Abstract
Nitrophorin 4 (NP4) is a heme protein that stores and delivers nitric oxide (NO) through pH sensitive conformational change. This protein uses the ferric state of a highly ruffled heme to bind NO tightly at low pH and release it at high pH. In this work, the rebinding kinetics of NO and CO to NP4 are investigated as a function of iron oxidation state and the acidity of the environment. The geminate recombination process of NO to ferrous NP4 at both pH 5 and pH 7 is dominated by a single ~7 ps kinetic phase that we attribute to the rebinding of NO directly from the distal pocket. The lack of pH dependence explains in part why NP4 cannot use the ferrous state to fulfill its function. The kinetic response of ferric NP4NO shows two distinct phases. The relative geminate amplitude of the slower phase increases dramatically as the pH is raised from 5 to 8. We assign the fast phase of NO rebinding to a conformation of the ferric protein with a closed hydrophobic pocket. The slow phase is assigned to the protein in an open conformation with a more hydrophilic heme pocket environment. Analysis of the ultrafast kinetics finds the equilibrium off-rate of NO to be proportional to the open state population as well as the pH-dependent amplitude of escape from the open pocket. When both factors are considered, the off-rate increases by more than an order of magnitude as the pH changes from 5 to 8. The recombination of CO to ferrous NP4 is observed to have a large non-exponential geminate amplitude with rebinding timescales of ~10−11–10−9 s at pH 5 and ~10−10–10−8 s at pH 7. The non-exponential CO rebinding kinetics at both pH 5 and pH 7 are accounted for using a simple model that has proven effective for understanding CO binding in a variety of other heme systems.
Introduction
Heme proteins are involved in many fundamental and widespread biological functions such as ligand transport, electron transfer, catalysis and signaling1–10. Nitrophorins (NPs) are nitric oxide (NO) carrier heme proteins found in the saliva of blood feeding insects2,11. The salivary gland of the “kissing bug” Rhodnuis Prolixus contains seven such proteins; four (NP1–4) in the adult insect and three additional proteins are found in the early stage of development12,13. Nitrophorins sequester NO produced by nitric oxide synthase (NOS) in the salivary glands of the insect14. They use the ferric state of the heme to store and deliver NO through a pH dependent protein conformational change. These proteins bind NO tightly in a low pH environment (pH~5 for insect saliva) and release it in the higher pH environment of the host tissue (pH ~7.5). The NO affinity of nitrophorins is regulated by off-rates, k off, which increase significantly when the pH changes from 5 to 815.
Releasing of NO initiates a cascade of signals that leads to a vasodilatation and reduced platelet aggregation, thereby increasing the blood supply for the insect2. Nitrophorins also sequester the histamine released by the host, which helps to displace NO and increase the efficiency of the vasodilatory activity. It also prevents inflammation associated with tissue damage and protects the insect from detection11,16.
Nitrophorin 4 (NP4), the subject of this work, has been extensively studied experimentally and theoretically15,17–30. The structure of NP4 has a lipocalin fold, consisting of β-barrel with a ruffled heme buried in one end of the barrel and covalently linked to the protein through the proximal histidine His5931. In addition to NO, small ligands such as H2O, CN− and CO can bind to the heme iron from the distal side. Binding of NO to ferric NP4 at low pH leads to reordering of the A–B and G–H loops which results in expulsion of water molecules and burying NO tightly inside the distal pocket19,21,31. The heme becomes highly ruffled due in part to its close contact with the distal leucine residues (Leu123 and Leu133)19,24. This leads to the stabilization of the ferric NP4NO complex11,19.
X-ray crystallography has revealed that the distal pocket of NP4 cycles between two conformations: a hydrophobic well organized closed conformer and a hydrophilic less ordered open conformer. Features of both conformers exist at all conditions, and the population ratio of the closed to the open conformer increases with decreasing the pH and/or the polarity of the ligand19,21,24. When compared to NO, the more polar and upright-bound CO molecule evidently leads to a smaller amount of ligand induced conformational change and a smaller population of the closed conformation23.
Site directed mutagenesis and kinetics of ligand binding to the heme following photolysis have been extensively used to investigate the structure, dynamics, and function of heme proteins and to explore the migration pathways of small molecules within them32–67. These studies have demonstrated that, in general, the kinetics of diatomic ligand (NO, O2 and CO) binding to heme proteins involve surprisingly complex processes 34,39,51,60,68,69. This is primarily due to the multidimensional energy landscape of the protein68 as revealed by kinetic hole burning64,70–72, structural relaxation of the protein39,64,73,74, and interconversion among its different conformational sub-states44,46. In some cases, these processes occur on the same time scale as ligand rebinding and migration between different docking sites within the protein matrix45,74–78. Despite the huge amount of theoretical and experimental work that has been carried out in this field over the last 50 years, the reactions of small diatomic molecules with the heme, and the kinetic mechanisms underlying recognition and discrimination, are still not fully understood79.
Nitrophorins are excellent examples of the class of proteins where function is controlled by environmental change. In this work we investigate how variations in the pH of the environment regulate the binding and release of small diatomic ligands (NO and CO) in NP4, focusing on the physiologically relevant case of nitric oxide release from ferric NP4.
Experimental Section
Sample preparation
Ferric NP4 was expressed and purified from E. coli as described previously17,80. The protein was dissolved in the appropriate buffers (100 mM sodium acetate buffer at pH 5.0, 100 mM phosphate buffer at pH 7.0, or 100 mM Tris-HCl buffer at pH 8.0 ). All solutions were continuously degassed with argon for at least 30 minutes. To prepare Ferric NP4NO samples, NO gas (99.0% pure, Mathenson) was bubbled successively through 0.1 M NaOH solution and the appropriate buffer, and then was introduced to the degassed ferric NP4 under 1 atm pressure for about 20 s. Ferrous samples were obtained by adding an excess of 1 molar sodium dithionite under anaerobic conditions. Ferrous NO adducts were prepared by further adding an equimolar quantity of degassed sodium nitrite solution to the ferrous samples, whereas CO adducts were prepared by flushing CO gas over the surface of the ferrous samples for about 1 h. In all cases the sample concentration was adjusted to have approximately an absorbance of 1 OD at the Soret peak in the 1 mm path length cell. The absorption spectra were recorded before and after each experiment using a spectrophotometer (U-4100 Hitachi). All measurements were carried out at room temperature (T = 22° C).
Experimental Setup
Two laser systems have been used in this study. The temporal kinetic traces were measured using a two color pump-probe instrument that has been described in detail elsewhere81. The instrument involves two Ti: sapphire regenerative amplifiers, both operating at 190 kHz, which are seeded by two synchronized Ti:sapphire oscillators: A master (Mira P, 76 MHz, 3 ps (FWHM)) and a slave (Mira F, 76 MHz, 100 fs (FWHM)). The output of the femtosecond regenerative amplifier (Coherent REGA9000) was focused in a 500 µm BBO crystal to generate the second harmonic at 403 nm, which used to pump the sample. The amplified picosecond pulses from a tunable picosecond regenerative amplifier81 were doubled using a 2 mm BBO crystal. This beam was tuned to 420 nm to probe ferric samples and to 438 nm when ferrous samples were under investigation. The two beams are then collinearly combined and focused in the sample using an achromatic lens with 2 in focal length. The time delay between the pump and probe pulses can be controlled electronically using a phase shifter in the synchrolock system (continuous scan from 0 to 16 ns) along with the femtosecond regenerative amplifier timing delay electronics that allow extending the scan window up to 5 µs by discrete steps of 26 ns. The timing jitter between the pump and probe pulses is ~800 fs. The polarization of the pump and the probe beam are set to the magic angle (~54.7°) to eliminate rotational relaxation artifacts.
The transient absorption spectra were measured using a laser system that has been described elsewhere49,82. A 580 nm pump beam is obtained from an optical parametric amplifier (OPA) which is pumped with an amplified Ti:Sapphire laser system (REGA 9050, Coherent. Inc.) operating at 200 kHz. Part of the 800 nm fundamental pulse train from the amplifier is focused onto a spinning fused silica disk to generate a white light continuum which is used as a probe. The time delay between the pump and the probe is controlled using a servo-motor translation stage with a full translation range of 5.6 ns. The pump and the probe are focused into a spinning sample cell (2 in diameter) rotating at 7000 rpm. After the sample, the pump is blocked using a short pass cutoff filter and the transmitted probe light is dispersed using a monochromator and detected by a photodiode array detector.
Kinetic Data Analysis for ferric NP4NO
A kinetic model has been recently put forward to describe the mechanism of NO escape from NP429. Here we propose a similar kinetic scheme, but without explicitly exposing the rates for opening and closing the heme pocket or for thermal dissociation of the heme-ligand bond.
Where the subscripts (c) and (o) represent the closed and open pockets, respectively. NPcL and NPoL represent the ligand bound state of NP4; NPc:L and NPo:L represent the ligand being dissociated from the heme, but still inside the protein matrix, whereas NPc+L and NPo+L are the states where the ligand has escaped into the solution. (Here we note that the “open state” is more accurately described as a dynamic distribution of open conformations.) The states NPL and NP:L correspond to the A and B states that are often used to describe ligand binding in heme systems, so the geminate rebinding rate constant is denoted as kBA. The quantities are the intrinsic geminate rates, the escape rates, and the entry rates in the open and closed conformers, respectively. The rates of closing and opening of the protein, kc and ko, are not shown explicitly because we assume these rates to be slower than the kinetic timescales under investigation here.
