Abstract
Aging is linked to loss of the self-renewal capacity of adult stem cells. Here, we observed that human multipotent stem cells (MSCs) underwent cellular senescence in vitro. Decreased expression of histone deacetylases (HDACs), followed by downregulation of polycomb group genes (PcGs), such as BMI1, EZH2 and SUZ12, and by upregulation of jumonji domain containing 3 (JMJD3), was observed in senescent MSCs. Similarly, HDAC inhibitors induced cellular senescence through downregulation of PcGs and upregulation of JMJD3. Regulation of PcGs was associated with HDAC inhibitor-induced hypophosphorylation of RB, which causes RB to bind to and decrease the transcriptional activity of E2F. JMJD3 expression regulation was dependant on histone acetylation status at its promoter regions. A histone acetyltransferase (HAT) inhibitor prevented replicative senescence of MSCs. These results suggest that HDAC activity might be important for MSC self-renewal by balancing PcGs and JMJD3 expression, which govern cellular senescence by p16INK4A regulation.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-009-0242-9) contains supplementary material, which is available to authorized users.
Keywords: Multipotent stem cell, Polycomb gene, JMJD3, Senescence, Epigenetic
Introduction
Modification of chromatin by epigenetic regulatory mechanisms, including acetylation, deacetylation and methylation of histones, is considered an important mechanism for stem cell pluripotency [1]. Nuclear transplantation remodels epigenetic marks to reprogram somatic cell by balancing between heterochromatic and euchromatic marks [2]. Transduction of exogenous “pluripotency genes,” such as OCT4 and SOX2, to somatic cells to induce pluripotency showed that epigenetic modulations for endogenous stemness genes rebound, and silence of exogenes are necessary for full reprogramming [3, 4]. Pan et al. [5] reported that localization of trimethylation of histon H3 at lysine 4 residue (H3K4Me3), an euchromatic mark of histone, is predominant in OCT4, SOX2 and NANOG genes in embryonic stem cells, and colocalization of both H3K4Me3 and trimethylation of histone H3 at lysine 27 residue (H3K27Me3) occurred during differentiation to repress these pluripotency genes. Taken together, the chromatin status of pluripotency genes is crucial for regulation of self-renewal and pluripotency of stem cells.
Aging of an organism is a complex process that involves the decline of multiple systems that are required to maintain the homeostasis of cells and tissues in addition to the impairment of tissue maintenance and repair [6]. Recent studies suggest that an age-associated decline in adult stem cell function might also be associated with these processes [7–14].
Replicative senescence of multipotent stem cells (MSCs) involves the “Hayflick phenomenon” [15], which is characterized by cell enlargement, changes in morphology and ultimate cessation of proliferation. Several lines of evidence show that the polycomb group genes (PcGs) and JMJD3 histone lysine demethylase are involved in the homeostasis of p16INK4A, a key regulator of premature cellular senescence. Polycomb repressor complex 2 (PRC2) initiates chromatin repression by binding to target genes and inducing methylation of histone proteins, primarily at lysine 27 of histone H3, by EZH2, a protein with histone methyltransferase activity. Thereafter, members of polycomb repressor complex 1 (PRC1), such as chromobox gene (CBX) and BMI1, are able to recognize trimethylated histone H3 lysine 27 (H3K27Me3) and bind to condensed nucleosomes [16]. Young cells have compacted chromosomes at the CDKN2 (p16INK4A–p19ARF) locus as a result of a high extent of H3K27Me3, which is caused by PcG interaction. On the other hand, replicative senescence hinders binding of PRC2 group proteins to the CDKN2 locus, thereby reducing the recruitment of the PRC1 maintenance complex to the p16INK4A promoter region. The collapse of polycomb protein complexes results in an increase of p16INK4A expression, which induces senescence [16, 17]. On the other hand, JMJD3 increases in oncogene- and stress-induced senescence to demethylate H3K27Me3 at the CDKN2 locus, leading to an increase of p16INK4A expression [18]. Modification of histones bound to lineage-specification genes is also a critical step for maintenance of multipotency [19]. In human adipose tissue-derived MSCs, H3K27Me3 and H3K4Me3 bivalent modifications repress adipogenic genes such as fatty acid-binding protein 4 (FABP4) and peroxisome proliferator activated receptor-γ2 (PPARγ2) by coexisting on their promoter region of adipogenic genes and repress the expression. Upon induction of adipogenesis, histone H3 bound to the promoters of adipogenic genes is acetylated at lysine 9 residues and is demethylated at lysine 27, activating transcription of adipogenic specific genes.
In contrast to its subordinate circuitry, the up-stream regulatory mechanism of PcGs is poorly understood. BMI1 possesses a c-MYC binding E-box motif in its proximal promoter, and its transcription has been demonstrated to be regulated by c-MYC [20]. EZH2, EED and SUZ12, members of the PRC2 complex, are induced by E2F overexpression in the U2OS human osteosarcoma cell line [21]. Indeed, the proximal promoters of EED and EZH2 are bound by the E2F transcription factor and are transcriptionally regulated by the RB-E2F interaction [22]. E2F target genes are known to be stably repressed by the RB tumor suppressor in senescent cells, but not in quiescent cells [23].
Taken together, key mechanisms in the regulation of the balance of PcGs and JMJD3 during replicative senescence, which may be crucial for maintenance of self-renewal in adult stem cells, remain unclear. Here, we examined the role of HDAC activity in PcGs and JMJD3 regulation to keep MSCs from undergoing cellular senescence.
Materials and methods
Isolation and culture of hMSCs
Human adipose tissue-derived MSCs (hAD-MSCs) [23] and human umbilical cord blood-derived MSCs (hUCB-MSCs) [24] were isolated and cultured as previously described. In brief, two clones of hAD-MSCs were established from freshly excised mammary fat tissue from Ba-Ram Plastic Surgery Hospital. Tissues were obtained from 20- to 30-year-old women during reduction mammoplasty with informed consent. The hAD-MSCs were maintained in K-NAC medium supplemented with 2 mM N-acetyl-l-cysteine (Sigma-Aldrich, St. Louis, MO) and l-ascorbic acid (0.2 mM, Sigma-Aldrich).
hUCB-MSCs were obtained from umbilical cord blood immediately after full-term delivery with consent from 20–30-year-old mothers and approval of the Boramae Hospital Institutional Review Board (IRB). Two hUCB-MSC clones were used in this experiment. hUCB-MSCs were maintained in DMEM (Invitrogen, Carlsbad, CA) containing 10% FBS. All procedures for preparation and utilization of hAD-MSCs (IRB no. 0611/001-001) and hUCB-MSCs (IRB no. 0603/001-002) for research purposes were approved by the institutional review board of Seoul National University.
Measurement of proliferation potential
The effects of VPA and NaBu on MSC proliferation were measured using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT, Sigma-Aldrich, St. Louis, MO) assay, as described previously [25]. In brief, cells were plated on 24-well plates at a density of 2.5 × 104/ml and cultured for 24 h. At the end of the incubation, 50 μl of MTT stock solution (5 mg/ml) was added, and the plates were further incubated for 4 h at 37°C. Formazan crystals were solublized with DMSO, and the absorbance was measured with an EL800 microplate reader (Bio Tek Instruments, Winooski, VT). Cell population doubling levels (CPDL) were also measured from passage 1 to passage 15. MSCs were seeded at a density of 4 × 105 cells per 10 cm plate and incubated for 4 days. Cell counting was performed at least three times, and the average values were used for the CPDL graph.
Senescence-associated beta-galactosidase staining
Senescence-associated beta-galactosidase (SA β-gal) staining was performed as described by Narita et al. [26] with modifications. MSCs were seeded on six-well plates at a density of 1 × 105 cells/well for late passage cells and 5 × 104 cells/well for early passage cells. Cells were incubated for 3 days until reaching appropriate confluency. For long-term HDAC inhibitor treatment, early passage cells were seeded at a density of 1–2 × 104 cells/well and treated for 1, 3, 5 or 7 days. Cells were washed twice with PBS and fixed with 0.5% glutaraldehyde in PBS (pH 7.2) for 5 min at room temperature. Cells were then washed with MgCl2-containing PBS (pH 7.2, 1 mM MgCl2) and stained in X-gal solution overnight at 37°C. Cells were washed twice with PBS, and the nuclei were stained with Hoechst 33258 (1 μg/ml; 10 min).
