Abstract
The ATP-utilizing chromatin assembly and remodelling factor (ACF) functions to generate regularly spaced nucleosomes, which are required for heritable gene silencing. The mechanism by which ACF mobilizes nucleosomes remains poorly understood. Here we report a single-molecule FRET study that monitors the remodelling of individual nucleosomes by ACF in real time, revealing previously unknown remodelling intermediates and dynamics. In the presence of ACF and ATP, the nucleosomes exhibit gradual translocation along DNA interrupted by well-defined kinetic pauses that occurred after approximately 7 or 3 – 4 base pairs of translocation. The binding of ACF, translocation of DNA, and exiting of translocation pauses are all ATP-dependent, revealing three distinct functional roles of ATP during remodelling. At equilibrium, a continuously bound ACF complex can move the nucleosome back-and-forth many times before dissociation, indicating that ACF is a highly processive and bidirectional nucleosome translocase.
The packaging of DNA into chromatin represses essential nucleic acid transactions, such as transcription, replication, repair and recombination. This repression is in part regulated by chromatin remodelling enzymes, which couple the energy of ATP hydrolysis to the assembly and mobilization of nucleosomes. ATP-dependent chromatin remodelling enzymes can be classified into several subfamilies, SWI/SNF, ISWI, CHD/Mi2 and INO80, depending on their composition and function1–5 . Despite possessing a conserved superfamily 2 ATPase subunit that facilitates DNA translocation6,7, different subfamilies exhibit divergent remodelling activities. For example, the ISWI enzymes have been shown to translocate the histone octamer along DNA and generate a repositioned nucleosome with a canonical structure8–11, whereas the SWI/SNF enzymes generate a variety of products including repositioned nucleosomes, alternative nucleosome structures containing DNA loops, and nucleosomes with altered histone composition1–5. The kinetic intermediates and pathways through which the nucleosome structure evolves during remodeling, however, remain largely elusive. Single molecule experiments are ideally suited to probe these dynamics. Recently, optical and magnetic tweezers have been used to study individual SWI/SNF remodelers, providing direct measurements of DNA translocation and loop formation by these enzymes12–14. In this work, we established a single-molecule Förster resonance energy transfer (FRET)15–17 assay to characterize the structural dynamics and kinetic intermediates of nucleosomes during remodelling. Human ACF18–22, a representative member of the ISWI family remodelers, was investigated using this approach.
Probing nucleosome translocation by FRET
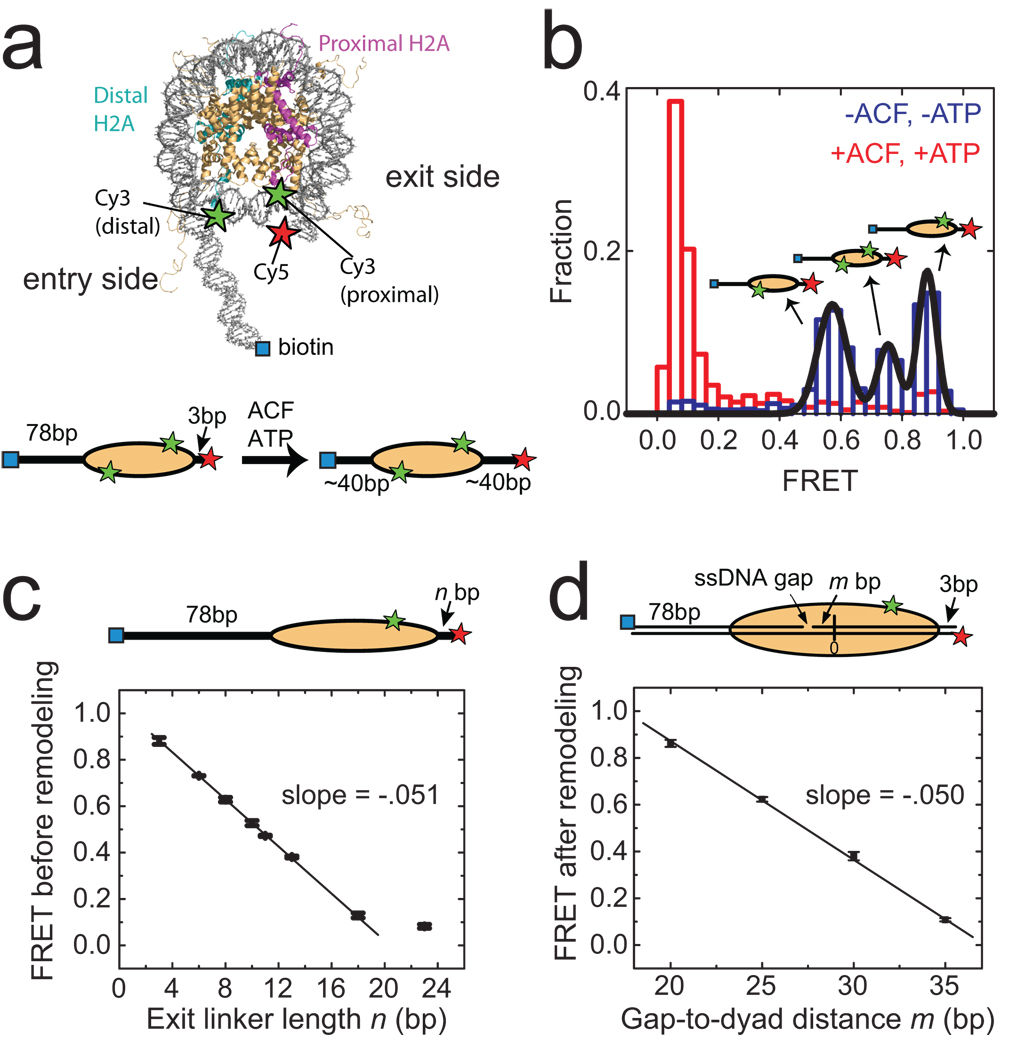
For FRET characterizations, we labelled histone octamers with a donor dye (Cy3) on histone H2A23 and reconstituted mononucleosomes with the Cy3-labeled octamer and a double-stranded DNA that contained an acceptor dye (Cy5) and a biotin at opposite ends. Unless otherwise indicated, we used the 601 nucleosome positioning sequence24 to place the octamer 3 base pairs (bp) away from the Cy5-labelled exit end of the DNA, leaving 78 bp of linker DNA on the entry side (Fig. 1a, Supplementary Fig. 1a, n = 3 bp). The nucleosomes were then anchored to a microscope slide via a biotin-streptavidin linkage and imaged by a total-internal-reflection-fluorescence (TIRF) microscope25. The presence of two H2A subunits in each octamer led to a heterogeneous population of nucleosomes with three different labelling configurations: 1) donor on the H2A subunit proximal to the acceptor, 2) donor on the H2A subunit distal to the acceptor, 3) donor on both H2A subunits. Single-molecule detection allowed these configurations to be discriminated. Three distinct peaks centred at FRET = 0. 88, 0.75 and 0.58 were observed in the FRET distribution (Fig. 1b). The assignment of these peaks to the three labelling configurations was further confirmed by individual FRET time traces, which showed one- or two-step photobleaching for nucleosomes bearing one or two donor dyes, respectively (Supplementary Fig. 2). In the following, we focus our analyses on nucleosomes containing a single donor on the proximal H2A (FRET = 0.88) to maximize the dynamic range in our experiments.