The interconversion NPo → NPc requires a large conformational change involving collapse of the two loops A–B and G–H, forming a hydrogen bond network and packing of hydrophobic side chains into the distal pocket. This process is expected to be slow23 (for example the rate of the well-known open/closed transition in myoglobin was found to be ~104 to 106 s−1 depending on temperature and solvent composition44,46). The geminate rebinding of the ligand in NP4, however, occurs on the picosecond time scale. Thus, for the sake of simplicity in Scheme 1, we assume that the interconversion NPo ↔ NPc is slow compared to ligand binding and escape. Under these conditions the equations that govern the rebinding kinetics in the closed and open conformations are decoupled and each one of these processes can be described by a simple three state model36,83:
| (1a) |
| (1b) |
where Nc (t) and No (t) are the normalized survival populations in the closed and open conformations, respectively. The overall rebinding process is then governed byN(t) = PcNc(t) +PoNo(t), where Pc and Po represent the closed and open populations in Scheme 1 (Pc + Po=1). Assuming that (i.e., geminate rebinding is much faster than bimolecular rebinding with i=c,o) then the temporal evolution of the total survival population on time scales shorter than the bimolecular kinetics can be written as
| (2) |
Within the three state model we have: with i=c,o If we consider that the ligand escapes only from the open conformer23,29, then in Eq. 2. Finally, we note that under this condition so the overall off-rate depends only on the “open” parameters and can be written as
| (3) |
where is the rate of thermal bond breaking in the open conformation.
Scheme 1.
Results
NO Rebinding kinetics to ferrous NP4
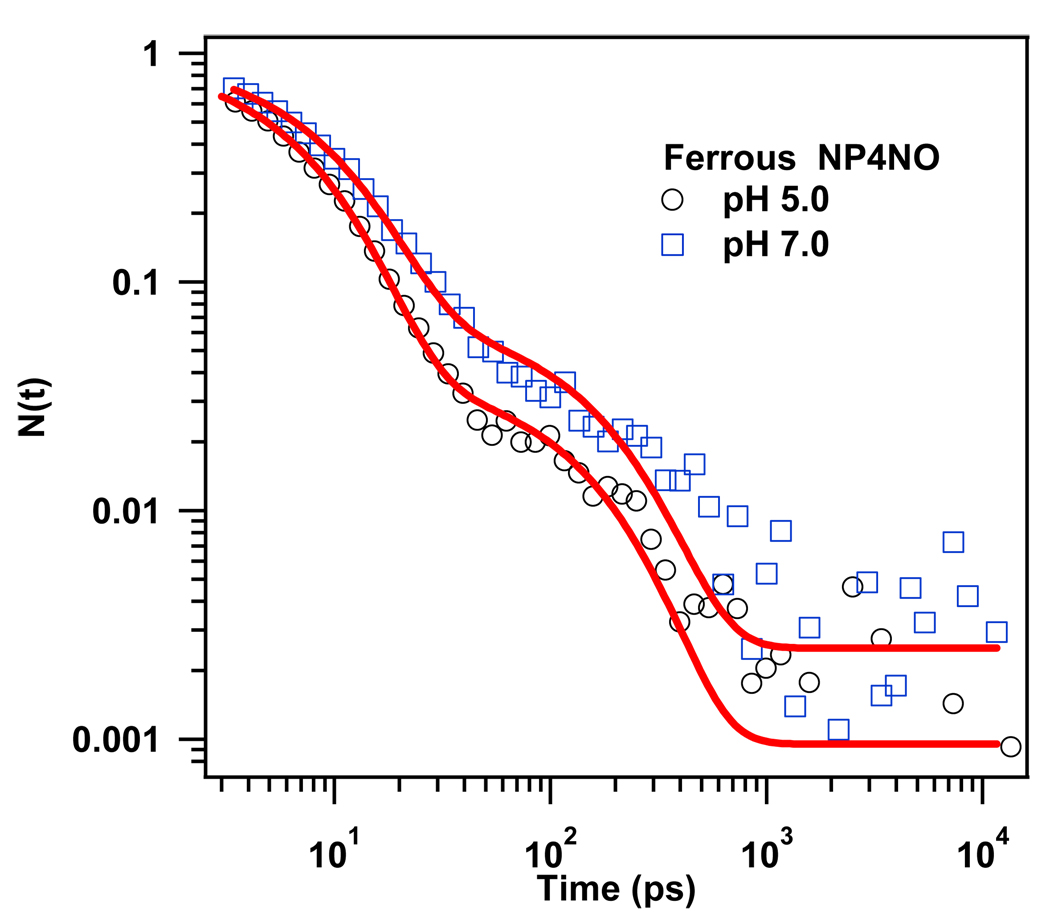
Figure 1 represents the kinetic traces of the geminate recombination of NO to Ferrous NP4 at pH 5.0 and pH 7.0. The rebinding kinetics in these two environments are very similar. In both cases NO rebinds very efficiently with a total geminate amplitude near unity and the amount of NO that escapes to the solvent is less than 1% and within the noise of our experiment. This means that the high affinity of the ferrous heme to NO is not significantly affected when the protein adopts a more open configuration at high pH. It also explains why NP4 cannot use its ferrous state to fulfill its function. The kinetic trace, in both cases, can be reasonably fit with two exponential decay functions (see Table 1). The first phase dominates and has a time constant ~7 ps. We assign this phase to the rebinding of NO directly from the distal pocket (state B). A second phase, around 100 ps was also detected. Its geminate amplitude is very small (<5%) and insensitive to the variation of the pH. As with NO binding to myoglobin51, this phase could reflect the rebinding of a very small fraction of the photolyzed NO molecules that have bifurcated or migrated to one of the xenon cavities that have been identified in the NP4 x-ray structure23 ( see discussion).
Figure 1.
Rebinding kinetics of NO to ferrous NP4 (pumped at 403 nm and probed at 438 nm) at pH 5.0 and pH7.0. The solid lines represent the fits of the data using a two exponential function. The kinetic traces are normalized to unity at time zero.
Table 1.
Fitting parameters of NO rebinding kinetics in ferrous NP4 extracted using a two exponential decay function (N(t) = A1 exp(−t /τ1)+A2 exp(−t /τ2)+A0).
| pH | τ1 (ps) | A1 | τ2 (ps) | A2 | A0 |
|---|---|---|---|---|---|
| 5.0 | 6.8 | 0.97 | 115 | 0.03 | 0.001 |
| 7.0 | 8.1 | 0.95 | 130 | 0.04 | 0.002 |
Dynamics of NO rebinding to Ferric NP4
Transient Absorption Spectra
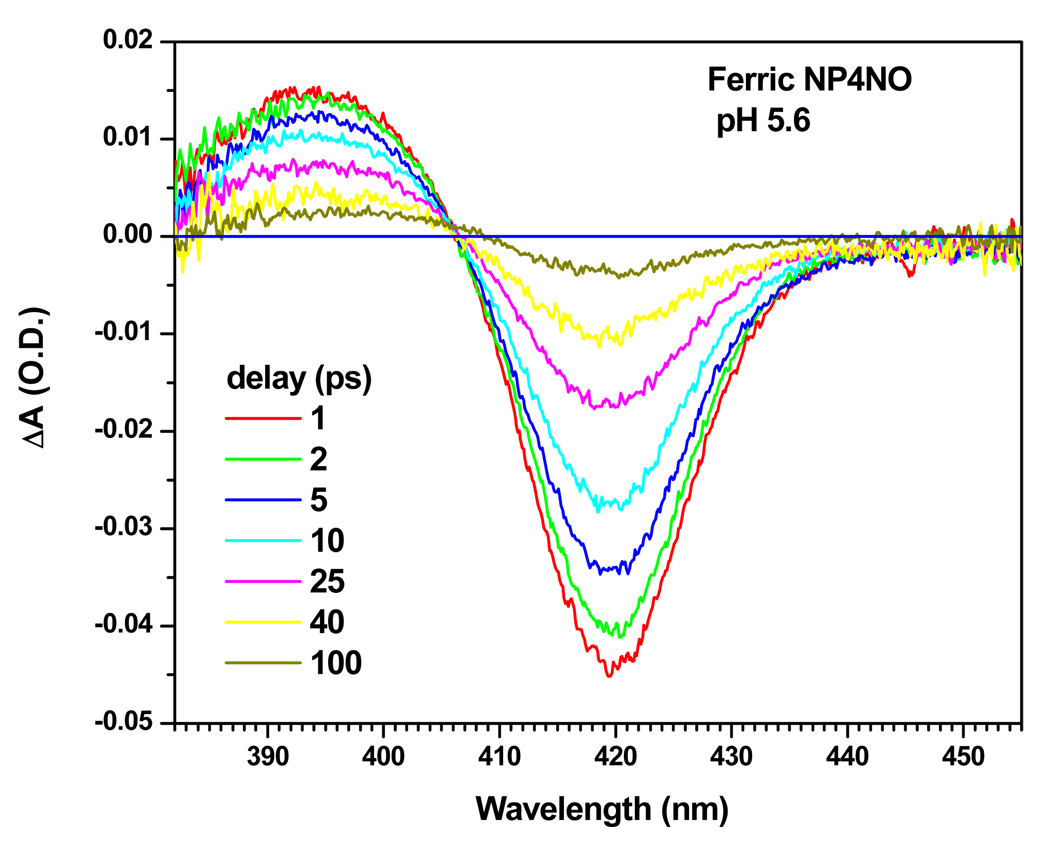
The equilibrium absorption spectra of NP4 and its different adducts has been documented previously 24. Ferric NP4 has a characteristic 6c high-spin absorption band where water is the 6 th ligand, whereas ferric NP4NO is a 6c low-spin species. Figure 2 shows the transient absorption spectra following the photo-dissociation of NO in ferric NP4 at pH 5.6 for different delays between the 580nm pump and the continuum probe. These spectra show the characteristic dynamics of simple two state rebinding with a fixed isosbestic point located at ~407 nm. The constructed absorption spectrum of the transient photoproduct, following the method described in the supporting materials section S1, has a Soret peak at ~395 nm and it is very similar to that of the ferric H64L mutant of myoglobin, which is known to be a 5c species 84. Thus, we assign the transient photoproduct created upon NO photolysis at pH 5.6 to a 5c ferric NP4 (i.e., the water ligand is not present following NO photolysis).