Immunocytochemistry
Immunocytochemical analyses of HDAC1, HDAC2, c-MYC, BMI1, SUZ12, p16INK4A, E2F-1 and RB were performed. Cells cultured with or without 10 mM VPA or 5 mM NaBu for 5 or 7 days were fixed in 4% paraformaldehyde and permeabilized with 0.2% Triton X-100 (Sigma-Aldrich, St Louis, MO). The cells were incubated with 10% normal goat serum (Zymed Laboratories Inc., San Francisco, CA) and then stained with antibodies against HDAC1 (1:200, Upstate Biotechnology, Lake Placid, NY), HDAC2 (1:200, Upstate Biotechnology), c-MYC (1:100, Santa Cruz, Santa Cruz, CA), BMI1 (1:200, Abcam, Cambridge, UK), SUZ12 (1:200, Abcam) and p16INK4A (1:200, Abcam), E2F-1 (1:200, Santa Cruz) and RB (Millipore, Billerica, MA) followed by 1 h incubation with Alexa 488- or Alexa 594-labeled secondary antibody (1:1,000; Molecular Probes, Eugene, OR). The nuclei were stained with Hoechst 33258 (1 μg/ml; 10 min). The images were captured with a confocal microscope (Nikon, Eclipse TE200, Japan).
Western blot analysis
Western blot analyses of HDAC1, HDAC2, c-MYC, JMJD3, BMI1, SUZ12, EZH2, p16INK4A, H3K4Me3, H3K27Me3 and RB were performed as described previously [25]. MSCs cultured with or without HDAC inhibitors for 1, 3, 5 or 7 days were lysed with 50 mM Tris–HCl buffer containing 0.1% Triton X100 freshly supplemented with protease/phosphatase inhibitor cocktail (1 mM phenylmethylsulfonyl fluoride, 1 mM aprotinin, 1 mM leupeptin, 1 mM antipain and 0.1 mM sodium orthovanadate). Protein samples were separated on 7.5–15% SDS-PAGE and then transferred to nitrocellulose membranes at 50 V, 350 mA for 5 h. All antibodies were used according to the manufacturer’s instructions. Antibodies used for Western blot analysis were HDAC1 (1:1,000, Upstate Biotechnology), HDAC2 (1:200, Upstate Biotechnology), c-MYC (1:100, R&D Systems, Minneapolis, MN), JMJD3 (1:500, Santa Cruz), BMI1 (1:200, Chemicon, Temecula, CA), SUZ12 (1:200, Abcam), EZH2 (1:200, Millipore), p16INK4A (1:200, Abcam), H3K4Me3 (1:2,500, Abcam), H3K27Me3 (1:2,500, Upstate Biotechnology) and RB (1:1,000, Millipore). Protein bands were detected using an enhanced chemiluminescence detection kit (Amersham Pharmacia Biotech, Buckinghamshire, UK) and X-ray film after recognition with the respective secondary antibodies conjugated with horseradish peroxidase.
RT-PCR
Total cellular RNA was extracted from cells with TRIzol reagentTM (Invitrogen) according to the manufacturer’s instructions. cDNA was synthesized by adding the purified RNA and oligo-dT primers to Accupower RT premix (Bioneer, Korea) according to the manufacturer’s instructions. PCR was conducted with Accupower PCR premix (Bioneer). The primer sets used for this study are listed in Table S1. All PCR products were analyzed by gel electrophoresis on 1.5% agarose gels with ethidium bromide staining, followed by fluorescence digitization using a Bio-Rad GelDoc XR system (Bio-Rad). Semi-quantitative RT-PCR was conducted by quantifying RT-PCR band using ImageJ image analysis software (National Institutes of Health, USA). Each gene was normalized with beta-actin or RPL13A as housekeeping control. At least three independent analyses were done for each gene.
siRNA inhibition study
For specific HDAC1 and HDAC2 inhibition, siRNA inhibition studies were performed using commercial siRNA targeting HDAC1 and HDAC2 along with a non-targeting siRNA (Dharmacon, ON Target plus SMART pool, cat. nos. L-003493-00, L-003495-00, D-001210-01-05). siRNA transfections were done according to manufacturer’s instructions. In brief, cells were seeded at a density of 5 × 104 cells/well, and siRNA-containing media (without addition of antibiotics) was added when cells reached 50% confluence. Cells were incubated with 50 or 100 nM siRNA for 48 or 96 h. To investigate the long-term effect of HDAC inhibition, cells were subcultured 48 h post-siRNA transfection. Subcultured cells were stabilized for 24 h and incubated with siRNA for 72 h at the same concentration. After inhibition, RNA, protein extraction or SA β-gal staining was performed for genetic or characteristic analyses.
Chromatin immunoprecipitation assays
hAD-MSC and hUCB-MSC were seeded in 10-cm plates at a density of 0.8–1 × 105 and cultured with or without 4 mM VPA or 2 mM NaBu for 7 days. Cells in late passage were seeded at a density of approximately 5 × 105 cells per 10-cm plate and incubated for 7 days. ChIP assays were performed according to the manufacturer’s protocol (ChIP assay kit, Upstate Biotechnology). Chromatin was immunoprecipitated by antibodies according to the manufacturer’s instructions. PCR reactions were performed in a final template dilution of 1:50, and PCR products were analyzed by gel electrophoresis on 1.5–2% agarose gels with ethidium bromide staining, followed by fluorescence digitization using a Bio-Rad GelDoc XR system (Bio-Rad). Semi-quantitative RT-PCR was conducted by quantifying RT-PCR bands using ImageJ image analysis software (National Institutes of Health, USA). Each value was expressed as a ratio of input value. The primer sets used for ChIP assays are listed in Table S2.
Statistical analysis
All experiments were conducted at least in triplicate (n = 3), and the results are expressed as the mean ± SD. Statistical analysis was conducted via analysis of variance (ANOVA), followed by Duncan’s multiple range tests or Student’s t test. A value of p < 0.05 was considered significant.
