Abstract
In this pilot study, we used primary human acute myeloid leukemia (AML) cell genomes as templates for exonic PCR amplification, followed by high-throughput resequencing, analyzing ≈7 million base pairs of DNA from 140 AML samples and 48 controls. We identified six previously described, and seven previously undescribed sequence changes that may be relevant for AML pathogenesis. Because the sequencing templates were generated from primary AML cells, the technique favors the detection of mutations from the most dominant clones within the tumor cell mixture. This strategy represents a viable approach for the detection of potentially relevant, nonrandom mutations in primary human cancer cell genomes.
Because many of the mutations relevant for the pathogenesis of cancer and other diseases can only be detected at the level of DNA sequence, many groups are now initiating sequence-based approaches for mutational screens (1). Reduced sequencing costs and improved high-throughput techniques have recently increased the plausibility of this approach. Many questions remain about the best ways to approach cancer genomics and it is clear that no single platform will detect all relevant mutations (2). Although sequence-based mutational profiling is the gold standard for detecting small mutations, it has been unclear whether the mutator phenotype associated with many cancer cell genomes would make resequencing data difficult to interpret. Bardelli et al. (3) and others (reviewed in ref. 1) have recently performed mutational profiling of genes from cell lines derived from tumors, or from cancer cells passaged in immunodeficient mice. This approach is attractive because the supply of DNA from the cell lines is virtually limitless, and because the cell lines are clonal. However, many cancer cells [acute myeloid leukemia (AML) cells included] do not readily adapt to tissue culture conditions (and not all can be passaged in mice), making the approach impractical for routine clinical application. In addition, it is possible that adaptation to tissue culture may require additional mutations for the immortalization of cells; subclones of cells from within a tumor may also be selected during the transition to in vitro growth conditions or during passage in mice. For these reasons, it is preferable to detect genetic changes in primary cancer cells that have not been manipulated. This issue presents a major technical challenge because many tumors are contaminated by nonmalignant cells that are often difficult to remove, and because the number of tumor cells available for analysis is often quite small.
We have attempted to address these issues in this study by examining the genomes of the readily available tumor cells from patients with acute myeloid leukemia. Although the bone marrow of overtly leukemic patients often contains some contaminating normal cells, we have learned that these populations generally do not obscure our ability to detect acquired mutations. By examining the frequency of sequence changes in a large number of AML genomes versus control genomes, we have learned that this resequencing strategy does not detect large numbers of irrelevant passenger mutations that occur randomly in AML genomes. By optimizing PCR techniques, we have been able to reduce the amount of starting material required to create a suitable amplicon for resequencing to 5 ng of genomic DNA, which markedly increases our ability to resequence large numbers of genes from individual patient samples. These data therefore suggest that sequence-based mutational profiling of the genomes of primary cancer cells will be a viable strategy for discovering sequence changes that are relevant for the pathogenesis of cancer.
Materials and Methods
Patient Materials. Appropriate consent for tissue banking was obtained from each patient after the nature and consequences of the study were explained, either at Washington University, or at a participating Cancer and Leukemia Group B (CALGB) institution, by using Institutional Review Board-approved protocols for the studies performed. Patient characteristics are listed in Tables 1 and 2, which are published as supporting information on the PNAS web site.
PCR and Sequencing. Gene sequences were extracted from the public draft human genome database, GenBank (www.ncbi.nlm.nih.gov) and used as reference sequences for assembly and primer construction. Exon boundaries were identified by LO-CUSLINK or by analyses of canonical domains of each gene. Primers were designed in CONSED (4) or by using EXON PRIMER (http://ihg.gsf.de/ihg/ExonPrimer.html) and were synthesized by Genosys (Sigma). Primers were designed to amplify the exon plus ≈100 base pairs of flanking intronic sequence. Sequences of all primers are available in Table 3, which is published as supporting information on the PNAS web site. Five nanograms of genomic DNA was amplified by using AccuTaq (Sigma) and MJ Research (Cambridge, MA) PTC-225 thermal cyclers with the following two-cycle parameters: hot start, 96°C for 2 min 30 sec; and 96°C for 20 sec, 60°C for 15 min, for 35 cycles. Amplicons were sequenced with Big Dye version 3.1 chemistry and analyzed by using ABI 3730 capillary sequencers. Exons (97.9%) were successfully amplified and 72.4% of the alterations were assessed from paired reads of single amplicons. Sequence traces were assembled in CONSED and analyzed to identify genomic alterations by using the POLYPHRED (5) software package.
