Abstract
Expression level, control, and intercoordination of 66 selected heart rhythm determinant (HRD) genes were compared in atria and ventricles of four male and four female adult mice. We found that genes encoding various adrenergic receptors, ankyrins, ion channels and transporters, connexins, cadherins, plakophilins, and other components of the intercalated discs form a complex network that is chamber dependent and differs between the two sexes. In addition, most HRD genes in atria had higher expression in males than in females, while in ventricles, expression levels were mostly higher in females than in males. Moreover, significant chamber differences were observed between the sexes, with higher expression in atria than ventricles for males and higher expression in ventricles than atria for females. We have ranked the selected genes according to their prominence (new concept) within the HRD gene web defined as extent of expression coordination with the other web genes and stability of expression. Interestingly, the prominence hierarchy was substantially different between the two sexes. Taken together, these findings indicate that the organizational principles of the heart rhythm transcriptome are sex dependent, with the newly introduced prominence analysis allowing identification of genes that are pivotal for the sexual dichotomy.
Keywords: Arrhythmia, Atria, Cardiac connexins, Intercalated discs, Ion channels, Ventricles
Introduction
There is now abundant evidence that suggests that sex is a potent modifier of the cardiovascular system (Pilote et al. 2007). In electrocardiographical studies, women display a plethora of electrophysiological variations from men, including faster resting heart rates (Bazett 1920; Adams 1936) and longer rate-corrected QT intervals (Merri et al. 1989; Rautaharju et al. 1992) among other significant differences (Pham and Rosen 2002). Retrospective analyses have revealed significant differences between males and females in terms of prevalence, presentation, management, and outcomes of arrhythmias (reviewed in Peters and Gold 2004). In general, malignant ventricular arrhythmias and sudden cardiac death are less common in women than in men (Albert et al. 1996; Kannel et al. 1998; Buxton et al. 1999a, b; AVID 1997). However, women are significantly more susceptible to drug-induced QT prolongation and torsades de pointes (Reinoehl et al. 1996; Drici et al. 1998; Lehman et al. 1999). Similarly, there is a greater preponderance in women for paroxysmal supraventricular tachycardia (Rodriguez et al. 1992). In striking contrast, male sex is a strong independent predictor of risk for atrial fibrillation, the most common sustained arrhythmia reported in clinical medicine (Benjamin et al. 1998; Peters and Gold 2004). Despite the growing appreciation of the importance of such variations, the mechanisms that underlie sex differences in cardiac function and dysfunction nonetheless remain largely unresolved.
Cardiac arrhythmia encompasses a large and heterogeneous group of heart conditions with complex pathogeneses arising from dynamic interactions among a wide range of structural, electrophysiological, inflammatory, hormonal, environmental, and genetic factors (Mohler and Wehrens 2007). In addition to adaptive or maladaptive morphological abnormalities (e.g., right/left atrial/ventricular hypertrophy), lethal cardiac arrhythmias can also result from altered expression or mutation of key genes (Nattel et al. 2007). Common inherited arrhythmia syndromes caused by variants in genes that encode cardiac ion channel α and β subunits include the congenital long QT syndrome, Brugada syndrome, short QT syndrome, and catecholaminergic polymorphic ventricular tachycardia (Mohler and Wehrens 2007). Moreover, inherited mutations in genes related to regulatory pathways of ion channels may also cause cardiac arrhythmias (Dostanic et al. 2004; Killeen et al. 2008; Lee et al. 2007; Mohler and Wehrens 2007; Teng et al. 2008). Such multiformity of arrhythmia forms suggests that normal cardiac rhythm requires appropriate co-expression of numerous and diverse molecular components.
Use of high throughput methods and of computational algorithms to process and interpret the large and unwieldy data sets obtained from comprehensive assessments of arrhythmia are critical for our understanding of this complex set of disease states and to generate hypotheses testable by more focused methods. Therefore, in order to identify the transcriptomic foundations of sex differences in cardiac arrhythmias, we have profiled the atrial and ventricular transcriptomes of male and female adult mice, with special emphasis on expression level, control, and intercoordination of 66 selected heart rhythm determinant (HRD) genes. In addition to the expected chamber-specific differences, results of our studies revealed prominent sex-specific differences that complement observations of other groups (e.g., Gilbert and Nijland 2008; Isensee et al. 2008; McCurley and Callard 2008; Silander et al. 2008; Witt et al. 2008). Through application of a variant of the principal component analysis, we have identified within the selected genes those proposed to be pivotal in chamber- and sex-specific differences.
Materials and methods
Animals
C57Bl/6j mice used in this study were purchased from Charles River Laboratories International, Inc. (Wilmington, MA, USA), housed in the Albert Einstein College of Medicine (AECOM) accredited animal facility and monitored daily by a trained veterinary personnel. The experimental protocol was approved by the AECOM Institutional Animal Care and Use Committee (IACUC). We have followed the IACUC program for prevention of disease, daily observation and surveillance for assessment of animal health, and the methods of animal handling, restraint, anesthesia, and analgesia. Animal euthanasia respected the guidelines of the Panel on Euthanasia of the American Veterinary Medical Association.
RNA extraction and hybridization
Four adult male (M) and four adult female (F) mice were decapitated, the hearts were removed, and atria (A) and ventricles (V) were collected in separate tubes. Total RNA (20 μg) extracted in Trizol from each of the 16 samples was reverse-transcribed in the presence of fluorescent Alexa Fluor® 647 and Alexa Fluor® 555-aha-dUTPs (Invitrogen, CA, USA) to obtain labeled cDNA. Red- and green-labeled samples of biological replicas were then co-hybridized (“multiple yellow” strategy; Iacobas et al. 2006) overnight at 50°C with mouse MO36k oligonucleotide arrays printed by the Duke University Microarray Facility with Operon Mouse Oligo Set, version 4.0 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GPL8928). After washing (0.1% SDS and 1% SSC) to remove the non-hybridized cDNA, each array was scanned with GenePix 4000B scanner (MDS, Toronto, Canada), and images were primarily analyzed with a GenePixPro 6.0 (Axon Instruments, CA, USA).