Results
Replicative senescence in hUCB- and hAD-MSCs downregulates HDAC, c-MYC and PcGs
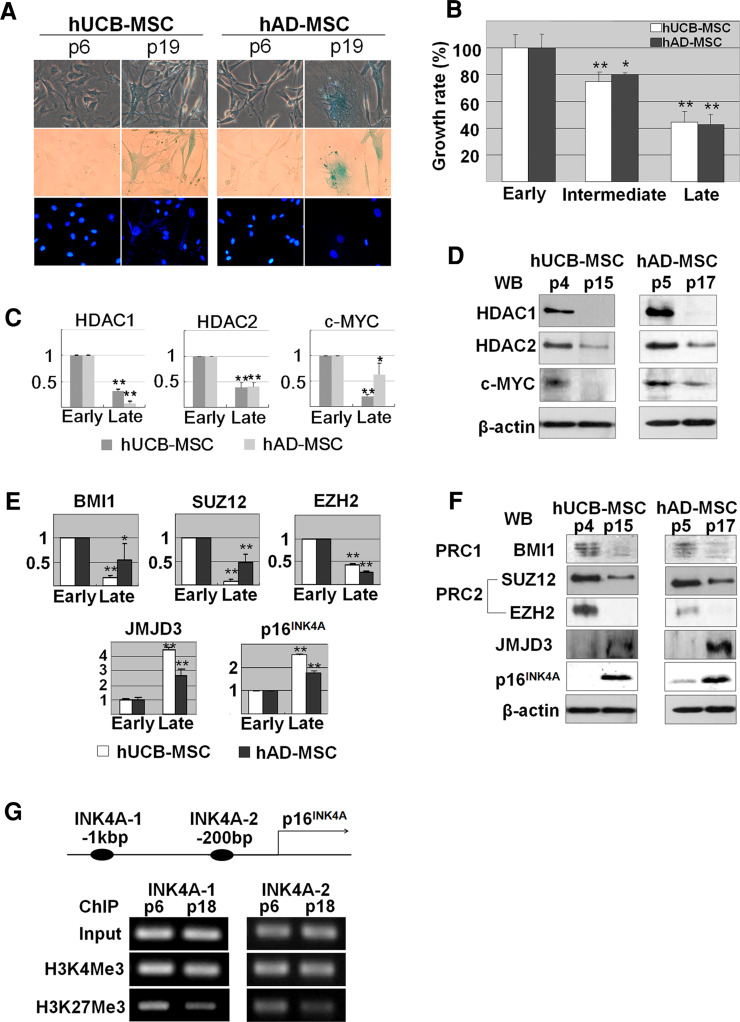
In order to induce replicative senescence, we repeatedly subcultured hUCB- and hAD-MSCs in vitro. Based on the cumulative population doubling level, it was assumed that human MSCs began to lose their proliferation potential at around the tenth passage (Figure S1). At the 19th passage, we confirmed that both hUCB- and hAD-MSCs completely lost their self-renewal capability, as evidenced by a substantial decrease of proliferation potential and the development of senescent phenotypes, which were revealed using senescence-associated beta-galactosidase (SA β-gal) staining (Fig. 1a). The senescence status of MSCs was divided into three grades based on passage numbers: early (fewer than 10 passages), intermediate (10 to 15 passages) and late (more than 16 passages). It was observed that senescent hUCB- and hAD-MSCs had reduced proliferative potential (less than 50% of early passage MSCs), as shown by the MTT cell proliferation assay (Fig. 1b). We speculated that the expression levels of HDAC1 and HDAC2, representative HDACs in eukaryotes, and c-MYC would decrease, based on our previous report indicating that HDAC activity is crucial for maintaining proliferative potential and the multi-lineage differentiation ability of MSCs in vitro. Indeed, mRNA and protein expression levels of HDAC1, HDAC2 and c-MYC were downregulated upon replicative senescence of MSCs (Fig. 1c; Figure S1A). In addition, in vitro cellular senescence altered the localization of these proteins from nuclear to cytosolic (Figure S1C).
Fig. 1.
Replicative senescence of hUCB- and hAD-MSCs. a MSCs undergo replicative senescence upon repeated (more than 15 passages) subculture in vitro, as visualized by SA β-gal staining. b MSCs begin to lose proliferation potential at around the tenth passage (intermediate), and in senescent stage (late), proliferation potential was reduced to 50% or less compared to early passage (fewer than seven passages) MSCs. Expression levels of HDAC1, HDAC2 and c-MYC are downregulated during repeated subculture-induced senescence of MSCs as shown by RT-PCR (c) and immunoblot analysis (d). Senescent MSCs show decreased expression levels of BMI1 (PRC1 member), SUZ12 and EZH2 (members of PRC2) and an increased expression level of JMJD3. p16INK4A expression levels also increased, as shown by RT-PCR (e) and immunoblot analysis (f). g Chromatin immunoprecipitation was performed to address whether upregulation of p16INK4A depends on demethylation of the H3K27 residue. Specific demethylation of H3K27, not of H3K4, was observed in late passage (p18) hUCB-MSCs. Two regions on the p16INK4A promoter targeted by specific primer sets are described schematically. Graphs represent semi-quantification of RT-PCR results from at least three independent experiments. * and ** represent statistical significance at the levels of p < 0.05 and p < 0.005, respectively
We observed that mRNA and protein expression levels of PcGs, such as BMI1, SUZ12 and EZH2, and JMJD3 histone lysine demethylase decreased in senescent hUCB- and hAD-MSCs. As a result, p16INK4A expression was increased in senescent MSCs. In contrast, the expression level of EED was not affected during senescence of MSCs, as shown by RT-PCR and immunoblot analyses (Fig. 1e, f; Figure S1B). In order to confirm whether the upregulation of p16INK4A depends on demethylation at lysine 27 of histone H3 (H3K27), we performed a ChIP assay and compared H3K27 to anti-trimethylated H3K4 (a repressive histone marker known as a target site for EZH2 histone methyltransferase) (Fig. 1g). It was observed that the promoter region was enriched with trimethylated H3K27 (H3K27Me3), not with the activated histone marker, H3K4Me3.
In order to confirm the alteration of HDACs, c-MYC and PcGs expression in vivo, we used an in vivo murine aging model to analyze expression levels of these genes by RT-PCR. Among several organs, liver and inguinal fat showed a drastic decrease in HDAC, c-MYC and PcG expression. In contrast, other organs, such as skin, did not show a significant difference between young and old mice. As shown in Figure S4, expression of HDAC1 and HDAC2 in 16-month-old (old) B6 mouse livers and inguinal fat pads were less than 50% of that of the 3-month-old (young) mice. In old animals, PcG expression levels were 20–30% of young animals, and expression levels of c-MYC in old animal livers were reduced to 10% of those in young animals. However, in the inguinal fat pad, the expression level of c-MYC was maintained at 60% of the young animal levels (Figure S4).
Inhibition of histone deacetylase activity causes MSC senescence
In a previous report, we observed that a 3-day treatment of HDAC inhibitors, such as sodium butyrate (NaBu) and valproic acid (VPA), induced p21CIP1/WAF1 expression and arrested MSCs in the G2/M phase of the cell cycle without affecting p16INK4A expression or cellular senescence [27]. It was suggested that abrupt expression of p21CIP1/WAF1 by high concentrations of HDAC inhibitors (10 and 5 mM for VPA and NaBu, respectively) placed cells into a quiescent stage before p16INK4A proteins could accumulate and the cells could senesce. In order to assess whether HDAC is involved in cellular senescence of MSCs, we extended the treatment period up to 7 days, where we observed that MSCs acquired a senescent phenotype based on SA β-gal staining (Fig. 2a). HDAC1, HDAC2, c-MYC (both total and phosphorylated form), BMI1 and SUZ12 showed similar cytosolic localization by HDAC inhibitor-induced cellular senescence of MSCs, as shown by immunocytochemistry (Fig. 2a; Figure S2). These observations suggest that inhibition of HDAC activity by chemical inhibitors, VPA and NaBu, results in MSC cellular senescence and, similar to replicative senescence of MSCs, downregulation of c-MYC.
Fig. 2.
The loss of HDAC activity is a crucial step in MSC senescence. a HDAC inhibitors, VPA and NaBu, induce cellular senescence in hUCB- and hAD-MSCs upon 7 days of treatment in culture, as shown by SA β-gal staining. Localization of HDAC1, HDAC2 and c-MYC proteins in HDAC inhibitor-treated MSCs were visualized by immunocytochemistry. b, c Protein expression levels of p16INK4A, JMJD3, c-MYC, BMI1 and SUZ12 were assessed using immunoblot analysis. Both hUCB- and hAD-MSCs were treated with 2 mM VPA or 1 mM NaBu for the indicated time periods. Twenty micrograms of whole cell lysates were loaded, and immunoblot analysis was performed as described in “Materials and methods.” d Time-course changes in global H3K27 and H3K4 methylation levels were assessed using immunoblot analysis upon treatment of HDAC inhibitors in hUCB-MSCs. e Cells were treated for 7 days, and ChIP assay was performed to analyze histone methylation patterns at the p16INK4A promoter region. A decrease of H3K27Me3 enrichment, not of H3K4Me3, was observed in both VPA- and NaBu-treated hUCB-MSCs. Two regions on p16INK4A promoter targeted by specific primer sets are described schematically
In order to further confirm whether molecular mechanisms of HDAC inhibitor-mediated cellular senescence are similar to that of replicative senescence, we conducted time-course RT-PCR (Figure S3) and immunoblot experiments (Fig. 2b, c) for the aforementioned markers, such as c-MYC, PcGs, JMJD3 and p16INK4A. hUCB- and hAD-MSCs showed decreased expression of c-MYC, BMI1 and SUZ12. JMJD3 was upregulated by 1-day treatment of HDAC inhibitors, and its expression level peaked at 3 days of treatment. p16INK4A was slightly increased from 3 days of treatment of HDAC inhibitors. Interestingly, a decrease of BMI1 protein expression was consistently followed by c-MYC downregulation, suggesting that BMI1 might be regulated by c-MYC. On the other hand, alteration of EZH2, SUZ12 and JMJD3 expression levels were observed immediately after VPA or NaBu treatment, indicating that those genes may have distinct regulatory mechanisms from that of BMI1. Upregulation of p16INK4A expression followed alteration of BMI1, EZH2, SUZ12 and JMJD3. To further confirm whether the alteration of EZH2 and JMJD3 expression level by HDAC inhibitor treatment causes histone lysine methylation, we performed immunoblot using anti-H3K27Me3 and anti-H3K4Me3 antibodies. Accordingly, it was observed that the protein level of H3K27Me3, not of H3K4Me3, was significantly reduced by treatment with 2 mM VPA or 1 mM NaBu, indicating that the alteration of EZH2 and JMJD3 expression level affected the balance between histone methyltransferase and demethylase activity, especially on lysine 27 residue of histone H3 (H3K27). Using ChIP, we further assessed whether this global alteration of methylation pattern on histone H3 by treatment of HDAC inhibitors affected the promoter region of p16INK4A. Indeed, the amount of H3K27Me3, but not of H3K4Me3, at the proximal promoter region of p16INK4A was decreased by treatment of HDAC inhibitors, again suggesting the specificity of EZH2 and JMJD3 on H3K27Me3 regulation (Fig. 2e). Taken together, treatment of HDAC inhibitor alters PcGs and JMJD3 expression levels, which are important for regulation of p16INK4A expression in young MSCs by erasing the H3K27Me3 closed chromatin structure in a substrate-specific manner.