Results
Strategy for Mutational Profiling. We tested an exonic resequencing strategy for the mutational profiling of genomes derived from primary AML cells. This strategy was chosen over a cDNA sequencing approach because many loss-of-function mutations that create RNA instability or loss could be missed with this technique. PCR techniques were carefully optimized so that only 5 ng of tumor cell DNA was required to create an amplicon for resequencing. One-hundred and ten exons from 12 target genes were PCR-amplified from the genomic DNA of primary AML cells by using primers that lie ≈100 base pairs upstream and downstream from the exon borders. The same primers (and/or additional primers from within the amplicon) were then used to sequence one or both strands by using robotic, high-throughput technologies. The sequence output was assembled in CONSED and screened for differences from the human National Center for Biotechnology Information reference sequence with POLY-PHRED. All potential changes were flagged and then verified by visual inspection, annotated for potential relevance, and assigned a priority score for further analysis. Priority 1 changes have the potential to change the function of a gene [e.g., changes that create amino acid substitutions (both nonconservative and conservative changes were included, because functional consequences are not always accurately predicted by this approach), stop codons, frameshifts, splicing abnormalities, etc.]. Priority 2 changes include all other differences from the reference sequence.
We selected a group of 46 previously banked primary AML samples for study (the pilot set). Thirty-one of the samples were derived from the bone marrow or peripheral blood of overtly leukemic patients with French-American-British subtype M2, and 15 patients had M3 AML (clinical characteristics of the samples are described in Table 1). The samples were originally frozen as cell pellets after red blood cell lysis, with no additional purification. Blast counts (or blast plus promyelocyte counts in the case of the M3 samples) ranged from 20% to 95%.
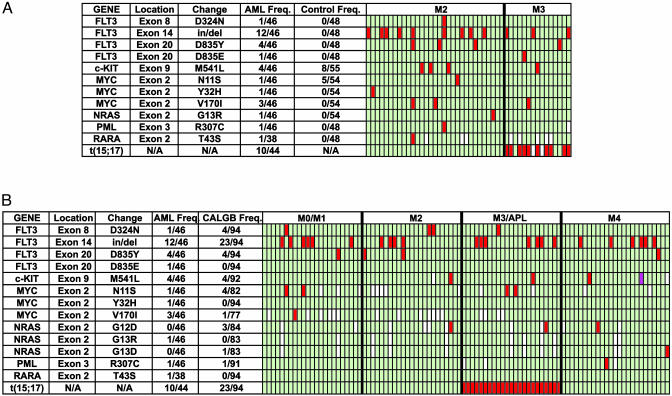
Defining the Sensitivity of the Resequencing Assay by Using the FLT3 Gene. Since acquired, activating mutations of the FLT3 receptor tyrosine kinase gene are currently the most commonly detected genetic changes in AML genomes [≈25-35% of AML patients (6-13)], we amplified and sequenced FLT3 exons 2-24 to test the sensitivity and specificity of the resequencing protocol (Fig. 1A). Priority 1 changes were found in exons 8, 14, and 20. The exon 8 change (D324N) was detected in one AML sample and had not previously been described. It occurs within one of the extracellular Ig domains of FLT3, and it may be relevant because the D residue at this position is conserved in c-KIT, a related tyrosine kinase receptor. Twelve samples had individually unique insertions within exon 14 (in some earlier papers, this exon was inaccurately numbered as 11) that represent internal tandem duplications of this region, which is the most common mutation previously detected in FLT3 (using PCR approaches). This mutation is almost certainly relevant for disease pathogenesis (14, 15). Five samples contained a base substitution in exon 20 that creates a constitutively activated tyrosine kinase domain (either D835Y or D835E). One patient had both an internal tandem duplication (ITD) and D835Y. Therefore, 16 of 46 samples (35%) had known activating mutations of FLT3, which agrees well with the frequency of FLT3 mutations detected in other studies of AML (6-12). Because germ-line DNA was not available for this set of AML samples, we resequenced the same 23 exons from DNA obtained from histologically normal, non-cancerous tissue derived from 48 patients with solid tumors (none of which had AML; see Table 2). These patients were similar for gender and ethnicity, but were older than the AML patients (67 ± 15 vs. 53 ± 24 years, P = 0.004). No priority 1 FLT3 sequence changes were detected in any control sample (Fig. 1 A).
Fig. 1.
Summary of sequencing results. (A) Data are from the genes that were resequenced in the AML pilot set. Each row represents one designated sequence change from the indicated gene. N/A, not available. Each column under the M2 and M3 headings represents one patient sample. Red indicates a sequence change that predicts altered gene function (priority 1), and green indicates that the indicated change was not found. White indicates that resequencing of a target exon was not completed for a particular sample. t(15;17) indicates the presence of the t(15;17) translocation that is found in >90% of patients with AML M3. (B) Data are from the genes that were resequenced from the 94 AML samples from the CALGB set. The color scheme is the same as for A. Purple indicates apparent homozygosity for a priority 1 sequence change.
