.5.
Fig. 2.
Ex vivo culture of mouse blastocysts. Photographs of representative Snf2h wild-type (+/+), heterozygous (+/-), and homozygous mutant (-/-) blastocysts at the time of their isolation at 3.5 dpc (Top, ×1260), after 1 day (Middle, ×1040), and after 6 days (Bottom, ×520) of culture in serumsupplemented media (n = 76). Freshly isolated Snf2h-/- blastocysts and those cultured for 1 day were viable and expanded similarly to their wild-type and heterozygous littermates (see fully developed ICM in all genotypes, Top and Middle). After 2–5 days of culture, ICM- and trophoectoderm-derived cells from all Snf2h-/- blastocysts (n = 20) underwent growth arrest and apoptosis (see the loss of cell structure and pycnotic nuclei indicated by the arrow and vacuolized cytoplasm indicated by the dot pointing to the few remaining cells that were undergoing apoptosis). In contrast, ICM- and trophoectoderm-derived cells from all wild-type (n = 21) and heterozygous (n = 35) blastocysts rapidly proliferated and developed into compact ES cell colonies (labeled ES in the figure), surrounded at the periphery by migrating trophoblast-like structures that morphologically resemble trophoblast giant cells (TS).
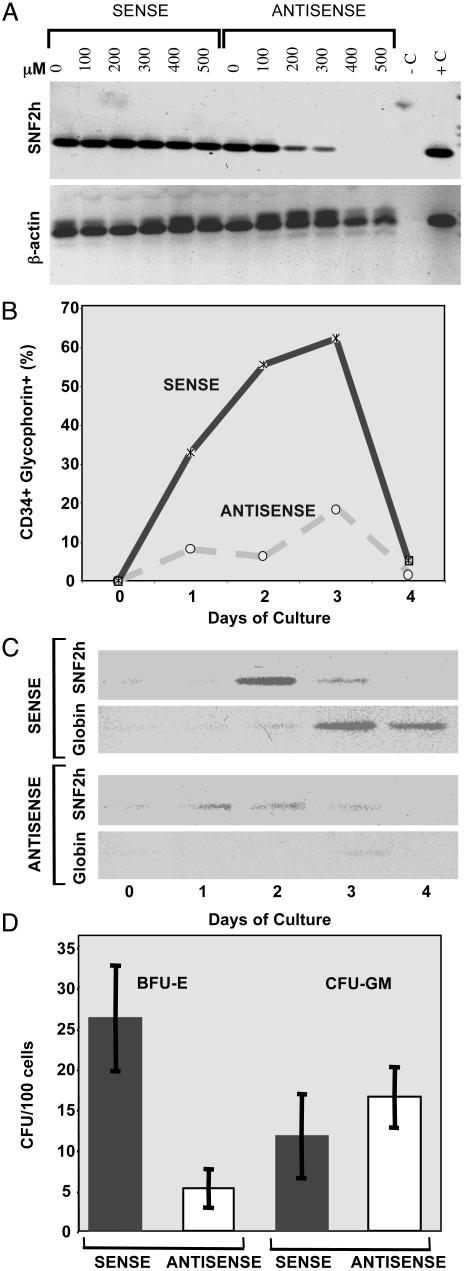
Inhibition of SNF2H Expression Blocks in Vitro Erythropoiesis. Because early lethality of Snf2h-null embryos prevented analysis of its role in later development, we investigated a requirement for SNF2h in adult erythropoiesis by antisense inhibition in primary human hematopoietic progenitors. We first determined the effectiveness of various concentrations of an antisense oligonucleotide directed against the translation-initiation region of SNF2h mRNA in K562 human erythroleukemia cells (Fig. 3A). SNF2h mRNA was reduced to very low levels by treating the cells with 400–500 μM antisense oligonucleotide, whereas no effect occurred in cells treated with the complementary sense oligonucleotide. CD34+GlyA- hematopoietic progenitors were obtained by magnetic sorting of normal human bone marrow cells. When these cells are cultured in serum-free conditions in the presence of 3 units/ml erythropoietin, they begin to proliferate and, after 1–2 days in culture, they develop into maturing erythroid cells that express GlyA (Fig. 3B). During this proliferative phase, SNF2h mRNA levels also rise and then decline as cells mature and cease to proliferate (Fig. 3C). Subsequently, β-globin mRNA levels rise, and the cells rapidly lose the CD34 antigen.
Fig. 3.