Figure 2.
Transient absorption spectra of the photolyzed ferric NP4NO at pH 5.6, for different time delays. The pump excitation wavelength was 580 nm.
Kinetics
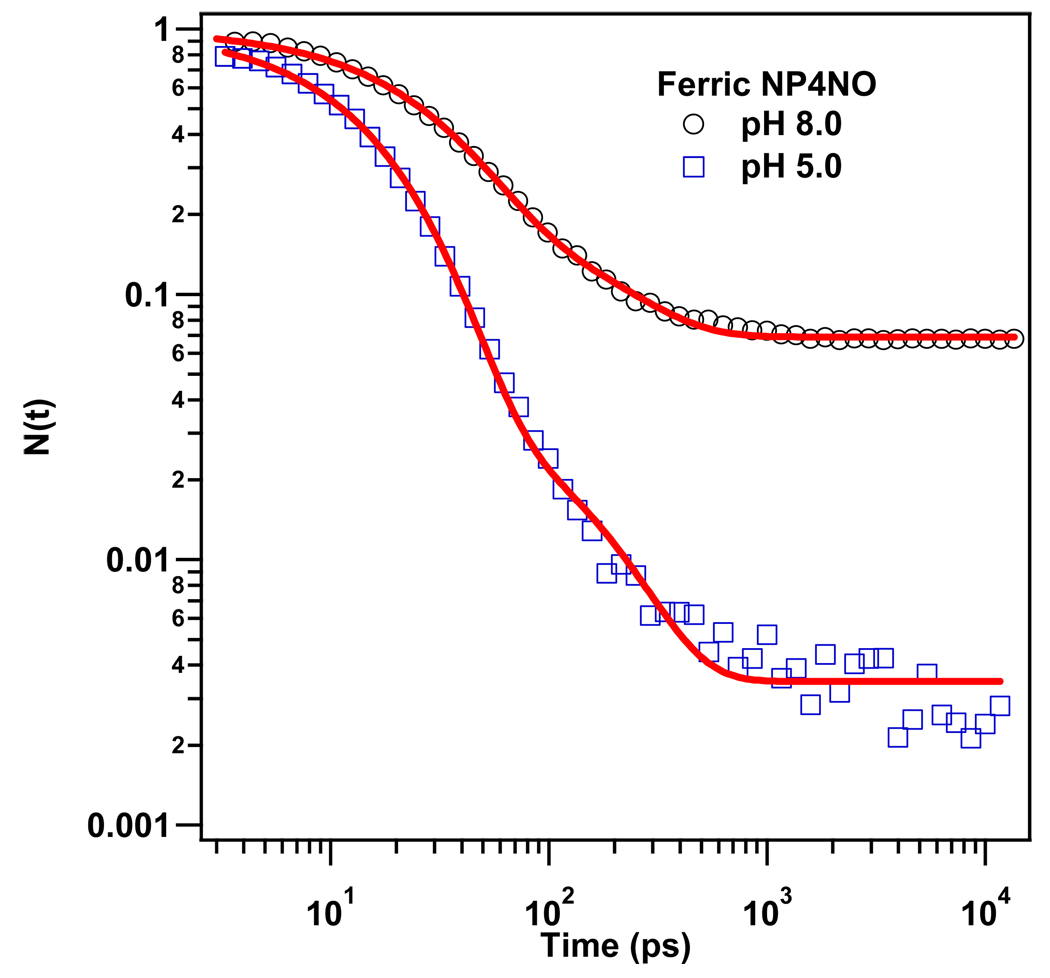
Figure 3 displays the geminate rebinding kinetics of ferric NP4NO at pH 5.0 and pH 8.0. We used the kinetic model developed in the previous section to fit the data and the results are displayed in Table. 2. The sample solution at pH 5.0 mimics the saliva of the insect where NO needs to be meticulously stored in order to protect the insect from the numerous harmful side reactions of the NO molecule. Whereas, the pH 8.0 solution is a mimic for the blood environment of the victim where NO needs to be released in order to trigger the cascade of chemical signals 2,11 that have been briefly described in the introduction.
Figure 3.
Rebinding kinetics of NO to ferric NP4 (pumped at 403 nm and probed at 420 nm) at pH 5.0 and pH8.0. The solid lines represent the fits to the data using the kinetic model developed in the text. The kinetic traces are normalized to unity at time zero.
Table 2.
Fitting parameters of NO rebinding kinetics in ferric NP4 extracted using the kinetic model developed in section 2. All rates are in units of 109 s−1. The equilibrium off rate, . The amplitude for geminate recombination in the closed state is taken to be unity.
| pH | Pc | kgc | kBAc | Po | kgo | Igo | kBAo | kouto | koff/kABo |
|---|---|---|---|---|---|---|---|---|---|
| 5.0 | 0.96 | 64 | 64 | 0.04 | 7.4 | 0.90 | 6.7 | 0.74 | 0.004 |
| 8.0 | 0.79 | 40 | 40 | 0.21 | 6.1 | 0.67 | 4.0 | 2.0 | 0.069 |
The kinetic traces at both pHs show two distinct geminate phases: a fast one with exponential decay (τ ~ 15ps) and a slow one, with a time constant around 150 ps. We assign the fast phase to the geminate rebinding of NO in the more homogeneous and organized closed conformer. The slower phase is assigned to recombination in the more heterogeneous open conformer. Since , we can identify the amplitude of the fast phase with Pc and find Po=1−Pc, which leads to the other parameters listed in Table 2. The geminate phase associated with the closed conformation of the protein is dominant at pH 5.0 and it shows a slightly faster rate than that at pH 8.0. At the higher pH, the open conformation increases its population to ~20% so that the overall off-rate of NO increases by more than an order-of-magnitude (see Discussion).
Photolysis Quantum Yield
The photolysis quantum yield is defined here as the probability for ligand dissociation following single photon excitation, prior to any geminate recombination. The method that we use to calculate the photolysis quantum yield of ferric NP4NO at time zero has been previously used to determine the quantum yields49 of NO and O2 in ferrous myoglobin (Mb), as well as methionine photolysis in ferrous cytochrome c82. A detailed description of the method can be found in the supporting information.
Figure 4 displays the transient absorption, the scaled equilibrium and the calculated photoproduct spectra of NP4NO at pH 5.6 (A) and MbCO (B), respectively. The measurements on these two samples are performed consecutively to ensure a minimum change in the experimental conditions. Since the quantum yield of MbCO is unity85,86, it is used as a reference to calculate the photolysis quantum yield of NP4NO using Eq.S5 in the supporting information materials. The fractional amount of MbCO photolyzed by the pump beam, η, is the ratio of the number of pump pulse photons absorbed to the number of MbCO molecules in the interaction region. This ratio can be calculated by comparing the transient spectrum, after vibrational cooling and prior to CO rebinding (e.g., at t = 20ps), to the equilibrium difference (Mb-MbCO) on an absolute scale. The ratio η for MbCO was found to be 11.5% and 10% in the two experiments reported here.
Figure 4.
Spectra used for calculation of photolysis quantum yield of ferric NP4NO. (A) Spectra of ferric NP4NO at pH 5.6, (B) Spectra of MbCO. In both figures (A) and (B) the red, black and blue lines represent, respectively, the transient absorption spectrum at t = 20 ps, the scaled equilibrium spectrum of the reactant, and the calculated spectrum of the 5-coordinate photoproduct at t = 20 ps.
In the case of NP4NO, the 5c spectrum of the ferric photoproduct is unknown because water is bound in the equilibrium state and the 5c species is not accessible in an equilibrium measurement. Thus η for NP4NO is derived by adding the transient spectrum at 20 ps to the scaled NO-bound equilibrium spectrum to recover a smooth spectrum that must represent the 5c photoproduct absorption (see Fig. 4). The scaling factor found by this process, η(20ps), is 2.1% and 2.2% in the two experiments reported in Table S1 (see supporting information). However, in order to calculate the quantum yield using Eq. S5, the normalized survival population N(t) of the unligated NP4 species at t =20 ps also needs to be taken into account because any rebinding during the first 20ps would reduce the value found for η(20ps) as discussed in the supporting information (Eq. S1). For MbCO, the value of N(20ps) is known to be unity. For NP4NO at pH 5.6, the value for N(20ps) is found to be 0.36 (data not shown, but similar to pH 5 in Fig. 3). Using the parameters displayed in Table S1, the photolysis quantum yield of ferric NP4NO at pH 5.6, , was found to be 91% and 108% in the two experiments. As a result we take the photolysis quantum yield of NP4NO at pH 5.6 to be .