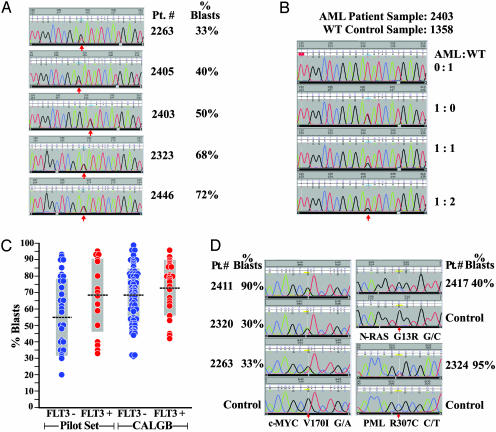
Because the leukemic cells within the AML samples used in this study were not enriched before banking, we wished to determine whether mutational detection was influenced by nonleukemic cells that could be contaminating the leukemic specimens. As noted above, five samples contained point mutations in FLT3 codon 835. The sequence traces identifying these mutations are shown in Fig. 2A, and the blast counts for each sample are indicated. For this study, we have assumed that each AML cell has one normal and one mutant allele, which predicts that a sample comprised of 100% clonal leukemia cells will have a 1:1 wild-type:mutant signal ratio for this FLT3 mutation. The intensity of the signal from the mutant allele is ≈75% that of wild type in the sample with 33% blasts (patient 2263), and ≈50% that of wild type in the sample with 40% blasts (patient 2405). These data suggest that a substantial number of nonleukemic cells were present in these samples; despite their presence, the mutations are clearly detected above the baseline of the sequence trace. The samples with 50%, 68%, and 72% blasts (patients 2403, 2303, and 2446) all demonstrated mutant signals that were virtually equivalent to that of wild type, which suggests that nearly all of the cells in the sample (regardless of morphology) were derived from the same clone and contained the heterozygous mutation. To test this idea, we performed serial dilutions of each of these AML samples with a different control sample containing two normal FLT3 alleles, and repeated the amplification and resequencing studies. A representative example is shown in Fig. 2B. Serial dilutions reduced the mutant signal proportional to the dilution factor in all cases. The mutant signal therefore directly reflects the proportion of cells that are part of the leukemic clone. Because the samples with 50%, 68%, and 72% blasts all had mutant:wild-type signals of 1:1, this finding suggests that the blast count provides a minimal estimate of the proportion of the sample that is part of the leukemic clone (16). To further assess the sensitivity of the sequencing assay, we plotted the presence or absence of FLT3 mutations against blast counts (Fig. 2C, pilot samples); FLT3 mutations were detected in samples with blast counts of 33-95%.
Fig. 2.
Analysis of mutations in the FLT3, c-MYC, and PML genes. (A) Sequence tracings from FLT3 exon 20 amplicons from five AML patients. Sample identifiers and blast counts are shown on the right. These patients were from the pilot set, so the samples were not enriched on Ficoll before they were banked. The positions of the mutations are shown with red arrows. G → T substitutions are present at position 199 in four patients (2263, 2405, 2323, and 2446), creating a D835Y substitution in FLT3. The other patient (2403) has a T → G substitution at position 201, creating a D835E substitution. (B) Sequence tracings from FLT3 exon 20 amplicons derived from an AML sample and a wild-type control sample at varying ratios. The two DNA samples were premixed at the designated ratios, and then exon 20 amplicons were created by PCR and sequenced. The position of the mutation that creates the D835E mutation is shown with a red arrow. The trace from control (WT) DNA is shown at the top, and the trace from the undiluted AML sample is shown next. The 1:1 dilution is shown next, and the 1:2 dilution is shown last. The signal from the mutant allele is detectable at all dilutions, but it decreases as the proportion of control DNA increases. (C) FLT3 mutations (either ITDs in exon 14, and/or activating point mutations at amino acid position 835) are plotted against the blast count of each sample. Data from the pilot set and from the CALGB set are indicated. Means are shown as black bars, and SDs are shown as gray boxes. (D) Sequence tracings from pilot-set AML samples with the c-MYC V170I change, the NRAS G13R change, and the PML R307C change. Note that the signal intensity from the mutant allele is less than that of wild type in the designated AML samples (except for one patient with c-MYC V170I), suggesting that the mutation is not present in all of the cells of the sample. Control tracings are shown for each region as indicated.