Requirement of SNF2h for in vitro erythropoiesis. (A) Antisense inhibition of SNF2h mRNA levels in human erythroleukemia cells. K562 cells were incubated in serum-free media for 18 h in the presence of the indicated concentrations (micromolar) of antisense and sense SNF2h oligonucleotides. Total cellular RNA was prepared and assayed for the levels of SNF2h and β-actin mRNAs by RT-PCR. For further details, see Materials and Methods. The lane marked -C contained no cDNA template, and the lane marked +C contained SNF2h cDNA as a template for PCR. (B and C) SNF2h inhibition blocks erythroid development of hematopoietic progenitors. Magnetically sorted CD34+GlyA- human bone marrow progenitors were incubated for the indicated times with 500 μM SNF2h antisense or sense oligonucleotides in serum-free medium containing 3 units/ml erythropoietin. (B) The fraction of cells expressing both CD34 antigen and GlyA was assayed by flow cytometry as described in Materials and Methods. (C) Levels of SNF2h andβ-globin mRNAs were assayed by RT-PCR of total cellular RNA. (D) SNF2h inhibition blocks development of committed erythroid progenitors. Cell populations were prepared and incubated for 4 days in the presence of antisense or sense oligonucleotides as described in B and C. The cells were then plated in semisolid cultures with 3 units/ml erythropoietin and incubated for 14 days. Colonies containing ≈500–2,000 cells were scored as either fully hemoglobinized BFU-E usually containing multiple clusters of erythroblasts in different stages of terminal differentiation, or unhemoglobinized concentric CFU-GM colonies mainly consisting of differentiating myeloblasts. BFU-E colonies were not observed in plates lacking erythropoietin, whereas the number of CFU-GM colonies was not influenced by the absence of erythropoietin.
Cultures treated with 500 μM SNF2h antisense oligonucleotide showed a marked reduction in development of GlyA+ cells and decreased β-globin mRNA accumulation (Fig. 3 B and C). The initial rise in SNF2h mRNA levels in response to culture with erythropoietin also was significantly reduced in antisense cultures (Fig. 3C). To determine whether the failure to develop differentiated cells in SNF2h inhibited cultures is due to inhibition of erythroid progenitor proliferation or simply inhibition of erythroid marker expression, we also examined production of erythroid BFU-E colonies in semisolid methylcellulose cultures. SNF2h antisense treatment of progenitor cells in liquid serum-free culture reduced the number of BFU-E colonies observed in semisolid media by ≈80% as compared with sense oligonucleotide-treated cultures (Fig. 3D) or with untreated cells (data not shown). The sorted CD34+GlyA- progenitor population treated with erythropoietin contains a low level of contaminating, committed granulocyte-macrophage progenitors that can spontaneously form CFU-GM colonies in the semisolid cultures. The number of these colonies was unaffected by SNF2h antisense oligonucleotide treatment, showing that the treatment with oligonucleotide in liquid culture is not toxic to cells and that multicellular colonies can develop subsequently in methylcellulose cultures. These results suggest that SNF2H is required for proliferation and/or differentiation of very immature progenitors with extensive cytokineinduced proliferative capacity. The observed down-regulation of Snf2h mRNA levels in differentiating cells after 3–4 days in liquid culture (Fig. 3C) and during mouse erythroleukemia cell differentiation (32) suggests that Snf2h may not be involved in the latter stages of erythroid differentiation.
Discussion
Mammals, like other organisms, contain multiple ATP-dependent complexes that remodel nucleosomes. Although some of the properties of these complexes overlap in in vitro assays, they contain one of three different families of ATPase subunits and a variety of different associated proteins, suggesting that these complexes have different functions in vitro (4). The most definitive approach to investigate their roles in mammalian development is to generate mice with mutations in the genes encoding their subunits. Inactivation of Brg1, encoding the core ATPase subunit of one of the major SWI/SNF complexes, showed that it is an essential gene in mice (24). Embryos homozygous for a Brg1-null mutation die during the periimplantation stage, some time between E3.5 and E6.5. Genotype analysis of embryos in utero from mutant intercrosses and blastocyst in vitro culture experiments indicate that Brg1 homozygotes die before implantation is complete. These studies also indicate that Brg1 is required for survival of both the inner cell mass and the trophectoderm. However, other experiments show that mouse embryo fibroblasts lacking BRG1 can proliferate, indicating that BRG1 is not required by all cells for survival and proliferation. SWI/SNF complexes contain either BRG1 or BRM ATPases. In contrast to the studies in which Brg1 was inactivated, Brm-null mice develop normally (25). The level of BRG1 protein is up-regulated in Brm-null mice, suggesting possible compensation of BRM functions by SWI/SNF complexes containing BRG1. However, the results with Brg1-null embryos show that BRM-containing complexes cannot compensate for complexes containing Brg1.
