Abstract
Human cytomegalovirus (HCMV), a ubiquitous herpesvirus, causes a lifelong subclinical infection in healthy adults but leads to significant morbidity and mortality in neonates and immunocompromised individuals. Its ability to grow in different cell types is responsible for HCMV-associated diseases, including mental retardation and retinitis, and vascular disorders. To globally assess viral gene function for replication in cells, we determined the genomic sequence of a bacterial artificial chromosome (BAC)-based clone of HCMV Towne strain and used this information to delete each of its 162 unique ORFs and generate a collection of viral mutants. The growth of these mutants in different cultured cells was examined to systematically investigate the necessity of each ORF for replication. Our results showed that 45 ORFs are essential for viral replication in fibroblasts and 117 are nonessential. Some genes were found to be required for viral replication in retinal pigment epithelial cells and microvascular endothelial cells, but not in fibroblasts, indicating their role as tropism factors. Interestingly, several viral mutants grew 10- to 500-fold better than the parental strain in different cell types, suggesting that the deleted ORFs encode replication temperance or repressing functions. Thus, HCMV encodes supportive and suppressive growth regulators for optimizing its replication in human fibroblasts, epithelial, and endothelial cells. Suppression of viral replication by virus-encoded temperance factors represents a novel mechanism for regulating the growth of an animal virus, and may contribute to HCMV's optimal infection of different tissues and successful proliferation among the human population.
Human cytomegalovirus (HCMV) is the leading viral cause of congenital abnormalities and mental retardation in newborns (1). This virus also causes mild or subclinical diseases in immunocompetent adults and leads to severe life-threatening complications in immunocompromised individuals, which include AIDS patients and transplant recipients (2). With a linear double-stranded DNA genome of >230 kb, HCMV is the largest member of the human herpesvirus family, which also includes the prototype herpes simplex virus 1 (HSV-1), varicella–zoster virus, Epstein–Barr virus, and Kaposi's sarcoma-associated herpesvirus (3–6). Within the family, HCMV is the prototype of the β-herpesvirus subgroup, which includes herpesviruses 6 and 7 (6). Genomic analysis of the HCMV AD169 strain genome and comparative analysis of other related herpesvirus genomes, combined with biochemical and functional studies, have led to the prediction of numerous viral ORFs and provided some insight into HCMV gene function (6–11). Yet, the roles of many ORFs in viral replication and pathogenesis remain unknown (12–16), partly because of the slow growth kinetics of HCMV and the lack of a suitable animal model.
Clinical data indicate that HCMV infects various tissue and cell types and, hence, is responsible for a myriad of complications including mental retardation, AIDS-associated retinitis, and vascular diseases (1, 17–19). Depending on the tissue type and the host's immune state, HCMV engages in three different modes of infection: acute infections with highly productive growth, persistent infections with low levels of replication, and latent infections where no viral progeny are produced (2, 6). In different cell types, HCMV exhibits various growth rates, suggesting that its replication in a particular cell type is tightly regulated and, thus, determines the outcome of diseases in specific tissues. Although there is evidence for a genetic basis of viral cell type-specific infection and growth regulation (20), many virus-encoded cell-tropism factors have not been identified, and their functional roles in viral replication are unclear.
In this study, we conducted a global functional analysis of HCMV genes by constructing virus gene-deletion mutants and examining their growth phenotypes in different natural HCMV host cells. We determined the genomic sequence of a bacterial artificial chromosome (BAC)-based clone (14, 21–29) of HCMV Towne strain that was previously constructed (14) and used this information to generate a collection of viral mutants with each of its 162 unique ORFs deleted. We provide direct evidence suggesting that 45 ORFs are essential for viral replication in fibroblasts and 117 are not. Moreover, some viral genes, though dispensable for replication in fibroblasts, are required for HCMV growth in human microvascular endothelial cells and retinal pigment epithelial cells, indicating that these ORFs encode cell-type tropism factors. Interestingly, viral mutants with deletions in distinct ORFs were found to replicate significantly better than the parental strain in different cell types, suggesting that these genes function as temperance factors to suppress viral growth in a cell type-specific fashion. This is, to our knowledge, the first observation in a herpesvirus that viral replication is suppressed by its own encoded ORFs. Identification of CMV tropism and temperance factors will provide insight into our understanding of CMV pathogenesis and significantly facilitate the development of novel approaches for controlling and preventing HCMV infections.
Materials and Methods
Virus and Cells. HCMV (Towne strain, American Type Culture Collection) and human cells were propagated as described (Supporting Materials and Methods, which is published as supporting information on the PNAS web site, and refs. 13 and 14). Human primary foreskin fibroblasts (HFF) (CC-2509) and human microvascular endothelial cells (HMVEC) (CC-2527) were obtained from Clonetics (San Diego), and human retinal pigment epithelial (RPE) cells (C4000-1) immortalized with human telomerase reverse transcriptase (hTERT) were purchased from Clontech.
Genomic Sequencing and Bioinformatic Analysis. TowneBAC DNAs were subjected to genomewide shotgun sequencing analysis at MWG Biotech (High Point, NC) and the Stanford Genome Technology Center (Stanford, CA). The sequence was determined to an average redundancy >10-fold.