To extend the data generated with the first set of samples, we analyzed a set of 94 AML samples that had been cryopreserved in the CALGB Leukemia Bank after Ficoll enrichment (the CALGB set). All samples were from pretreatment bone marrow obtained from patients older than 18 years with de novo AML. All had two or fewer cytogenetic abnormalities, and all had at least 30% blasts (or promyelocytes, in the case of M3 AML). We amplified and resequenced exons 8, 14, and 20 from the FLT3 gene. Four samples had the D342N change in exon 8. Twenty-six of 94 (28%) samples contained either FLT3 ITD and/or D835Y (Fig. 1B); 23 of the mutations were ITDs in exon 14, and 4 were D835Y mutations. One patient had both mutations. All of the mutations were apparently heterozygous. Seven of 23 patients with M0/M1, 5 of 23 patients with M2, 7 of 23 patients with M3, and 7 of 25 patients with M4 AML had activating mutations of FLT3. The mean blast count in samples with the FLT3 mutation was 73 ± 16%, vs. 68 ± 17% without the FLT3 mutation, which is not statistically significant (P = 0.21). The frequency of FLT3 mutations in samples with 30-49% blasts (3 of 14, 21%) was not different from the frequency of FLT3 mutations in samples containing 50-96% blasts (23 of 78, 29%, P = 0.64; blast counts were not available for two samples).
Defining Sequence Changes in 11 Other AML-Associated Genes. After defining the utility of the exonic resequencing strategy with FLT3, we rapidly resequenced all of the exons of 11 other genes that could potentially be relevant for the pathogenesis of AML‡ (6, 11, 17-35), by using the pilot-set samples. Priority 1 changes are summarized in Fig. 1 A; all exons containing these changes were also resequenced in at least 48 control samples. A change predicting M541L in the c-KIT gene was detected in four AML samples and eight controls. One AML sample contained a priority 1 change in c-MYC that predicted N11S; this change was also present in 5 of 54 controls. Because these two changes also occurred in the control samples, they may either represent functionally inconsequential polymorphisms, or they may be germ-line changes that predispose to cancer susceptibility (because our control population patients had all developed solid tumors).
Several additional changes only occurred in AML samples and could therefore be relevant for disease pathogenesis (although the power to make this prediction is small without germ-line DNA from the same patients). A change in c-MYC (Y32H) was detected in one AML sample, and a second c-MYC change (V170I) was detected in three samples. A previously described change (11) that predicts G13R in exon 2 of NRAS was detected in one AML sample. Changes that predict R307C in the PML gene and T43S in the RARA gene were detected in one M2 patient each. No priority 1 changes were found in the genes encoding CBFB, AML-1/RUNX1, CEBPA, HRAS, KRAS, or SPI1/PU.1.
The exons containing priority 1 changes were then resequenced in most of the CALGB samples (Fig. 1B). c-KIT M541L was present in 4 of 92 samples. c-MYC N11S was present in 4 of 82, Y32H in 0 of 94, and V170I in 1 of 77 CALGB samples. NRAS G13R was not detected in the CALGB set, but two other previously described NRAS mutations [G12D (3 of 84) and G13D (1 of 83) were detected (25)]. The PML R307C mutation was detected in 1 of 91 samples, but RARA T43S was not detected. All of the exons of the PU.1 gene were also resequenced in the CALGB set, but no priority 1 changes were identified.
Assessing Somatic vs. Germ-Line Sequence Changes. Direct evaluation of the sequence traces suggested that many of these changes were acquired (i.e., somatic). When two bases are present at a given position, and the polymorphic signal is less than that of the reference signal (except in the case of A residues, which are always favored with our sequencing chemistry), it is highly likely that the base change is present in only a fraction of the total cells, which means that it must be acquired. For example, the c-MYC V170I variant signal was less than that of the reference signal in 2 of 3 pilot-set samples (and it was not detected in controls) strongly suggesting that it was acquired (Fig. 2D); the mutant signal in one of these samples (patient 2263) was similar to the mutant signal for the FLT3 D835Y mutation that occurred in the same patient (Fig. 2 A). Likewise, the NRAS G13R and PML R307C signals were less than that of wild type in the same position, and were not found in control samples (Fig. 2D). All other base changes described in Fig. 1 A yielded variant signals that were equivalent to that of wild type, suggesting that they were either germline changes, or if acquired, they were present in virtually all cells in the sample.