Geminate Rebinding Kinetics of NP4CO
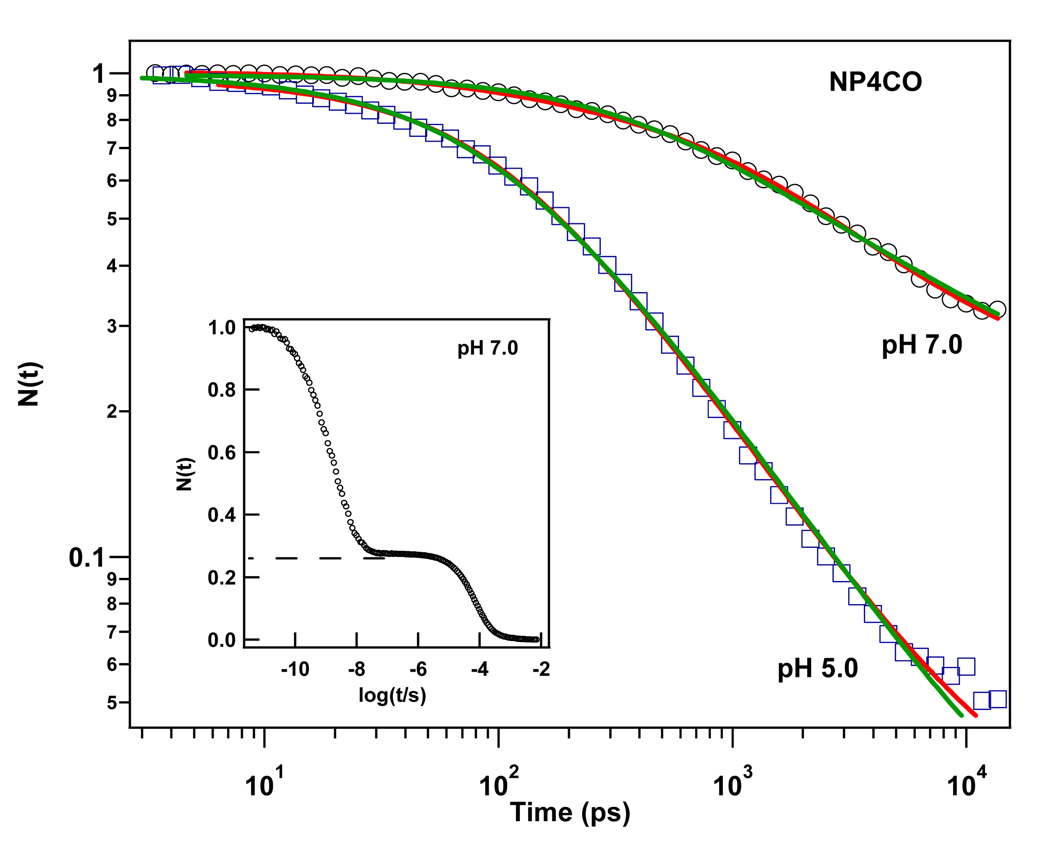
Figure 5 displays the semi-log plot comparing the geminate rebinding dynamics of CO to NP4 at pH 5.0 and pH 7.0. In contrast to myoglobin36,39, the geminate recombination process of CO in NP4 takes place on the 10−11–10−8 s time scale with high geminate yield. The kinetic response exhibits a highly non-exponential behavior at both pHs over the observed time window. This indicates that the geminate rebinding process of CO in NP4 is non-exponential in both conformations. The kinetic traces were fit using a simple model that accounts for the distribution of heme out-of-plane displacements that affect CO, but not NO51 rebinding, and works well to describe CO kinetics heme systems87,88. We could successfully fit the kinetic data using either a single distribution, or we could allow for two distributions in order to account for the rebinding of CO in each of the protein conformations (closed and open). As for NP4NO, we suppose that CO escapes into the solvent only from the open conformer (i.e., ). The details of the fitting procedure are described in the supporting information and discussed below. The fitting parameters are displayed in Table 3 for the case of separate open and closed conformations. For the single “average” distribution, the fitting parameters are given in Table 4. It should be noted that the closed conformer is predominant at pH 5 but at pH 7 it is reduced to ~25% of the population. These findings are in agreement with the x-ray data24, which demonstrate that the population of the closed conformer is dominant at pH 5 and the population of the open conformer is dominant at pH 7.5.
Figure 5.
Kinetic traces of CO rebinding to NP4 at pH 7.0 and pH 5.0. (pumped at 403 nm and probed at 438 nm). The solid lines are fits using the heme doming distribution model: The red line represents a fit function that assumes a separate distribution for each of the open and closed states. The green line represents a fit function with only a single “average” distribution. The data are normalized to unity at time zero. The inset represents the rebinding kinetics of CO to NP4 at pH 7.0 over a wider time window. The slower bimolecular phase has a ~25% amplitude.
Table 3.
Kinetic parameters for NP4CO rebinding using the two distribution model.
| pH | Pc | Po | Igo | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 5.0 | 0.32 | 0.12 | 20 | 0.27 | 0.12 | 1.33 | 0.93 | 0.06 | 0.68 | ||||||
| 7.0 | 0.33 | 0.12 | 18 | 0.28 | 0.12 | 1.25 | 0.23 | 0.77 | 0.72 |
Table 4.
Kinetic parameters for NP4CO rebinding using a single distribution model.
| pH | Ig | ||||||
|---|---|---|---|---|---|---|---|
| 5.0 | 0.32 | 0.13 | 21 | 0.97 | |||
| 7.0 | 0.33 | 0.15 | 4.7 | 0.82 |
We note from Table 3 that the fitting parameter, (with i = c, o) is more than an order of magnitude larger in the closed conformation than in the open conformation . When only a single distribution is used (Table 4), a weighted average of the parameters in Table 3 is obtained for k1. The fitting parameter is a function of both the standard Arrhenius prefactor and the enthalpic barrier assigned to the distal pocket :
| (4) |
In order to extract and independently, temperature dependent kinetic measurements are needed88.
Discussion
Secondary docking sites and ligand migration in NP4
X-ray structure has revealed that NP4 has two xenon cavities labeled Xe1 and Xe2. Xe1 is a major cavity and Xe2 is a minor cavity and their xenon occupancies were 1 and 0.3, respectively, in the crystal structure 23. Temperature derivative spectroscopy at cryogenic temperatures showed that NO and CO rebind to the heme iron exclusively from the distal pocket 23. No rebinding from distinct secondary sites was detected 23. Furthermore, molecular dynamics simulations on ferric NP4NO showed that the residues on the internal side of the distal pocket in NP4 remain tightly packed regardless of the pH, and NO does not migrate into the protein interior during more than 100 ns of simulations 29. Our results support these findings and show that even at room temperature the xenon cavities do not play a crucial role in modulating the geminate recombination of small diatomic ligands in NP4. The kinetic traces of both NP4CO and ferric NP4NO show two different geminate phases with amplitudes that are highly dependent on the pH. We therefore assign the two kinetic phases to the rebinding of the ligand in the open or closed protein conformations.
In the case of ferrous NP4NO, the rebinding kinetics at both pH 5 and pH 7 are dominated by a 7 ps phase (see Table 1). A similar phase for the geminate recombination of NO has been detected in many ferrous heme proteins51,59 and model compounds51 and attributed to the rebinding of NO to the heme iron directly from the (B-state) distal pocket51. The temperature dependence of this phase has been investigated in the case of myoglobin51 and for the bare heme (FePPIX) in a glycerol water mixture51,88. It was found that the enthalpic barrier for NO binding is zero with a prefactor on the order of 1011 s−1. For ferrous MbNO, a slower phase (~200 ps) was also detected49,51,53,54,89 and it was attributed51 to the transition of NO back into the distal pocket from a more remote docking site (probably the Xe4 pocket). The kinetic response of ferrous NP4NO shows a second phase around 100 ps with a very small geminate amplitude (<5%). This phase cannot be assigned to the rebinding of NO exclusively in one of the (closed or open) protein conformations because its amplitude is insensitive to variation of the pH. In analogy to the case of MbNO, this phase probably reflects a very small fraction of the photolyzed NO molecules that bifurcate or migrate to one of the xenon cavities of NP4, most probably to the minor cavity Xe2. Such molecules might then encounter a small enthalpic barrier before escaping from the xenon cavity, returning to the pocket, and rebinding.
Mechanism of NO binding and release in NP4
NP4 stores and delivers NO through a pH dependent conformational change. The protein traps and protects NO in a low pH environment (saliva of the insect, pH~5) by packing two loops A–B and G–H into the distal pocket, and then releases it in the high pH environment of the victim’s tissue (pH~7.5) where the loops are repelled from each other18,22. It is thought that the opening of the protein is mainly triggered by the deprotonation of residue Asp3022,90. We should emphasize that both conformations exist in thermodynamic equilibrium, but the population of the closed conformer is dominant at low pH21. Furthermore, FTIR spectroscopy studies23 showed that the NO-bound ferric NP4 at pH 7.5 displays two infrared absorption bands, a large and sharp band at 1908 cm−1, and a smaller and broader one at 1922 cm−1, representing two distinct heme bound conformations labeled A1908 and A1922. These two conformations were associated23 with the closed and open conformers of the protein, respectively. Analogous pH dependent IR states were also previously assigned to the open and closed states of Mb91, where CO was used as the molecular reporter group for the protein conformational distribution.
Previous measurements22 revealed that the dramatic increase of NO dissociation constant Kd at higher pH in NP4 is in fact governed by the off rate, koff. These measurements also showed that the off rate is biphasic under all conditions with koff1=1.8s−1, koff2= 0.6s−1 at pH 8; and koff1=0.15 s−1, koff2= 0.02 s−1 at pH 5. The normalized amplitudes of the faster off-rates are 0.53 and 0.59 at pH 5.0 and pH8.0, respectively. Upon mutation of key residues in the A–B and G–H loops, the protein loses the pH dependence of NO release but still displays the multiphasic kinetic behavior22.