Construction of Deletion and Rescued Mutants. The mutagenesis procedures were carried out by using a PCR-based approach (30). To construct the deletion cassettes, two oligonucleotide primers (up1 and dn1) were constructed and contained the following components (from 3′ to 5′): 19 homologous nucleotides to the antibiotic resistance cassette KanMX4 (31), a 20-nt unique barcode tag, a common 19-nt primer, and a 25-nt region homologous to the first 25 nt adjacent to either the start or stop codon of the ORF being targeted for deletion.
All predicted ORFs that potentially encode proteins >100 aa were initially selected for deletion. The deletion cassette was designed to remove the entire coding sequence for a given ORF. Although ≈10% of HCMV ORFs overlapped with each other, the position of the deletions was not adjusted, nor were there any attempts made to avoid essential genes, genes in which a previous deletion had been constructed, or genes with a well defined function.
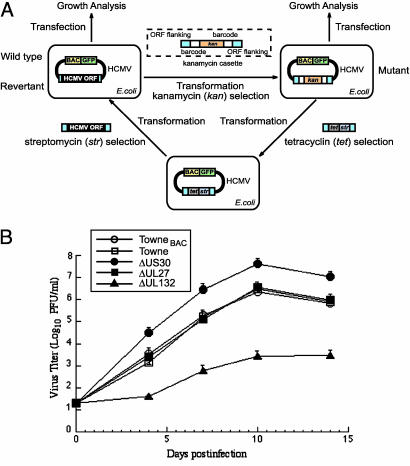
Construction of rescued BAC mutants was carried out by adapting a two-step homologous recombination approach in Escherichia coli (Fig. 2A) (Supporting Materials and Methods and ref. 32).
Fig. 2.
(A) Procedures for constructing deletion and rescued mutants. Detailed procedures are described in Materials and Methods and Supporting Materials and Methods. (B) Multiple-step growth [multiplicity of infection (moi) = 0.05] of HCMV mutants in HFF. Cells were infected with each virus and at different time points postinfection, and cells and culture media were harvested and sonicated. The viral titers were determined by plaque assays on HFF (13). The values of the viral titer represent the average obtained from triplicate experiments. The standard deviation is indicated by the error bars.
Growth Analysis of Viral Mutants in Cells. HFF were electroporated with the BAC DNAs, then plated onto six-well plates and observed for 3–15 weeks for GFP expression and cytopathic effect (CPE). When constructs yielded a growth phenotype different from the parental TowneBAC on initial transfection, two additional independent DNA preparations and transfections were performed to verify the growth phenotype of the constructs. No viral progeny were produced from TowneBAC DNAs containing deletions of essential genes.
Results
Genomic Sequencing of a BAC-Based Clone of HCMV Towne Strain. A construct, TowneBAC, was produced by inserting a BAC sequence into the HCMV genome (Towne strain) and replacing the dispensable, 10-kb US1–US12 region (US, unique short) (14). The TowneBAC DNA, though maintained as a BAC-based plasmid in E. coli, produces infectious progeny in human fibroblasts and retains wild-type growth characteristic in vitro (14).
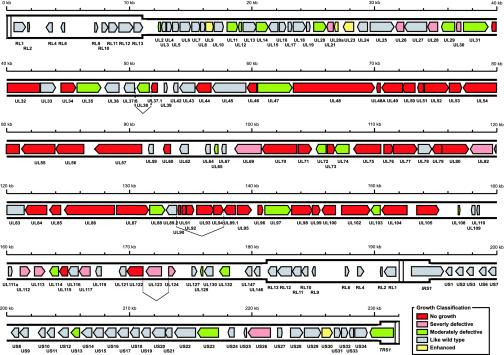
The cloned HCMV Towne sequence in the TowneBAC construct was determined (GenBank accession no. AY315197) by using the shotgun sequencing approach. Similar to the sequence of HCMV AD169 strain, which is the only other complete HCMV genome that has been sequenced (10), the Towne genome is composed of a unique long (UL) region and a US region, both flanked by inverted repeat regions (RL and RS) (Fig. 1). Except for the four Towne strain-specific genes (UL146, UL147, RL12, and RL13) that were reported (9), the overall sequences of the two strains exhibited 98% identity. The Towne sequence present in the TowneBAC construct is predicted to encode 152 unique ORFs, with 9 of these present in two copies in the RL elements (Fig. 1). Taking into account the 10 putative ORFs within the deleted US1–US12 region, the Towne strain potentially encodes at least 162 unique ORFs. Many of these genes have homologues in the AD169 strain, which was recently reanalyzed and predicted to encode 145 unique ORFs (7). The ORFs identified in this study correspond to those identified by Chee et al. (10) and Cha et al. (9) with respect to the ORF designation and sequence identity. Some minor variations in sequence length and identity were observed and expected as the strain used in our work differed from those used in the previous studies. A detailed comparative sequence analysis will be described elsewhere.
Fig. 1.