Frequency of Sequence Changes in Control vs. AML Genomes. Of the 46 AML samples from the pilot set, 30 had one or more priority 1 change. Five of the M3 samples had one priority 1 change and also t(15;17), a change that is relevant for the initiation of AML M3 (29). One 69-year-old patient with de novo M2 AML had four priority 1 changes (FLT3 ITD and D835Y, RARA T43S, and c-MYC V170I) and a possible deletion of one allele of CBFB (three polymorphisms in this gene were apparently homozygous, suggesting that either one allele was deleted, or that the patient was homozygous for a rare haplotype). The overall frequency of sequence changes in this sample was 0.69 per kb (see below). Another 63-year-old M2 patient in first relapse had FLT3 ITD, FLT3 D324N, and the PML R307C change. The overall frequency of changes in this patient was 0.36 per kb, which was less than the average for the AML patients (0.47 per kb). The significance of the described changes for AML pathogenesis will require validation with additional sequencing studies, and in experimental model systems.
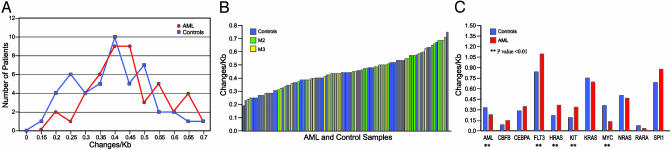
We resequenced a total of 3,675,997 base pairs of DNA from the pilot-set AML samples and 2,598,122 base pairs of DNA from the controls (for some genes, we resequenced control exons only when priority 1 changes were identified in the AML set). An average of 79,913 base pairs of sequence data were obtained for each pilot-set AML sample, and 50,961 base pairs for each control sample. From these data, we were able to define overall frequencies of sequence differences from the NCBI human reference sequence. The pilot AML set contained 1,762 differences from the reference sequence; 30 of these (1.70%) were priority 1 changes. The control group had 1,115 total differences, of which 13 (1.17%) were priority 1. The overall base change frequency was 0.47 change per kb for the AML group, and 0.42 for the controls; this difference is statistically significant (P = 0.0041 with χ2 analysis) because of the large amount of sequence data obtained; the biologic significance of this small numerical difference is unclear. The frequencies of sequence differences (displayed as Changes/Kb) are distributed in a normal fashion for both sets (Fig. 3A). The frequencies of sequence changes for each sample are rank ordered in Fig. 3B. M2 and M3 samples were scattered throughout the controls and there were no striking outliers in either group. Finally, we plotted the frequency of sequence changes by gene (Fig. 3C). For three genes (HRAS, FLT3, and c-KIT), the frequency was significantly higher in the AML pilot set, but for two others (c-MYC and AML-1/RUNX1), the frequency was higher in the control set. When the changes in all 12 genes were compared, there was no difference between the pilot AML set and control set (P = 0.5781, through a paired t test analysis).
Fig. 3.
Analyses of the frequency of base changes per kilobase of sequenced DNA from the AML pilot set and the control set. (A) Total sequence changes per kilobase of sequenced DNA plotted as a function of the numbers of samples with the designated frequencies. More than 98% of sequence changes detected in both sets were priority 2. (B) Total sequence changes per kilobase of sequenced DNA plotted for each pilot set AML sample and control sample arranged in rank order. (C) Total sequence changes per kilobase of sequenced DNA, plotted for each of the 12 genes analyzed. More changes were noted in the AML set for HRAS, FLT3, and c-KIT, whereas more were in the control set for c-MYC and AML-1/RUNX1 (P < 0.01). The other genes had insignificant differences between the sets. When all genes were compared there was no significant difference between the AML set and the control set.
Discussion
Our results from this study suggest the feasibility and plausibility of large-scale mutational screens of primary cancer cell genomes with an exon-based DNA sequencing approach, which is well suited to high-throughput technologies. One important concern that has been expressed about this approach is that the genomic instability of cancer cells would create a high background of unimportant genetic changes (passenger mutations) that could make the analysis of mutational relevancy a virtually impossible task (1). However, by creating the resequencing templates from the genomes of large numbers of primary tumor cells through PCR, random mutations occurring in individual cells do not appear to obscure the analysis. In fact, the design of this strategy favors the detection of mutations that are biologically relevant because they confer proliferation and/or survival advantages to the dominant clone(s) in these primary samples. A better understanding of the relevancy of tumor cell sequence changes will be facilitated in future studies by having matched germ-line samples available for all tumor samples (3) so that all changes can immediately be assigned as germ line vs. acquired. To address this issue, we are now prospectively banking our AML samples with matched germ-line DNA samples obtained from skin biopsies obtained at the time of bone marrow aspiration.