We have measured the kinetics of the geminate recombination of NO in ferric NP4 (Fig. 3) and presented a simple model to describe our data (Scheme 1). This model takes into account that NP4 has two conformers, closed and open, or A1908 and A1922 in the FTIR notation23. We assume that the interconversion between these two substates requires the rearrangement of two large portions of the protein (loops A–B and G–H)18,21 and is slow compared to the geminate rebinding process, which takes place on the picosecond time scale. This assumption is motivated by our previous work46,91 where we investigated the time scale for the open and closed interconversion [A0↔(A1+A3)] in MbCO. We found that the rate for this transition ranges between 104 and 106 s−1 depending on temperature and solvent composition46. This is consistent with the assumption that the open and closed transition rates in NP4 are slow compared to the geminate rebinding and escape rates. It allows us to decouple the rebinding processes of the two protein conformers and analyze each of them using a standard three state model36,83. We also assume that the ligand escapes to the solvent only in the open conformation , which is consistent with recent molecular dynamic simulations29. A similar situation exists for MbCO, but because of the much slower CO geminate rebinding rate in Mb, there is time for the closed to open transition to take place before the CO rebinds to the heme. As a result, in Mb most of the CO escapes from the distal pocket into solution.
The results of the analysis for NO binding to ferric NP4 are displayed in Table 2. Two distinct exponential phases are sufficient to fit the kinetic traces of ferric NP4NO at both pHs. One is fast (~15 ps) and dominant at pH 5.0, while the other is ~10 fold slower. We assign the fast phase to the rebinding of NO in the closed conformer and the slower phase to NO rebinding in the open conformer. The model (with ) allows us to estimate the populations in the closed (Pc) and the open (Po) conformers (see Table 2) Since independent measurements indicate that the photolysis quantum yield of ferric NP4NO is equal to unity, these two quantities represent the relative populations of the two conformers in the NO bound state. Thus, our estimate of the relative bound-state populations in the closed (Pc) and open (Po) conformers are: Pc =0.96 and Po = 0.04 at pH 5.0; and Pc =0.79 and Po = 0.21 at pH 8.0. The later result is in good agreement with the populations measured using the FTIR technique at pH 7.5 and T =3 K, which found A1908 = 0.80 and A1922 = 0.20.
It should be noted that the intrinsic geminate rebinding rate, kBA, of NO in the closed conformer is ~10 fold larger than in the open conformer. On one hand, this result may be considered surprising because one might expect that in the open conformation any steric constraints imposed on the ligand binding would be relaxed and therefore the rate of the geminate recombination would be enhanced. On the other hand, the larger geminate rate in the closed conformation could arise from residues that form a well-organized distal pocket so that the translational motion of the ligand is restricted and it is retained in a favorable orientation for rebinding (near 180°). This latter effect would reduce the entropic barrier for rebinding and increase the geminate recombination rate. A similar fast geminate phase (~15 ps) has been recently detected for NO rebinding in ferric horseradish peroxidase (HRP)52 and nitric oxide reductase (NOR)92. It is nearly as fast as the ~10 ps phase of the enthalpically barrierless NO binding in ferrous heme proteins51 (see previous section). Therefore, we conclude that NO rebinds to the heme iron in the closed conformation of NP4 directly from the distal pocket with a minimal enthalpic and entropic barrier.
Using Eq. 3, and assuming that the rate of thermal bond breaking kAB is not dependent on pH, we find that the overall off-rate of NO in NP4 increases by 17-fold when the pH varies from 5.0 to 8.0 (see Table 2). This is in good agreement with the ratio of the fast phase off-rates, koff1(pH5)/koff1(pH8) =12, that was previously measured22. Except for a small (factor of ~3) change in , the rates appearing on the right hand side of Eq. 3 do not significantly change with the pH (see Table 2). Thus, the simple model developed here indicates that the ratio of the overall off-rates, and therefore the ratio of dissociation constants Kd of NP4NO at pH 5 and 8, is governed by the ratio of the corresponding populations of the open conformation (factor of ~5) and the pH-dependent escape rate (factor of ~3). This is consistent with the bimolecular kinetics of NP4 mutants D30A, D30N (A–B loop) and D129A/L130A (G–H loop)22. In these mutants, the NO binding does not induce a conformational change as it does in the wild type. As a result, the pH dependence of the off-rate is eliminated for D129A/L130A and considerably reduced for D30A and D30N22. Clearly, the pH dependent open/closed equilibrium plays a key role in determining the NO off-rate in nitrophorin systems.
The existence of the secondary off-rate (i.e., the biphasic, or non-exponential, nature of the dissociation process) remains somewhat mysterious. This could arise in several scenarios: (i) The rate of thermal bond-breaking, kAB, could be biphasic (e.g., due to differing NO interactions with surrounding residues), (ii) The escape rate in the closed conformation could be non-zero, (iii) kAB and the interconversion rate between the two conformers could be comparable, leading to a genuinely non-exponential (rather than bi-exponential) behavior of the off-rate. Unfortunately, such scenarios cannot be differentiated based on the available data.
Role of water
Internal water molecules are crucial for protein function and structural stability. In Mb water molecules enter into the distal pocket and bind to the heme in its ferric form. Water does not bind to the ferrous heme in Mb but it is hydrogen bonded to the nearby distal histidine, His6493. This water molecule blocks ligand access to the distal pocket and needs to be displaced in order for heme to bind small diatomic molecules (O2, NO, CO). Recently, a spectrokinetic assay for water entry into the distal pocket of deoxy myoglobin and its mutants was developed65 and demonstrated that heme pocket hydration is one of the primary factors in controlling the overall bimolecular association rate for CO binding65. Previous work84 also studied the kinetics of water entry and binding to ferric myoglobin following the photodissociation of NO and it was found that entry of water to the distal pocket of myoglobin is correlated with displacement of the distal histidine His64 into the solvent. The rate of water entry was found to be 5.7×106 s−1. It was also observed that the NO bimolecular rebinding rate was enhanced by 3 orders of magnitude for both the H64L mutant, which does not bind water in the ferric state, and for the H64G mutant, where the bound water is no longer stabilized by hydrogen bonding with His6484. This result indicates that in Mb the coordinated water significantly alters NO bimolecular binding when it is stabilized by hydrogen bonding with His64.
Nienhaus et al.23 hypothesized that the release of NO from the open conformation of NP4, especially at higher pH, is enhanced by blockage of the active site by water which hinders immediate NO recombination23. Recent molecular dynamic simulations revealed that, in the high pH conformation, water molecules are more abundant and closer to the iron90. Monitoring the Fe-OH2 distance of the closest water molecule in this conformation following NO dissociation showed that that water comes close to the iron atom quite rapidly (at about 0.3 ns) when NO is only 5 Å away from the heme90.
NP4 gives a unique opportunity to investigate the binding rate of distal pocket water molecules to the heme. In contrast to myoglobin, water molecules coexist with the exogenous ligand in the heme pocket. In the open conformation of ferric NP4NO several waters co-exist in the distal pocket. Upon NO photolysis, if one of these water molecules replaces NO and binds to the heme, it should leave a spectroscopic imprint. The NO rebinding kinetics displayed in Fig. 3 were monitored at λ = 420 nm. Figure 6 is similar to Fig. 4A and displays the transient absorption spectrum of NP4NO at t = 20 ps, the scaled equilibrium spectrum of NP4NO, the calculated spectrum of the photoproduct, and, in addition, the scaled equilibrium spectrum of NP4-H2O. As can be seen from this figure, the signals arising from the binding of NO or water have different signs when probed between λ = 417–440 nm. If water starts to bind to the 5c NP4NO photoproduct, the optical transmission signal should rise. However, the kinetic response shows a monotonic decay of −ΔA over the fist 500 ps followed by a plateau (e.g., see Fig. 3). Therefore we conclude that in NP4 water does not bind to the heme iron, at least during the first 16 ns following NO photodissociation.
Figure 6.
Transient absorption spectrum of ferric NP4NO at t =20 ps (red), the scaled equilibrium spectrum of NP4NO (black), the calculated spectrum of the 5-coordinate photoproduct (blue) and the scaled equilibrium spectrum of 6-coordinate NP4-H2O (magenta).
We propose the following scenario for NO binding and release in the hydrophilic open conformation. In this conformation, the volume accessible to ligand in the distal pocket is larger than that of the closed conformation. Therefore, geminate recombination of NO is retarded due to the development of an entropic rebinding barrier. The NO ligand can more easily diffuse away from the heme iron and water molecules can collapse into the vacancy, rendering the region around the iron more hydrophilic and forming an effective barrier against the reassociation of the apolar NO ligand. However, the water does not bind to the iron, at least during the first 15 ns. Overall, this enhances the escape of NO into solution. Table 2 shows that the escape rate of NO from the open form is ~3 fold faster at pH 8 than at pH 5. We attribute the increased escape rate at pH 8 to the larger abundance of water molecules in the distal pocket of the protein.
Ultrafast rebinding kinetics of CO in NP4
As can be seen from Fig 5, the geminate recombination of CO to NP4 is highly non-exponential and takes place on the picosecond and early nanosecond time scale with high geminate yield. This ultrafast CO rebinding behavior was first observed for heme model compounds 83,88,94–97 and it has been recently reported for a variety of other heme proteins including CooA 98,99, CBS 100, carboxymethyl cytochrome c (CM-cytc) 101,102 and methionine substituted mutants of cythochrome c 103.