Genome organization and ORFs of HCMV (Towne strain) based on the genomewide shotgun sequencing of the viral sequence cloned in a BAC. Similar to the HCMV AD169 genome (7, 10), the Towne genome is composed of a UL region and a US region, both flanked by inverted repeat regions (RL and RS). RL and RS are shown in a thicker format than UL and US. Each of the ORFs (RL1–RL13, UL2–UL147, IRS1, US1–US34, and TRS1) is color-coded according to the growth properties of their corresponding virus gene-deletion mutants in HFF (Table 1). The vertical dashed lines represent the splicing junctions.
Generation of Virus ORF-Deletion Mutants and Rescued Mutants. To systematically analyze the function of each ORF in viral replication, we used a rapid bacterial homologous recombination system and generated a collection of mutants in E. coli by deleting each of the predicted ORFs from TowneBAC, using a PCR-based approach (Fig. 2A) (30). Each gene was precisely deleted from the start to stop codons and replaced with a kanamycin resistance cassette, which was PCR-amplified by using primers containing 50-nt sequences flanking the regions of the targeted ORF (Fig. 2A and Supporting Materials and Methods) (30). Each deletion was verified by using PCR screening, restriction digest profiling, and Southern analysis (Fig. 4, which is published as supporting information on the PNAS web site). All of the predicted 152 genes were deleted (Table 1).
Table 1. A list of HCMV Towne strain ORFs categorized by the growth properties of their respective deletion mutants in cultured human foreskin HFFs.
| ORFs | Conservation | Function | Growth |
|---|---|---|---|
| No growth (45 mutants) | |||
| UL32 | β-herpes | Tegument | Essential* |
| UL34 | CMV | Unknown (transcription) | Essential* |
| UL37.1 | β-herpes/CMV | Antiapoptotic | Essential* |
| UL44 | Core | DNA replication | Essential* |
| UL46 | Core | Capsid | Essential* |
| UL48 | Core | Tegument | Essential* |
| UL48.5 | Core | Capsid protein | Essential* |
| UL49 | Core | Unknown | Essential* |
| UL50 | Core | Egress | Essential* |
| UL51 | Core | DNA packaging/cleavage | Essential* |
| UL52 | Core | DNA packaging/cleavage | Essential* |
| UL53 | Core | Egress | Essential* |
| UL54 | Core | DNA polymerase | Essential* |
| UL55 | Core | Glycoprotein B | Essential† |
| UL56 | Core | DNA packaging/cleavage | Essential* |
| UL57 | Core | ssDNA binding protein | Essential* |
| UL60 | CMV | Unknown (OriLyt?) | Essential* |
| UL70 | Core | Helicase/primase | Essential* |
| UL71 | Core | Unknown | Essential* |
| UL73 | Core | Glycoprotein N | Essential† |
| UL75 | Core | Glycoprotein H | Essential† |
| UL76 | Core | Unknown | Essential* |
| UL77 | Core | DNA packaging/cleavage | Essential* |
| UL79 | Core | Unknown | Essential* |
| UL80 | Core | Capsid assembly | Essential† |
| UL84 | β-herpes | DNA replication | Essential* |
| UL85 | Core | Capsid | Essential* |
| UL86 | Core | Capsid | Essential* |
| UL87 | Core | Unknown | Essential* |
| UL89.1 | Core | DNA packaging/cleavage | Essential* |
| UL90 | CMV | Unknown | Essential* |
| UL91 | β-herpes | Unknown | Essential* |
| UL92 | β-herpes | Unknown | Essential* |
| UL93 | Core | Unknown | Essential* |
| UL94 | Core | Unknown (tegument) | Essential* |
| UL95 | Core | Unknown | Essential* |
| UL96 | β-herpes | Unknown | Essential* |
| UL98 | Core | Alkaline nuclease | Essential* |
| UL99 | Core | Tegument | Essential* |
| UL100 | Core | Glycoprotein M | Essential† |
| UL102 | Core | Helicase/primase | Essential* |
| UL104 | Core | DNA packaging/cleavage | Essential* |
| UL105 | Core | Helicase/primase | Essential* |
| UL115 | Core | Glycoprotein L | Essential† |
| UL122 | β-herpes | IE2 (transcription) | Essential† |
| Severe growth defect (12 mutants) | |||
| UL21 | CMV | Unknown | <2 × 10-4* |
| UL26 | CMV | Tegument (transcription) | <2 × 10-4* |
| UL28 | β-herpes | Unknown | <2 × 10-4* |
| UL30 | CMV | Unknown | <2 × 10-4* |
| UL69 | Core | Tegument (transcription) | <2 × 10-4† |
| UL82 | β-herpes | Tegument (transcription) | <2 × 10-4† |
| UL112 | β-herpes | Major early protein | <2 × 10-4* |
| UL113 | β-herpes | Major early protein | <2 × 10-4* |
| UL117 | β-herpes | Unknown | <2 × 10-4* |
| UL123 | CMV | IE1 | <2 × 10-4† |
| UL124 | CMV | Latent transcript (ORF152) | <2 × 10-4‡ |