Histone deacetylase activity is specific for regulation of MSC senescence
To confirm whether the regulation of PcGs and JMJD3, which leads to upregulation of p16INK4A, is specific to HDAC activity, inhibition assays using short interference RNAs that specifically targeted either HDAC1 or HDAC2 were conducted. Both siHDAC1 and siHDAC2 inhibited c-MYC expression in hUCB- and hAD-MSCs to less than 50% of control groups (Fig. 3a, b). The expression levels of PcGs, such as BMI1, EZH2 and SUZ12, were also reduced by specific inhibition of HDACs using siRNA. On the other hand, JMJD3 and p16INK4A were upregulated by siRNA-mediated inhibition of HDACs. To determine whether the alteration of PcGs, JMJD3 and p16INK4A expression levels by siRNA-mediated specific inhibition of HDAC1 or HDAC2 is enough to trigger cellular senescence in MSCs, we performed SA β-gal staining of siRNA transfectants. It was observed that siHDAC1 was more efficient in causing cellular senescence, as evidenced by slightly elevated SA β-gal activity in siHDAC1-transfected groups (Fig. 3c). Taken together, these data demonstrate that HDAC activity is essential for PcG and JMJD3 expression levels and the repression of p16INK4A, all of which maintain the self-renewal capability of MSCs.
Fig. 3.
HDAC1 and HDAC2 inhibition regulates histone-modulating genes. a, b Specific targeting of HDAC1 and HDAC2 by using siRNA induced increased expression of JMJD3 and repression of c-MYC and polycomb genes, as shown by RT-PCR analysis. Graphs represent semi-quantification of RT-PCR results from at least three independent experiments. c Specific downregulation of HDAC1 and HDAC2 is enough to cause cellular senescence in MSCs. siHDAC1 caused an increase in SA β-gal activity
Regulatory mechanism of HDACs; RB hypophosphorylation by HDAC inhibitors causes downregulation of PcGs
To clarify how HDAC activity governs JMJD3 expression, we performed ChIP for the JMJD3 proximal promoter region using specific antibodies against acetylated histone H3 (AcetylH3) and H4 (AcetylH4) in hUCB-MSCs. As shown in Fig. 4, the promoter region of JMJD3 was enriched with AcetylH3 by treatment of HDAC inhibitors for 12 or 24 h, indicating that HDAC activity is important in JMJD3 expression because of chromatin modification in the JMJD3 promoter region.
Fig. 4.
HDAC inhibitors regulate JMJD3 expression by histone H3 acetylation. After 12- or 24-h treatment of HDAC inhibitors on hUCB-MSCs, ChIP assays were performed to investigate acetylation of histone H3 and H4 on JMJD3 promoter regions. In the diagram, four target regions on the promoter of JMJD3 that were investigated using specific primer sets are described schematically
In regard to PcGs, we looked into RB and E2F interaction. As reported, c-MYC has a binding site for the E2F transcription factor in its P2 promoter region, and hence, it is transcriptionally activated by E2F [28]. The retinoblastoma protein (RB) interacts with DNA-binding proteins such as E2F to regulate transcriptional activity or to serve as a binding dock for transcriptional silencers, such as HDACs [29], HMTases [30] and DNA methyltransferases (DNMTs), to suppress gene expression [31, 32]. The phosphorylation status of the RB protein is important for determining the activity of RB as a transcriptional repressor. Hyperphosphorylation, induced by the cyclin-CDK complex, for example, detaches RB from E2F, allowing non-bound free E2F to have transcriptional activity [28, 33]. SUZ12 has an E2F binding site on its upstream promoter and is regulated by E2F [34]. EZH2, however, lacks E2F binding sequences, but is still regulated by E2F transcriptional activity, as reported by Bracken et al. [22]. Several lines of evidence indicate that HDAC activity correlates to RB phosphorylation status. Inhibition of HDAC activity using a HDAC inhibitor, trapoxin, results in hypophosphorylation of RB [35].
To assess whether RB phosphorylation status is modulated during HDAC inhibitor-induced MSC senescence, immunoblot analysis was performed. As shown in Fig. 5a, hyperphosphorylated RB in control MSCs was significantly dephosphorylated by 12 h and more significantly by 24 h after treatment with VPA and NaBu. The significant dephosphorylation 24 h post-treatment occurred concomitantly with the initiation of EZH2, SUZ12 and c-MYC downregulation, as shown in Fig. 2b and c. Immunocytochemical analysis showed that RB localization at the nucleus was slightly increased by treatment of HDAC inhibitors (Figure S5A). Moreover, HDAC inhibitor treatment increased E2F bound to RB as shown by immunoprecipitation (Figure S5B). ChIP analysis data showed that the RB protein is enriched in c-MYC, EZH2 and SUZ12 promoter regions, suggesting that HDAC inhibitor-mediated hypophosphorylated RB binds to the E2F transcription factor and suppresses expression of target genes, including EZH2, SUZ12 (Fig. 5b, c) and c-MYC (Figure S6A).
Fig. 5.
HDAC inhibitors regulate c-MYC, EZH2 and SUZ12 by RB hypophosphorylation. a HDAC inhibitors induced RB hypophosphorylation within 24 h as shown by immunoblot assay. b, c After 12 or 24 h treatment of HDAC inhibitors on hUCB-MSCs, ChIP assays were performed to investigate binding of RB and HDAC1 protein on EZH2 (b) and SUZ12 (c) promoter regions. In the diagram, two target regions on the promoter of each gene investigated using specific primer sets and E2F binding site are described schematically
Taken together, these data demonstrate that HDAC regulates JMJD3 by affecting the histone acetylation status of the JMJD3 promoter region. On the other hand, c-MYC, EZH2 and SUZ12 expression regulated the phosphorylation status of the RB protein upon treatment of HDAC inhibitors. In turn, changes of JMJD3 and PcGs expression levels during HDAC inhibitor-mediated cellular senescence of MSCs decreased expression of the repressive chromatin marker, H3K27Me3, on the p16INK4A promoter region, thereby causing an increase of p16INK4A expression.