We chose the 12 genes used in this study based on previous information that had implicated all of these genes in the pathogenesis of cancer and/or AML. A variety of genes were chosen from several classes, including that of receptor tyrosine kinases (i.e., FLT3 and c-KIT), targets of MAP kinases (NRAS, HRAS, and KRAS), transcription factors (c-MYC, SPI1/PU.1, and CEBPA), and genes involved in AML-associated translocations (CBFB, AML1/RUNX1, PML, and RARA). Our studies with FLT3 verified the high frequency of acquired mutations in this gene in AML (6-13), thus validating the sensitivity of the PCR/resequencing approach for mutational detection in non-purified bone marrow samples with >30% blasts. Mutations in c-MYC, c-KIT, PML, and RARA were also identified, but their relevance for AML pathogenesis awaits functional studies of the mutant forms of these genes.
NRAS mutations were found in one sample in the pilot set and in four CALGB samples. Therefore, 5 of 125 sequenced AML samples (4%) had previously described activating mutations of NRAS. This result is lower than the 13% frequency of NRAS mutations detected in other studies of AML (P = 0.0031, Fisher's exact test; refs. 6, 11, and 25). The difference is unlikely to reflect mutations that were missed due to contaminating cells because we were able to detect NRAS mutations in samples with blast counts ranging from 40% to 64%, which is clearly adequate for the detection of FLT3 mutations. The frequency difference may be due to the exclusion of M3 samples in other studies, which accounted for only 9 of 414 samples evaluated. M3 samples in our study accounted for 34 of 125 samples (P < 0.0001, Fisher's exact test). No mutations in PU.1 (0 of 140) or AML1 (0 of 46) were found in our sample sets, which is not significantly different from other reports for PU.1 (7 of 263, 2.6%) or AML1 (17 of 232, 7.3%) (P > 0.05 for both; refs. 26, 27, 31, 36, and 37). These reported frequencies fall to 0% for PU.1 and 4.2% for AML1 if only M2 and M3 samples are considered (26, 27, 31, 36, 37). Similarly, 0 of 46 pilot set samples contained mutations in CEBPA, which previously had been reported to have a mutation frequency of 7.4% (16 of 215; refs, 20 and 28), not statistically different from 0 of 46 (P > 0.05). Therefore, the apparently low frequencies of mutations for these three genes are most likely accounted for by sample selection or small sample numbers, rather than a low sensitivity of the PCR-based sequencing strategy used in this study.
The current PCR-based approach to template generation efficiently utilizes the limited supply of DNA available from tumor samples. In our experience, bone marrow samples from patients with overt AML yield an average of 1-10 × 107 cells for banking. Because 106 AML cells yield ≈4 μg of genomic DNA in our experience, most banked samples yield 40-400 μg of total DNA. Because 5 ng of DNA is used to generate an amplicon, 8,000-80,000 exonic amplicons can be generated per sample (representing 800-8,000 genes, assuming 10 exons per gene). Multiplexing of PCR amplicons can potentially extend this yield up to 5-fold. Typical samples of banked AML bone marrow cells will therefore yield adequate DNA to resequence hundreds to thousands of genes with the approaches described here.
These data create a plausibility structure for using sequencing and robotic technologies pioneered by large genome sequencing centers for the mutational profiling of complex genetic diseases like cancer. Although this approach will not identify all of the genetic and epigenetic changes that may be relevant for cancer pathogenesis (e.g., translocations, large deletions, mutations in distant regulatory regions, methylation alterations, etc.), it will provide a wealth of information regarding the structure and sequence of cancer cell genomes. By coupling sequence-based mutational profiling with high-resolution comparative genomic hybridization and RNA profiling, many genetic changes relevant for cancer susceptibility, initiation, progression, and relapse/ resistance will surely be discovered, and new targets for therapy should soon follow.
Supplementary Material
Acknowledgments
This work was supported in part by National Institutes of Health Grants CA83962 and CA101937, the Bakewell Cancer Research Fund, the Buder Charitable Foundation, the Siteman Cancer Center, the Washington University Department of Medicine (T.J.L.), a grant from the Danforth Foundation (R.K.W.), U.S. Public Health Service Grants CA31946 and CA101140 (Cancer and Leukemia Group B), the Coleman Leukemia Research Foundation (C.D.B.), and a Fellowship grant from the Leukemia Research Foundation (M.J.W.).
Abbreviations: AML, acute myeloid leukemia; ITD, internal tandem duplication; CALGB, Cancer and Leukemia Group B.
Footnotes
Gurrieri, C., Nafa, K., Taha, M., Jain, V., Douer, D., Biondi, A., Nimer, S., Gallagher, R. & Pandolfi, P. (2001) Blood 98, 835a.