| Us26 | β-herpes | Unknown | <2 × 10-4* |
| Moderate growth defect (23 mutants) | |||
| UL2 | CMV | Unknown | 10-1-10-2† |
| UL11 | CMV | Glycoprotein | 10-2-10-3* |
| UL12 | CMV | Unknown | 10-1-10-2* |
| UL14 | CMV | Unknown | 10-2-10-3* |
| UL20 | CMV | TCR homolog | 10-2-10-3† |
| UL29 | β-herpes | Unknown | 10-2-10-3* |
| UL31 | β-herpes | Transcription | 10-2-10-3* |
| UL35 | β-herpes | Tegument/transcription | 10-2-10-3* |
| UL38 | β-herpes | Unknown | 10-2-10-3* |
| UL47 | Core | Tegument-DNA release | 10-3-10-4† |
| UL65 | CMV | Unknown (pp67 virion protein) | 10-2-10-3* |
| UL72 | Core | dUTPase | 10-3-10-4* |
| UL74 | β-herpes | Glycoprotein O | 10-3-10-4† |
| UL88 | β-herpes | Tegument | 10-2-10-3* |
| UL97 | Core | Protein kinase | 10-2-10-3† |
| UL103 | Core | Unknown | 10-2-10-3* |
| UL108 | CMV | Unknown | 10-2-10-3* |
| UL114 | Core | Uracil DNA glycosylase | 10-3-10-4† |
| UL129 | CMV | Unknown | 10-2-10-3* |
| UL132 | CMV | Unknown | 10-2-10-3* |
| US13 | CMV | Unknown | 10-1-10-2‡ |
| US23 | β-herpes | Unknown | 10-2-10-3* |
| TRS1 | CMV | Transcription/egress | 10-2-10-3† |
| Growth like wild type (68 mutants, 78 ORFs) | |||
| UL3 | CMV | Unknown | Dispensable† |
| UL4 | CMV | Glycoprotein | Dispensable† |
| UL5 | CMV | Unknown | Dispensable† |
| UL6 | CMV | Unknown | Dispensable† |
| UL7 | CMV | Unknown | Dispensable† |
| UL8 | CMV | Unknown | Dispensable† |
| UL10 | CMV | Unknown | Dispensable† |
| UL13 | CMV | Unknown | Dispensable* |
| UL15 | CMV | Unknown | Dispensable* |
| UL16 | CMV | Immunomodulation | Dispensable† |
| UL17 | CMV | Unknown | Dispensable* |
| UL18 | CMV | MHC homolog | Dispensable† |
| UL19 | CMV | Unknown | Dispensable* |
| UL24 | β-herpes | Tegument | Dispensable* |
| UL25 | β-herpes | Tegument | Dispensable* |
| UL27 | β-herpes | Unknown | Dispensable* |
| UL33 | β-herpes | G protein receptor | Dispensable† |
| UL36 | β-herpes | Antiapoptotic | Dispensable† |
| UL37.3 | β-herpes | Unknown | Dispensable† |
| UL39 | CMV | Unknown | Dispensable* |
| UL42 | CMV | Unknown | Dispensable† |
| UL43 | β-herpes | Tegument | Dispensable† |
| UL45 | Core | Ribonucleotide reductase | Dispensable† |
| UL59 | CMV | Unknown | Dispensable* |
| UL62 | CMV | Unknown | Dispensable* |
| UL64 | CMV | Unknown | Dispensable* |
| UL67 | CMV | Unknown | Dispensable* |
| UL78 | CMV | G protein receptor | Dispensable† |
| UL83 | β-herpes | Tegument | Dispensable† |
| UL89.2 | Core | DNA packaging/cleavage | Dispensable* |
| UL109 | CMV | Unknown | Dispensable* |
| UL110 | CMV | Unknown | Dispensable* |
| UL111a | CMV | IL-10 homolog | Dispensable* |
| UL116 | CMV | Unknown | Dispensable* |
| UL119 | CMV | Fc receptor | Dispensable* |
| UL121 | CMV | Unknown | Dispensable* |
| UL127 | CMV | Unknown | Dispensable† |
| UL130 | CMV | Unknown | Dispensable* |
| UL146 | CMV | Chemokine | Dispensable* |
| UL147 | CMV | Chemokine homolog | Dispensable* |
| IRS | CMV | Transcription | Dispensable† |
| (US1) | CMV | Unknown | Dispensable† |
| (US2) | CMV | Immunomodulation | Dispensable† |
| (US3) | CMV | Immunomodulation | Dispensable† |
| (US6) | CMV | Immunomodulation | Dispensable† |
| (US7) | CMV | Unknown | Dispensable† |
| (US8) | CMV | Immunomodulation | Dispensable† |
| (US9) | CMV | Unknown | Dispensable† |
| (US10) | CMV | Immunomodulation | Dispensable† |
| (US11) | CMV | Immunomodulation | Dispensable† |
| (US12) | CMV | Unknown | Dispensable† |
| US14 | CMV | Unknown | Dispensable† |
| US15 | CMV | Unknown | Dispensable* |
| US16 | CMV | Unknown | Dispensable* |
| US17 | CMV | Unknown | Dispensable* |
| US18 | CMV | Unknown | Dispensable* |
| US19 | CMV | Unknown | Dispensable* |
| US20 | CMV | Unknown | Dispensable* |
| US21 | CMV | Unknown | Dispensable* |
| US22 | β-herpes | Unknown | Dispensable* |
| US24 | CMV | Unknown | Dispensable* |
| US25 | CMV | Unknown | Dispensable* |
| US27 | CMV | G protein receptor | Dispensable† |
| US28 | β-herpes | G protein receptor | Dispensable† |
| US29 | CMV | Unknown | Dispensable* |
| US31 | CMV | Unknown | Dispensable* |
| US32 | CMV | Unknown | Dispensable* |
| US33 | CMV | Unknown | Dispensable* |
| US34 | CMV | Unknown | Dispensable* |
| RL1 | CMV | Unknown | Dispensable* |
| RL2 | CMV | Unknown | Dispensable* |
| RL4 | CMV | Early protein | Dispensable† |