Figure 1. Monitoring ACF-catalyzed nucleosome remodelling by single-molecule FRET.
a, (upper panel) The nucleosome structure33 with labelling sites for Cy3 and Cy5 indicated by green and red stars, respectively. Additional B-form DNA is modelled onto the entry and exit sides of the nucleosome to show the flanking DNA linkers. (lower panel) A linear nucleosome scheme showing the footprint of the histone octamer (yellow oval) on the DNA (black line) before and after ACF-catalyzed remodelling. b, The FRET distribution of the n = 3 bp nucleosomes before (blue bars) and after (red bars) remodelling. The three initial peaks centred at FRET = 0.88, 0.75 and 0.58 (derived from Gaussian fit, black line) result from the three distinct Cy3-labeling configurations. After equilibration with ACF and ATP, the FRET values reduce to below 0.1. c, The initial FRET value as a function of the exit linker DNA length (n). The data were fit to a line with a slope of −0.051 ± 0.002 (black line). The last point near zero FRET is excluded from the linear fit. In this and subsequent figures, data from nucleosomes with a single Cy3 dye on the proximal H2A subunit are presented. The selection criteria for these nucleosomes are described in Online Methods. d, The final FRET values after remodelling by ACF as a function of m, the number of base pairs between the ssDNA gap and the nucleosome dyad (denoted as 0). The linear fit (black line) gives a slope of −0.050 ± 0.002. Error bars are ± s. e.m.
Recombinant ACF, comprised of a catalytic ATPase subunit, SNF2h, and an accessory subunit, Acf119–22, was added to the surface-anchored nucleosomes to induce remodelling. FRET decreased substantially upon addition of ACF and ATP (Fig. 1b), whereas incubation with ACF alone resulted in no significant change in FRET (data not shown). The observed decrease in FRET is consistent with the ability of ACF to centre mononucleosomes on DNA10,11,23,26,27 (Fig. 1a). The average remodelling rate measured from nucleosomes anchored to the surface was quantitatively similar to that determined from measurements of nucleosomes in solution, indicating that surface-anchoring of nucleosomes did not inhibit the activity of ACF (Supplementary Fig. 3).
In order to correlate the observed FRET value to the octamer position quantitatively, we measured FRET for a series of nucleosome constructs with different linker DNA lengths (n) on the exit side (Fig. 1c). The FRET value decreased monotonically with increasing exit linker length in a manner similar to the distance-dependence of FRET observed between donor and acceptor dyes attached to a DNA duplex (Supplementary Fig. 4). To further test whether the ACF-induced FRET change was indeed due to translocation of the histone octamer on DNA, we designed nucleosomes with stall sites defined by single-stranded (ss) DNA gaps. It has been shown that the ATPase domain of ISWI remodellers contacts a DNA region two helical turns (~20 bp) from the dyad axis of the nucleosome, and that ssDNA gaps located in this region inhibit nucleosome translocation28–30. We thus prepared a series of nucleosomes with the same linker DNA lengths (78 bp on the entry side and 3 bp on the exit side), each possessing a two-nucleotide ssDNA gap at a specified distance (m bp) away from the dyad axis (Fig. 1d). While the initial FRET values of these constructs were similar to that observed for the construct without the ssDNA gap, the final FRET values after remodelling showed a strong dependence on the position of the ssDNA gap (Fig. 1d), with little FRET change for the construct with m = 20 bp and a FRET versus m slope identical to that observed for the exit linker length dependence shown in Fig 1c. These results demonstrate that the observed ACF-induced FRET changes can be quantitatively interpreted in terms of nucleosome translocation along DNA, though we cannot formally exclude the possibility that other alterations in nucleosome structure could also make a minor contribution. An interesting consideration is the spontaneous site exposure due to fraying DNA ends previously reported to occur in the 0.01 – 0.05 s time scale31, which should not cause significant fluctuations in FRET observed here with 0.1 – 2 s time resolution.