Role of c-MYC in cellular senescence of MSCs
We have described that HDAC activity is indispensable for MSCs to maintain their self-renewal capability by keeping them in the stem cell compartment and preventing them from acquiring a senescent phenotype. In this context, expression levels of c-MYC and BMI1 were diminished during replicative senescence of MSCs (Fig. 1; Figure S1), and, indeed, c-MYC expression was specifically regulated by HDAC-mediated modulation of RB hypophosphorylation, as shown in Fig. 5a and Figure S6A. To further assess the role of c-MYC in MSC senescence, a chemical inhibitor of c-MYC, 10058-F4, was used in hUBC- and hAD-MSCs. A decrease in cell proliferation in a dose- and time-dependent fashion was observed (Figure S6B). Moreover, treatment of cells with 10058-F4 increased SA β-gal activity, indicating that c-MYC activity is a pivotal regulator of MSC senescence (Figure S6C). Additionally, BMI1 expression was decreased by 10058-F4 treatment in hUCB- and hAD-MSCs in a time-dependent fashion (Figure S6D), suggesting that inhibition of c-MYC activity precedes the downregulation of BMI1 expression in MSC senescence. Indeed, c-MYC activity is important in the self-renewal capability of MSC.
Histone acetyltransferase inhibitor prevents MSCs from replicative senescence by sustaining PcG and repressing JMJD3 expression levels
The aforementioned results strongly suggest that HDAC activity, via regulation of PcGs, is pivotal for maintenance of self-renewal and prevention of senescence of MSCs. In this context, HDAC activity might regulate chromatin structure in the proximity of the JMJD3 promoter. In addition, the phosphorylation status of RB might directly regulate EZH2 and SUZ12, or indirectly affect BMI1 through c-MYC modulation. Since histone acetyltransferases (HAT) are counteractive to HDACs, we assessed the effects of a chemical inhibitor of HATs, anacardic acid (AA), on the replicative senescence of MSCs. Treatment of cells with AA slowed regression of the doubling level of hUCB-MSCs, and moreover, increased the proliferation potential of hAD-MSCs, as shown by cumulative population doubling levels (Fig. 6a, b). In order to confirm whether the molecular mechanisms of MSC senescence that lead to loss of self-renewal potential are reversed by the HAT inhibitor, we compared the expression levels of HDAC, c-MYC, PcGs and p16INK4A in AA-treated and control MSCs that displayed the same doubling level. Ten micromolar anacardic acid effectively prevented the replicative senescence-mediated downregulation of HDAC2, BMI1 and SUZ12 in both hUCB- and hAD-MSCs. Additionally, HDAC1, c-MYC and EZH2 expression levels were upregulated in hUCB-MSCs treated with 10 μM AA compared to the control group (Fig. 6c). However, upregulation of those genes in hAD-MSCs treated with AA treatment was not observed, presumably because cells in the control group did not fall into a senescent stage, as shown in the CPDL curve (Fig. 6a). The expression level of JMJD3 was downregulated by treatment of AA in both hUCB- and hAD-MSCs. Likewise, p16INK4A expression levels were downregulated by AA treatment in hUCB-MSCs, but not in hAD-MSCs, compared to control group cells. These results suggest that inhibition of HAT activity is counteractive to replicative senescence-mediated inactivation of HDACs and that the HAT inhibitor, anacardic acid, might extend the self-renewal potential of MSCs.
Fig. 6.
A histone acetyltransferase inhibitor, anacardic acid (AA), prevented MSCs from replicative senescence. a Cumulative population doubling level was measured. The doubling level of the AA-treated group was higher than that of control group. b Anacardic acid prevented replicative senescence by sustaining proliferation ability and early state morphology. c RNA was extracted during CPDL experiments. Gene expression levels of AA treated and non-treated cells were compared at the same doubling level by RT-PCR and PCR. d Semi-quantification of PCR product was performed by Image J analysis. Graph shows relative gene expression levels of AA-treated cells compared with control cells
Discussion
Replicative senescence is considered the cellular counterpart of in vivo aging [36, 37], which is accompanied by gene expression changes that might be regulated by epigenetic mechanisms resulting in permanent arrest of cell proliferation [15]. Bypassing replicative senescence is a crucial step for maintenance of stem cell self-renewal [36, 37]. Loss of self-renewal leads to breakdown of cell and tissue homeostasis and the impairment of tissue maintenance and repair [6].
Self-renewal of a stem cell, which can be described as resulting from asymmetrical cell division by which a stem cell gives rise to an identical stem cell and a daughter cell with less self-renewal capability than its mother, is pivotal for the maintenance of stem cell clonality and functionality. In contrast, symmetrical division occurs in culture dishes where stem cell populations robustly increase. The increase of cell number by symmetrical cell division appears to cause cells to lose stemness and eventually leads to cellular senescence. It has been speculated that the loss of a somatic stem cell population, as well as its functionality, is involved in age-related diseases [38].
Here, we report that hUCB- and hAD-MSCs senesce and lose their self-renewal capability as a result of repetitive subculture in vitro, as demonstrated by elevated p16INK4A expression and SA β-gal activity. Our results indicate that MSC senescence in vitro initiates at around the 10th passage (i.e., 30 cumulative doublings), and cells completely undergo cellular senescence at around passage 15, although differences exist depending on the sources and donors. In this context, a novel role of HDACs in the regulation of polycomb group genes (PcGs), such as BMI1, EZH2 and SUZ12, was crucial to keep MSCs in the stem cell compartment. In order to extend our observations to an in vivo aging model, we adopted an aged mouse model and assessed mRNA expression of cellular senescence genes by RT-PCR within several solid organs, as well as bone marrow. Among these, liver and inguinal fat showed significant regulation of target genes, such as HDACs, c-MYC, BMI1 and SUZ12.
Our previous report suggests that HDACs play a pivotal role in the proliferation and controlled differentiation of MSCs. In brief, we reported that HDAC inhibitors reduce proliferation potential of human MSCs by upregulating p21CIP1/WAF1, a cyclin-dependent kinase (CDK) inhibitor, and diminish the differentiation efficiency of MSCs into adipocytes, chondrocytes and neuronal cells. In contrast, HDAC inhibitors increase the osteogenic differentiation capability of MSCs. Results from the previous study also show that p16INK4A is not upregulated in the same conditions that increase p21CIP1/WAF, suggesting a profound difference in the regulatory mechanisms of both CDK inhibitors. Cells were placed into a quiescent stage by high concentrations of HDAC inhibitors, which mediate abrupt expression of p21CIP1/WAF1 before p16INK4A protein can accumulate and cells acquire senescent phenotypes. In addition, it is well known that the upregulation of p21CIP1/WAF1 is mediated by hyperacetylation of core histones upon inhibition of HDAC activity [39]; on the other hand, the regulatory mechanism of p16INK4A involves histone methyltransferase (HMTase) activity [17]. Given these data, the roles of p21CIP1/WAF1 and p16INK4A are likely to differ in terms of stem cell self-renewal. It has been suggested that the upregulation of p21CIP1/WAF1 is transient and strongly effective on cell cycle progression, but not cellular senescence. The transient cell cycle arrest by p21CIP1/WAF1 may be reversed when causative factors, such as the HDAC inhibitors used in our previous study, are removed. On the other hand, expression of p16INK4A shows an accumulative effect on cellular senescence, which is not likely reversed by removal of initiation factors.
As previously reported by Bracken et al. [17], p16INK4A, which is adjacent to p19ARF at the CDKN2A locus, is regulated by HMTase-induced methylation of H3 core histones on lysine 27 (H3K27Me3; a marker for transcriptional silencing). In young fibroblasts, the promoter of p16INK4A is bound by members of the PRC2-initiating complex, such as SUZ12, which in turn recruits EZH2, a member of the PRC2 complex that has HMTase activity and specifically trimethylates the H3K27 residue. The PRC1 maintenance complex can recognize and bind to the PRC2 complex, which stabilizes the complexes and permanently silences transcription. Either the downregulation of PRC1 or PRC2 causes somatic cellular senescence. More recently, JMJD3, a histone lysine demethylase, was identified as a transcriptional activator of p16INK4A by demethylating H3K27Me3 residues at CDKN2A locus [18]. Histone modification by methylation of lysine residues is also involved in lineage-specific differentiation of MSCs [19]. Noer et al. have reported that H3K27Me3 and H3K4Me3 bivalent marks of histone repress adipogenic genes in undifferentiated hAD-MSCs. Upon adipogenic stimulation, histones bound to adipogenic gene promoters develop permissive transcription by acetylating H3K9 residues and by demethylating H3K27 residues. Indeed, histone modification enzymes affect MSC self-renewal by regulating senescence and differentiation capabilities.
