Abstract
Overproduction of nitric oxide (NO) by inducible NO synthase (iNOS) has been implicated in the pathogenesis of many diseases. iNOS is active only as a homodimer. Dimerization of iNOS represents a potentially critical target for therapeutic intervention. In this study, we show that intracellular iNOS forms dimers that are ”undisruptable” by boiling, denaturants, or reducing agents. Undisruptable (UD) dimers are clearly distinguishable from the easily dissociated dimers formed by iNOS in vitro. UD dimers do not form in Escherichia coli-expressed iNOS and could not be assembled in vitro, which suggests that an in vivo cellular process is required for their formation. iNOS UD dimers are not affected by intracellular depletion of H4B. However, the mutation of Cys-115 (critical for zinc binding) greatly affects the formation of UD dimers. This study reveals insight into the mechanisms of in vivo iNOS dimer formation. UD dimers represent a class of iNOS dimers that had not been suspected. This unanticipated finding revises our understanding of the mechanisms of iNOS dimerization and lays the groundwork for future studies aimed at modulating iNOS activity in vivo.
Nitric oxide (NO) is an important signaling and cytotoxic molecule that is synthesized from l-Arg by isoforms of NO synthase (NOS) (1-3). As a signaling molecule, NO is produced by two constitutive calcium (Ca2+)-dependent isoforms: neuronal NOS and endothelial NOS. Ca2+-activated calmodulin binds to and transiently activates constitutive NOS dimers. Due to the transient nature of elevated Ca2+ levels, the activity of produced NO is short-lived. As an agent of inflammation and cell-mediated immunity, NO is generated by a Ca2+-independent cytokine-inducible NOS (iNOS) that is widely expressed in diverse cell types under transcriptional regulation by inflammatory mediators (2-4). Even at basal Ca2+ levels, calmodulin is tightly bound to iNOS. For this reason, iNOS is notably distinguished from the constitutive isoforms for its production of relatively large amounts of NO (5). iNOS has been implicated in the pathogenesis of many diseases, some of which include Alzheimer's disease, tuberculosis, asthma, glaucoma, inflammatory bowel disease, arthritis, stroke, and septic shock (6, 7). Such wide-based implication has in turn produced an interest in understanding the regulation of NO synthesis by iNOS with the intrinsic goal of developing therapeutic strategies aimed at selective modulation of iNOS activity (6-8).
The human iNOS gene contains 26 exons and encodes a protein of 131 kDa (9, 10). Human iNOS has three domains: (i) an amino-terminal oxygenase domain that binds heme, tetrahydrobiopterin (H4B) and l-Arg; (ii) a carboxyl-terminal reductase domain that binds FMN, FAD, and NADPH; and (iii) an intervening calmodulin-binding domain that regulates electron transfer between the oxygenase and reductase domains (3, 9, 11). iNOS, similar to other NOSs, is active only as a homodimer in which the subunits align in a head-to-head manner, the aminoterminal oxygenase domains forming the dimer interface (12).
Posttranslational subunit dimerization of iNOS represents a potentially critical locus for therapeutic interventions aimed at regulating its activity. There have been a large number of studies addressing the mechanisms of iNOS dimerization (3, 12-15), but most of these studies were performed in vitro under dictated experimental conditions and by using either recombinant protein or partial domains. Very few studies have addressed iNOS dimerization in vivo using cultured or primary cells. In this study, we show that intracellular iNOS forms dimers that are ”undisruptable” by heat, SDS, strong denaturants, and/or reducing agents. These dimers are clearly distinguishable from the easily dissociated dimers formed by iNOS in vitro. This unexpected finding revises our understanding of the mechanisms of iNOS dimerization.
Materials and Methods
Cell culture, transfection, iNOS activity assays, DNA mutagenesis, gel permeation chromatography, and recombinant human iNOS production in Escherichia coli were done as described (16-18). Mass spectrometry was done by using an Applied Biosystems Voyager-DE STR Biospectrometry Workstation.
Cell Lysis. The cell layer was lysed on ice for 45 min in 40 mM Bis-Tris propane buffer (pH 7.7), 150 mM NaCl, and 10% glycerol with 25 mM sodium taurocholate containing protease inhibitors (19, 20).
Standard Western Analysis (i.e., Under Fully Denaturing Conditions). Cell lysates were mixed with one-third volume of 4× Laemmli sample buffer (200 mM Tris·HCl, pH 6.8/8% SDS/40% glycerol/400 mM DTT), heated at 100°C for 5 min. Proteins were resolved on SDS/PAGE by using 4% gels, transferred, and probed with specific antibody (19, 20).
Low-Temperature Partially Denaturing Western Analysis. The procedure was performed as above except that, before electrophoresis, cell lysates were incubated at 37°C for 30 min in the presence of 2 mM l-Arg and 0.1 mM H4B, and electrophoresis was performed at 4°C (16, 17, 21).
iNOS Production in SF9 Insect Cells. Human iNOS cDNA was inserted into baculovirus transfer vectors (BD Biosciences). Production and amplification of baculovirus vectors were performed according to the manufacturer's instructions.
All data are shown as representatives of at least three independent experiments.