| RL6 | CMV | Unknown | Dispensable† |
| RL9 | CMV | Unknown | Dispensable† |
| RL10 | CMV | Glycoprotein | Dispensable† |
| RL11 | CMV | Unknown | Dispensable† |
| RL12 | CMV | Unknown | Dispensable† |
| RL13 | CMV | Unknown | Dispensable† |
| Enhanced growth (4 mutants) | |||
| UL9 | CMV | Unknown | 1 × 10* |
| UL20a | CMV | Unknown | 1 × 10* |
| UL23 | β-herpes | Tegument | 1 × 10* |
| US30 | CMV | Unknown | 1 × 10* |
Also shown are the sequence conservations of these ORFs with those in HCMV AD169 strain and other herpesviruses, the genome sequences of which are currently available (7, 9-11), and their functions and the functions of their homologues in other herpesviruses that have been shown or implicated from previous studies (ref. 6 and references therein). Although virus mutants with a deletion in each of the 10 ORFs in the US1—US12 region (marked with parentheses) were not individually constructed, these ORFs are listed as dispensable because they were collectively deleted and were not present in TowneBAC (14).
Results from this study only.
Results from this study consistent with those from previous studies (6).
Results from this study not consistent with those from previous studies (6).
The mutant BAC DNAs were isolated from bacteria and transfected into cultured HFF. Of the 152 constructed mutants, 107 produced viral progeny, indicating that the mutated genes are not essential for HCMV replication in HFF. In contrast, 45 mutants did not yield infectious progeny even after repeated transfection and extensive incubation (Table 1). In our experiments, when constructs did not yield viral progeny on the initial transfection, two additional independent DNA preparations and transfections were performed to verify the no-growth phenotype of the construct. To further confirm their no-growth phenotype, revertant BAC clones were constructed for several mutants (e.g., ΔUL32) by restoring the deletion with the intact ORF sequence (Figs. 2A and 4). The rescued mutant (e.g., rescued-UL32) produced progeny and grew as well as the TowneBAC, thereby confirming that deleting the ORF sequence causes the no-growth phenotype (Fig. 5, which is published as supporting information on the PNAS web site).
Analysis of Viral Mutants with Deletion of Essential and Nonessential Genes. We identified 45 ORFs that are essential for viral replication in HFF (Table 1). Thirty-seven of these ORFs had not been previously reported to be essential, of which 15 had not even been suggested to be essential based on the studies of other herpesviruses. Over 90% of the essential genes are conserved among all herpesviruses (core genes) or β-herpesviruses (Table 1). In contrast, ≈70% of the nonessential genes are HCMV-specific and are not conserved among β-herpesviruses.
Based on their growth properties in fibroblasts, viral mutants carrying deletions in nonessential genes were further categorized into four groups: severe growth defect, moderate growth defect, growth like the wild type, and enhanced growth (Table 1). Twelve mutants were classified to have a severe growth defect in HFF, thereby precluding the generation of sufficient titers for growth studies. Five of these ORFs have unknown functions, and the remaining seven genes are involved in regulating transcription or genome replication (6). Moderate growth defect mutants reached a peak titer of 10–10,000 times less than TowneBAC after 14 days in a multiple-step growth analysis (e.g., ΔUL132; Fig. 2B). This group contains 23 viral mutants of which 11 of the deleted ORFs have not been characterized, and their functions are currently unknown.
Sixty-eight mutants retained growth properties that ranged from wild-type levels to <10-fold fewer plaque-forming units at 14 days postinfection (e.g., ΔUL27; Fig. 2B). These “growth like wild type” mutants (Table 1) are considered to have deletions in dispensable genes, the majority of which are HCMV-specific ORFs.
The mutant group that showed enhanced growth reached a 10-fold greater peak titer than the parental Towne strain or TowneBAC virus during a 14-day infection (e.g., ΔUS30; Fig. 2B). Although their functions are currently unknown, recent bioinformatic analyses suggest that these ORFs are all either β-herpesvirus or HCMV-specific transmembrane proteins (8).