X-ray crystallography showed that upon CO binding to NP4 the protein undergoes a conformational change similar to that of the NO complex24. However it was suggested that the formation of the hydrophobic trap is less efficient in the case of the CO complex because the CO molecule is more polar than NO23. The linear (upright) orientation of bound CO compared to the bent NO bound form may also reduce the population of the closed form. In fact, the FTIR spectrum of NP4CO at pH7.5 shows a broad band at ~1960 cm−1 resulting from multiple overlapping absorbance bands23. This indicates that the active site is significantly heterogeneous at pH 7.5, which is consistent with the presence of a larger fraction of the NP4 open conformation. This is also indicated from the analysis of the CO rebinding kinetics (see Table 3). However, as the pH decreases, the distal pocket population with CO bound still favors the more organized hydrophobic closed conformer24. As with ferric NP4NO, we assume that CO rebinds to NP4 following Scheme I. However, because CO (unlike NO) encounters a significant barrier due to heme doming51,88, we fit the non-exponential geminate recombination of CO using a fine-grained distribution of heme doming configurations as discussed previously87,88. This was done using either two distributions (one each for the open and closed states), as shown in Table 3, or with a single “averaged” distribution (Table 4). When the two distributions are used, we let the interconversion between the protein conformations be slow compared to the timescales (10−11–10−8 s) for CO rebinding and escape to solvent. As for NP4NO and MbCO41,46, the escape is assumed to take place only from the open conformer.
The model87,88 used to describe the heme doming conformations, and related barrier distributions, is presented in the supporting information materials. It is based on a distribution of heme doming equilibrium positions, so that the total enthalpic barrier, H, for CO binding can be separated into two parts
| (5) |
where Hp(a) represents the proximal barrier due to the heme doming and a is the Gaussian distributed equilibrium position of the generalized iron out-of-plane doming coordinate. The quantity K is an effective force constant along the doming coordinate and H0 represents the remaining (mostly distal) contributions to the enthalpic barrier, which contains energies involving ligand docking sites and steric constraints associated with the distal pocket along with a small a-independent term from the linearly coupled heme potential surfaces87. For the sake of simplicity we suppose here that the distal barrier (i = o, c) is constant in both conformations. However on these time scales (ps to ns), ligand binding can be coupled to the structural relaxation of the protein, which gives rise to time dependent enthalpic and entropic barriers. Also, in the heterogeneous open conformation, the ensemble of open proteins is more likely to lead to a distributed distal barrier, i.e., P(H0). When escape from the closed form is not allowed, the survival population at time t after photolysis, (see supporting information), can be written as
| (6a) |
with
| (6b) |
Pc and Po are the normalized (Pc + Po = 1) initial photolyzed populations in the unbound states (NP:CO)o and (NP:CO)c, respectively. The parameter represents the geminate amplitude in the open conformation and because there is no ligand escape in the closed form. The last term in Eq. 6a is the total amount of ligand that escapes to the solvent, given that escape is allowed only in the open conformation. The generalized force constant K is taken as 13.8Nm−1, as found for other heme systems87,100 and from the frequency (~40cm−1) of the heme doming mode104. As a result, the parameters , that describe the heme out-of-plane distribution (i = o, c), and the distal pocket barrier ( in Eq. 5), can be found from the temperature dependent fitting parameters {Ai, Ci, } in Eq. 6b as,
| (7) |
We used the following procedure to fit the data when the model with two distributions was employed. Because the crystal structure shows24 that the closed conformer is dominant at pH 5, we first fitted the kinetic trace at this pH value using only a single distribution for the dominant closed form. From this we got an estimate for the closed distribution parameters . We found , which are typical for heme systems. As a result, we fixed these two parameters and fit the kinetics using Eqs. 6a and 6b assuming that both conformers have the same heme distribution . Using these results as initial input, we relaxed the condition that the open and closed distributions be equal and found that only small changes in the distributions were observed. In order to limit the number of free parameters, we ended up restricting the width of the distributions, σa = 0.12 Å while allowing the mean doming displacement to vary. As can be seen from Table 3, the mean out-of-plane displacement for the open and closed states only varied between 0.27–0.33Å.
When a single distribution was used, the parameters that resulted are given in Table 4. The value for k1 turns out to be a weighted average of what is found in the two distribution model. This is not too surprising and it provides an independent check of the robustness of the fitting procedures.
As can be seen in Table 3 the parameter is ~15 fold smaller in the open conformation compared to that of the closed conformation. As with NP4NO, we suggest that the larger volume accessible to the CO ligand in the open conformation retards its geminate recombination due to the increase of the entropic barrier. Following the diffusion of CO away from the heme iron, water molecules may collapse and help block access to the heme active site. This clustering and reorganization of water molecules around the heme iron may create an effective barrier against the immediate recombination of CO in the open form.
As can be seen from Table 3, the extracted populations Pc and Po for NP4CO at high pH are very different from those of NP4NO (Table 2). The population of the open conformation in the CO bound state becomes dominant at pH 7. This can be confirmed by examining the data in Fig. 5 where the insert reveals a CO escape amplitude of ~0.25 at pH 7.0. Using Eq. 6a we find that the overall escape amplitude is consistent with the product of the open population (~0.8) and its individual escape amplitude (~0.3). This result is evidently due to the fact that the closure of the distal pocket (hydrophobic trapping) is less efficient in the case of CO at pH>7 than it is for NO. This has also been previously suggested as an explanation for the difference in the heterogeneity of the IR bands of NP4CO and NP4NO at pH 7.5.23 The FTIR spectrum of NP4CO shows multiple absorbance bands denoted A1939, A1953, A1963 and A1972 23. Each one of these bands is associated with a different conformation of the protein; however, it remains unclear how these conformations are divided between the open and closed states.
Conclusions
In summary, we have shown in this work that binding and release of small diatomic ligands (NO and CO) in NP4 are highly modulated by pH changes. The geminate recombination process of NO to ferrous NP4 at both pH 5 and pH 7 is dominated by a single ~7 ps phase that is attributed to the rebinding of NO directly from the heme distal pocket. The kinetic response of ferric NP4NO exhibits two exponential phases. The relative geminate amplitudes associated with these two phases depend on the pH. We assign the fast phase to the rebinding of NO within the hydrophobic closed conformation of the protein and the slow phase to its recombination from the hydrophilic open conformation. The overall off-rate of NO was found to increase by more than one order of magnitude when the pH changes from 5.0 to 8.0, assuming that the open conformer allows ligand escape but the closed conformer does not. The geminate recombination of CO in NP4 takes place on the picosecond and early nanosecond time scale with high geminate yield and exhibits a highly non-exponential behavior. The kinetic traces at both pH 5 and pH 7 were analyzed by using a simple model that includes both the heme doming distribution and the distal pocket contributions to the enthalpic barrier. Secondary distal pocket docking sites do not seem to play a significant role in modulating the enthalpic barrier for geminate rebinding of either NO or CO in NP4. Within the 15 ns experimental window, water does not immediately bind to the heme iron following NO photodissociation from ferric NP4. However, we suggest that in the open conformation of NP4 there is an increased entropic barrier against NO (and CO) recombination due to increased distal pocket volume and to clustering and reorganization of water in the vicinity of the heme.
Supplementary Material
Acknowledgements
This work is supported by grants from the NIH (DK35090 to P.M.C. and HL62969 to W.R.M.) and NSF (MCB-0744738 to P.M.C.). We thank Ms. Jacqueline L. Brailey for her help in expression and purification of NP4.