Results and Discussion
iNOS Forms ”Undisruptable” (UD) Dimers in Cultured Cells but Not in E. coli. Previous studies have shown that neuronal NOS can form SDS-resistant dimers, which are detected by using low temperature partially denaturing SDS/PAGE (21-23). In that procedure, the samples were preincubated in the presence of l-Arg and H4B and were not heated before gel electrophoreses, which was performed at 4-15°C. These SDS-resistant dimers seemed to be stabilized by the presence of increasing concentrations of l-Arg and H4B but were completely dissociated at the temperature of 40-50°C. Subsequent studies failed to demonstrate the presence of SDS-resistant dimers for recombinant iNOS (24) even after incubation with 1 mM l-Arg and 100 μM H4B, conditions that are known to induce iNOS dimer formation (13, 14).
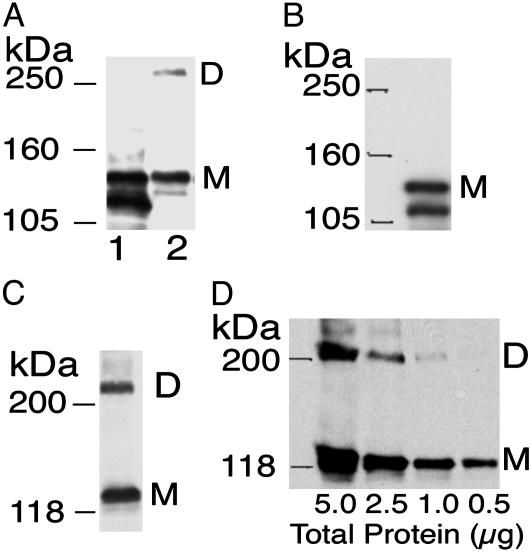
To investigate the presence of SDS-resistant iNOS dimers, partially denaturing Western analysis was performed on lysates of HEK293 cells transfected with human iNOS cDNA and on E. coli lysates expressing recombinant human iNOS. The same human iNOS cDNA was used to generate both mammalian and E. coli expression vectors (16, 18). E. coli-generated iNOS was shown to be active and primarily dimeric on gel permeation chromatography (18). On partially denaturing SDS/PAGE, recombinant iNOS migrated only as a monomer (expected size of 131 kDa) whereas iNOS in HEK293 cell lysates was detected as both a monomer and a dimer (Fig. 1A). The experiments were then conducted by using standard Western analysis (i.e., under fully denaturing conditions and without prior incubation of the samples with l-Arg or H4B). As anticipated, recombinant iNOS was detected only as a monomer (Fig. 1B). Surprisingly, iNOS in HEK293 cell lysates was detected as both a monomer and a dimer (Fig. 1C). This finding indicated that iNOS in HEK293 cells formed dimers that could survive boiling in 2% SDS and 100 mM DTT. Neither increasing the SDS concentration in Laemmli buffer to 5% nor replacing DTT with up to 10% 2-mercaptoethanol had any effect on the observed iNOS dimers (data not shown). We designated this species using the terminology ”undisruptable (UD) dimers.”
Fig. 1.
iNOS Forms UD dimers in cultured cells but not in E. coli. (A) iNOS expression was evaluated under partially denaturing conditions in lysates of E. coli expressing human iNOS (lane 1) and in HEK293 cells transfected with human iNOS cDNA (lane 2). Lysates were incubated at 37°C for 30 min in the presence of 2 mM l-Arg and 0.1 mM H4B and subjected to SDS/PAGE in 4% gels at 4°C and immunoblotting with anti-iNOS Ab. In addition, lysates of E. coli expressing iNOS (B) and HEK293 cells transfected with iNOS cDNA (C) were subjected to standard Western analysis (i.e., under fully denaturing conditions and without prior incubation with l-Arg and H4B) by using anti-iNOS Ab. (D) Lysates of HEK293 expressing iNOS were subjected to standard Western analysis for iNOS by using decreasing amounts of total protein. Letters D and M denote dimer and monomer, respectively. The lower molecular mass band seen in some of the blots represents an iNOS degradation product resulting from aminoterminal proteolytic cleavage (25).
We hypothesized that the reason for not detecting iNOS UD dimers thus far may be related to the possibility that, on SDS/PAGE, monomers represent the majority of iNOS species and therefore are easily detected, whereas the detection of UD dimers requires a more sensitive analysis. To test this hypothesis, we performed standard Western analysis on lysates of HEK293 cells expressing iNOS (Fig. 1D). With decreasing amounts of cell lysates, detection of both iNOS monomers and dimers gradually decreased. Although the Western analysis used was highly sensitive, only iNOS monomers were detected when 0.5 μg of cell lysates total proteins was used. These results indicate that UD dimers detected by Western analysis represent a minor part of total iNOS species and are likely to be overlooked if a sensitive method of detection is not used. In this context, it should be noted that UD dimers are likely to be underestimated by Western analysis because of the lower blotting transfer efficiency of the 262-kDa dimer compared with that of the monomer (23).
Intracellular iNOS exists in both the soluble cytosolic fraction and the particulate membrane fraction (16, 26). To determine whether iNOS UD dimers are predominant in either fraction, we analyzed soluble and particulate fractions of HEK293 cells expressing human iNOS. After cell lysis by sonication, lysates were fractionated into soluble and particulate fractions by centrifugation at 100,000 × g for 1 h. iNOS UD dimers were detected in both fractions, indicating that the formation of these dimers is not limited to a specific cytosolic or membrane component (Fig. 8, which is published as supporting information on the PNAS web site).
In an attempt to reproduce UD dimers in vitro, recombinant iNOS was incubated with l-Arg and H4B in concentrations previously shown to induce iNOS dimerization (13, 14). Moreover, following the reasoning that factors in eukaryotic cell lysates may be required, recombinant iNOS was also incubated with HEK293 cell lysates and evaluated for the presence of UD dimers. All in vitro attempts failed to reproduce the results found with the cultured cells (data not shown), which suggests that an active biological process in eukaryotic cells may be required for such dimers to form.