Data processing
1. Filtering and normalization
All spots affected by local corruption, or with saturated pixels, or with foreground red or green fluorescence less than twice the background level on any slide were removed from the analysis. The background subtracted signals were normalized iteratively (Iacobas et al. 2005b), alternating red/green, interblock, lowess and scale, and intra- and interslide normalization until the average residual error of the ratio between the spot median net fluorescence and that of the corresponding block (i.e., the 784 spots printed by the same pin) median of valid spots fell below 5% in subsequent steps. Normalized relative expression levels were then organized into redundancy groups composed of all spots probing the same gene, and each group is then represented by the weighted average of the individual spot values. With this reduction of redundancy, after eliminating also the spots targeting not yet well-annotated genes, we arrived at 6,512 fully annotated distinct genes that were appropriately quantified in all 16 RNA samples.
2. Selection of HRD genes
Ank2, Ank3, Cacna1c, Kcne2, Kcnh2, Kcnq1, Lamp2, Lmna, Myh7, Pkp2, Ryr2, Scn5a, Slc25a20, Tpm1, and Ttn were selected from the Online Mendelian Inheritance in Man database as respondent to the keyword “arrhythmia”. The list of potassium channels was completed with Kcnd3, Kcnj11, Kcnj12, Kcnj8, Kcnj9, Abbc8, and Abcc9, with the last two genes also known as Sur1 and Sur2. Other genes were selected from the NCBI Gene Ontology categories (www.godatabase.org): regulation of heart rate (Epas1), regulation of heart contraction (Dmpk, Gnao1, and Thra), cardiac inotropy (Adrbk1, Atp1a1, Atp1a2, Csrp3, Gaa, Prkaca, and Slc8a1), decreased strength of heart contraction during baroreceptor response to increased blood pressure (Adra1d), positive regulation of heart contraction rate by epinephrine–norepinephrine (Adra1b and Adrb1). Casq2 was included because its mutations together with those of Ryr2 were associated with catecholaminergic polymorphic ventricular tachycardia (Györke 2009).
From the five connexins that form gap junction channels between cardiomyocytes, we have analyzed Cx37, Cx40, Cx43, and Cx45, encoded by the genes Gja4, Gja5, Gja1, and Gjc1. Cx43 is the primary connexin in ventricles but also abundantly expressed in atria and in smaller amounts in the distal conduction system. Cx40 is expressed in atria, endothelial cells, and nodal tissue. Cx45 is confined to the conduction system in the adult heart, being present in the sinoatrial and atrioventricular nodes as well as in the atrioventricular conduction bundle, while Cx37 is an endothelial connexin found throughout the vascular tree, including the endocardial vessels (Duffy et al. 2006). Cx30.2, which forms gap junction channels with very low conductance (~9 pS; Kreuzberg et al. 2005) in nodal and conduction systems, was not quantified in this experiment.
In addition to the above connexins, we have also considered other components of the intercalated discs (ID) required for the integration of the heart mechanical and electrical properties. Thus, we have analyzed several adherens (Jup and Vezt), cadherins (Cdh11, Cdh13, Cdh2, Cdh3, Cdh4, and Cdh5), protocadherins (Pcdh12, Pcdh7, Pcdha11, Pcdha12, and Pcdha2) and cadherin associated proteins (the catenins: Ctnna1, Ctnna2, Ctnb1, and Ctnd1), desmosomal proteins (Dsc2, Dsg2, Pkp2, and Pkp4), and tight junction proteins (Cxadr, Jam2, Jam3, Tjap1, and Tjp1). Most of the ID genes were reported as related to various heart diseases (e.g., Dalal et al. 2006; Fischer et al. 2009; Herren et al. 2009; Li et al. 2006; Perriard et al. 2003).
3. Detection of differentially expressed genes
relied on both absolute >1.5×-fold change and <0.05 p-value of the heteroscedastic t test applied to the means of the background subtracted normalized fluorescence values in the four biological replicas of the compared transcriptomes. The p-values (two samples, unequal variance) were computed with a Bonferroni type correction applied to the redundancy group (Iacobas et al. 2005a). Moreover, by using the same criteria, we have determined whether the entire HRD gene cohort was differentially expressed in male heart when compared to female atria/ventricles.
4. Analysis of transcription control
The Relative Expression Variability (REV) was determined as the midrange of the χ2 estimate of the coefficient of variability of the transcript abundance among biological replicas. Then, the genes were ordered with respect to decreasing REV so that the first percentile or gene expression stability score (GES < 1) contained the most unstably expressed and the 100th percentile the most stably expressed genes (Iacobas et al. 2007a).