Interestingly, as shown by Guney et al. [20], BMI1 has a c-MYC binding E-box motif in its proximal promoter, and hence it is transcriptionally regulated by c-MYC. In the present report, we demonstrated that binding of c-MYC to the BMI1 promoter region decreases during MSC senescence. Additionally, when MSCs were exposed to an inhibitor of c-MYC, 10058-F4, the expression level of BMI1 was decreased in a time-dependent fashion, suggesting that c-MYC directly relates BMI1 regulation. Since the protooncogene c-MYC has an E2F-binding motif in its P2 promoter, it is possible to speculate that BMI1 is indirectly regulated by RB-E2F via c-MYC. In addition, the PRC2 genes, EZH2 and SUZ12, are target genes of the E2F transcriptional factor as shown in this report and by others [22, 34], suggesting that there might be a common regulatory mechanism of PcGs. Hypophosphorylated RB binds to and disrupts transcriptional activity of E2F, whereas hyperphosphorylation of RB detaches RB from E2F, which results in E2F target gene transcription [28, 33]. Therefore, the phosphorylation status of the RB protein plays an important intermediate role for the transcription of E2F target genes, including PcGs. One of the most important observations in this report is that the alteration of HDAC, c-MYC, PcG and p16INK4A genes by replicative senescence was completely prevented by treatment of cells with a HAT inhibitor, anacardic acid, during subculture in vitro. This phenomenon showed that the loss of HDAC activity is a crucial initiation step for MSC senescence and the failure of self-renewal. As mentioned above, one of the biggest shortcomings of adult stem cells is premature senescence, which might limit the number of cells for transplantation. To overcome premature senescence and enhance the usage of MSCs, inhibition of HAT activity, which can be achieved not by exogenous genetic modulation but by relatively safe chemicals, might be a good strategy.
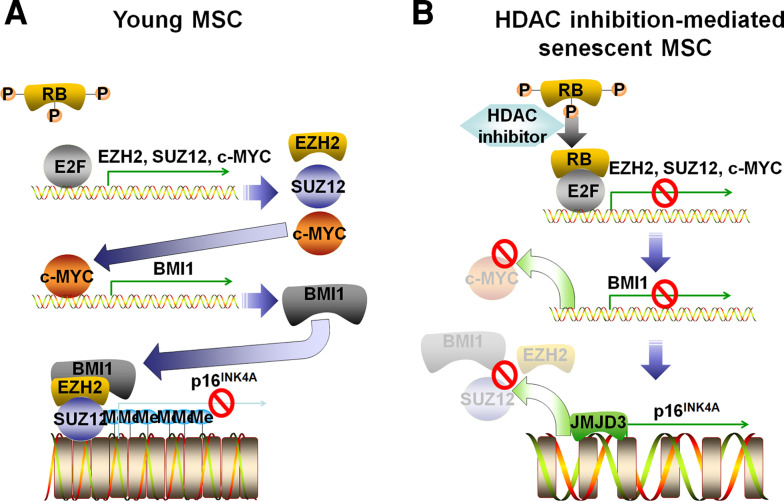
Taken together, the data in this report show that replicative senescence and loss of MSC self-renewal involve downregulation of HDACs, c-MYC and PcGs, which eventually causes an accumulation of p16INK4A expression and an upregulation of SA β-gal activity as summarized in Fig. 7. HDAC activity is crucial for the maintenance MSC self-renewal because it regulates c-MYC expression, as shown by experiments using HDAC chemical inhibitors and specific siRNA constructs. A novel role of HDACs is to regulate BMI1 indirectly by modulating the RB phosphorylation status and hence regulating binding to the E2F binding site on the c-MYC P2 promoter. In turn, c-MYC binds to the BMI1 promoter and regulates expression of BMI1. The other essential role of HDACs is to regulate expression of EZH2 and SUZ12, members of the PRC2 complex, directly through the RB-E2F pathway. E2F directly binds to the promoter regions of EZH2 and SUZ12, and therefore, hypophosphorylation of RB, mediated by HDAC inhibition, represses their expression level. Consequently, PcGs, including BMI1 and SUZ12, bind to the p16INK4A promoter and the trimethylated lysine 27 residue on histone H3 to transcriptionally repress this gene. Interestingly, the senescence-associated regulation of HDACs, c-MYC, PcGs and p16INK4A was completely reversed by treatment of MSCs with a HAT inhibitor. In conclusion, HDAC activity is pivotal for MSC self-renewal capability, and development of methodologies for maintaining HDAC activity during ex vivo expansion of MSCs while preserving stem cell capacity should be pursued.
Fig. 7.
Schematic diagram of senescence regulation in young MSCs and HDAC inhibitor-mediated senescent MSCs. a In young MSCs, RB is hyperphosphorylated, and expression of EZH2, SUZ12 and c-MYC is controlled by free E2F bound to their promoter region. HDAC inhibitors dephosphorylate RB to bind to E2F transcription factor to repress its transcriptional activity and silence EZH2, SUZ12 and c-MYC genes. Because c-MYC is an upstream regulator of BMI1, HDAC inhibition results in BMI1 downregulation. b p16INK4A is repressed in young MSCs because of PcG-induced H3K27Me3 enrichment. Repression of PcG expression level and increase of JMJD3 expression level caused by HDAC inhibition lead to demethylation of H3K27 and p16INK4A expression to senesce MSCs
Electronic supplementary material
Below is the link to the electronic supplementary material.
Figure S1. Gene expression level and cellular localization change during cellular senescence. Gene expression levels of HDAC1, HDAC2 and c-MYC (a) and PcGs, JMJD3 and p16INK4A (b) were shown using RT-PCR analysis. (c) Immunocytochemistry was performed to confirm cellular localization of HDAC1, HDAC2 and c-MYC, and the localization was altered from nuclear to cytosolic during cellular senescence. (TIFF 2927 kb)
Figure S2. Localization of BMI1, SUZ12, p16INK4A and phosphorylated c-MYC protein in HDAC inhibitor-treated hUCB-MSCs was visualized by immunocytochemistry. After treatment of HDAC inhibitor, BMI1 and SUZ12 proteins were localized from nucleus to cytosol. White arrows indicate the nuclei that showed a decreased level of target protein. p16INK4A and phosphorylated c-MYC at residues of tyrosin 58 and serine 62, a pre-degradation form of c-MYC, increased, suggesting that the HDAC inhibitor may affect c-MYC activity not only by expression regulation, but also by post-translational regulation. (TIFF 1577 kb)
Figure S3. HDAC inhibitor decreases c-MYC and PcGs expression and increases JMJD3 expression in hUCB-, hAD-MSC in time- and dose-dependent manners. After treatement of 2mM VPA or 1mM NaBu, RT-PCR analysis was performed in (a) hUCB- and (b) hAD-MSC. c-MYC, SUZ12 and BMI1 mRNA expression levels were decreased in a time-dependent manner. JMJD3 was significantly increased in hUCB-MSC treated by HDAC inhibitors for 1 day. (c) After treatment of VPA and NaBu at various concentrations on hUCB-MSC, RT-PCR analysis was performed. Data show that c-MYC, SUZ12 and BMI1 mRNA expression levels were decreased in a dose-dependent manner by treatment of HDAC inhibitors. (TIFF 2395 kb)
Figure S4. Regulation of HDAC1, HDAC2, c-MYC, BMI1 and SUZ12 expressions in an in vivo aging model. RNA was isolated from indicated organs of young and old mice, and RT-PCR analysis was performed. Graphs represent semi-quantification results from at least three independent RT-PCR experiments. Relative expression levels of genes were calculated as percent of the corresponding genes in the same organ from young animal. * and ** represent statistical significance at the levels of p<0.05 and p<0.005, respectively. (TIFF 1493 kb)
Figure S5. E2F binds to RB protein after HDAC inhibitor treatment. (a) Localization of E2F and RB protein in HDAC inhibitor-treated hUCB-MSCs was visualized by immunocytochemistry. After treatment of HDAC inhibitor for 48 h, E2F was localized from cytoplasm to nucleus. (b) Immunoprecipitation was performed to confirm binding of E2F to RB protein. After treatment of HDAC inhibitor for 24 h, E2F binding to RB protein was increased. (TIFF 2004 kb)
Figure S6. Cellular senescence induced by 10058-F4, a pharmacological inhibitor of c-MYC. (a) 10058-F4 decreases proliferation potential of hUCB- (upper panel) and hAD-MSCs (lower panel) in time- and dose-dependent manners. (b) Inhibition of c-MYC using 10058-F4-induced cellular senescence in MSCs as visualized by SA β-gal staining. (c) 10058-F4 downregulates c-MYC and its downstream BMI1 protein expression levels as shown by immunoblot assay. (d) Cells were treated with HDAC inhibitors for 12 h or 24 h, and ChIP assay was performed as described in Materials and Methods to detect the RB-bound region on c-MYC promoter. In the diagram, two target regions on c-MYC promoter investigated using specific primer sets and E2F binding site were described schematically. * and ** represent statistical significance at the levels of p < 0.05 and p < 0.005, respectively. (TIFF 3150 kb)
Acknowledgment
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea Ministry of Education, Science and Technology (MEST no. M10841000119-08N4100-11910).