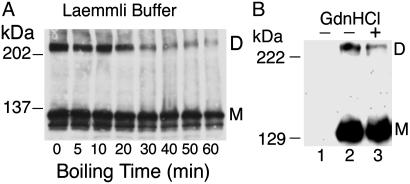
iNOS UD Dimers Can Survive Prolonged Boiling in Laemmli Buffer and Denaturation with Urea or Gdn·HCl. To determine whether the observed iNOS UD dimers were merely residual dimers detected due to inadequate denaturation or reducing treatment, we examined the effects of treating cell lysates with progressively harsher denaturation conditions. To determine whether longer boiling time in Laemmli buffer would be necessary to disrupt iNOS dimers, we boiled HEK293 cell lysates expressing iNOS up to 1 h in Laemmli buffer before Western analysis (Fig. 2A). Importantly, iNOS dimers could still be detected even after 1 h of boiling. Boiling led to a progressive reduction in both iNOS monomers and dimers. This effect was likely due to observed formation of crude precipitates in sample solution, thus reducing amounts of proteins resolved on SDS/PAGE. In addition, there is a potential for peptide bond cleavage with prolonged boiling (27). Densitometry analysis showed that the ratio of dimers as a percentage of total iNOS decreased from 45% to 36% after the standard 5 min of boiling, and to 18% after 1 h of boiling. Considering the deteriorating effect of boiling on sample resolution and the knowledge that Western analysis is inherently not quantitative, we concluded that boiling had no significant effect on dissociation of the iNOS UD dimers. Similar results were obtained in additional experiments that included the presence of 4M urea in sample buffer (Fig. 9, which is published as supporting information on the PNAS web site).
Fig. 2.
iNOS UD dimers survive prolonged boiling in Laemmli buffer and denaturation with Gdn·HCl. (A) Lysates of HEK293 cells transfected with human iNOS cDNA were boiled for various time points in the presence of Laemmli sample buffer containing 2% SDS and 100 mM DTT before Western analysis with anti-iNOS Ab. (B) Western analysis with anti-iNOS Ab of lysates of untransfected HEK293 cells (lane 1) or HEK293 cells stably expressing human iNOS (lanes 2 and 3). In lane 3, cells were lysed by direct addition of 100 mM Tris·HCl (pH 8.0) buffer containing6MGdn·HCl and 100 mM DTT. Lysates were incubated in the same buffer for 20 min at room temperature and subsequently at 100°C for 10 min. Immediately before SDS/PAGE, Gdn·HCl was removed from cell lysates by using PAGEprep columns (Pierce).
Gdn·HCl is used widely as a strong protein denaturant (28). To further ensure iNOS denaturation during Western analysis, we incubated lysates of HEK293 expressing iNOS with a buffer containing 6 M Gdn·HCl and 100 mM DTT for 20 min at room temperature and then at 100°C for 10 min. Gdn·HCl was removed from samples immediately before SDS/PAGE so it would not interfere with the procedure. This precaution was done by coupling removal of Gdn·HCl to eluting lysates in Laemmli buffer. Western analysis showed that iNOS UD dimers in cell lysates survived the Gdn·HCl treatment (Fig. 2B).
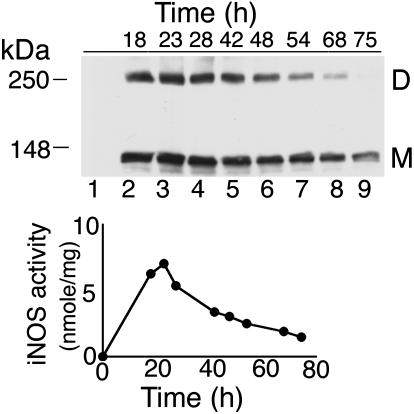
Time Course for Formation of Intracellular UD Dimers of iNOS. Maximum detection of iNOS UD dimers by Western analysis occurred at 23 h after transfection of iNOS cDNA in HEK293 cells (Fig. 3). UD dimers progressively decreased over time, but they were still detectable up to 68 h. At 75 h, only iNOS monomers could be detected. Significantly, maximum levels of iNOS activity coincided with maximal UD dimers detection at 23 h and mirrored changes in UD dimers. This correlation suggests that UD dimers are temporally associated with iNOS activity intracellularly.
Fig. 3.
Time course for formation of UD dimers of iNOS. HEK293 cells were transfected with cDNA of vector only (lane 1) or of human iNOS (lanes 2-9) and lysed at different time points after transfection. Lysates were evaluated by standard Western analysis with anti-iNOS Ab. iNOS activity of cell lysates is expressed as nanomoles of nitrite per mg of total cell protein.