References
- 1.Strausberg, R., Simpson, A. & Wooster, R. (2003) Nat. Rev. 4, 409-418. [DOI] [PubMed] [Google Scholar]
- 2.Weber, B. L. (2002) Cancer Cell 1, 37-47. [DOI] [PubMed] [Google Scholar]
- 3.Bardelli, A., Parsons, D., Silliman, N., Ptak, J., Szabo, S., Saha, S., Markowitz, S., Willson, J., Parmigiani, G., Kinzler, K., et al. (2003) Science 300, 949. [DOI] [PubMed] [Google Scholar]
- 4.Gordon, D., Abajian, C. & Green, P. (1998) Genome Res. 8, 195-202. [DOI] [PubMed] [Google Scholar]
- 5.Nickerson, D., Tobe, V. & Taylor, S. (1997) Nucleic Acids Res. 25, 2745-2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kiyoi, H., Naoe, T., Nakano, Y., Yokota, S., Minami, S., Miyawaki, S., Asou, N., Kuriyama, K., Jinnai, I., Shimazaki, C., et al. (1999) Blood 93, 3074-3080. [PubMed] [Google Scholar]
- 7.Kottaridis, P., Gale, R., Frew, M., Harrison, G., Langabeer, S., Belton, A., Walker, H., Wheatley, K., Bowen, D., Burnett, A., et al. (2001) Blood 98, 1752-1759. [DOI] [PubMed] [Google Scholar]
- 8.Kottaridis, P., Gale, R., Langabeer, S., Frew, M., Bowen, D. & Linch, D. (2002) Blood 100, 2393-2398. [DOI] [PubMed] [Google Scholar]
- 9.Nakao, M., Yokota, S., Iwai, T., Kaneko, H., Horiike, S., Kashima, K., Sonoda, Y., Fujimoto, T. & Misawa, S. (1996) Leukemia 10, 1911-1918. [PubMed] [Google Scholar]
- 10.Schnittger, S., Schoch, C., Dugas, M., Kern, W., Staib, P., Wuchter, C., Loffler, H., Sauerland, C., Serve, H., Buchner, T., et al. (2002) Blood 100, 59-66. [DOI] [PubMed] [Google Scholar]
- 11.Stirewalt, D., Kopecky, K., Meshinchi, S., Appelbaum, F., Slovak, M., Willman, C. & Radich, J. (2001) Blood 97, 3589-3595. [DOI] [PubMed] [Google Scholar]
- 12.Yamamoto, Y., Kiyoi, H., Nakano, Y., Suzuki, R., Kodera, Y., Miyawaki, S., Asou, N., Kuriyama, K., Yagasaki, F., Shimazaki, C., et al. (2001) Blood 97, 2434-2439. [DOI] [PubMed] [Google Scholar]
- 13.Gilliland, D. G. & Griffin, J. D. (2002) Blood 100, 1532-1542. [DOI] [PubMed] [Google Scholar]
- 14.Kelly, L. M., Kutok, J. L., Williams, I. R., Boulton, C. L., Amaral, S. M., Curley, D. P., Ley, T. J. & Gilliland, D. G. (2002) Proc. Natl. Acad. Sci. USA 99, 8283-8288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sohal, J., Phan, V., Chan, P., Davis, E., Patel, B., Kelly, L., Abrams, T., O'Farrell, A., Gilliland, D., LeBeau, M. & Kogan, S. (2003) Blood 101, 3188-3197. [DOI] [PubMed] [Google Scholar]
- 16.Haferlach, T., Loffler, H., Nickenig, C., Ramm-Peterson, L., Meeder, M., Schoch, R., Schlegelberger, B., Schnittger, S., Schoch, C. & Hiddemann, W. (1998) Br. J. Haematol. 103, 93-99. [DOI] [PubMed] [Google Scholar]
- 17.Beghini, A., Peterlongo, P., Ripamonti, C., Larizza, L., Cairoli, R., Morra, E. & Mecucci, C. (2000) Blood 95, 726-727. [PubMed] [Google Scholar]
- 18.de Souza Fernandez, T., Silva, M., de Souza, J., de Paula, M. & Abdelhay, E. (1996) Cancer Genet. Cytogenet. 86, 183-184. [DOI] [PubMed] [Google Scholar]
- 19.Ding, W., Li, Y.-P., Nobile, L., Grills, G., Carrera, I., Paietta, E., Tallman, M., Wiernik, P. & Gallagher, R. (1998) Blood 92, 1172-1183. [PubMed] [Google Scholar]