ISWI complexes are a second major category of ATP-dependent chromatin remodeling complexes, and SNF2h is the predominate ATPase subunit present in these complexes. The results reported here show that homozygous inactivation of Snf2h produces a set of phenotypes that are very similar to those observed in Brg1-null embryos. Both types of null embryos die during early mouse development. Furthermore, in both cases, blastocyst culture experiments indicate that loss of either of these two ATPases leads to growth arrest and cell death of both the trophectoderm and inner cell mass. On the other hand, we found evidence that Snf2h-/- embryos underwent implantation, whereas Brg1-/- embryos appear to die before implantation. The difference could suggest that requirements for BRG1- versus SNF2h-containing complexes are manifested at different times in early development. It is also possible that the difference reflects a difference in the rates of loss of the maternal mRNA or protein stores for these two factors.
The finding that Snf2h-null embryos die early in development is consistent with the expression pattern of the gene. Snf2h is widely expressed during both fetal and postnatal development. Snf2h mRNA is detectable in embryos as early as E9.5 (35). We and others have found that Snf2h protein is detectable in ES cells (Fig. 1C) (16). This finding suggests that Snf2h also plays an essential role at a very early stage in development, a role that is not revealed in Snf2h-null embryos because of the maternal contribution.
Similar to SWI/SNF complexes that contain either BRG1 or BRM ATPases, at least two homologous ATPases can occupy ISWI complexes. Snf2h and Snf2l are two highly homologous ISWI-type ATPases that share 81% amino acid sequence identity and 75% DNA-coding sequence identity (35). Expression of Snf2l is upregulated in terminally differentiated cells in adult brain, ovary, testis, and lymphocytes, whereas expression of Snf2h is generally associated with highly proliferating cells of the same organs (35). However, both proteins are present in remodeling complexes obtained from ES cells (16) and both mRNAs are detectable in E9.5 mouse embryos (35). Nevertheless, our experiments indicate that Snf2l is not able to compensate for loss of Snf2h during early embryogenesis. Most likely, these two ISWI-type ATPases play distinct roles in development. This view is supported by the finding that recessive mutations in Snf2l are associated with Schimke immunoosseous dysplasia, a human disorder with specific postnatal phenotypes (36).
The studies of both Brg1 and Brm mutant mice suggest that mammalian SWI/SNF complexes participate in control of cell proliferation. Brm-null mice are heavier than controls and exhibit an increased mitotic index in liver tissue (25). Brm-null mouse embryo fibroblasts were deficient in their ability to undergo G0/G1 arrest in response to stimuli that normally induce quiescence (25). Brg1 heterozygous mutants were found to be susceptible to developing spontaneous tumors (24). These results are consistent with the view that SWI/SNF complexes are important in negative regulation of cell-cycle progression. Indeed, germ-line mutations of INI1, a noncatalytic component of SWI/SNF complexes, are associated with development of rhabdoid tumors in human patients (37) and mice heterozygous for INI1 develop such tumors (38–40). In contrast, we did not find any evidence of increased neoplasia in Snf2h heterozygotes. We have monitored 70 heterozygous mutants and 70 wild-type littermates of both sexes for up to 12 months, and we have not observed tumor formation in any of the mice. In fact, expression of Snf2h is generally associated with active cell proliferation, and we and others have found that Snf2h is expressed in all tested immortal or tumor-derived cell lines (15, 26, 32). The studies of Snf2h-null embryos reported here certainly support the view that Snf2h is required for cell proliferation. As shown in Fig. 2, the ICM is clearly intact in Snf2h-null blastocysts, but in vitro culture of such blastocysts failed to show any evidence of ES cell colony formation, whereas culture of wild-type and heterozygous blastocysts consistently produced colonies with ES cell characteristics. Furthermore, we made numerous attempts to derive Snf2h-null ES cell lines and all such attempts failed, whereas ES cell lines were readily established from wild-type or heterozygous littermate blastocysts. We also found by antisense inhibition experiments that Snf2h is required for the development of erythroid colonies from early hematopoietic progenitors (Fig. 3). Complexes containing Snf2h localize to foci of replicating heterochromatin and depletion of Snf2h in cultured cell lines reduces the rate of DNA replication throughout S phase (15). The Snf2h gene also has been identified as a target for regulation by the E2F transcription factors, which are key regulators of other genes required for cell proliferation (41). These observations may explain, at least in part, the proposed requirement for Snf2h in cycling cells but further studies will be needed to understand the roles of Snf2h in proliferation of normal cells. The gene-inactivated mouse strains described here should be very useful for such work, as well as for elucidating the role of Snf2h in specific cell types during differentiation and development.