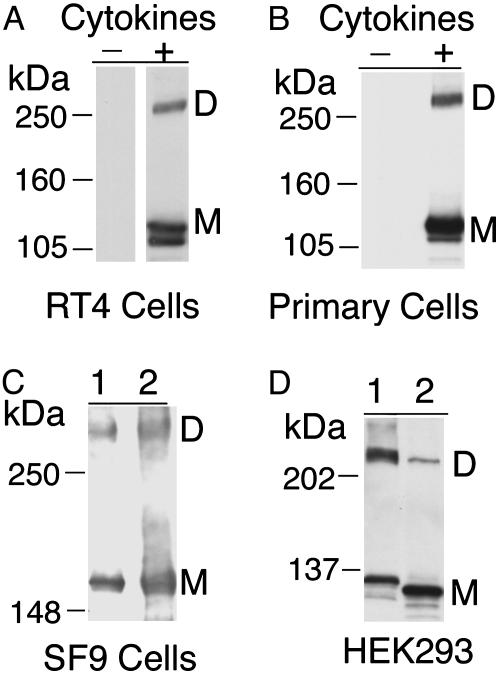
Cytokine-Induced iNOS Forms UD Dimers. iNOS is known to be up-regulated in vivo in response to cytokines and inflammatory mediators (2, 4, 7). We therefore sought to examine endogenous iNOS UD dimers after cytokine induction. By cytokine induction, we further imitated the in vivo state and eliminated concerns over the consequences of exogenous expression of iNOS in HEK293 cells. To this end, we used RT4, which is a human urinary bladder transitional cell papilloma cell line (19). RT4 cells were stimulated for 24 h with a cytokine mixture of IFN-γ, tumor necrosis factor-α, IL-1β, and IL-6. iNOS UD dimers were detected by Western analysis of cell lysates (Fig. 4A). Time course studies revealed that, after cytokine addition, iNOS UD dimers can be detected as early as 8 h and as late as 48 h, with the majority detected at 24 h (data not shown). These results indicate that the presence of UD dimers of iNOS is neither unique to HEK293 cells nor the result of overexpression.
Fig. 4.
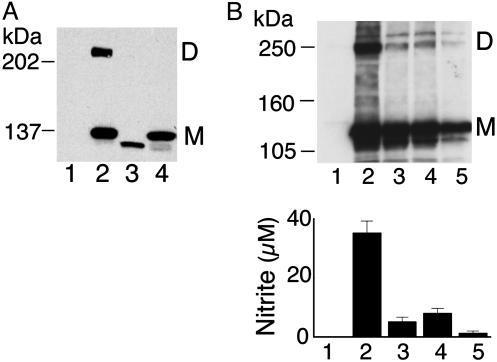
iNOS UD dimers form in various cell types. Shown are representative standard Western analyses using anti-iNOS Ab of RT4 cells incubated for 24 h in the presence or absence of a cytokine mixture of IFN-γ (100 units/ml), IL-1β (0.5 ng/ml), tumor necrosis factor α, (10 ng/ml), and IL-6 (200 units/ml) (A); primary bronchial epithelial cells cultured for 24 h in the presence or absence of a cytokine mixture similar to that of A except that IL-6 was replaced with IL-4 (100 ng/ml) (B); crude cell lysates (lane 1) and partially purified product (lane 2) of GST-iNOS fusion protein expressed in SF9 insect cells (C); and HEK293 cells transfected with cDNA of human iNOS (lane 1) or murine iNOS (lane 2) (D).
iNOS in Primary Human Cells Forms UD Dimers. In the human lung, bronchial epithelial cells are the principal cell type responsible for iNOS production in response to cytokines and inflammatory mediators. Up-regulation of iNOS in bronchial epithelial cells accounts for the increased exhaled NO in asthma patients and has been implicated in the pathogenesis of airway inflammation, the hallmark of asthma (4, 7). For these reasons, it is vital to understand the mechanisms of iNOS regulation in these cells. Primary bronchial epithelial cells, freshly isolated by bronchial brushing of normal subjects, were cultured for 24 h in the presence of a cytokine mixture of IFN-γ, IL-1β, tumor necrosis factor α, and IL-4 (4, 20). iNOS UD dimers were clearly detected in cell lysates after cytokine induction (Fig. 4B).
iNOS Forms UD Dimers in Insect Cells. Because iNOS UD dimers were observed in cultured mammalian cells but not in E. coli, we sought to determine whether iNOS UD dimers would form in a nonmammalian eukaryotic cell type. We used a baculovirus system to express human iNOS in SF9 insect cells, with GST fused to its amino terminus. Western analysis of SF9 crude cell lysates and glutathione affinity-purified GST-iNOS revealed the presence of GST-iNOS as monomers and UD dimers (Fig. 4C). Because GST adds 26 kDa to the 131-kDa molecular mass of iNOS, the expected sizes for GST-iNOS monomer and GST-iNOS dimer are 157 and 314 kDa, respectively. These estimates are consistent with the Western analysis shown in Fig. 4C. These data confirm that iNOS can form UD dimers in insect cells. In addition, they provide further evidence that the observed slower-migrating iNOS band represents an iNOS dimer and not a heterodimer between iNOS and another protein of a similar molecular mass. In the latter case, the fusion of GST would be expected to add only 26 kDa to the molecular mass of both iNOS monomer and the heterodimer. We also expressed human iNOS without GST fusion in SF9 cells and observed the presence of iNOS UD dimer (data not shown).
Murine iNOS Forms UD Dimers Intracellularly. To investigate whether the formation of UD dimers is a unique feature of human iNOS or shared with the murine isoform, HEK293 cells were transfected with human or murine iNOS cDNA, and Western analysis was subsequently performed on cell lysates. Similar to the data obtained with human iNOS, UD dimers were detected for murine iNOS (Fig. 4D). Murine iNOS is 1 kDa smaller than human iNOS (2, 9). The difference in molecular mass between the two isoforms was clearly detected for both monomers and UD dimers. Additional experiments confirmed the presence of iNOS UD dimers in the murine macrophage cell line RAW264.6 after their stimulation with IFN-γ and lipopolysaccharide (Fig. 10, which is published as supporting information on the PNAS web site).