- 20.Gomart, A., Hofmann, W.-K., Kawano, S., Takeuchi, S., Krug, U., Kwok, S., Larsen, R., Asou, H., Miller, C., Hoelzer, D. & Koeffler, H. (2002) Blood 99, 1332-1340. [DOI] [PubMed] [Google Scholar]
- 21.Harada, H., Harada, Y., Tanaka, H., Kimura, A. & Inaba, T. (2003) Blood 101, 673-680. [DOI] [PubMed] [Google Scholar]
- 22.Langabeer, S., Gale, R., Rollinson, S., Morgan, G. & Linch, D. (2002) Genes Chromosomes Cancer 34, 24-32. [DOI] [PubMed] [Google Scholar]
- 23.Ma, Y., Zeng, S., Metcalfe, D., Akin, C., Dimitrijevic, S., Butterfield, J., McMahon, G. & Longley, B. (2002) Blood 99, 1741-1744. [DOI] [PubMed] [Google Scholar]
- 24.Michaud, J., Wu, F., Osato, M., Cottles, G., Yanagida, M., Asou, N., Shigesada, K., Ito, Y., Benson, K., Raskind, W., et al. (2002) Blood 99, 1364-1372. [DOI] [PubMed] [Google Scholar]
- 25.Misawa, S., Horiike, S., Kaneko, H., Sasai, Y., Ueda, Y., Nakao, M., Yokota, S., Taniwaki, M., Fujii, H., Nakagawa, H., et al. (1998) Leuk. Res. 22, 631-637. [DOI] [PubMed] [Google Scholar]
- 26.Mueller, B., Pabst, T., Osato, M., Asou, N., Johansen, L., Minden, M., Behre, G., Hiddemann, W., Ito, Y. & Tenen, D. (2002) Blood 100, 998-1007. [DOI] [PubMed] [Google Scholar]
- 27.Osato, M., Asou, N., Abdalla, E., Hoshino, K., Yamasaki, H., Okubo, T., Suzushima, H., Takatsuki, K., Kanno, T., Shigesada, K. & Ito, Y. (1999) Blood 93, 1817-1824. [PubMed] [Google Scholar]
- 28.Pabst, T., Mueller, B., Zhang, P., Radomska, H., Narravula, S., Schnittger, S., Behre, G., Hiddemann, W. & Tenen, D. (2001) Nat. Genet. 27, 263-270. [DOI] [PubMed] [Google Scholar]
- 29.Pollock, J. L., Westervelt, P., Walter, M. J., Lane, A. A. & Ley, T. J. (2001) Curr. Opin. Hematol. 8, 206-211. [DOI] [PubMed] [Google Scholar]
- 30.Preudhomme, C., Warot-Loze, D., Roumier, C., Grardel-Duflos, N., Garand, R., Lai, J., Dastugue, N., Macintyre, E., Denis, C., Bauters, F., et al. (2000) Blood 96, 2862-2869. [PubMed] [Google Scholar]
- 31.Preudhomme, C., Sagot, C., Boissel, N., Cayuela, J.-M., Tigaud, I., de Botton, S., Thomas, X., Raffoux, E., Lamandin, C., Castaigne, S., et al. (2002) Blood 100, 2717-2723. [DOI] [PubMed] [Google Scholar]
- 32.Schaich, M., Ritter, M., Illmer, T., Lisske, P., Thiede, C., Schakel, U., Mohr, B., Ehninger, G. & Neubauer, A. (2001) Br. J. Haematol. 112, 300-307. [DOI] [PubMed] [Google Scholar]
- 33.Song, W.-J., Sullivan, M., Legare, R., Hutchings, S., Tan, X., Kufrin, D., Ratajczak, J., Resende, I., Haworth, C., Hock, R., et al. (1999) Nat. Genet. 23, 166-175. [DOI] [PubMed] [Google Scholar]
- 34.Walker, L., Stevens, J., Campbell, H., Corbett, R., Spearing, R., Heaton, D., Macdonald, D., Morris, C. & Ganly, P. (2002) Br. J. Haematol. 117, 878-881. [DOI] [PubMed] [Google Scholar]
- 35.Zhou, D.-C., Kim, S., Ding, W., Schultz, C., Warrell, R., Jr., & Gallagher, R. (2002) Blood 99, 1356-1363. [DOI] [PubMed] [Google Scholar]
- 36.Lamandin, C., Sagot, C., Roumier, C., Lepelley, P., DeBotton, S., Cosson, A., Fenaux, P. & Preudhomme, C. (2002) Blood 100, 4680-4681. [DOI] [PubMed] [Google Scholar]
- 37.Vegesna, V., Takeuchi, S., Hofmann, W. K., Ikezoe, T., Tavor, S., Krug, U., Fermin, A. C., Heaney, A., Miller, C. W. & Koeffler, H. P. (2002) Leuk. Res. 26, 451-457. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.