Multiple ATP-dependent remodelling steps
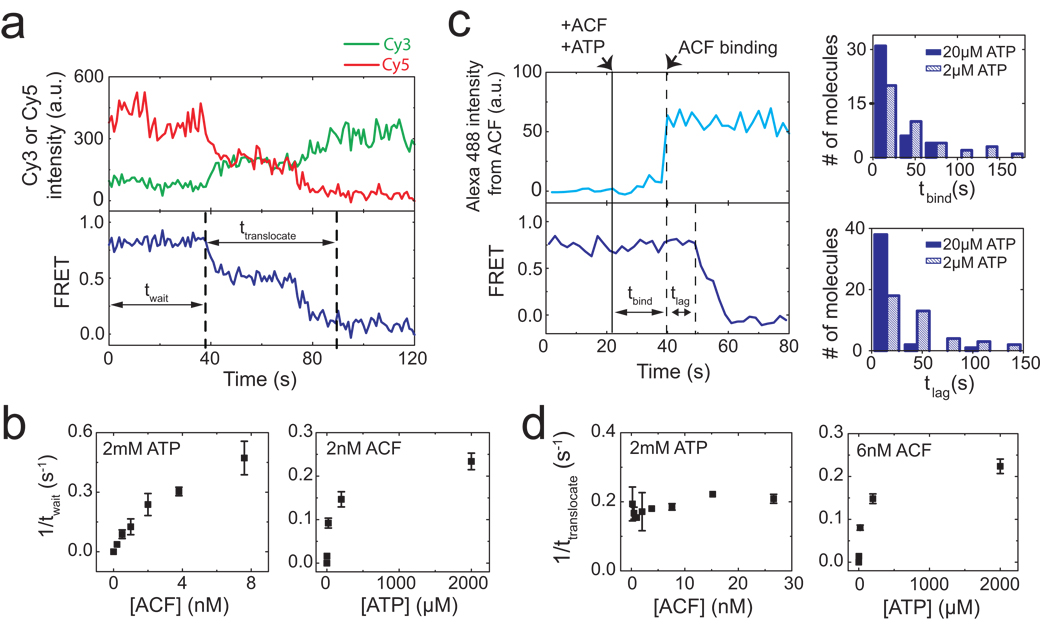
Next we characterized the remodelling kinetics by adding ACF and ATP to the nucleosomes in situ during data acquisition. After the addition of ACF and ATP, individual nucleosomes exhibited a “waiting” period prior to any detectable change in FRET, followed by a “translocation” period, during which FRET decreased to the background level (Fig. 2a). The duration of the waiting period (twait) depended on both ACF and ATP concentrations (Fig. 2b). The distributions of twait obtained at various ACF and ATP conditions suggest that the waiting phase included at least two steps, one depending on the ACF concentration and the other on ATP (Supplementary Fig. 5). To determine the order of these two steps, we performed a three-colour experiment with dye-labelled ACF, in which signal from the Alexa 488 dye on ACF directly reported the binding of the enzyme, while the FRET pair on the nucleosome reported the nucleosome position on the DNA. Notably, the binding of ACF preceded the onset of FRET decrease (Fig. 2c). Both the time before ACF binding (tbind) and the time lag (tlag) from ACF binding to the onset of FRET decrease depended on the ATP concentration (Fig. 2c), indicating that the waiting phase consisted of an ATP-dependent ACF binding step followed by an additional ATP-dependent step after the enzyme bound.
Figure 2. Real-time dynamics of ACF-catalyzed nucleosome translocation.
a, Donor fluorescence (green), acceptor fluorescence (red), and FRET (blue) traces showing the ACF-induced remodelling of a single nucleosome (n = 3 bp). ACF (6 nM) and ATP (2 µM) were added at time zero. The durations of the waiting phase and the translocation phase are denoted as twait and ttranslocate, respectively b, Dependence of the mean twait value on ACF and ATP concentrations. c, Simultaneous monitoring of the binding of ACF and the remodelling of nucleosomes. (left panels) The upper trace shows the fluorescence signal from the Alexa 488-labelled ACF. The lower trace shows FRET between Cy3 and Cy5 on the nucleosome. ACF (4 nM) and ATP (20 µM) were added at the time indicated by the solid black line. The binding event of ACF (indicated by the first dashed line) further divides twait into two phases, tbind and tlag. (right panels) The distributions of tbind and tlag at two different ATP concentrations and 4 nM ACF. The distributions at different ATP concentrations are statistically distinct with 95% confidence for tbind and more than 99% confidence for tlag according to the Kolmogorov-Smirnov test. d, Dependence of the mean ttranslocate value on ACF and ATP concentrations. Error bars are ± s.e.m.
In contrast to the waiting phase, the duration of the translocation phase (ttranslocate) was only dependent on ATP, but not on ACF, concentration (Fig. 2d and Supplementary Fig. 6), suggesting that binding of additional ACF molecules was not required during this phase. Consistent with this notion, when we prebound nucleosomes with ACF and then removed unbound ACF with a buffer containing ATP to initiate remodeling, the majority (86%) of the remodelled nucleosomes showed a complete decrease in FRET to below 0.1, indicating that the translocation phase did not require binding of additional ACF from the solution.