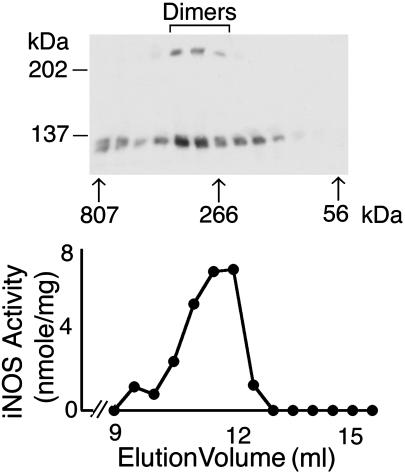
Size Analysis of iNOS UD Dimers by Gel Permeation Chromatography. To confirm the native molecular mass of iNOS UD dimers, lysates of HEK293 cells transfected with human iNOS were subjected to size exclusion chromatography (16, 18). As expected, maximum iNOS activity was detected in fractions eluting near 262 kDa, corresponding to the expected size of human iNOS dimers (Fig. 5). The corresponding Western analysis of those fractions revealed the presence of two species of iNOS dimers with regard to the SDS/PAGE migration patterns. Both species eluted at approximately the same fractions, indicating that they have the same native molecular mass. Yet when resolved by SDS/PAGE, they exhibited two distinct migration patterns, which strongly suggests that iNOS dimers that were formed intracellularly and subsequently retrieved from cell lysates contain two subsets. The first subset is comprised of dimers that are easily dissociated on SDS/PAGE and migrate as monomeric iNOS. This subset accounts for the dimers previously and extensively studied in vitro. These dimers can be easily induced to dissociate, and they reassembled in vitro by altering the cofactors and the substrate concentrations in their solvent environment. The second subset is a subset of dimers that we describe in this study. These particular dimers do not dissociate after full denaturation, and they migrate as dimers on SDS/ PAGE, suggesting they are either covalently linked or very tightly associated. The UD dimers in the gel permeation eluted fractions were estimated by densitometry to be ≈34% of total iNOS detected. The higher structural stability of iNOS UD dimers and their association with maximum iNOS activity suggest that they represent higher order intracellular complexes than dissociable dimers.
Fig. 5.
Analysis of iNOS UD dimers by gel permeation chromatography. Lysates of HEK293 cells transfected with human iNOS cDNA were subjected to FPLC by using a Superdex 200 column in the presence of 1 mM l-Arg, 4 μMH4B, 4 μM FAD, and 3 mM DTT. Fractions (eluted between 9 and 15.5 ml) were assayed for iNOS activity and examined by standard Western analysis with anti-iNOS Ab. Arrows indicate the calculated molecular masses of the corresponding fractions.
Analysis of iNOS UD Dimers by Mass Spectrometry. We aimed to directly confirm the identity of the UD dimers of iNOS by mass spectrometry. Murine iNOS, induced in RAW264.7 cells by using IFN-γ and lipopolysaccharide, was partially purified by using 2′, 5′ADP-Sepharose before performing SDS/PAGE. A band that corresponds to iNOS UD dimers was excised from the gel and subjected to trypsin digest. The generated peptides were analyzed by a matrix-assisted laser desorption ionization/time of flight (Fig. 10). The peptide map was then compared with the GenBank database, which confirmed the product to be iNOS. Despite several repetitions of the purification and identification procedures, no other proteins were coidentified in the excised band. The direct identification of iNOS as the sole protein in the UD dimer band effectively rules out the alternative possibility of iNOS forming a heterodimer with another protein of equal molecular mass.
H4B Depletion in Cultured Cells or Inhibition of NO Synthesis by iNOS Has No Significant Effect on iNOS UD Dimers. Although the cofactor H4B is required for iNOS activity, its exact role in NO synthesis is not entirely clear. A much more controversial issue is the role of H4B in promoting and/or maintaining iNOS dimerization (15, 29). Although H4B is not required for dimerization of iNOS expressed in E. coli, it promoted dimerization of iNOS monomers in vitro and of iNOS expressed in NIH 3T3 cells (13, 14, 30). Further discrepancies recently became evident from interpretations of the crystal structures of the iNOS oxygenase domain. The first structure analysis proposed a role for H4B in favoring protein conformational changes that help form and stabilize the dimer (31). However, a subsequent study of the crystal structures of both endothelial NOS and iNOS oxygenase domains suggests that H4B may not have such a critical role in forming a stable dimer (32, 33).
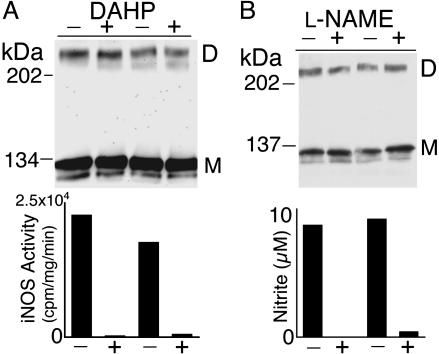
In our study, the addition of H4B during cell lysis or in a subsequent incubation with cell lysates had no significant effect on the detection of iNOS UD dimers (data not shown). Thus, we questioned whether reducing the intracellular levels of H4B would affect the formation or stability of UD dimers of iNOS. Intracellular H4B synthesis can be inhibited by 2,4-diamino-6-hydroxypyrimidine (DAHP), which is a selective inhibitor of GTP cyclohydrolase I, the rate-limiting enzyme for de novo H4B synthesis (34). Incubation of HEK293 cells expressing iNOS with 10 mM DAHP for 24 h resulted in virtual elimination of iNOS activity but did not have detectable effect on iNOS UD dimers (Fig. 6A). Similar results were obtained by using the murine macrophage RAW264.7 cells after their stimulation with lipopolysaccharide and IFN-γ (data not shown). Although we did not directly measure H4B levels, the virtual elimination of NO synthesis by iNOS indicates that H4B levels were critically reduced by DAHP, as previously shown (23, 34). Furthermore, iNOS activity was restored by incubating cells with sepiapterin, an agent that circumvents the inhibition of H4B synthesis by DAHP (data not shown), confirming that iNOS inhibition was due to a lack of H4B. These results suggest that H4B does not play a major role in forming or stabilizing iNOS UD dimers.