Acknowledgments
We thank Jana Cmejlova (Institute of Hematology, Prague) for help with sequencing analyses of the Snf2h locus, Petr Kobylka (Cord Blood Bank, Eurocord Project) for providing bone marrow and cord blood samples, and Jan Zivny (First Medical Faculty, Charles University) for help with flow cytometry analyses. Injections of Snf2h+/- ES cells into pseudopregnant females were done by Harry Hou, Jr., and Marcus Vargas (Gene Targeting Facility, Albert Einstein College of Medicine). This work was supported by National Institutes of Health Grant CA16368. T.S. was supported by the SASS Foundation for Leukemia Research, the Leukemia Research Foundation, and the Grant Agency of the Czech Republic (301-00-1061). A.I.S. also receives support from National Cancer Institute Cancer Center Grant 2P30CA13330.
Abbreviations: BRM, brahma; dpc, days postcoitum; ES, embryonic stem; BRG1, BRM-related gene 1; ICM, inner cell mass; PGK, phosphoglycerate kinase; En, embryonic day n; GlyA, glycophorin A.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. AF375046 (Mus musculus ATP-dependent chromatin remodeling protein Snf2h mRNA, complete cds) and NT_039467 (M. musculus chromosome 8 genomic contig, strain C57BL/6J)].
References
- 1.Richmond, T. J. & Davey, C. A. (2003) Nature 423, 145–150. [DOI] [PubMed] [Google Scholar]
- 2.Felsenfeld, G. & Groudine, M. (2003) Nature 421, 448–453. [DOI] [PubMed] [Google Scholar]
- 3.Narlikar, G. J., Fan, H. Y. & Kingston, R. E. (2002) Cell 108, 475–487. [DOI] [PubMed] [Google Scholar]
- 4.Fan, H. Y., He, X., Kingston, R. E. & Narlikar, G. J. (2003) Mol. Cell 11, 1311–1322. [DOI] [PubMed] [Google Scholar]
- 5.Emerson, B. M. (2002) Cell 109, 267–270. [DOI] [PubMed] [Google Scholar]
- 6.Kadam, S. & Emerson, B. M. (2002) Curr. Opin. Cell Biol. 14, 262–268. [DOI] [PubMed] [Google Scholar]
- 7.Peterson, C. L. & Workman, J. L. (2000) Curr. Opin. Genet. Dev. 10, 187–192. [DOI] [PubMed] [Google Scholar]
- 8.Peterson, C. L. (2003) Curr. Biol. 13, R195–R197. [DOI] [PubMed] [Google Scholar]
- 9.Kadam, S. & Emerson, B. M. (2003) Mol. Cell 11, 377–389. [DOI] [PubMed] [Google Scholar]
- 10.Liu, R., Liu, H., Chen, X., Kirby, M., Brown, P. O. & Zhao, K. (2001) Cell 106, 309–318. [DOI] [PubMed] [Google Scholar]
- 11.Strobeck, M. W., Knudsen, K. E., Fribourg, A. F., DeCristofaro, M. F., Weissman, B. E., Imbalzano, A. N. & Knudsen, E. S. (2000) Proc. Natl. Acad. Sci. USA 97, 7748–7753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang, H. S., Gavin, M., Dahiya, A., Postigo, A. A., Ma, D., Luo, R. X., Harbour, J. W. & Dean, D. C. (2000) Cell 101, 79–89. [DOI] [PubMed] [Google Scholar]
- 13.Zhou, Y., Santoro, R. & Grummt, I. (2002) EMBO J. 21, 4632–4640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Santoro, R., Li, J. & Grummt, I. (2002) Nat. Genet. 32, 393–396. [DOI] [PubMed] [Google Scholar]
- 15.Collins, N., Poot, R. A., Kukimoto, I., Garcia-Jimenez, C., Dellaire, G. & Varga-Weisz, P. D. (2002) Nat. Genet. 32, 627–632. [DOI] [PubMed] [Google Scholar]
- 16.Bozhenok, L., Wade, P. A. & Varga-Weisz, P. (2002) EMBO J. 21, 2231–2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fyodorov, D. V. & Kadonaga, J. T. (2001) Cell 106, 523–525. [DOI] [PubMed] [Google Scholar]
- 18.Peterson, C. L. & Herskowitz, I. (1992) Cell 68, 573–583. [DOI] [PubMed] [Google Scholar]
- 19.Wilson, C. J., Chao, D. M., Imbalzano, A. N., Schnitzler, G. R., Kingston, R. E. & Young, R. A. (1996) Cell 84, 235–244. [DOI] [PubMed] [Google Scholar]