Translocation pauses during remodelling
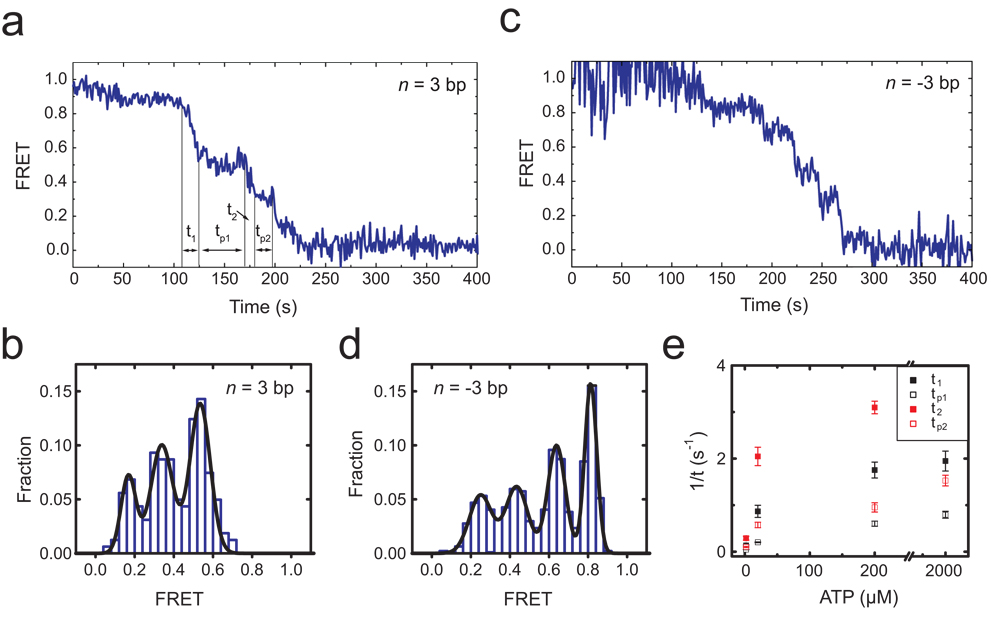
Notably, translocation of the nucleosome did not proceed at a constant rate. Instead, the translocation phase exhibited periods of gradual decrease in FRET interrupted by translocation pauses (Fig. 3). For nucleosomes with the initial exit linker length n = 3 bp, the first pause occurred at a FRET value of 0.53 ± 0.03 (Fig. 3a, b), corresponding to an increase of linker length to 9.9 ± 0.6 bp and thus nucleosome translocation by 6.9 ± 0.6 bp. The pause position appeared to be independent of the initial linker length: for nucleosome constructs with four different linker DNA lengths (n = −3, 0, 3 and 6 bp), the first pause all occurred after approximately 7 bp of DNA translocation (Supplementary Fig. 7a).
Figure 3. ACF-catalyzed nucleosome translocation is interrupted by well defined kinetic pauses.
a, FRET time trace of a nucleosome (n = 3 bp) showing kinetic pauses that divide the entire translocation phase into several translocation and pause sub-phases: t1, tp1, t2, tp2…. ACF (6 nM) and ATP (2 µM) were added at time zero. b, FRET distribution of the pauses observed for the n = 3 bp nucleosomes. The peak FRET values, 0.53, 0.34, and 0.17 (obtained from Gaussian fit, black line), corresponds to 6.9 bp of translocation between the initial position and the first pause, 3.8 bp between the first and second pauses, and 3.3 bp between the second and third pauses. c, FRET time trace of a n = −3 bp nucleosome after addition of ACF (6 nM) and ATP (2 µM) at time zero. The proximity of the initial donor and acceptor positions causes partial quenching of their fluorescence and thus relatively large fluctuations in initial FRET. d, FRET distribution of the pauses observed for the n = −3 bp nucleosomes. The peaks correspond to 7.3 bp of translocation between the initial position and the first pause, 3.4 bp between the first and second pauses, 4.0 bp between the second and third pauses, and 3.6 bp between the third and fourth pauses. e, ATP-dependence of the mean t1, tp1, t2, and tp2 values. Error bars are ± s.e.m.
In addition, we tested the dependence of the pause position on DNA sequence using a weaker positioning sequence “A-100” (Supplementary Fig. 1b), which has ~100 fold lower affinity than the 601 sequence32. The first pause of these nucleosomes again occurred after approximately 7 bp of translocation (Supplementary Fig. 7b). While we cannot formally rule out the possibility that the positioning sequences contributed to the position of this initial pause, the observation that nucleosomes with two substantially different DNA sequences exhibit the same initial pause position suggests a potentially general feature of remodelling by ACF.
In addition to the first pause, subsequent translocation pauses were observed at lower FRET values (Fig. 3a, c). For nucleosomes with initial exit linker length n = 3 bp, a second and third pause preferentially occurred at FRET = 0.34 ± 0.03 and 0.17 ± 0.03, corresponding to 3.8 ± 0.6 bp and 3.3 ± 0.6 bp of translocation prior to pausing, respectively (Fig. 3a, b). Similar pauses were also observed for the n = −3 bp nucleosomes, except that the shorter exit linker length after the third pause allowed detection of a fourth pause, which occurred after 3.6 ± 0.8 bp of translocation from the third pause (Figs. 3c, d). Taken together, these results indicate that the nucleosomes were translocated by a shorter distance (3 – 4 bp) between the subsequent pauses. Both the dwell time of the pauses and the duration of the translocation phases in between pauses depended on the concentration of ATP, indicating that ATP binding was required in both phases (Fig. 3e and Supplementary Fig. 8). The dwell times of the subsequent pauses were similar to each other but substantially shorter than that of the first pause.