Fig. 6.
Effect of H4B depletion and iNOS inhibition on UD dimers of iNOS. (A) HEK293 expressing iNOS were incubated for 24 h in the presence or absence of 10 mM of the H4B synthesis inhibitor DAHP. Cell lysates were evaluated by standard Western analysis with anti-iNOS Ab. iNOS activity in cell lysates is determined by measuring the conversion of l-[3H]Arg to l-[3H]citrulline. (B) HEK293 cells expressing iNOS were incubated for 48 h in the presence or absence of 3 mM of the NOS inhibitor l-NAME. Cell lysates were evaluated by standard Western analysis with anti-iNOS Ab. The iNOS activity of cells is shown by the measurements of nitrite accumulation in the culture media. Results are shown in duplicate.
To determine whether NO synthesis by iNOS was required for UD dimer formation, HEK293 cells expressing iNOS were incubated with the substrate competitive inhibitor l-NAME (Nω-nitro-l-Arg methyl ester) to inhibit NO synthesis (Fig. 6B). Compared with untreated cells, there was no significant difference detected in the formation of iNOS UD dimers in cells treated with l-NAME, despite marked reduction of NO production. These data suggest that the formation of UD dimers by iNOS is not linked to NO production.
iNOS UD Dimers Are Not Detected in Cells Expressing Either an iNOS Splice Variant or an iNOS Mutant, both of Which Are Unable to Dimerize. To confirm that prevention of iNOS dimerization would prevent the formation of UD dimers, we used a splice variant of human iNOS that lacks exons 8 and 9 (iNOS8-9-) and cannot form dimers, which we previously cloned and characterized. In addition, we have generated and characterized several iNOS mutants, including a deletion mutant iNOSΔ241-248 that cannot form dimers (4, 16-18). In contrast to full-length human iNOS, neither iNOS8-9- nor iNOSΔ241-248 could form UD dimers when transfected in HEK293 cells (Fig. 7A). In addition, we examined the consequences of preventing dimerization of wild-type, full-length human iNOS on UD dimer formation. Recently, a class of imidazole-related compounds was found to be allosteric, potent dimerization inhibitors of NOS (35). The NOS dimerization inhibitor BBS1 prevented iNOS UD dimer formation in HEK293 cells expressing iNOS (data not shown). These results confirm that a lower order of dimer formation is required for UD dimer formation.
Fig. 7.
Requirements for formation of iNOS UD dimers. (A) HEK293 cells were transfected with plasmids containing only the vector (lane 1), human iNOS (lane 2), iNOS splice variant iNOS8-9- (lane 3), or iNOS deletion mutant Δ241-248 (lane 4). (B) HEK 293 cells were transfected with plasmids containing the vector only (lane 1), human iNOS (lane 2), or iNOS mutants in which Cys-115 was replaced with Ala (lane 3), His (lane 4), or Ser (lane 5). Twenty-four hours after transfection, cell lysates were evaluated by standard Western analysis with anti-iNOS Ab. (Lower) Nitrite accumulation in the culture medium for experiments of B is shown, mean ± SD (n = 3).
Zinc Binding Is Required for Maximal Formation of UD-iNOS Dimers. Recently, the crystal structure of the oxygenase domain of all NOSs revealed a conserved zinc ion center tetrahedrally coordinated to pairs of symmetry-related Cys residues at the dimer interface. The conserved motif and its strategic location suggest a structural role in maintaining the intersubunit contacts (32, 33). Cys-115 of human iNOS forms one of the ligands for zinc binding. Mutation of its cognate residue in neuronal NOS to Ala resulted in an inability of the mutant to bind zinc (36). We mutated Cys-115 of human iNOS to Ala, His, or Ser and characterized the resulting mutants after their transfection in HEK293 cells. All iNOS mutants had marked reduction in their NO synthesis ability (Fig. 7B). The formation of UD dimers was also markedly reduced. More importantly, there was a correlation between the magnitude of reduction of UD dimers and the loss of activity. Interestingly, the Cys-115 → His mutant was the least affected. Because His is known to serve as a zinc ligand in other proteins, the loss of zinc binding in this mutant may be less severe (37). These results indicate that the metal zinc center in iNOS is important for the formation and/or the stability of the UD dimers.