5. Analysis of expression coordination
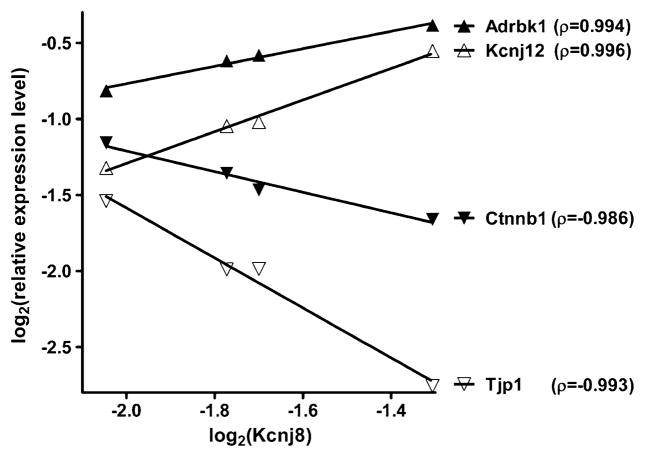
Two genes were considered as synergistically expressed if their expression levels had a positive covariance within biological replicas, as antagonistically expressed when they manifested opposite tendencies (i.e., negative covariance) and as independently expressed when their transcription levels were not correlated (close to zero covariance). In the case of four biological replicas, the (<0.05) cutoff for synergism is pair-wise Pearson correlation coefficient ρ>0.90, for antagonism ρ<−0.90, and for independence |ρ| < 0.05 (Iacobas et al. 2008a). Figure 1 illustrates this concept, using as examples genes that are synergistically or antagonistically expressed with the gene encoding the inwardly rectifying potassium channel, subfamily J, and member 8 (Kcnj8) in male ventricles. For comparison, ankyrin 2, brain (Ank2, not illustrated) was found as independently expressed (ρ=0.025) with Kcnj8.
Fig. 1.
Examples of synergistically (adrbk1adrenergic receptor kinase, beta 1; Kcnj12 potassium inwardly rectifying channel, subfamily J, member 12) and antagonistically (Ctnnb1 catenin (cadherin associated protein), beta 1; Tjp1 tight junction protein 1) expressed genes with Kcnj8 in the male ventricles. Numbers in parentheses are the pair-wise Pearson correlation coefficients between the sets of log2 expression levels in the biological replicas of the compared samples
For each gene i, we have determined all synergistically (SYN), antagonistically (ANT), and independently (IND) expressed partners within the selected HRD gene webs, as well as the (significant) coordination degree (COORD). In order to have a more comprehensive measure of the coordination degree of gene i within the gene web W, we here define the connectivity percentage (CON) as:
| (1) |
where {W}, {SYN}i, and {ANT}i are, respectively, the numbers of genes in the web, synergistic, and antagonist partners of gene i. Both COORD and CON values range from 0 (absolute independent expression, i.e., ρ=0 with respect to each other gene) to 100 (absolute synergistically or antagonistically expressed, i.e., |ρ|=1 with each other gene). For instance, the COORD and CON values of the illustrated genes in Fig. 1 were 29% and 51% (Adrbk1), 28% and 50% (Ctnnb1), 25% and 52% (Kcnj12), 28% and 52% (Kcnj8), and 28% and 52% (Tjp1), all of them outscoring the 44% average for the male ventricles.
6. Analysis of gene prominence
We here introduce the prominent gene analysis to select the most relevant HRD genes and build the atrial and ventricular HRD gene webs of male and female mice. Expression of a prominent gene is assumed to be (1) coordinated with that of numerous other genes to control the web and (2) resistant to the variation of the local conditions to protect the web. Therefore, we define the prominence of gene gi within the web W as percentile locating the score:
| (2) |
GP takes values from 0 to 100 and indicates the percentile of that gene in the prominence hierarchy, with GP=0 the most prominent and GP=100 the least prominent gene.
Results
Data complying with the “minimum information about microarray experiments” were deposited in the National Center for Biotechnology Information Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo) as GSE17324.
1. Analysis of differential expression