Footnotes
Supporting information available
The method used for calculation of the absolute quantum yield of NP4NO photolysis. The complete analysis of NP4CO ligand binding kinetics. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Antonini E, Brunori M. Hemoglobin and Myoglobin in their Reactions with Ligands. Amsterdam-London, The Netherlands: North-Holland Publishing Co; 1971. [Google Scholar]
- 2.Ribeiro JM, Hazzard JM, Nussenzveig RH, Champagne DE, Walker FA. Science. 1993;260:539–541. doi: 10.1126/science.8386393. [DOI] [PubMed] [Google Scholar]
- 3.Ortiz de Montellano PR. CYTOCHROME P450 : Structure, Mechanism and Biochemistry. 3rd Edition. New York: Kluwer Academic/Plenum Publishers; 2005. [Google Scholar]
- 4.Li H, Poulos TL. J. Inorg. Biochem. 2005;99:293–305. doi: 10.1016/j.jinorgbio.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 5.Rousseau DL, Li D, Couture M, Yeh SR. J. Inorg. Biochem. 2005;99:306–323. doi: 10.1016/j.jinorgbio.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 6.Bredt DS, Snyder SH. Annu. Rev. Biochem. 1994;63:175–195. doi: 10.1146/annurev.bi.63.070194.001135. [DOI] [PubMed] [Google Scholar]
- 7.Gilles-Gonzalez MA, Ditta GS, Helinski DR. Nature. 1991;350:170–172. doi: 10.1038/350170a0. [DOI] [PubMed] [Google Scholar]
- 8.Hou SB, Larsen RW, Boudko D, Riley CW, Karatan E, Zimmer M, Ordal GW, Alam M. Nature. 2000;403:540–544. doi: 10.1038/35000570. [DOI] [PubMed] [Google Scholar]
- 9.Aono S, Nakajima H. Coord. Chem. Rev. 1999;192:267–282. [Google Scholar]
- 10.Dioum EM, Rutter J, Tuckerman JR, Gonzalez G, Gilles-Gonzalez MA, McKnight SL. Science. 2002;298:2385–2387. doi: 10.1126/science.1078456. [DOI] [PubMed] [Google Scholar]
- 11.Walker FA. J. Inorg. Biochem. 2005;99:216–236. doi: 10.1016/j.jinorgbio.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 12.Champagne DE, Nussenzveig RH, Ribeiro JMC. J. Biol. Chem. 1995;270:8691–8695. doi: 10.1074/jbc.270.15.8691. [DOI] [PubMed] [Google Scholar]
- 13.Andersen JF, Gudderra NP, Francischetti IMB, Valenzuela JG, Ribeiro JMC. Biochemistry. 2004;43:6987–6994. doi: 10.1021/bi049655t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ribeiro JMC, Nussenzveig RH. Febs Letters. 1993;330:165–168. doi: 10.1016/0014-5793(93)80265-v. [DOI] [PubMed] [Google Scholar]
- 15.Andersen JF, Ding XD, Balfour C, Shokhireva TK, Champagne DE, Walker FA, Montfort WR. Biochemistry. 2000;39:10118–10131. doi: 10.1021/bi000766b. [DOI] [PubMed] [Google Scholar]
- 16.Ribeiro JMC, Walker FA. J. Exp. Med. 1994;180:2251–2257. doi: 10.1084/jem.180.6.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andersen JF, Weichsel A, Balfour CA, Champagne DE, Montfort WR. Structure. 1998;6:1315–1327. doi: 10.1016/s0969-2126(98)00131-2. [DOI] [PubMed] [Google Scholar]
- 18.Weichsel A, Andersen JF, Roberts SA, Montfort WR. Nat. Struct. Biol. 2000;7:551–554. doi: 10.1038/76769. [DOI] [PubMed] [Google Scholar]
- 19.Roberts SA, Weichsel A, Qiu Y, Shelnutt JA, Walker FA, Montfort WR. Biochemistry. 2001;40:11327–11337. doi: 10.1021/bi0109257. [DOI] [PubMed] [Google Scholar]
- 20.Berry RE, Ding XD, Shokhireva TK, Weichsel A, Montfort WR, Walker FA. J. Biol. Inorg. Chem. 2004;9:135–144. doi: 10.1007/s00775-003-0505-0. [DOI] [PubMed] [Google Scholar]
- 21.Kondrashov DA, Roberts SA, Weichsel A, Montfort WR. Biochemistry. 2004;43:13637–13647. doi: 10.1021/bi0483155. [DOI] [PubMed] [Google Scholar]
- 22.Maes EM, Weichsel A, Andersen JF, Shepley D, Montfort WR. Biochemistry. 2004;43:6679–6690. doi: 10.1021/bi049748a. [DOI] [PubMed] [Google Scholar]
- 23.Nienhaus K, Maes EM, Weichsel A, Montfort WR, Nienhaus GU. J. Biol. Chem. 2004;279:39401–39407. doi: 10.1074/jbc.M406178200. [DOI] [PubMed] [Google Scholar]
- 24.Maes EM, Roberts SA, Weichsel A, Montfort WR. Biochemistry. 2005;44:12690–12699. doi: 10.1021/bi0506573. [DOI] [PubMed] [Google Scholar]
- 25.Amoia AM, Montfort WR. Protein. Sci. 2007;16:2076–2081. doi: 10.1110/ps.072981907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kubo M, Gruia F, Benabbas A, Barabanschikov A, Montfort WR, Maes EM, Champion PM. J. Am. Chem. Soc. 2008;130:9800–9811. doi: 10.1021/ja800916d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wondimagegn T, Ghosh A. J. Am. Chem. Soc. 2001;123:5680–5683. doi: 10.1021/ja004314y. [DOI] [PubMed] [Google Scholar]
- 28.Menyhard DK, Keseru GM. Febs Letters. 2005;579:5392–5398. doi: 10.1016/j.febslet.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 29.Kondrashov DA, Montfort WR. J. Phys. Chem. B. 2007;111:9244–9252. doi: 10.1021/jp071136n. [DOI] [PubMed] [Google Scholar]
- 30.Marti MA, Estrin DA, Roitberg AE. J. Phys. Chem. B. 2009;113:2135–2142. doi: 10.1021/jp808055e. [DOI] [PubMed] [Google Scholar]
- 31.Weichsel A, Andersen JF, Champagne DE, Walker FA, Montfort WR. Nat. Struct. Biol. 1998;5:304–309. doi: 10.1038/nsb0498-304. [DOI] [PubMed] [Google Scholar]
- 32.Scott EE, Gibson QH, Olson JS. J. Biol. Chem. 2001;276:5177–5188. doi: 10.1074/jbc.M008282200. [DOI] [PubMed] [Google Scholar]
- 33.Gibson QH. J.Physiol. 1956;134:112–122. doi: 10.1113/jphysiol.1956.sp005627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Austin RH, Beeson KW, Eisenstein L, Frauenfelder H, Gunsalus IC. Biochemistry. 1975;14:5355–5373. doi: 10.1021/bi00695a021. [DOI] [PubMed] [Google Scholar]
- 35.Greene BI, Hochstrasser RM, Weisman RB, Eaton WA. Proc. Natl. Acad. Sci. USA. 1978;75:5255–5259. doi: 10.1073/pnas.75.11.5255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Henry ER, Sommer JH, Hofrichter J, Eaton WA. J. Mol. Biol. 1983;166:443–451. doi: 10.1016/s0022-2836(83)80094-1. [DOI] [PubMed] [Google Scholar]
- 37.Jongeward KA, Magde D, Taube DJ, Marsters JC, Traylor TG, Sharma VS. J. Am. Chem. Soc. 1988;110:380–387. [Google Scholar]
- 38.Anfinrud PA, Han C, Hochstrasser RM. Proc. Natl. Acad. Sci. USA. 1989;86:8387–8391. doi: 10.1073/pnas.86.21.8387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tian WD, Sage JT, Srajer V, Champion PM. Phys. Rev. Lett. 1992;68:408–411. doi: 10.1103/PhysRevLett.68.408. [DOI] [PubMed] [Google Scholar]
- 40.Balasubramanian S, Lambright DG, Marden MC, Boxer SG. Biochemistry. 1993;32:2202–2212. doi: 10.1021/bi00060a011. [DOI] [PubMed] [Google Scholar]
- 41.Tian WD, Sage JT, Champion PM. J. Mol. Biol. 1993;233:155–166. doi: 10.1006/jmbi.1993.1491. [DOI] [PubMed] [Google Scholar]
- 42.Walda KN, Liu XY, Sharma VS, Magde D. Biochemistry. 1994;33:2198–2209. doi: 10.1021/bi00174a029. [DOI] [PubMed] [Google Scholar]
- 43.Olson JS, Phillips GN., Jr. J. Biol. Chem. 1996;271:17593–17596. doi: 10.1074/jbc.271.30.17593. [DOI] [PubMed] [Google Scholar]
- 44.Johnson JB, Lamb DC, Frauenfelder H, Muller JD, McMahon B, Nienhaus GU, Young RD. Biophys. J. 1996;71:1563–1573. doi: 10.1016/S0006-3495(96)79359-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Srajer V, Teng TY, Ursby T, Pradervand C, Ren Z, Adachi S, Schildkamp W, Bourgeois D, Wulff M, Moffat K. Science. 1996;274:1726–1729. doi: 10.1126/science.274.5293.1726. [DOI] [PubMed] [Google Scholar]
- 46.Tian WD, Sage JT, Champion PM, Chien E, Sligar SG. Biochemistry. 1996;35:3487–3502. doi: 10.1021/bi952474u. [DOI] [PubMed] [Google Scholar]
- 47.Scott EE, Gibson QH. Biochemistry. 1997;36:11909–11917. doi: 10.1021/bi970719s. [DOI] [PubMed] [Google Scholar]
- 48.Dadusc G, Ogilvie JP, Schulenberg P, Marvet U, Miller RJD. Proc. Natl. Acad. Sci. USA. 2001;98:6110–6115. doi: 10.1073/pnas.101130298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ye X, Demidov AA, Champion PM. J. Am. Chem. Soc. 2002;124:5914–5924. doi: 10.1021/ja017359n. [DOI] [PubMed] [Google Scholar]
- 50.Cao W, Ye X, Sjodin T, Christian JF, Demidov AA, Berezhna S, Wang W, Barrick D, Sage JT, Champion PM. Biochemistry. 2004;43:11109–11117. doi: 10.1021/bi049077g. [DOI] [PubMed] [Google Scholar]
- 51.Ionascu D, Gruia F, Ye X, Yu AC, Rosca F, Beck C, Demidov A, Olson JS, Champion PM. J. Am. Chem. Soc. 2005;127:16921–16934. doi: 10.1021/ja054249y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ye X, Yu AC, Champion PM. J. Am. Chem. Soc. 2006;128:1444–1445. doi: 10.1021/ja057172m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Petrich JW, Poyart C, Martin JL. Biochemistry. 1988;27:4049–4060. doi: 10.1021/bi00411a022. [DOI] [PubMed] [Google Scholar]