In summary, we report that intracellular iNOS forms a class of dimers that are tightly associated and clearly distinguishable from the easily dissociated dimers that were previously and extensively studied. These UD dimers do not form when dimerization is prevented by either a dimerization inhibitor or in mutants of iNOS that cannot dimerize. This finding signifies that UD dimers most likely represent a higher order of dimers. UD dimers do not form in E. coli-expressed iNOS, and they could not be assembled in vitro. This result suggests that an intracellular process (possibly chaperon interaction) is required for the assembly of these dimers. UD dimers are not affected by lack of H4B in the cell, and they are not dependent on NO production by iNOS. However, the mutation of Cys-115, which is critical for zinc binding, strongly affects the formation of UD dimers. This finding implies that the metal center is an integral part to maintaining the integrity and the stability of the UD dimer.
The finding that intracellular iNOS UD dimers are undisruptable under strong denaturation conditions suggests that, unlike the widely studied in vitro dimers, the former are either covalently linked or have very strong association. Because iNOS UD dimers survive reducing agents, any covalent linkage that they might have must be a non-disulfide type. Dityrosine cross linkage is a particularly intriguing possibility because it is known to be induced by peroxynitrite (38). The latter is produced by a reaction between NO and superoxide. However, the apparent lack of effect on iNOS UD dimers after inhibition of NO production by l-NAME makes that possibility less likely. In the absence of a covalent linkage, an alternative mechanism for iNOS UD dimers would be through a very tight association between iNOS subunits. Our data suggest that the zinc metal center plays a role in either forming or stabilizing iNOS UD dimers. Zinc atoms have been shown to have stabilization structural roles in other enzymes such as alcohol dehydrogenase, aspartate transcarbamylase, and protein kinase C (39).
This study reveals insights into the mechanisms of in vivo iNOS dimer formation. The UD dimers represent a previously unsuspected class of iNOS dimers. The direct correlation between UD dimer formation and iNOS activity suggests that modulation of iNOS activity in vivo may involve control of UD dimer formation and/or dissociation, which may offer future avenues of therapeutic intervention in diseases associated with overproduction of NO by iNOS. In this context, the use of selective inhibitors of iNOS has been advocated as a novel therapeutic approach for several inflammatory diseases (6, 8). A major difficulty in the use of the current iNOS inhibitors is their lack of specificity for the iNOS isoform (3, 35). The discovery of iNOS UD dimers as a possible regulator of iNOS activity offers a potential means for iNOS inhibition that could provide a more selective approach than current inhibitors. This possibility is further strengthened by the observation that iNOS UD dimers seem to be distinct from dimers previously reported of other NOS isoforms.
Supplementary Material
Acknowledgments
We thank W. Patton, M. Moczygemba, D. Ghosh, Y. Osawa, and C. S. Raman for critical review of the manuscript; T. Cabello for editorial assistance; R. Cook for assistance with mass spectrometry; and J. Parkinson (Berlex) for the gift of BBS1 inhibitor. This work was supported by the American Lung Association, the American Heart Association, the National Heart, Lung, and Blood Institute, and the National Institute of Allergy and Infectious Diseases.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: iNOS, inducible NO synthase; l-NAME, Nω-nitro-l-Arg methyl ester; H4B, tetrahydrobiopterin; DAHP, 2,4-diamino-6-hydroxypyrimidine; UD, undisruptable.
References
- 1.Ignarro, L. J., Buga, G. M., Wood, K. S., Byrns, R. E. & Chaudhuri, G. (1987) Proc. Natl. Acad. Sci. USA 84, 9265-9269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xie, Q.-W., Cho, H. J., Calaycay, J., Mumford, R. A., Swiderek, K. M., Lee, T. D., Ding, A., Troso, T. & Nathan, C. (1992) Science 256, 225-228. [DOI] [PubMed] [Google Scholar]
- 3.Stuehr, D. J. (1999) Biochim. Biophys. Acta 1411, 217-230. [DOI] [PubMed] [Google Scholar]
- 4.Eissa, N. T., Strauss, A. J., Haggerty, C. M., Choo, E. K., Chu, S. C. & Moss, J. (1996) J. Biol. Chem. 271, 27184-27187. [DOI] [PubMed] [Google Scholar]
- 5.Cho, H. J., Xie, Q.-w., Calaycay, J., Mumford, R. A., Swiderek, K. M., Lee, T. D. & Nathan, C. (1992) J. Exp. Med. 176, 599-604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nathan, C. (1997) J. Clin. Invest. 100, 2417-2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo, F. H., Comhair, S. A. A., Zheng, S., Dweik, R. A., Eissa, N. T., Thomassen, M. J., Calhoun, W. & Erzurum, S. C. (2000) J. Immunol. 164, 5970-5980. [DOI] [PubMed] [Google Scholar]