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Footnotes
J.-W. Jung and S. Lee contributed equally.
References
- 1.Bibikova M, Laurent LC, Ren B, Loring JF, Fan JB. Unraveling epigenetic regulation in embryonic stem cells. Cell Stem Cell. 2008;2:123–134. doi: 10.1016/j.stem.2008.01.005. [DOI] [PubMed] [Google Scholar]
- 2.Bian Y, Alberio R, Allegrucci C, Campbell KH, Johnson AD. Epigenetic marks in somatic chromatin are remodelled to resemble pluripotent nuclei by amphibian oocyte extracts. Epigenetics. 2009;4:194–202. doi: 10.4161/epi.4.3.8787. [DOI] [PubMed] [Google Scholar]
- 3.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 4.Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, Slukvin II, Thomson JA. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 5.Pan G, Tian S, Nie J, Yang C, Ruotti V, Wei H, Jonsdottir GA, Stewart R, Thomson JA. Whole-genome analysis of histone H3 lysine 4 and lysine 27 methylation in human embryonic stem cells. Cell Stem Cell. 2007;1:299–312. doi: 10.1016/j.stem.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 6.Zhao B, Benson EK, Qiao R, Wang X, Kim S, Manfredi JJ, Lee SW, Aaronson SA. Cellular senescence and organismal ageing in the absence of p21CIP1/WAF1 in ku80(−/−) mice. EMBO Rep. 2009;10:71–78. doi: 10.1038/embor.2008.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Janzen V, Forkert R, Fleming HE, Saito Y, Waring MT, Dombkowski DM, Cheng T, DePinho RA, Sharpless NE, Scadden DT. Stem-cell ageing modified by the cyclin-dependent kinase inhibitor p16INK4a . Nature. 2006;443:421–426. doi: 10.1038/nature05159. [DOI] [PubMed] [Google Scholar]
- 8.Molofsky AV, Slutsky SG, Joseph NM, He S, Pardal R, Krishnamurthy J, Sharpless NE, Morrison SJ. Increasing p16INK4a expression decreases forebrain progenitors and neurogenesis during ageing. Nature. 2006;443:448–452. doi: 10.1038/nature05091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Noda S, Horiguchi K, Ichikawa H, Miyoshi H. Repopulating activity of ex vivo-expanded murine hematopoietic stem cells resides in the CD48-c-Kit+Sca-1+lineage marker-cell population. Stem Cells. 2008;26:646–655. doi: 10.1634/stemcells.2007-0623. [DOI] [PubMed] [Google Scholar]
- 10.Meshorer E, Gruenbaum Y. Gone with the Wnt/Notch: stem cells in laminopathies, progeria, and aging. J Cell Biol. 2008;181:9–13. doi: 10.1083/jcb.200802155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rao MS, Mattson MP. Stem cells and aging: expanding the possibilities. Mech Ageing Dev. 2001;122:713–734. doi: 10.1016/S0047-6374(01)00224-X. [DOI] [PubMed] [Google Scholar]
- 12.Scaffidi P, Misteli T. Lamin A-dependent misregulation of adult stem cells associated with accelerated ageing. Nat Cell Biol. 2008;10:452–459. doi: 10.1038/ncb1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Snyder EY, Loring JF. A role for stem cell biology in the physiological and pathological aspects of aging. J Am Geriatr Soc. 2005;53:S287–S291. doi: 10.1111/j.1532-5415.2005.53491.x. [DOI] [PubMed] [Google Scholar]
- 14.Trosko JE. Role of diet and nutrition on the alteration of the quality and quantity of stem cells in human aging and the diseases of aging. Curr Pharm Des. 2008;14:2707–2718. doi: 10.2174/138161208786264106. [DOI] [PubMed] [Google Scholar]
- 15.Hayflick L. The limited in vitro lifetime of human diploid cell strains. Exp Cell Res. 1965;37:614–636. doi: 10.1016/0014-4827(65)90211-9. [DOI] [PubMed] [Google Scholar]
- 16.Sparmann A, van Lohuizen M. Polycomb silencers control cell fate, development and cancer. Nat Rev Cancer. 2006;6:846–856. doi: 10.1038/nrc1991. [DOI] [PubMed] [Google Scholar]
- 17.Bracken AP, Kleine-Kohlbrecher D, Dietrich N, Pasini D, Gargiulo G, Beekman C, Theilgaard-Monch K, Minucci S, Porse BT, Marine JC, Hansen KH, Helin K. The polycomb group proteins bind throughout the INK4A-ARF locus and are disassociated in senescent cells. Genes Dev. 2007;21:525–530. doi: 10.1101/gad.415507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Agger K, Cloos PA, Rudkjaer L, Williams K, Andersen G, Christensen J, Helin K. The H3K27me3 demethylase JMJD3 contributes to the activation of the INK4A-ARF locus in response to oncogene- and stress-induced senescence. Genes Dev. 2009;23:1171–1176. doi: 10.1101/gad.510809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Noer A, Lindeman LC, Collas P. Histone H3 modifications associated with differentiation and long-term culture of mesenchymal adipose stem cells. Stem Cells Dev. 2009;18:725–736. doi: 10.1089/scd.2008.0189. [DOI] [PubMed] [Google Scholar]
- 20.Guney I, Wu S, Sedivy JM. Reduced c-Myc signaling triggers telomere-independent senescence by regulating Bmi-1 and p16INK4a . Proc Natl Acad Sci USA. 2006;103:3645–3650. doi: 10.1073/pnas.0600069103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muller H, Bracken AP, Vernell R, Moroni MC, Christians F, Grassilli E, Prosperini E, Vigo E, Oliner JD, Helin K. E2Fs regulate the expression of genes involved in differentiation, development, proliferation, and apoptosis. Genes Dev. 2001;15:267–285. doi: 10.1101/gad.864201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bracken AP, Pasini D, Capra M, Prosperini E, Colli E, Helin K. EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer. EMBO J. 2003;22:5323–5335. doi: 10.1093/emboj/cdg542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Narita M, Nunez S, Heard E, Lin AW, Hearn SA, Spector DL, Hannon GJ, Lowe SW. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell. 2003;113:703–716. doi: 10.1016/S0092-8674(03)00401-X. [DOI] [PubMed] [Google Scholar]
- 24.Seo KW, Lee SR, Bhandari DR, Roh KH, Park SB, So AY, Jung JW, Seo MS, Kang SK, Lee YS, Kang KS. OCT4A contributes to the stemness and multi-potency of human umbilical cord blood-derived multipotent stem cells (hUCB-MSCs) Biochem Biophys Res Commun. 2009;384:120–125. doi: 10.1016/j.bbrc.2009.04.094. [DOI] [PubMed] [Google Scholar]
- 25.Jung JW, Cho SD, Ahn NS, Yang SR, Park JS, Jo EH, Hwang JW, Jung JY, Kim SH, Kang KS, Lee YS. Ras/MAP kinase pathways are involved in Ras specific apoptosis induced by sodium butyrate. Cancer Lett. 2005;225:199–206. doi: 10.1016/j.canlet.2004.11.029. [DOI] [PubMed] [Google Scholar]
- 26.Narita M, Krizhanovsky V, Nunez S, Chicas A, Hearn SA, Myers MP, Lowe SW. A novel role for high-mobility group a proteins in cellular senescence and heterochromatin formation. Cell. 2006;126:503–514. doi: 10.1016/j.cell.2006.05.052. [DOI] [PubMed] [Google Scholar]