Ratios of expression levels in atria and ventricles of male and female mice are provided in Table 1 for genes whose differential expression between the compared samples was statistically significant. Fold change in up- or down-regulation is denoted by a positive or negative number, while a missing value indicates no significant difference. For example, the entry for Adrbk1 demonstrates that this gene is expressed at lower levels in male than in female (both atria and ventricles) and at higher levels in atria than in ventricles (both male and female).
Table 1.
Statistically significant expression ratios (negative for down-regulations) of HRD genes when comparing male (M)/female (F) atria (A)/ventricles (V)
| Gene | Symbol | Cytoband | MA/FA | MV/FV | MA/MV | FA/FV |
|---|---|---|---|---|---|---|
| Adrenergic receptor kinase, beta 1 | Adrbk1 | 19 A|19 3.0 cM | −4.64 | −1.89 | 1.51 | 3.71 |
| Adrenergic receptor, alpha 1b | Adra1b | 11 B1.1|11 19.0 cM | 2.46 | −4.01 | ||
| Adrenergic receptor, alpha 1d | Adra1d | 2 F1|2 74.9 cM | ||||
| Adrenergic receptor, beta 1 | Adrb1 | 19 D2|19 51.0 cM | 1.66 | 4.09 | ||
| Ankyrin 2, brain | Ank2 | 3 G2|3 62.5 cM | −1.95 | |||
| Ankyrin 3, epithelial | Ank3 | 10 B5.3|10 38.0 cM | −1.96 | −1.79 | ||
| ATPase, Na+/K+ transporting, alpha 1 polypeptide | Atp1a1 | 3 F3|3 48.4 cM | −2.17 | −2.67 | ||
| ATPase, Na+/K+ transporting, alpha 2 polypeptide | Atp1a2 | 1 H3|1 94.2 cM | −2.14 | −4.71 | ||
| ATP-binding cassette, subfamily C (CFTR/MRP), member 8 | Abcc8 | 7 B4|7 41.0 cM | −2.99 | −2.05 | ||
| ATP-binding cassette, subfamily C (CFTR/MRP), member 9 | Abcc9 | 6 G2|6 70.0 cM | −1.88 | −3.93 | −15.11 | |
| Cadherin 11 | Cdh11 | 8 D2|8 46.5 cM | 3.65 | −5.25 | ||
| Cadherin 13 | Cdh13 | 8 E1|8 64.0 cM | 2.69 | −4.93 | ||
| Cadherin 2 | Cdh2 | 18 A1|18 6.0 cM | 2.21 | 1.58 | 2.77 | |
| Cadherin 3 | Cdh3 | 8 D3|8 53.3 cM | 3.15 | |||
| Cadherin 4 | Cdh4 | 2 H4|2 106.0 cM | 2.11 | −1.65 | −3.08 | |
| Cadherin 5 | Cdh5 | 8 D3|8 51.0 cM | −2.04 | 2.43 | ||
| Calcium channel, voltage-dependent, L type, alpha 1C subunit | Cacna1c | 6 F1|6 56.0 cM | −2.46 | −3.91 | ||
| Calsequestrin 2 | Casq2 | 3 F3 | 3.97 | −7.11 | ||
| Catenin (cadherin associated protein), alpha 1 | Ctnna1 | 18 B1|18 11.0 cM | 1.78 | −2.01 | −3.04 | |
| Catenin (cadherin associated protein), alpha 2 | Ctnna2 | 6 B3–D|6 34.2 cM | −1.89 | |||
| Catenin (cadherin associated protein), beta 1 | Ctnnb1 | 9 F4|9 72.0 cM | −1.51 | |||
| Catenin (cadherin associated protein), delta 1 | Ctnnd1 | 2 D|2 47.8 cM | −1.93 | −2.03 | ||
| Coxsackievirus and adenovirus receptor | Cxadr | 16 C | ||||
| Cysteine and glycine-rich protein 3 | Csrp3 | 7 B4 | −5.36 | |||
| Desmocollin 2 | Dsc2 | 18 A2|18 7.0 cM | 4.02 | 3.43 | ||
| Desmoglein 2 | Dsg2 | 18 A2|18 7.05 cM | −1.57 | −2.00 | ||
| Dystrophia myotonica-protein kinase | Dmpk | 7 A3|7 4.0 cM | 2.43 | |||
| Endothelial PAS domain protein 1 | Epas1 | 17 E4 | −1.93 | −2.31 | ||
| Gap junction protein, alpha 1 | Gja1 | 10 B4|10 29.0 cM | −2.08 | −4.26 | ||
| Gap junction protein, alpha 4 | Gja4 | 4 D2.2|4 57.6 cM | 1.71 | −1.62 | 2.83 | |
| Gap junction protein, alpha 5 | Gja5 | 3 F2.1|3 45.6 cM | 9.64 | 5.44 | ||
| Gap junction protein, gamma 1 | Gjc1 | 11 E1 | −1.71 | |||
| Glucosidase, alpha, acid | Gaa | 11 D–E|11 74.0 cM | −1.76 | 6.15 | ||
| Guanine nucleotide binding protein, alpha o | Gnao1 | 8 C5|8 45.0 cM | 2.71 | −3.05 | 2.53 | −3.27 |
| Junction adhesion molecule 2 | Jam2 | 16 C3.3 | 2.59 | −2.85 | 2.82 | −2.61 |
| Junction adhesion molecule 3 | Jam3 | 9 A4 | −2.60 | −1.76 | 2.23 | 3.30 |
| Junction plakoglobin | Jup | 11 D|11 60.0 cM | −1.82 | |||
| Lamin A | Lmna | 3 F1|3 42.6 cM | −2.07 | −5.99 | ||