- 8.Hobbs, A. J., Higgs, A. & Moncada, S. (1999) Annu. Rev. Pharamacol. Toxicol. 39, 191-220. [DOI] [PubMed] [Google Scholar]
- 9.Geller, D. A., Lowenstein, C. J., Shapiro, R. A., Nussler, A. K., Di Silvio, M., Wang, S. C., Nakayama, D. K., Simmons, R. L., Snyder, S. H. & Billiar, T. R. (1993) Proc. Natl. Acad. Sci. USA 90, 3491-3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chartrain, N. A., Geller, D. A., Koty, P. P., Sitrin, N. F., Nussler, A. K., Hoffman, E. P., Billiar, T. R., Hutchinson, N. I. & Mudgett, J. S. (1994) J. Biol. Chem. 269, 6765-6772. [PubMed] [Google Scholar]
- 11.Abu-Soud, H. M. & Stuehr, D. J. (1993) Proc. Natl. Acad. Sci. USA 90, 10769-10772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ghosh, D. K. & Stuehr, D. J. (1995) Biochemistry 34, 801-807. [DOI] [PubMed] [Google Scholar]
- 13.Baek, J. K., Thiel, B. A., Lucas, S. & Stuehr, D. J. (1993) J. Biol. Chem. 268, 21120-21129. [PubMed] [Google Scholar]
- 14.Ghosh, D. K., Wu, C., Pitters, E., Moloney, M., Werner, E. R., Mayer, B. & Stuehr, D. J. (1997) Biochemistry 36, 10609-10619. [DOI] [PubMed] [Google Scholar]
- 15.Panda, K., Rosenfeld, R. J., Ghosh, S., Meade, A. L., Getzoff, E. D. & Stuehr, D. J. (2002) J. Biol. Chem. 277, 31020-31030. [DOI] [PubMed] [Google Scholar]
- 16.Eissa, N. T., Yuan, J., Haggerty, C. M., Choo, E. K. & Moss, J. (1998) Proc. Natl. Acad. Sci. USA 95, 7625-7630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eissa, N. T., Haggerty, C. M., Palmer, C. D., Patton, W. & Moss, J. (2001) Am. J. Respir. Cell Mol. Biol. 24, 616-620. [DOI] [PubMed] [Google Scholar]
- 18.Ghosh, D. K., Rashid, M. B., Crane, B., Taskar, V., Mast, M., Misukonis, M., Weinberg, J. B. & Eissa, N. T. (2001) Proc. Natl. Acad. Sci. USA 98, 10392-10397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Musial, A. & Eissa, N. T. (2001) J. Biol. Chem. 276, 24268-24273. [DOI] [PubMed] [Google Scholar]
- 20.Kolodziejski, P. J., Musial, A., Koo, J. S. & Eissa, N. T. (2002) Proc. Natl. Acad. Sci. USA 99, 12315-12320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klatt, P., Schmidt, K., Lehner, D., Glatter, O., Bächinger, H. P. & Mayer, B. (1995) EMBO J. 14, 3687-3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bender, A. T., Nakatsuka, M. & Osawa, Y. (2000) J. Biol. Chem. 275, 26018-26023. [DOI] [PubMed] [Google Scholar]
- 23.Habisch, H. J., Gorren, A. C., Liang, H., Venema, R. C., Parkinson, J. F., Schmidt, K. & Mayer, B. (2003) Mol. Pharmacol. 63, 682-689. [DOI] [PubMed] [Google Scholar]
- 24.Venema, R. C., Ju, H., Zou, R., Ryan, J. W. & Venema, V. J. (1997) J. Biol. Chem. 272, 1276-1282. [DOI] [PubMed] [Google Scholar]
- 25.Vodovotz, Y., Russell, D., Xie, Q. W., Bogdan, C. & Nathan C. (1995) J. Immunol. 154, 2914-2925. [PubMed] [Google Scholar]
- 26.Wheeler, M. A., Smith, S. D., García-Cardena, G., Nathan, C. F., Weiss, R. M. & Sessa, W. C. (1997) J. Clin. Invest. 99, 110-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kowit, J. D. & Maloney, J. (1982) Anal. Biochem. 123, 86-93. [DOI] [PubMed] [Google Scholar]
- 28.Pace, C. N. (1986) Methods Enzymol. 131, 266-280. [DOI] [PubMed] [Google Scholar]
- 29.Hurshman, A. R., Krebs, C., Edmondson, D. E., Huynh, B. H. & Marletta, M. A. (1999) Biochemistry 38, 15689-15696. [DOI] [PubMed] [Google Scholar]
- 30.Tzeng, E., Billiar, T. R., Robbins, P. D., Loftus, M. & Stuehr, D. J. (1995) Proc. Natl. Acad. Sci. USA 92, 11771-11775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Crane, B. R., Arvai, A. S., Ghosh, D. K., Wu, C., Getzoff, E. D., Stuehr, D. J. & Tainer, J. A. (1998) Science 279, 2121-2126. [DOI] [PubMed] [Google Scholar]
- 32.Raman, C. S., Li, H., Martasek, P., Kral, V., Masters, B. S. & Poulos, T. L. (1998) Cell 95, 939-950. [DOI] [PubMed] [Google Scholar]
- 33.Li, H., Raman, C. S., Glaser, C. B., Blasko, E., Young, T. A., Parkinson, J. F., Whitlow, M. & Poulos T. L. J. Biol. Chem. (1999) 274, 21276-21284. [DOI] [PubMed] [Google Scholar]
- 34.Gross, S. S. & Levi, R. (1992) J. Biol. Chem. 267, 25722-25729. [PubMed] [Google Scholar]
- 35.McMillan, K., Adler, M., Auld, D. S., Baldwin, J. J., Blasko, E., Browne, L. J., Chelsky, D., Davey, D., Dolle, R. E., Eagen, K. A., et al. (2000) Proc. Natl. Acad. Sci. USA 97, 1506-1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller, R. T., Martasek, P., Raman, C. S. & Masters, B. S. (1999) J. Biol. Chem. 274, 14537-14540. [DOI] [PubMed] [Google Scholar]
- 37.Vallee, B. L. & Auld, D. S. (1990) Biochemistry 29, 5647-5659. [DOI] [PubMed] [Google Scholar]
- 38.Giulivi, C. & Davies, K. J. (1994) Methods Enzymol. 233, 363-271. [DOI] [PubMed] [Google Scholar]
- 39.Vallee, B. L. & Falchuk, K. H. (1993) Physiol. Rev. 73, 79-118. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.