- 54.Petrich JW, Lambry JC, Balasubramanian S, Lambright DG, Boxer SG, Martin JL. J. Mol. Biol. 1994;238:437–444. doi: 10.1006/jmbi.1994.1302. [DOI] [PubMed] [Google Scholar]
- 55.Treuffet J, Kubarych KJ, Lambry JC, Pilet E, Masson JB, Martin JL, Vos MH, Joffre M, Alexandrou A. Proc. Natl. Acad. Sci. USA. 2007;104:15705–15710. doi: 10.1073/pnas.0703279104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kruglik SG, Jasaitis A, Hola K, Yamashita T, Liebl U, Martin JL, Vos MH. Proc. Natl. Acad. Sci. USA. 2007;104:7408–7413. doi: 10.1073/pnas.0700445104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liebl U, Bouzhir-Sima L, Negrerie M, Martin JL, Vos MH. Proc. Natl. Acad. Sci. USA. 2002;99:12771–12776. doi: 10.1073/pnas.192311699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Negrerie M, Berka V, Vos MH, Liebl U, Lambry JC, Tsai AL, Martin JL. J. Biol. Chem. 1999;274:24694–24702. doi: 10.1074/jbc.274.35.24694. [DOI] [PubMed] [Google Scholar]
- 59.Vos MH. Biochim. Biophys. Acta. 2008;1777:15–31. doi: 10.1016/j.bbabio.2007.10.004. [DOI] [PubMed] [Google Scholar]
- 60.Bredenbeck J, Helbing J, Nienhaus K, Nienhaus GU, Hamm P. Proc. Natl. Acad. Sci. USA. 2007;104:14243–14248. doi: 10.1073/pnas.0607758104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Finkelstein IJ, Ishikawa H, Kim S, Massari AM, Fayer MD. Proc. Natl. Acad. Sci. USA. 2007;104:2637–2642. doi: 10.1073/pnas.0610027104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ishikawa H, Kwak K, Chung JK, Kim S, Fayer MD. Proc. Natl. Acad. Sci. USA. 2008;105:8619–8624. doi: 10.1073/pnas.0803764105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang Y, Baskin JS, Xia T, Zewail AH. Proc. Natl. Acad. Sci. USA. 2004;101:18000–18005. doi: 10.1073/pnas.0408379102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Levantino M, Cupane A, Zimanyi L, Ormos P. Proc. Natl. Acad. Sci. USA. 2004;101:14402–14407. doi: 10.1073/pnas.0406062101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Goldbeck RA, Bhaskaran S, Ortega C, Mendoza JL, Olson JS, Soman J, Kliger DS, Esquerra RM. Proc. Natl. Acad. Sci. USA. 2006;103:1254–1259. doi: 10.1073/pnas.0507840103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Olson JS, Soman J, Phillips GN. Iubmb Life. 2007;59:552–562. doi: 10.1080/15216540701230495. [DOI] [PubMed] [Google Scholar]
- 67.Olson JS, Phillips GN. J. Biol. Inor. Chem. 1997;2:544–552. [Google Scholar]
- 68.Frauenfelder H, Sligar SG, Wolynes PG. Science. 1991;254:1598–1603. doi: 10.1126/science.1749933. [DOI] [PubMed] [Google Scholar]
- 69.Frauenfelder H, Wolynes PG. Science. 1985;229:337–345. doi: 10.1126/science.4012322. [DOI] [PubMed] [Google Scholar]
- 70.Srajer V, Champion PM. Biochemistry. 1991;30:7390–7402. doi: 10.1021/bi00244a005. [DOI] [PubMed] [Google Scholar]
- 71.Campbell BF, Chance MR, Friedman JM. Science. 1987;238:373–376. doi: 10.1126/science.3659921. [DOI] [PubMed] [Google Scholar]
- 72.Ormos P, Szaraz S, Cupane A, Nienhaus GU. Proc. Natl. Acad. Sci. USA. 1998;95:6762–6767. doi: 10.1073/pnas.95.12.6762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schmidt M, Nienhaus K, Pahl R, Krasselt A, Anderson S, Parak F, Nienhaus GU, Srajer V. Proc. Natl. Acad. Sci. USA. 2005;102:11704–11709. doi: 10.1073/pnas.0504932102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Srajer V, Ren Z, Teng TY, Schmidt M, Ursby T, Bourgeois D, Pradervand C, Schildkamp W, Wulff M, Moffat K. Biochemistry. 2001;40:13802–13815. doi: 10.1021/bi010715u. [DOI] [PubMed] [Google Scholar]
- 75.Ostermann A, Waschipky R, Parak FG, Nienhaus GU. Nature. 2000;404:205–208. doi: 10.1038/35004622. [DOI] [PubMed] [Google Scholar]
- 76.Bourgeois D, Vallone B, Schotte F, Arcovito A, Miele AE, Sciara G, Wulff M, Anfinrud P, Brunori M. Proc. Natl. Acad. Sci. USA. 2003;100:8704–8709. doi: 10.1073/pnas.1430900100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schotte F, Lim MH, Jackson TA, Smirnov AV, Soman J, Olson JS, Phillips GN, Wulff M, Anfinrud PA. Science. 2003;300:1944–1947. doi: 10.1126/science.1078797. [DOI] [PubMed] [Google Scholar]
- 78.Tomita A, Sato T, Ichiyanagi K, Nozawa S, Ichikawa H, Chollet M, Kawai F, Park SY, Tsuduki T, Yamato T, Koshihara S, Adachi S. Proc. Natl. Acad. Sci. USA. 2009;106:2612–2616. doi: 10.1073/pnas.0807774106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang ZY, Benabbas A, Ye X, Yu AC, Champion PM. J. Phys. Chem. B. 2009;113:10923–10933. doi: 10.1021/jp9031805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Andersen JF, Champagne DE, Weichsel A, Ribeiro JMC, Balfour CA, Dress V, Montfort WR. Biochemistry. 1997;36:4423–4428. doi: 10.1021/bi9628883. [DOI] [PubMed] [Google Scholar]
- 81.Yu A, Ye X, Ionascu D, Cao W, Champion PM. Rev. Sci. Instr. 2005;75:114301. [Google Scholar]
- 82.Wang W, Ye X, Demidov AA, Rosca F, Sjodin T, Cao WX, Sheeran M, Champion PM. J. Phys. Chem. B. 2000;104:10789–10801. [Google Scholar]
- 83.Ye X, Yu AC, Georgiev GY, Gruia F, Ionascu D, Cao WX, Sage JT, Champion PM. J. Am. Chem. Soc. 2005;127:5854–5861. doi: 10.1021/ja042365f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cao W, Christian JF, Champion PM, Rosca F, Sage JT. Biochemistry. 2001;40:5728–5737. doi: 10.1021/bi010067e. [DOI] [PubMed] [Google Scholar]
- 85.Bucher T, Kaspers J. Biochim. Biophys. Acta. 1947;1:21–34. [Google Scholar]
- 86.Schuresko DD, Webb WW. Biophys. J. 1978;24:382–383. doi: 10.1016/S0006-3495(78)85390-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Srajer V, Reinish L, Champion PM. J. Am. Chem. Soc. 1988;110:6656–6670. [Google Scholar]
- 88.Ye X, Ionascu D, Gruia F, Yu A, Benabbas A, Champion PM. Proc. Natl. Acad. Sci. USA. 2007;104:14682–14687. doi: 10.1073/pnas.0702622104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Shreve AP, Franzen S, Simpson MC, Dyer RB. J. Phys. Chem. B. 1999;37:7969–7975. [Google Scholar]
- 90.Swails JM, Meng Y, Walker FA, Marti MA, Estrin DA, Roitberg AE. J. Phys. Chem. B. 2009;113:1192–1201. doi: 10.1021/jp806906x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Morikis D, Champion PM, Springer BA, Sligar SG. Biochemistry. 1989;28:4791–4800. doi: 10.1021/bi00437a041. [DOI] [PubMed] [Google Scholar]
- 92.Kapetanaki SM, Field SJ, Hughes RJL, Watmough NJ, Liebl U, Vos MH. BBA-Bioenergetics. 2008;1777:919–924. doi: 10.1016/j.bbabio.2008.03.012. [DOI] [PubMed] [Google Scholar]
- 93.Quillin ML, Arduini RM, Olson JS, Phillips GN., Jr. J Mol Biol. 1993;234:140–155. doi: 10.1006/jmbi.1993.1569. [DOI] [PubMed] [Google Scholar]
- 94.Postlewaite JC, Miers JB, Dlott DD. J. Am. Chem. Soc. 1989;111:1248–1255. [Google Scholar]
- 95.Cao W, Ye X, Georgiev GY, Berezhna S, Sjodin T, Demidov AA, Wang W, Sage JT, Champion PM. Biochemistry. 2004;43:7017–7027. doi: 10.1021/bi0497291. [DOI] [PubMed] [Google Scholar]
- 96.Lim M, Jackson TA, Anfinrud PA. J. Biol. Inor. Chem. 1997;2:531–536. [Google Scholar]
- 97.Traylor TG, Magde D, Taube DJ, Jongeward KA, Bandyopadhyay D, Luo JK, Walda KN. J. Am. Chem. Soc. 1992;114:417–429. [Google Scholar]
- 98.Rubtsov IV, Zhang TQ, Nakajima H, Aono S, Rubtsov GI, Kumazaki S, Yoshihara K. J. Am. Chem. Soc. 2001;123:10056–10062. doi: 10.1021/ja011023w. [DOI] [PubMed] [Google Scholar]
- 99.Kumazaki S, Nakajima H, Sakaguchi T, Nakagawa E, Shinohara H, Yoshihara K, Aono S. J. Biol. Chem. 2000;275:38378–38383. doi: 10.1074/jbc.M005533200. [DOI] [PubMed] [Google Scholar]
- 100.Venugopal K, Benabbas A, Sun Y, Zhang Z, Singh S, Benerjee R, Champion PM. Submitted. [Google Scholar]
- 101.Silkstone G, Jasaitis A, Vos MH, Wilson MT. Dalton Trans. 2005:3489–3494. doi: 10.1039/b508183c. [DOI] [PubMed] [Google Scholar]
- 102.Kim J, Park J, Lee T, Lim M. J. Phys. Chem. B. 2009;113:260–266. doi: 10.1021/jp804656t. [DOI] [PubMed] [Google Scholar]
- 103.Silkstone G, Jasaitis A, Wilson MT, Vos MH. J. Biol. Chem. 2007;282:1638–1649. doi: 10.1074/jbc.M605760200. [DOI] [PubMed] [Google Scholar]
- 104.Gruia F, Ye X, Ionascu D, Kubo M, Champion PM. Biophys. J. 2007;93:4404–4413. doi: 10.1529/biophysj.107.114736. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.