We note that the sum of a 7 bp and a 3 – 4 bp step and the sum of three 3 – 4 bp steps are both close to the 10 bp periodicity of DNA-histone contacts within the nucleosome33. Interestingly, the remodelling intermediates at a fraction of the periodicity (7 bp and 3 – 4 bp) were not stable in the absence of the remodelling enzyme: upon removal of ACF, these intermediates collapsed to nucleosomal states in which the histone octamer was repositioned by a multiple of ~10 bp from the pre-remodelling position (Supplementary Fig. 9). These collapsed states, consistent with the previously observed accumulation of remodelling products at ~10 bp intervals of nucleosome translocation28,29, are likely imposed by structural constraints of the nucleosome.
Processive and bidirectional translocation
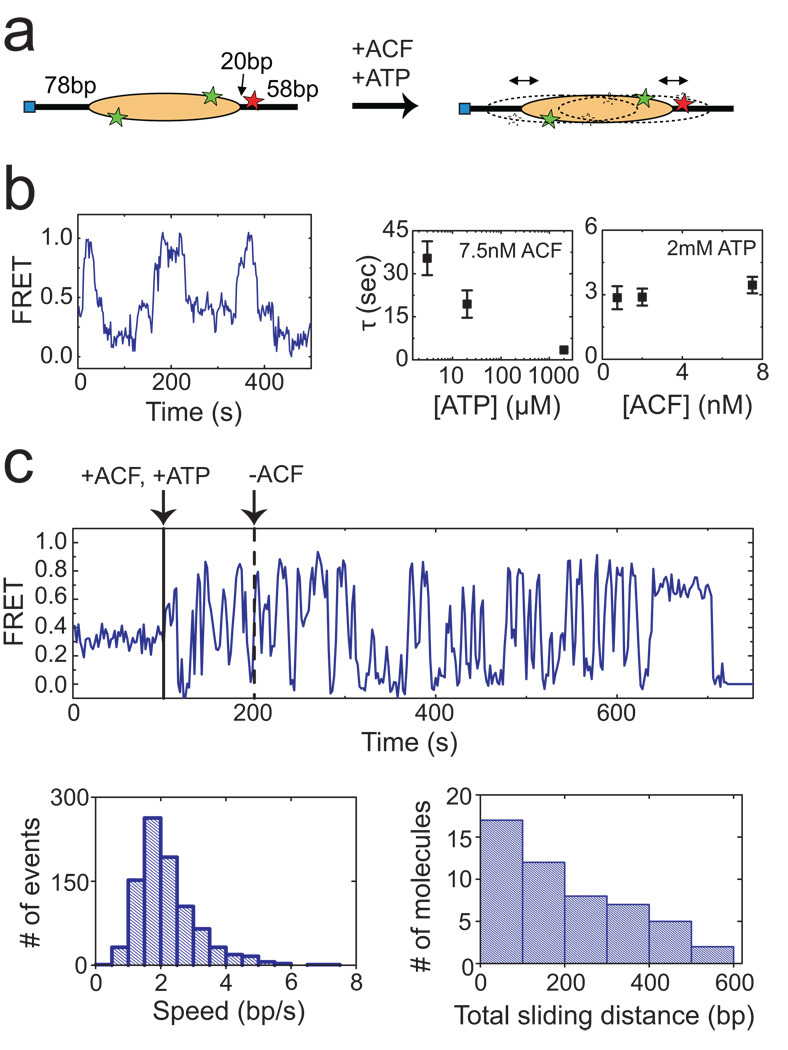
The above experiments with end-positioned nucleosomes provide quantitative analyses of remodelling kinetics and intermediates. The limited dynamic range of FRET, however, made it difficult to characterize the equilibrium state(s) after remodelling using these substrates. Considering that ACF tends to centre the nucleosome on the DNA, we reasoned that a centre-positioned nucleosome with an initial FRET value within the dynamic range of FRET would facilitate the analysis of equilibrium remodelling dynamics. To this end we constructed a centre-positioned mononucleosome with the 601 sequence flanked by 78 bp of DNA on each side and an internal acceptor label (Fig. 4a, Supplementary Fig. 1a)). The initial FRET distribution showed a narrow peak at FRET = 0.3 (Supplementary Fig. 10). After equilibration with ACF and ATP, the FRET distribution broadened substantially (Supplementary Fig. 10) and the time traces of individual nucleosomes exhibited large-amplitude oscillations in FRET (Fig. 4b), indicating that the histone octamer was translocated back-and-forth along the DNA by the remodelling enzyme. Bidirectional remodelling was observed to be the predominant behaviour (> 70% of remodelled nucleosomes), even at sub-saturating conditions in which a low concentration (1 nM) of ACF was added to induce remodelling of only a small fraction (<10%) of the nucleosomes. Autocorrelation analysis of these FRET time traces revealed a characteristic oscillation time that was dependent on the ATP concentration but independent of the ACF concentration (Fig. 4b), suggesting that the observed bidirectional translocation was accomplished by continuously bound ACF without requiring dissociation and rebinding of ACF from the solution. To further test this notion, we performed three-colour experiments with Alexa 488-labelled ACF and FRET-labelled nucleosomes, in which signal from the Alexa 488 dye directly reported the binding of ACF. Repeated back-and-forth movement of the nucleosomes was observed within individual ACF binding events (Supplementary Fig. 11), further confirming that the bidirectional nucleosome translocation was accomplished by a continuously bound ACF complex.
Figure 4. ACF catalyzes processive and bidirectional nucleosome translocation.
a, A centre-positioned nucleosome flanked by 78 bp of linker DNA on both sides is subject to remodelling. b, (left panel) FRET trace of a nucleosome in equilibrium with 7.5 nM ACF and 3 µM ATP showing back-and-forth translocation on DNA. (right panel) The characteristic time of the FRET fluctuations (τ) depends on ATP, but not on ACF, concentration. The τ values were derived from autocorrelation analysis as described in Online Methods. c, (upper panel) A FRET trace showing processive and bidirectional nucleosome translocation by a continuously bound ACF complex. ACF (3 nM) and ATP (2 mM) were added at the time indicated by the solid black line. Unbound ACF (but not ATP) was then removed from the solution at the time indicated by the dashed black line. (lower left panel) Distribution of the translocation speed within each segment of unidirectional translocation. (lower right panel) Distribution of the cumulative distance travelled by individual nucleosomes after removal of unbound ACF. Estimate of the travelling speed and distance is described in Online Methods. Error bars are ± s.e.m.