| Lysosomal-associated membrane protein 2 | Lamp2 | X A3.3|X 13.0 cM | −1.87 | −3.76 | ||
| Myosin, heavy polypeptide 7, cardiac muscle, beta | Myh7 | 14 C3|14 20.0 cM | 1.89 | 1.55 | −1.51 | |
| Plakophilin 2 | Pkp2 | 16 B1 | −3.65 | |||
| Plakophilin 4 | Pkp4 | 2 C3 | −2.08 | 2.65 | ||
| Potassium inwardly rectifying channel, subfamily J, member 11 | Kcnj11 | 7 B4|7 41.0 cM | −1.87 | −2.14 | ||
| Potassium inwardly rectifying channel, subfamily J, member 12 | Kcnj12 | 11 B2|11 34.15 cM | 2.75 | −1.74 | −4.24 | |
| Potassium inwardly rectifying channel, subfamily J, member 8 | Kcnj8 | 6 G2|6 70.0 cM | 1.96 | |||
| Potassium inwardly rectifying channel, subfamily J, member 9 | Kcnj9 | 1 H3|1 94.2 cM | 2.66 | 3.87 | ||
| Potassium voltage-gated channel, Isk-related subfamily, gene 2 | Kcne2 | 16 C4 | −2.08 | |||
| Potassium voltage-gated channel, Shal-related family, member 3 | Kcnd3 | 3 F3 | 2.06 | 2.77 | ||
| Potassium voltage-gated channel, subfamily H (eag-related), member 2 | Kcnh2 | 5 A3|5 12.0 cM | 2.18 | −2.19 | 3.32 | |
| Potassium voltage-gated channel, subfamily Q, member 1 | Kcnq1 | 7 F5|7 69.3 cM | ||||
| Protein kinase, cAMP dependent, catalytic, alpha | Prkaca | 8 C3 | −2.28 | 1.80 | ||
| Protocadherin 12 | Pcdh12 | 18 B3 | 4.33 | 2.10 | ||
| Protocadherin 7 | Pcdh7 | 5 C3–D | 2.18 | −1.84 | −2.42 | |
| Protocadherin alpha 11 | Pcdha11 | 18 B3 | 1.98 | 2.18 | ||
| Protocadherin gamma subfamily A, 12 | Pcdhga12 | 18 C | 4.96 | |||
| Protocadherin gamma subfamily A, 2 | Pcdhga2 | 18 B3 | −2.01 | 3.98 | 1.96 | |
| Ryanodine receptor 2, cardiac | Ryr2 | 13 A1–A2|13 7.0 cM | 2.06 | −2.10 | 1.87 | −2.31 |
| Sodium channel, voltage-gated, type V, alpha | Scn5a | 9 F3–F4|9 70.0 cM | −1.75 | −2.72 | −6.46 | |
| Solute carrier family 25 (mitochondrial carnitine/acylcarnitine translocase), member 20 | Slc25a20 | 9 F2 | 2.59 | −1.95 | −1.68 | −8.52 |
| Solute carrier family 8 (sodium/calcium exchanger), member 1 | Slc8a1 | 17 E3|17 48.0 cM | −2.56 | 5.09 | ||
| Thyroid hormone receptor alpha | Thra | 11 D–E|11 57.0 cM | −1.76 | 2.87 | 1.67 | |
| Tight junction associated protein 1 | Tjap1 | 17 C | −2.47 | 2.43 | ||
| Tight junction protein 1 | Tjp1 | 7 C|7 28.5 cM | −1.98 | 1.77 | ||
| Titin | Ttn | 2 D|2 44.0 cM | 1.77 | |||
| Tropomyosin 1, alpha | Tpm1 | 9 C|9 40.0 cM | −2.60 | 2.07 | ||
| Vezatin, adherens junctions transmembrane protein | Vezt | 10 C2 | −2.48 | −1.69 |
Note the diversity of the cytoband locations of the selected genes, the bias toward positive expression ratios in male vs female atria and toward negative ratios (down-regulation) in ventricles, and the shift in atria/ventricles ratios in female with respect to male
Overall, this analysis revealed that sex dichotomy (found for 54 genes) is almost as substantial as the chamber dichotomy (59 genes) for the expression of HRD genes. Except for the delta-1 adrenergic receptor (Adra1d), coxsackievirus and adenovirus receptor (Cxadr), and the QT-associated potassium channel (Kcnq1), all 63 other genes showed statistically significant bias toward one sex or/and one chamber. Interestingly, four ID genes (the catenins Ctnna2, Ctnnb1, Ctnnd1 and Jup, and one potassium channel (Kcne2)) showed only sex bias, while nine genes (cadherin 3 (Cdh3), cysteine and glycine-rich protein 3 (Csrp3), dystrophia myotonica-protein kinase (Dmpk), connexin40 (Gja5), connexin45 (Gjc1), potassium inwardly rectifying channel, subfamily J, member 8 (Kcnj8), protocadherin gamma subfamily A, 12 (Pcdhga12), plakophilin 2 (Pkp2), and the sarcomeric protein itin (Ttn)) showed only chamber bias.