- 20.Sudarsanam, P., Iyer, V. R., Brown, P. O. & Winston, F. (2000) Proc. Natl. Acad. Sci. USA 97, 3364–3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsukiyama, T., Palmer, J., Landel, C. C., Shiloach, J. & Wu, C. (1999) Genes Dev. 13, 686–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deuring, R., Fanti, L., Armstrong, J. A., Sarte, M., Papoulas, O., Prestel, M., Daubresse, G., Verardo, M., Moseley, S. L., Berloco, M., et al. (2000) Mol. Cell 5, 355–365. [DOI] [PubMed] [Google Scholar]
- 23.Elfring, L. K., Daniel, C., Papoulas, O., Deuring, R., Sarte, M., Moseley, S., Beek, S. J., Waldrip, W. R., Daubresse, G., DePace, A., et al. (1998) Genetics 148, 251–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bultman, S., Gebuhr, T., Yee, D., La Mantia, C., Nicholson, J., Gilliam, A., Randazzo, F., Metzger, D., Chambon, P., Crabtree, G., et al. (2000) Mol. Cell 6, 1287–1295. [DOI] [PubMed] [Google Scholar]
- 25.Reyes, J. C., Barra, J., Muchardt, C., Camus, A., Babinet, C. & Yaniv, M. (1998) EMBO J. 17, 6979–6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aihara, T., Miyoshi, Y., Koyama, K., Suzuki, M., Takahashi, E., Monden, M. & Nakamura, Y. (1998) Cytogenet. Cell Genet. 81, 191–193. [DOI] [PubMed] [Google Scholar]
- 27.Masuoka, H. C. & Townes, T. M. (2002) Blood 99, 736–745. [DOI] [PubMed] [Google Scholar]
- 28.Fan, Y., Sirotkin, A., Russell, R. G., Ayala, J. & Skoultchi, A. I. (2001) Mol. Cell. Biol. 21, 7933–7943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matushansky, I., Radparvar, F. & Skoultchi, A. I. (2000) Proc. Natl. Acad. Sci. USA 97, 14317–14322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lewandoski, M., Wassarman, K. M. & Martin, G. R. (1997) Curr. Biol. 7, 148–151. [DOI] [PubMed] [Google Scholar]
- 31.Miller, C. L. & Eaves, C. J. (1997) Proc. Natl. Acad. Sci. USA 94, 13648–13653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stopka, T., Zakova, D., Fuchs, O., Kubrova, O., Blafkova, J., Jelinek, J., Necas, E. & Zivny, J. (2000) Leukemia 14, 1247–1252. [DOI] [PubMed] [Google Scholar]
- 33.Plasilova, M., Zivny, J., Jelinek, J., Neuwirtova, R., Cermak, J., Necas, E., Andera, L. & Stopka, T. (2002) Leukemia 16, 67–73. [DOI] [PubMed] [Google Scholar]
- 34.Prchal, J. F. & Axelrad, A. A. (1974) N. Engl. J. Med. 290, 1382. [DOI] [PubMed] [Google Scholar]
- 35.Lazzaro, M. A. & Picketts, D. J. (2001) J. Neurochem. 77, 1145–1156. [DOI] [PubMed] [Google Scholar]
- 36.Boerkoel, C. F., Takashima, H., John, J., Yan, J., Stankiewicz, P., Rosenbarker, L., Andre, J. L., Bogdanovic, R., Burguet, A., Cockfield, S., et al. (2002) Nat. Genet. 30, 215–220. [DOI] [PubMed] [Google Scholar]
- 37.Versteege, I., Sevenet, N., Lange, J., Rousseau-Merck, M. F., Ambros, P., Handgretinger, R., Aurias, A. & Delattre, O. (1998) Nature 394, 203–206. [DOI] [PubMed] [Google Scholar]
- 38.Guidi, C. J., Sands, A. T., Zambrowicz, B. P., Turner, T. K., Demers, D. A., Webster, W., Smith, T. W., Imbalzano, A. N. & Jones, S. N. (2001) Mol. Cell. Biol. 21, 3598–3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roberts, C. W., Leroux, M. M., Fleming, M. D. & Orkin, S. H. (2002) Cancer Cell 2, 415–425. [DOI] [PubMed] [Google Scholar]
- 40.Klochendler-Yeivin, A., Fiette, L., Barra, J., Muchardt, C., Babinet, C. & Yaniv, M. (2000) EMBO Rep. 1, 500–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weinmann, A. S., Yan, P. S., Oberley, M. J., Huang, T. H. & Farnham, P. J. (2002) Genes Dev. 16, 235–244. [DOI] [PMC free article] [PubMed] [Google Scholar]