To further quantify the processivity of ACF, we performed buffer exchange experiments in which ACF and ATP were added and unbound ACF (but not ATP) was subsequently removed in situ as the position of individual nucleosomes was monitored. Remarkably, the ACF-induced bidirectional movement persisted for a long period of time after unbound ACF was removed from the solution (Fig. 4c). The nucleosomes were translocated with an average speed of approximately 2 bp/s. The lower-bound estimate of the cumulative distance travelled by the nucleosome after removal of unbound ACF exhibits a broad distribution with a mean of 200 bp (Fig. 4c). Taken together, these results indicate that ACF is a highly processive and bidirectional nucleosome translocase. The observed processivity is consistent with the strong commitment of ISWI enzymes to nucleosomal templates once chromatin assembly and remodelling are initiated34,35.
It is striking that an ACF complex remaining bound to the nucleosome could cause such a highly processive, back-and-forth nucleosome movement. Such a demanding task could be accomplished if ACF preferentially binds the nucleosome as a dimer, in which two ACF monomers, particularly their corresponding ATPase domains, are bound on opposite sides of the nucleosome and oriented for translocation in opposing directions. Coordinated action of the two monomers would then allow processive back-and-forth translocation of the nucleosome. This hypothesis is supported by our three-colour experiments with Alexa 488-labelled ACF and FRET-labelled nucleosomes. To determine the number of ACF bound to the nucleosome, we performed statistical analyses of the Alexa 488 intensity and the number of Alexa 488 photobleaching steps associated with each ACF binding event. These analyses suggest that the binding events leading to bidirectional nucleosome remodelling preferentially contained two ACF monomers, whereas the binding events leading to unidirectional remodelling preferentially contained a single ACF monomer (Supplementary Fig. 12). Further supporting this model, electron microscopy and biochemical data showed cooperative binding of two SNF2h proteins to a single nucleosome with each SNF2h occupying one side of the nucleosome in an activated ATP state (Racki et al manuscript). The diffusion coefficient of ACF bound to DNA is also consistent with a complex of two Acf1 and two SNF2h subunits36. Interestingly, the SWI/SNF subfamily enzymes can also reversibly create and retract DNA loops12,13, but it is unclear whether the bidirectional nucleosome translocation by ACF and the reversible DNA loop formation by SWI/SNF share a common mechanism.
Discussion
We have developed a single-molecule assay to monitor the remodelling of individual nucleosomes by chromatin remodelling enzymes in real time. This assay allowed quantitative characterization of the structural dynamics and kinetic intermediates of nucleosomes during remodelling. Using this approach, we showed that the human ACF enzyme induced gradual translocation of nucleosomes along DNA interrupted by well-defined kinetic pauses. ATP plays multiple functional roles in the remodelling process. The three distinct steps during remodelling, namely binding of ACF, translocation of the nucleosome, and translocation pauses, were all ATP dependent, revealing a versatile usage of ATP by an enzyme with only one type of ATP binding site.
Quantification of the FRET traces of end-positioned nucleosomes showed that the first kinetic pause occurred after approximately 7 bp of nucleosome translocation whereas subsequent pauses were separated by only 3 – 4 bp. Although it is currently unclear whether these remodelling intermediates occur only at the beginning of remodelling or continue into the processive remodelling phase, similar translocation pauses were also observed during the continuous remodelling process of centre-positioned nucleosomes (Fig. 4b) and thus may represent a fundamental property of ACF-induced remodelling. One possible origin of these intermediates is an ATP-dependent conformational change of the remodelling enzyme that prepares the nucleosome for the next round of DNA translocation (for example, by forming a DNA loop for subsequent propagation around the nucleosome)30,37,38. The unique properties of the first pause, as compared to the subsequent pauses, may imply a complex initiation phase of remodelling.
On centre-positioned nucleosomes, ACF was observed to exhibit remarkable processivity and bidirectionality: an ACF complex continuously bound to a nucleosome could translocate the histone octamer back-and-forth by a total distance of more than 200 bp and switch directions more than 20 times on average before dissociation. Statistical analyses suggest that the bidirectional remodelling is most probably caused by ACF dimers. The processive and bidirectional translocation of nucleosomes potentially allows ACF to rapidly sample the DNA on both sides of the nucleosome to generate regular inter-nucleosomal spacing.
METHODS SUMMARY
Detailed description of sample preparation and single-molecule FRET measurements are given in Online Methods. Briefly, various mononucleosome constructs, with different DNA sequences, DNA linker lengths, and ssDNA gap locations, were reconstituted using histone octamers that were labelled with a green FRET donor dye (Cy3) and double stranded DNA that was labeled with a red FRET acceptor dye (Cy5) and a biotin. The nucleosomes were then anchored to a microscope slide via a biotin-streptavidin linkage. Unlabelled ACF or ACF labelled with a blue dye (Alexa 488) were added to the surface-anchored nucleosomes together with ATP to induce remodelling. The fluorescence signals from Alexa 488, Cy3, and Cy5 were detected by a TIRF microscope, separated by dichroic mirrors, and imaged onto separate areas of an amplified-CCD camera after passing through various fluorescence emission filters. Custom-written software was used to identify single nucleosomes on the slide and to monitor the Alexa 488, Cy3 and Cy5 fluorescence at these positions for extended periods of time. The FRET value was defined as IA / (ID + IA), where ID and IA represent the fluorescence signals detected in the Cy3 and Cy5 channels, respectively.