In atria, 20 genes had higher expression in male and only four genes: Adrbk1, delta 1 catenin (Ctnnd1), junction adhesion molecule 3 (Jam3), and junction plakoglobin (Jup) were more highly expressed in female. Interestingly, the situation was reversed in ventricles, with only cadherin 2 (Cdh2) more expressed in male and 40 genes more expressed in female. We found also sex differences when comparing the expressions in the two chambers of the heart. Thus, in male, 31 genes were biased toward atria and four genes (ATP-binding cassette, subfamily C (CFTR/MRP), member 9 (Abcc9), lysosomal-associated membrane protein 2 (Lamp2), sodium channel, voltage-gated, type V, alpha (Scn5a), and mitochondrial carnitine/acylcarnitine translocase (Slc25a20)) toward ventricles. This 31:4 ratio in male is transformed into 7:30 ratio in female.
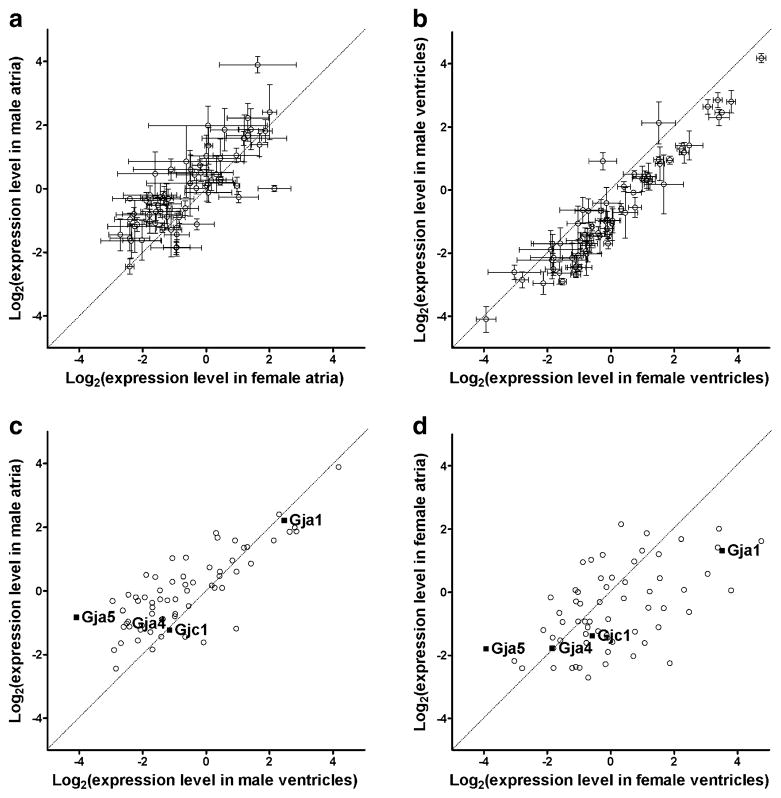
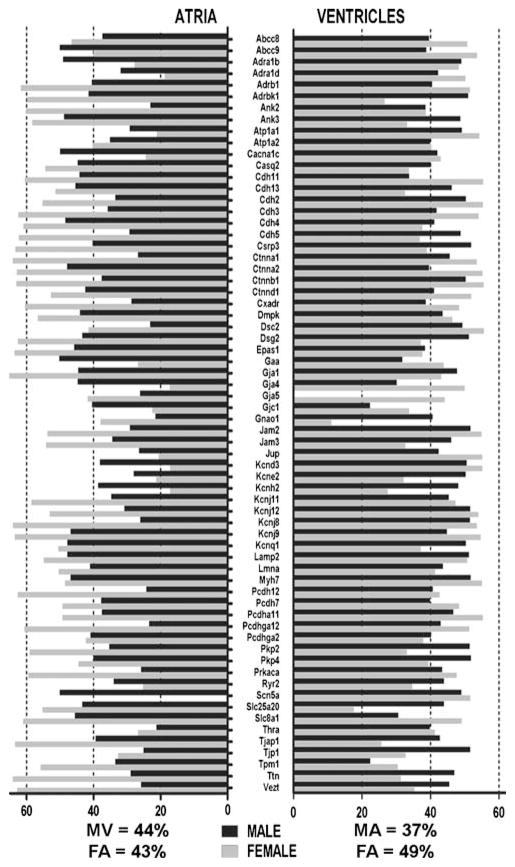
Figure 2 plots the 95% confidence intervals of the relative expression levels of the selected genes in male against the corresponding values in female atria (A) and ventricles (B) and the average levels in atria against ventricles in males (C) and females (D). The plots highlight the differences in gene expression between the sexes, reiterating the male bias in atria but female bias in ventricle, as well the shift in atria–ventricle ratios in female with respect to male.
Fig. 2.
Comparisons of relative expression levels of the selected genes in male/female atria/ventricles. The diagonals of the graphs represent equal expression levels in the compared samples, and the error bars indicate the 95% confidence intervals in each set of biological replicas. Note the male bias in a and female bias in b, the atria bias in c, and ventricles bias in d, as well as the nonuniform variability of the expression levels in a and b. The expression levels of the four connexins quantified in this experiment are indicated by black squares in c and d
2. Control of transcript abundance of HRD genes
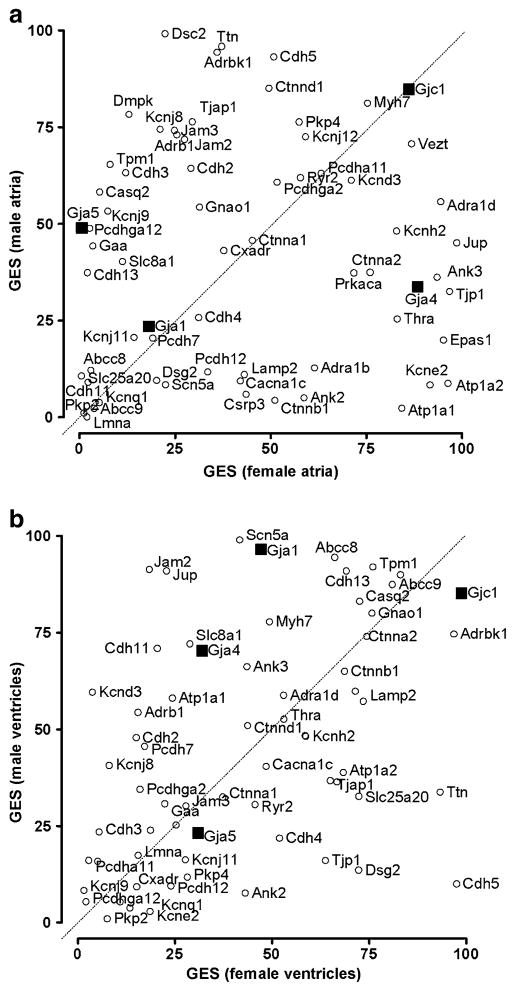
One may also note in Fig. 2 the non-uniform chamber and sex-dependent expression variability of the HRD genes from the extent of the 95% confidence intervals of their expressions. The differences in the hierarchy of genes with regard to the control strength are illustrated in Fig. 3 by plotting the GES percentiles in male atria/ventricles against the corresponding values in female atria/ventricles. GES percentiles were determined for all 6,512 distinct well-annotated unigenes quantified in all 16 RNA samples. Focusing only on the HRD cohort, we found that most of the selected genes displayed a wide range of differences in expression control when comparing the two sexes or the two chambers, without exhibiting any statistically significant bias toward one sex or chamber. On average, the most stably expressed genes in all samples were those encoding Cx45 (Gja1, average GES = 89), the adherens junction protein vezatin (Vezt, 83), and Adrbk1 (75), while the least stably expressed were two of the genes not showing sex differences: Pkp2 (3) and Kcnq1 (6). Of particular note is the variation in expression control between the sexes for the genes encoding the connexins (Cx). Thus, while in atria, Cx40 (Gja5) is more controlled in male and Cx37 (Gja4) in female and Cx43 (Gja1) and Cx45 (Gjc1) exhibit similar stability in both sexes, in ventricles, Gja1 and Gja4 are more controlled in males, with Gjc1 and Gja5 being similarly controlled in both sexes.
Fig. 3.