- 27.Lee S, Park JR, Seo MS, Roh KH, Park SB, Hwang JW, Sun B, Seo K, Lee YS, Kang SK, Jung JW, Kang KS. Histone deacetylase inhibitors decrease proliferation potential and multilineage differentiation capability of human mesenchymal stem cells. Cell Prolif. 2009;42:711–720. doi: 10.1111/j.1365-2184.2009.00633.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ishida S, Shudo K, Takada S, Koike K. A direct role of transcription factor E2F in c-myc gene expression during granulocytic and macrophage-like differentiation of HL60 cells. Cell Growth Differ. 1995;6:229–237. [PubMed] [Google Scholar]
- 29.Ferreira R, Naguibneva I, Mathieu M, Ait-Si-Ali S, Robin P, Pritchard LL, Harel-Bellan A. Cell cycle-dependent recruitment of HDAC-1 correlates with deacetylation of histone H4 on an Rb-E2F target promoter. EMBO Rep. 2001;2:794–799. doi: 10.1093/embo-reports/kve173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nicolas E, Roumillac C, Trouche D. Balance between acetylation and methylation of histone H3 lysine 9 on the E2F-responsive dihydrofolate reductase promoter. Mol Cell Biol. 2003;23:1614–1622. doi: 10.1128/MCB.23.5.1614-1622.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morris EJ, Dyson NJ. Retinoblastoma protein partners. Adv Cancer Res. 2001;82:1–54. doi: 10.1016/S0065-230X(01)82001-7. [DOI] [PubMed] [Google Scholar]
- 32.Fanciulli M (2006) Rb and tumorigenesis. Landes Bioscience/Eureka.com, Springer, Georgetown, New York
- 33.Park IK, Morrison SJ, Clarke MF. Bmi1, stem cells, and senescence regulation. J Clin Invest. 2004;113:175–179. doi: 10.1172/JCI20800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kirmizis A, Bartley SM, Farnham PJ. Identification of the polycomb group protein SU(Z)12 as a potential molecular target for human cancer therapy. Mol Cancer Ther. 2003;2:113–121. [PubMed] [Google Scholar]
- 35.Sambucetti LC, Fischer DD, Zabludoff S, Kwon PO, Chamberlin H, Trogani N, Xu H, Cohen D. Histone deacetylase inhibition selectively alters the activity and expression of cell cycle proteins leading to specific chromatin acetylation and antiproliferative effects. J Biol Chem. 1999;274:34940–34947. doi: 10.1074/jbc.274.49.34940. [DOI] [PubMed] [Google Scholar]
- 36.Alcorta DA, Xiong Y, Phelps D, Hannon G, Beach D, Barrett JC. Involvement of the cyclin-dependent kinase inhibitor p16INK4a in replicative senescence of normal human fibroblasts. Proc Natl Acad Sci USA. 1996;93:13742–13747. doi: 10.1073/pnas.93.24.13742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maurelli R, Zambruno G, Guerra L, Abbruzzese C, Dimri G, Gellini M, Bondanza S, Dellambra E. Inactivation of p16INK4a (inhibitor of cyclin-dependent kinase 4A) immortalizes primary human keratinocytes by maintaining cells in the stem cell compartment. Faseb J. 2006;20:1516–1518. doi: 10.1096/fj.05-4480fje. [DOI] [PubMed] [Google Scholar]
- 38.Trosko JE. Human stem cells as targets for the aging and diseases of aging processes. Med Hypotheses. 2003;60:439–447. doi: 10.1016/S0306-9877(02)00446-2. [DOI] [PubMed] [Google Scholar]
- 39.Ocker M, Schneider-Stock R. Histone deacetylase inhibitors: signalling towards p21cip1/waf1 . Int J Biochem Cell Biol. 2007;39:1367–1374. doi: 10.1016/j.biocel.2007.03.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Gene expression level and cellular localization change during cellular senescence. Gene expression levels of HDAC1, HDAC2 and c-MYC (a) and PcGs, JMJD3 and p16INK4A (b) were shown using RT-PCR analysis. (c) Immunocytochemistry was performed to confirm cellular localization of HDAC1, HDAC2 and c-MYC, and the localization was altered from nuclear to cytosolic during cellular senescence. (TIFF 2927 kb)
Figure S2. Localization of BMI1, SUZ12, p16INK4A and phosphorylated c-MYC protein in HDAC inhibitor-treated hUCB-MSCs was visualized by immunocytochemistry. After treatment of HDAC inhibitor, BMI1 and SUZ12 proteins were localized from nucleus to cytosol. White arrows indicate the nuclei that showed a decreased level of target protein. p16INK4A and phosphorylated c-MYC at residues of tyrosin 58 and serine 62, a pre-degradation form of c-MYC, increased, suggesting that the HDAC inhibitor may affect c-MYC activity not only by expression regulation, but also by post-translational regulation. (TIFF 1577 kb)
Figure S3. HDAC inhibitor decreases c-MYC and PcGs expression and increases JMJD3 expression in hUCB-, hAD-MSC in time- and dose-dependent manners. After treatement of 2mM VPA or 1mM NaBu, RT-PCR analysis was performed in (a) hUCB- and (b) hAD-MSC. c-MYC, SUZ12 and BMI1 mRNA expression levels were decreased in a time-dependent manner. JMJD3 was significantly increased in hUCB-MSC treated by HDAC inhibitors for 1 day. (c) After treatment of VPA and NaBu at various concentrations on hUCB-MSC, RT-PCR analysis was performed. Data show that c-MYC, SUZ12 and BMI1 mRNA expression levels were decreased in a dose-dependent manner by treatment of HDAC inhibitors. (TIFF 2395 kb)
Figure S4. Regulation of HDAC1, HDAC2, c-MYC, BMI1 and SUZ12 expressions in an in vivo aging model. RNA was isolated from indicated organs of young and old mice, and RT-PCR analysis was performed. Graphs represent semi-quantification results from at least three independent RT-PCR experiments. Relative expression levels of genes were calculated as percent of the corresponding genes in the same organ from young animal. * and ** represent statistical significance at the levels of p<0.05 and p<0.005, respectively. (TIFF 1493 kb)
Figure S5. E2F binds to RB protein after HDAC inhibitor treatment. (a) Localization of E2F and RB protein in HDAC inhibitor-treated hUCB-MSCs was visualized by immunocytochemistry. After treatment of HDAC inhibitor for 48 h, E2F was localized from cytoplasm to nucleus. (b) Immunoprecipitation was performed to confirm binding of E2F to RB protein. After treatment of HDAC inhibitor for 24 h, E2F binding to RB protein was increased. (TIFF 2004 kb)
Figure S6. Cellular senescence induced by 10058-F4, a pharmacological inhibitor of c-MYC. (a) 10058-F4 decreases proliferation potential of hUCB- (upper panel) and hAD-MSCs (lower panel) in time- and dose-dependent manners. (b) Inhibition of c-MYC using 10058-F4-induced cellular senescence in MSCs as visualized by SA β-gal staining. (c) 10058-F4 downregulates c-MYC and its downstream BMI1 protein expression levels as shown by immunoblot assay. (d) Cells were treated with HDAC inhibitors for 12 h or 24 h, and ChIP assay was performed as described in Materials and Methods to detect the RB-bound region on c-MYC promoter. In the diagram, two target regions on c-MYC promoter investigated using specific primer sets and E2F binding site were described schematically. * and ** represent statistical significance at the levels of p < 0.05 and p < 0.005, respectively. (TIFF 3150 kb)