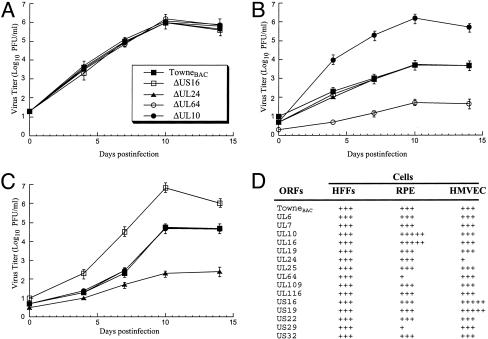
Identification of Viral Mutants with Deletion of ORFs Important for Infections in Specific Cell Types. Although 68 ORFs are found to be dispensable for viral replication in HFF, it is possible that these ORFs encode important functions for HCMV infection in vivo, including those involved in immunomodulation (6, 33). Because of the lack of an animal model for study of HCMV pathogenesis, cultured natural host cells have been used. In vivo, HCMV infects human RPE cells and HMVEC, leading to viral-associated retinitis and vascular diseases, respectively (17–19). It is conceivable that some of the ORFs, though dispensable for HCMV growth in fibroblasts, are important for supporting viral replication in other cell types. To test this hypothesis, HMVEC and RPE cells were individually infected with a collection of 15 viral mutants that grew as well as the parental virus in HFF. The growth of each virus in HMVEC and RPE cells was compared with the result found in HFF. Diverse growth phenotypes of these mutants were observed in HMVEC and RPE cells (Fig. 3). For instance, the UL24-deletion mutant grew as well as the TowneBAC in HFF and RPE cells but was significantly defective in growth in HMVEC. Another mutant with a UL64 deletion replicated normally in HMVEC and HFF but barely produced viral progeny in RPE cells (Fig. 3). Our results suggest that UL24 and UL64 are important for viral replication in HMVEC and RPE, respectively. Interestingly, a UL10 deletion mutant grew normally in HFF and HMVEC but reached a 500-fold higher titer than TowneBAC in RPE cells. Another mutant with a deletion of ORF US16 replicated as well as the TowneBAC in HFF and RPE cells but grew 100-fold better in HMVEC (Fig. 3). These observations imply that UL10 and US16 encode cell type-specific functions for virus-growth inhibition.
Fig. 3.
Analysis of multiple-step growth of different mutants and TowneBAC in HFF (A) [multiplicity of infection (moi) = 0.05], RPE cells (B) (moi = 0.25), and HMVEC (C) (moi = 0.05). (D) Comparison of the growth properties of 15 mutants in these three cell types with those of TowneBAC. +++, Peak titer similar to that of TowneBAC; +++++, peak titer at least 100 times higher than that of TowneBAC; +, peak titer at least 100 times lower than that of TowneBAC. Cells were infected with each virus, and at different time points postinfection, cells and culture media were harvested and sonicated. The viral titers were determined by plaque assays on HFF (13). The values of the viral titer represent the average obtained from triplicate experiments. The standard deviation is indicated by the error bars.
Discussion
In this study, we reported the global analysis of the HCMV genome with an emphasis on the identification of viral genes important for growth in cultured cells. The constructed mutants exhibited diverse growth properties from no growth to enhanced growth in HFF, thereby suggesting that the function of viral genes ranges from absolutely supportive to suppressive in viral replication. All of the predicted 162 unique ORFs encoded by HCMV Towne strain were deleted. Our data provide direct evidence to suggest that 45 ORFs encode essential genes, 15 of which have unknown function. Moreover, these results also indicate that 117 encode nonessential ORFs, with >70 genes of unknown function. The functional profiling of HCMV ORFs reported here is only an initial step toward elucidating the role of each gene in viral infection. It is possible that some of the observed phenotypes may have resulted from the impact of deleting the ORF on surrounding genes, which may have overlapping regions with the targeted ORF. Meanwhile, the absence of the US1–US12 region, which was deleted in all of the mutants, may influence some mutant phenotypes.
Research during the last two decades has collectively shown that the prototype herpesvirus, herpes simplex virus 1, encodes 37 essential genes and 48 nonessential genes (3). Our results show that the majority (78%) of the 45 HCMV genes that are essential for replication in HFF are highly conserved across all herpesviruses, suggesting that these core ORFs may represent the minimal ancestral genome of all herpesviruses. HCMV may have evolved from the progenitor genome through the acquisition of nonessential genes that are responsible for its infection and pathogenesis in various tissues. Indeed, this notion is consistent with our observations that deletions of nonessential genes have diverse effects on viral growth in different cell types including HCMV natural host cells, HMVEC, and RPE cells. This hypothesis is further supported by the identification of Epstein–Barr virus and Kaposi's sarcoma-associated herpesvirus-specific genes that have a role in their unique latent infections (4, 5).
The ability of HCMV to replicate in many cell types and tissues is responsible for the variety of sequelae associated with HCMV infection. Phenotypic screening for growth properties in a particular cell type or tissue by using the entire collection of HCMV mutants generated in this study should lead to the identification of viral genes and determinants that are responsible for HCMV's cell or tissue tropism. Given the fact that there are no animal models suitable for HCMV infection, such a global analysis of the HCMV genome, as described in our study, may prove to be a valuable alternative for the investigation of viral pathogenesis.