Plot of GES scores of HRD genes in male atria or ventricles against the corresponding values in female atria or ventricles. Genes located above/below the diagonal are more/less controlled in male than in female. Positions of connexin genes are marked by black squares to make their locations more obvious. Note that although the expression stability is largely different in male and female, there is no overall bias of the GES scores toward one sex
3. Analysis of HRD genes’ networking
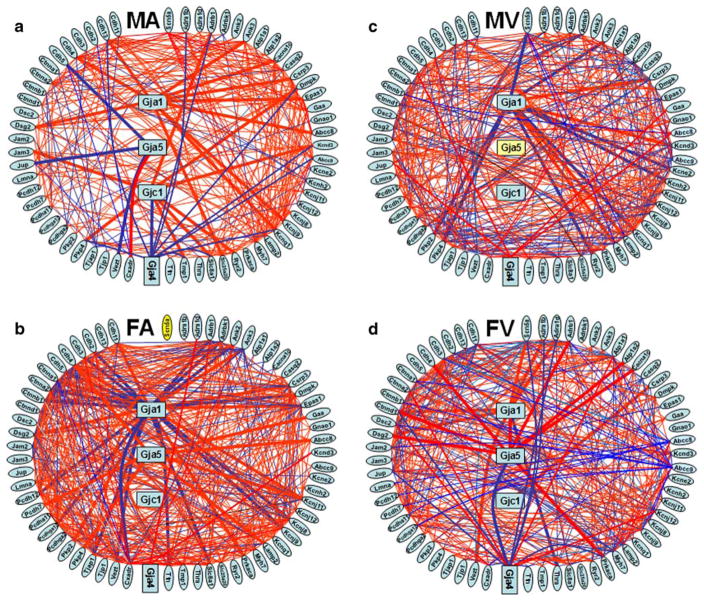
Figure 4 presents the statistically significant synergisms and antagonisms among the expressions of the HRD genes in the atria and ventricles of male and female mice. We found that not only was the gene networking profoundly different between males and females but also the densities of significant expression coordination. Thus, while in males the significant interlinkages of HRD genes were more abundant in ventricles (19% vs 12% in atria), in females, they were more abundant in atria (22% vs 18% in ventricles). The percentage of significantly coordinated partners within the HRD gene web ranged from 0% (for Gjc1 in FA and MV, guanine nucleotide binding protein, alpha o (Gnao1) and Slc25a20 in FV and alpha-tropomyosin 1 (Tmp1) in MV) to over 40%: alpha (E)-catenin (Ctnna1), tight junction associated protein 1 (Tjap1), Ctnna2, Gja1, and Vezst in FA. Again, some of the most striking sex and chamber differences were observed among the gap junction proteins. Hence, Gja1 is coordinated with 43% of the selected genes in female atria but it has substantially fewer coordinations in male atria (17%) and ventricles (18%), a percentage that is even further diminished in female ventricles (15%). The male atria had the largest number of independently expressed gene pairs, genes exceeding 10% independence: protocadherin gamma subfamily A, 12 (Pcdhga12, {IND} = 12%), Kcnj12 (11%), protein kinase, cAMP dependent, catalytic, alpha (Prkaca, 11%), and thyroid hormone receptor alpha (Thra, 11%).
Fig. 4.
Expression coordination of HRD genes in atria and ventricles of male and female adult mouse. a Male atria, b female atria, c male ventricles, d female ventricles. Red/blue lines (thicker for connexin partners) indicate synergistic/antagonistic expression of the linked genes. Note that in male, HRD genes are more interconnected in ventricles than in atria, while in female there are more connections in atria than in ventricles. The components of the intercalated disk complex were grouped in the left side to optimize visualization of their interlinkages. The yellow background of Gja5 in male ventricles and of Scn5a in female atria indicates that these genes were not adequately quantifiable in all four biological replicas owing to printing errors and therefore not suitable for the coordination analysis
Analysis of expression independence (Supplementary Figure 1) revealed the limits of the composing transcriptomic networks within the HRD gene web. Interestingly, both Gja1 and Gja5 have more significant independences in males than in females. Thus, Gja1 is independently expressed with four genes (endothelial PAS domain protein 1 (Epas1), Gnao1, and the protocadherins Pcdh12 and Pcdhga2) in male ventricles, while in female ventricles, it is independent with respect to only Pcdhga2 and Plakophilin 4 (Pkp4). Gja5 is independently expressed with regard to five genes: calcium channel, voltage-dependent, L type, alpha 1C subunit (Cacna1c), solute carrier family 8 (sodium/calcium exchanger), member 1 (Slc8a1), Thra, junction adhesion molecule 2 (Jam2), and Tjap1 in male atria and with regard to only Thra in female atria.
Figure 5 illustrates the sex differences in connectivity among HRD genes in atria and ventricles. On average, in atria, the genes exhibited lower connectivity in males than in females (37% vs 49%), while in ventricles, the percentages were very close (44% vs 43%). The largest connectivity (65%) was shown by Gja1 in female atria and the smallest (11%) by Gnao1in female ventricles.
Fig. 5.