Supplementary Material
Acknowledgements
We thank J. Widom (Northwestern University) for providing the plasmid containing the 601 positioning sequence and R.E. Kingston (Harvard Medical School) for the plasmids containing SNF2h and Acf1 genes. We also thank Lisa Racki and Elio Abbondanzieri for helpful discussions, and William Huang and Bryan Harada for help with some experiments. This work is supported in part by Howard Hughes Medical Institute (to X.Z.) and the National Institutes of Health (GM073767) and the Beckman Foundation (to G.J.N). X.Z. is a Howard Hughes Medical Institute investigator. M.D.S. was a NIH Ruth L. Kirschstein NSRA Fellow, G.J.N is a Leukemia and Lymphoma Society Scholar.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author information T.R.B. performed the experiments and analysis with help from M.D.S. J.G.Y. made the enzymes and histone proteins. T.R.B., G.J.N, and X.Z. designed the experiments. X.Z. oversaw the project.
References
- 1.Becker PB, Horz W. ATP-dependent nucleosome remodeling. Annu Rev Biochem. 2002;71:247–273. doi: 10.1146/annurev.biochem.71.110601.135400. [DOI] [PubMed] [Google Scholar]
- 2.Narlikar GJ, Fan HY, Kingston RE. Cooperation between complexes that regulate chromatin structure and transcription. Cell. 2002;108:475–487. doi: 10.1016/s0092-8674(02)00654-2. [DOI] [PubMed] [Google Scholar]
- 3.Flaus A, Owen-Hughes T. Mechanisms for ATP-dependent chromatin remodelling: farewell to the tuna-can octamer? Curr Opin Genet Dev. 2004;14:165–173. doi: 10.1016/j.gde.2004.01.007. [DOI] [PubMed] [Google Scholar]
- 4.Smith CL, Peterson CL. ATP-dependent chromatin remodeling. Curr Top Dev Biol. 2005;65:115–147. doi: 10.1016/S0070-2153(04)65004-6. [DOI] [PubMed] [Google Scholar]
- 5.Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu Rev Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- 6.Saha A, Wittmeyer J, Cairns BR. Chromatin remodeling by RSC involves ATP-dependent DNA translocation. Genes Dev. 2002;16:2120–2134. doi: 10.1101/gad.995002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Whitehouse I, Stockdale C, Flaus A, Szczelkun MD, Owen-Hughes T. Evidence for DNA translocation by the ISWI chromatin-remodeling enzyme. Mol Cell Biol. 2003;23:1935–1945. doi: 10.1128/MCB.23.6.1935-1945.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsukiyama T, Palmer J, Landel CC, Shiloach J, Wu C. Characterization of the Imitation Switch subfamily of ATP-dependent chromatin-remodeling factors in Saccharomyces cerevisiae. Genes Dev. 1999;13:686–697. doi: 10.1101/gad.13.6.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hamiche A, Sandaltzopoulos R, Gdula DA, Wu C. ATP-dependent histone octamer sliding mediated by the chromatin remodeling complex NURF. Cell. 1999;97:833–842. doi: 10.1016/s0092-8674(00)80796-5. [DOI] [PubMed] [Google Scholar]
- 10.Langst G, Bonte EJ, Corona DF, Becker PB. Nucleosome movement by CHRAC and ISWI without disruption or trans-displacement of the histone octamer. Cell. 1999;97:843–852. doi: 10.1016/s0092-8674(00)80797-7. [DOI] [PubMed] [Google Scholar]
- 11.Kassabov SR, Henry NM, Zofall M, Tsukiyama T, Bartholomew B. High-resolution mapping of changes in histone-DNA contacts of nucleosomes remodeled by ISW2. Mol Cell Biol. 2002;22:7524–7534. doi: 10.1128/MCB.22.21.7524-7534.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Y, Smith CL, Saha A, Grill SW, Mihardja S, Smith SB, Cairns BR, Peterson CL, Bustamante C. DNA translocation and loop formation mechanism of chromatin remodeling by SWI/SNF and RSC. Mol Cell. 2006;24:559–568. doi: 10.1016/j.molcel.2006.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lia G, Praly E, Ferreira H, Stockdale C, Tse-Dinh YC, Dunlap D, Croquette V, Bensimon D, Owen-Hughes T. Direct observation of DNA distortion by the RSC complex. Mol Cell. 2006;21:417–425. doi: 10.1016/j.molcel.2005.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shundrovsky A, Smith CL, Lis JT, Peterson CL, Wang MD. Probing SWI/SNF remodeling of the nucleosome by unzipping single DNA molecules. Nat Struct Mol Biol. 2006;13:549–554. doi: 10.1038/nsmb1102. [DOI] [PubMed] [Google Scholar]
- 15.Stryer L, Haugland RP. Energy transfer: a spectroscopic ruler. Proc Natl Acad Sci U S A. 1967;58:719–726. doi: 10.1073/pnas.58.2.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ha T, Enderle T, Ogletree DF, Chemla DS, Selvin PR, Weiss S. Probing the interaction between two single molecules: fluorescence resonance energy transfer between a single donor and a single acceptor. Proc Natl Acad Sci U S A. 1996;93:6264–6268. doi: 10.1073/pnas.93.13.6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhuang X, Bartley L, Babcock H, Russell R, Ha T, Herschlag D, Chu S. A single molecule study of RNA catalysis and folding. Science. 2000;288:2048–2051. doi: 10.1126/science.288.5473.2048. [DOI] [PubMed] [Google Scholar]