Our analysis of the mutant library suggests the presence of viral encoded factors that regulate viral growth in different cell types. The identification of viral genes that support viral replication in various cell types was not entirely unexpected. However, the discovery of HCMV-encoded factors that repress viral replication on a cell type-specific basis represents a discovery in the field of animal viruses. Deletion of distinct ORFs resulted in mutant viruses with enhanced growth in specific cell types (e.g., ΔUS30 in HFF, ΔUL10 in RPE cells, and ΔUS16 in HMVEC). The notion that these ORFs function in a cell type-specific manner is further supported by our observation that although the UL23 deletion mutant exhibited enhanced growth in HFF (Table 1), it retained wild-type growth kinetics in HMVEC and RPE cells (data not shown). Although the mechanism by which these genes repress viral replication is currently unknown, we speculate that the genes may either directly block CMV growth or activate cellular antiviral machinery to suppress viral replication.
The presence of these growth-repressor factors may initially seem counterproductive from the perspective of the virus; however, their existence is consistent with the observations that HCMV exhibits different growth rates in various cell types (2, 6). In vivo, these inhibitors may moderate viral loads to levels optimal for transmission but prevent viral replication from reaching levels that may result in severe tissue damage or host death. Furthermore, they may suppress productive lytic replication to low levels or cease viral replication, thereby facilitating persistent and latent infections. This notion is consistent with the recent observation that a retroviral spumavirus with a deletion of a specific gene is more easily activated from latency than the wild-type virus (34). Therefore, these repressor factors may in fact have the effect of enhancing virus survival. We believe that this strategy of pathogen temperance, whereby a pathogen achieves optimal coexistence with the host through self-moderation of its replication or virulence, is a fundamental component in a pathogen's repertoire of factors that function to enhance its long-term existence.
The discovery of such temperance genes in an animal virus suggests that pathogen temperance is a prevalent survival strategy and present in other higher order organisms with greater genome content. This hypothesis is consistent with recent observations in infectious organisms where deletion of certain pathogen-encoded factors resulted in a hypervirulent infection in the host (35, 36). The recognition of pathogen temperance may radically alter the way we perceive the emergence of hypergrowth virulent variants from benign pathogens. Arguably, the underlying mechanism for hypervirulence may, in some cases, be the loss of these temperance factors as opposed to the acquisition of virulence genes. Accordingly, drugs that mimic or activate temperance factors may lead to effective therapies against infectious diseases. Further studies of pathogen temperance will provide insight into the evolution of new and emerging virulent pathogens and facilitate the development of novel approaches for controlling future epidemics caused by these virulent strains.
Supplementary Material
Acknowledgments
We thank Edward Mocarski for generous help and invaluable advice; Neal Copeland and Craig A. Strathdee for DY380 strain and str/tet constructs; and Karen Chan, Kihoon Kim, Paul Rider, Hua Zou, Waraporn Tongprasit, Sergey Smirnov, Tong Toung, Jen Chen, Manfred Lee, and Alice Chu for technical assistance. This work was supported by a Chancellor's Initiative Grant from the University of California, Berkeley, and by the National Institutes of Health.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: BAC, bacterial artificial chromosome; HCMV, human cytomegalovirus; HFF, human primary foreskin fibroblasts; HMVEC, human microvascular endothelial cells; RPE, retinal pigment epithelial; UL, unique long; US, unique short.
Data deposition: Sequences of the TowneBAC isolate have been deposited in the GenBank database (accession no. AY315197).
References
- 1.Britt, W. J. (1999) in Sexually Transmitted Diseases and Adverse Outcomes of Pregnancy, eds. Hitchcock, H., MacKay, T. & Wasserheit, J. N. (Am. Soc. Microbiol., Washington, DC), pp. 269–281.