- 18.Ito T, Bulger M, Pazin MJ, Kobayashi R, Kadonaga JT. ACF, an ISWI-containing and ATP-utilizing chromatin assembly and remodeling factor. Cell. 1997;90:145–155. doi: 10.1016/s0092-8674(00)80321-9. [DOI] [PubMed] [Google Scholar]
- 19.Ito T, Levenstein ME, Fyodorov DV, Kutach AK, Kobayashi R, Kadonaga JT. ACF consists of two subunits, Acf1 and ISWI, that function cooperatively in the ATP-dependent catalysis of chromatin assembly. Genes Dev. 1999;13:1529–1539. doi: 10.1101/gad.13.12.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bochar DA, Savard J, Wang W, Lafleur DW, Moore P, Cote J, Shiekhattar R. A family of chromatin remodeling factors related to Williams syndrome transcription factor. Proc Natl Acad Sci U S A. 2000;97:1038–1043. doi: 10.1073/pnas.97.3.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leroy G, Loyola A, Lane WS, Reinberg D. Purification and characterization of a human factor that assembles chromatin. J Biol Chem. 2000;275:14787–14790. doi: 10.1074/jbc.C000093200. [DOI] [PubMed] [Google Scholar]
- 22.Poot RA, Dellaire G, Hulsmann BB, Grimaldi MA, Corona DF, Becker PB, Bickmore WA, Varga-Weisz PD. HuCHRAC, a human ISWI chromatin remodelling complex contains hACF1 and two novel histone-fold proteins. Embo J. 2000;19:3377–3387. doi: 10.1093/emboj/19.13.3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang JG, Madrid TS, Sevastopoulos E, Narlikar GJ. The chromatin-remodeling enzyme ACF is an ATP-dependent DNA length sensor that regulates nucleosome spacing. Nat Struct Mol Biol. 2006;13:1078–1083. doi: 10.1038/nsmb1170. [DOI] [PubMed] [Google Scholar]
- 24.Lowary PT, Widom J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J Mol Biol. 1998;276:19–42. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- 25.Abbondanzieri EA, Bokinsky G, Rausch JW, Zhang JX, Le Grice SF, Zhuang X. Dynamic binding orientations direct activity of HIV reverse transcriptase. Nature. 2008;453:184–189. doi: 10.1038/nature06941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He X, Fan HY, Narlikar GJ, Kingston RE. Human ACF1 alters the remodeling strategy of SNF2h. J Biol Chem. 2006;281:28636–28647. doi: 10.1074/jbc.M603008200. [DOI] [PubMed] [Google Scholar]
- 27.Stockdale C, Flaus A, Ferreira H, Owen-Hughes T. Analysis of nucleosome repositioning by yeast ISWI and Chd1 chromatin remodeling complexes. J Biol Chem. 2006;281:16279–16288. doi: 10.1074/jbc.M600682200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schwanbeck R, Xiao H, Wu C. Spatial contacts and nucleosome step movements induced by the NURF chromatin remodeling complex. J Biol Chem. 2004;279:39933–39941. doi: 10.1074/jbc.M406060200. [DOI] [PubMed] [Google Scholar]
- 29.Zofall M, Persinger J, Kassabov SR, Bartholomew B. Chromatin remodeling by ISW2 and SWI/SNF requires DNA translocation inside the nucleosome. Nat Struct Mol Biol. 2006;13:399–346. doi: 10.1038/nsmb1071. [DOI] [PubMed] [Google Scholar]
- 30.Dang W, Bartholomew B. Domain architecture of the catalytic subunit in the ISW2-nucleosome complex. Mol Cell Biol. 2007;27:8306–8317. doi: 10.1128/MCB.01351-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li G, Levitus M, Bustamante C, Widom J. Rapid spontaneous accessibility of nucleosomal DNA. Nat Struct Mol Biol. 2005;12:46–53. doi: 10.1038/nsmb869. [DOI] [PubMed] [Google Scholar]
- 32.Partensky PD, Narlikar GJ. Chromatin remodelers act globally, sequence positions nucleosomes locally. J Mol Biol. 2009;391:12–25. doi: 10.1016/j.jmb.2009.04.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 34.Fyodorov DV, Kadonaga JT. Dynamics of ATP-dependent chromatin assembly by ACF. Nature. 2002;418:897–900. doi: 10.1038/nature00929. [DOI] [PubMed] [Google Scholar]
- 35.Gangaraju VK, Prasad P, Srour A, Kagalwala MN, Bartholomew B. Conformational changes associated with template commitment in ATP-dependent chromatin remodeling by ISW2. Mol Cell. 2009;35:58–69. doi: 10.1016/j.molcel.2009.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Strohner R, Wachsmuth M, Dachauer K, Mazurkiewicz J, Hochstatter J, Rippe K, Langst G. A 'loop recapture' mechanism for ACF-dependent nucleosome remodeling. Nat Struct Mol Biol. 2005;12:683–690. doi: 10.1038/nsmb966. [DOI] [PubMed] [Google Scholar]
- 37.Fitzgerald DJ, DeLuca C, Berger I, Gaillard H, Sigrist R, Schimmele K, Richmond TJ. Reaction cycle of the yeast Isw2 chromatin remodeling complex. Embo J. 2004;23:3836–3843. doi: 10.1038/sj.emboj.7600364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cairns BR. Chromatin remodeling: insights and intrigue from single-molecule studies. Nat Struct Mol Biol. 2007;14:989–996. doi: 10.1038/nsmb1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.