- 2.Pass, R. F. (2001) in Fields Virology, eds. Knipe, D. M. & Howley, P. M. (Lippincott William & Wilkins, Philadelphia), Vol. 2, pp. 2675–2706. [Google Scholar]
- 3.Roizman, B. & Knipe, D. M. (2001) in Fields Virology, eds. Knipe, D. M. & Howley, P. M. (Lippincott William & Wilkins, Philadelphia), Vol. 2, pp. 2399–2460. [Google Scholar]
- 4.Kieff, E. & Rickinson, A. B. (2001) in Fields Virology, eds. Knipe, D. M. & Howley, P. M. (Lippincott William & Wilkins, Philadelphia), Vol. 2, pp. 2511–2574. [Google Scholar]
- 5.Moore, P. S. & Chang, Y. (2001) in Fields Virology, eds. Knipe, D. M. & Howley, P. M. (Lippincott William & Wilkins, Philadelphia), Vol. 2, pp. 2803–2834. [Google Scholar]
- 6.Mocarski, E. S. & Courcelle, C. T. (2001) in Fields Virology, eds. Knipe, D. M. & Howley, P. M. (Lippincott William & Wilkins, Philadelphia), Vol. 2, pp. 2629–2673. [Google Scholar]
- 7.Davison, A. J., Dolan, A., Akter, P., Addison, C., Dargan, D. J., Alcendor, D. J., McGeoch, D. J. & Hayward, G. S. (2003) J. Gen. Virol. 84, 17–28. [DOI] [PubMed] [Google Scholar]
- 8.Rigoutsos, I., Novotny, J., Huynh, T., Chin-Bow, S. T., Parida, L., Platt, D., Coleman, D. & Shenk, T. (2003) J. Virol. 77, 4326–4344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cha, T. A., Tom, E., Kemble, G. W., Duke, G. M., Mocarski, E. S. & Spaete, R. R. (1996) J. Virol. 70, 78–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chee, M. S., Bankier, A. T., Beck, S., Bohni, R., Brown, C. M., Cerny, R., Horsnell, T., Hutchison, C. A., 3rd, Kouzarides, T., Martignetti, J. A. & et al. (1990) Curr. Top. Microbiol. Immunol. 154, 125–169. [DOI] [PubMed] [Google Scholar]
- 11.McGeoch, D. J., Dolan, A. & Ralph, A. C. (2000) J. Virol. 74, 10401–10406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hobom, U., Brune, W., Messerle, M., Hahn, G. & Koszinowski, U. H. (2000) J. Virol. 74, 7720–7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dunn, W., Trang, P., Khan, U., Zhu, J. & Liu, F. (2001) Proc. Natl. Acad. Sci. USA 98, 14831–14836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marchini, A., Liu, H. & Zhu, H. (2001) J. Virol. 75, 1870–1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meyer, H. H., Ripalti, A., Landini, M. P., Radsak, K., Kern, H. F. & Hensel, G. M. (1997) J. Gen. Virol. 78, 2621–2631. [DOI] [PubMed] [Google Scholar]
- 16.Azad, R. F., Driver, V. B., Tanaka, K., Crooke, R. M. & Anderson, K. P. (1993) Antimicrob. Agents Chemother. 37, 1945–1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Epstein, S. E., Speir, E., Zhou, Y. F., Guetta, E., Leon, M. & Finkel, T. (1996) Lancet 348, Suppl. 1, s13–s17. [DOI] [PubMed] [Google Scholar]
- 18.Scholz, M., Doerr, H. W. & Cinatl, J. (2003) Trends Microbiol. 11, 171–178. [DOI] [PubMed] [Google Scholar]
- 19.Streblow, D. N., Orloff, S. L. & Nelson, J. A. (2001) J. Nutr. 131, 2798S–2804S. [DOI] [PubMed] [Google Scholar]
- 20.Gerna, G., Percivalle, E., Sarasini, A., Baldanti, F., Campanini, G. & Revello, M. G. (2003) J. Gen. Virol. 84, 1431–1436. [DOI] [PubMed] [Google Scholar]
- 21.Mocarski, E. S., Post, L. E. & Roizman, B. (1980) Cell 22, 243–255. [DOI] [PubMed] [Google Scholar]
- 22.Roizman, B. & Jenkins, F. J. (1985) Science 229, 1208–1214. [DOI] [PubMed] [Google Scholar]
- 23.Weber, P. C., Levine, M. & Glorioso, J. C. (1987) Science 236, 576–579. [DOI] [PubMed] [Google Scholar]
- 24.Messerle, M., Crnkovic, I., Hammerschmidt, W., Ziegler, H. & Koszinowski, U. H. (1997) Proc. Natl. Acad. Sci. USA 94, 14759–14763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith, G. A. & Enquist, L. W. (2000) Proc. Natl. Acad. Sci. USA 97, 4873–4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brune, W., Menard, C., Heesemann, J. & Koszinowski, U. H. (2001) Science 291, 303–305. [DOI] [PubMed] [Google Scholar]
- 27.Borst, E. M., Hahn, G., Koszinowski, U. H. & Messerle, M. (1999) J. Virol. 73, 8320–8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hahn, G., Rose, D., Wagner, M., Rhiel, S. & McVoy, M. A. (2003) Virology 307, 164–177. [DOI] [PubMed] [Google Scholar]
- 29.Yu, D., Smith, G. A., Enquist, L. W. & Shenk, T. (2002) J. Virol. 76, 2316–2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee, E. C., Yu, D., Martinez de Velasco, J., Tessarollo, L., Swing, D. A., Court, D. L., Jenkins, N. A. & Copeland, N. G. (2001) Genomics 73, 56–65. [DOI] [PubMed] [Google Scholar]
- 31.Wach, A., Brachat, A., Pohlmann, R. & Philippsen, P. (1994) Yeast 10, 1793–1808. [DOI] [PubMed] [Google Scholar]
- 32.Stavropoulos, T. A. & Strathdee, C. A. (2001) Genomics 72, 99–104. [DOI] [PubMed] [Google Scholar]
- 33.Ploegh, H. L. (1998) Science 280, 248–253. [DOI] [PubMed] [Google Scholar]
- 34.Meiering, C. D. & Linial, M. L. (2002) Proc. Natl. Acad. Sci. USA 99, 15130–15135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parish, T., Smith, D. A., Kendall, S., Casali, N., Bancroft, G. J. & Stoker, N. G. (2003) Infect. Immun. 71, 1134–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cunningham, M. L., Titus, R. G., Turco, S. J. & Beverley, S. M. (2001) Science 292, 285–287. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.