HRD genes have different connectivity scores in male/female atria/ventricles. The numbers indicate the average CON scores in male (M) and female (F) atria (A) and ventricles (V). In agreement with the statistically significant partnerships presented in Fig. 4, the average connectivity was higher in female than in male for atria and larger in male than in female in the case of ventricles. Note the larger relative variability (CV=SD/average) among HRD genes in female atria and ventricles (CVFA=32%, CVFV=24%) compared to the male counterparts (CVMA=23%, CVFV=15%)
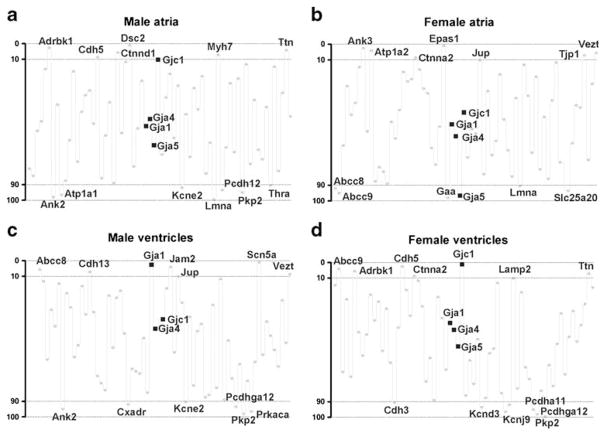
4. Analysis of gene prominence
By combining the connectivity and stability features of the selected genes, we have been able to discern their respective prominence scores using the novel analytical technique of prominent gene analysis. Figure 6 illustrates the results obtained for the selected HRD genes. Of note are the substantial differences between the chambers for the same sex and between sexes for the same chamber. The most prominent genes (GP<5 percentile) in each sample were those encoding: desmocollin 2 (Dsc2), Adrbk1, and Ttn in male atria; Epas1, ankyrin 3 epithelial (Ank3) and ATPase, Na+/K+ transporting, and alpha 2 polypeptide (Atp1a2) in female atria; Snc5a, Gja1, and Jam2 in male ventricles, and Gjc1, cadherin 5 (Cdh5), and Abcc9 in female ventricles. The least prominent genes (GP>95 percentile) in each sample were those encoding lamin-A (Lmna), Ank2 and ATPase, Na+/K+ transporting, alpha 1 polypeptide (Atp1a1) in male atria; glucosidase, alpha, acid (Gaa), Gja5, and Abcc9 in female atria; Pkp2, Prkaca, and Ank2 in male ventricles; and Pkp2, Pcdhga12, and potassium inwardly rectifying channel, subfamily J, member 9 (Kcnj9) in female ventricles. On average, the most prominent genes were myosin, heavy polypeptide 7, cardiac muscle, beta (Myh7, <GP>=13), Adrbk1 (15), and Vezst (19), and the least prominent ones were Pkp2 (95), Lmna (88), and Kcnq1 (86).
Fig. 6.
Prominence of HRD genes in atria and ventricles of adult male and female mice. In order to optimize the readability, only the symbols of connexins and of the most (GP<10%) and the least (GP>10%) prominent genes are presented. Note the chamber and sex differences
Discussion
Approach
Our results suggest that the list of disease biomarkers should be extended beyond the significantly regulated genes by including those emphasizing also significant changes in the expression control and intercoordination within functional pathways. Such broader approach might explain the diversity of phenotypic consequences when individual genes are similarly altered in various animal models of human diseases, knockouts, or/and stressed animals (e.g., Boden and Kennaway 2006; Boles et al. 2009; Chugh et al. 2003; Harris 2008; Donnelly-Roberts et al. 2009; Iacobas et al. 2005a, 2007a, b, 2008a, b; Isensee et al. 2008; Joshi-Mukherjee et al. 2008; Kamiński et al. 2007; Wilhelm et al. 2006). It might also explain why different gene and protein profiles are associated with similar phenotypes as in patients with chronic atrial fibrillation that presented either elevated levels of Cx40 protein (Polontchouk et al. 2001), reduced Cx40 transcript and protein expression (Nao et al. 2003), or no detectable change in the amounts of Cx40 (Kanagaratnam et al. 2004). The flexibility of the biological systems in preserving the vital functions regardless of the environment change (including variation in the gonadal hormones) indicates that several reserve gene networks may be on place.
Our findings support the hypothesis that alteration of expression coordination might induce phenotypic changes even without significant regulation of expression levels. Thus, we found that none of Gja1, Gja5, or Gjc1 was differently expressed in atria despite the higher prevalence of atrial fibrillation (which has been proposed to involve atrial connexins) in males than in females. However, we found large differences in the coordination degrees of these connexins: Gja1 (17% in males vs 43% in females), Gja5 (6% vs 11%), and Gjc1 (14% vs 0%). There are also substantial differences in the expression control of Gja1 and Gja5 in atria: Gja1 (GES=24 in male vs GES=18 in female), Gja5 (49 vs 1). The results regarding the connexins are important because altered expression or mutation of their encoding genes have been reported as causing or accompanying cardiac arrhythmias (Danik et al. 2008; Fishman 2005; Hervé et al. 2008; Wilhelm et al. 2006). In addition to ensuring synchronous contraction of the myocytes by providing low-resistance pathways for impulse propagation and cytoplasmic continuity for Ca2+ and second messengers, the connexins are involved in the control of cell growth (Kardami et al. 2007), regulation of numerous functional pathways (Iacobas et al. 2005a), and networking of many transcription factors (e.g., Iacobas et al. 2007b; Spray and Iacobas 2007) and polymerase III gene cluster (Iacobas et al. 2008b).
Expression of HRD genes is chamber and sex dependent
While chamber differences of HRD genes were expected given the divergent structures and functional behaviors of atria and ventricles, the extent of sex-associated differences was surprising. Not only was expression of most HRD genes in atria higher in males than in females while in ventricles the situation was reversed, but also the atrial/ventricular expression ratios were practically opposite in the two sexes. On average, the sequence of expression levels of HRD genes was female ventricles>male atria> female atria≈male ventricles.
Our results support the findings of other groups. For instance, the higher expression of the adrenergic receptors Adra1b and Adrb1 in males vs females may explain why females are less affected than males by Adra1a–Adra1b double KO (O’Connell et al. 2006) and the influence of gonadal hormones on Adrb1 blockade (Lujan and Dicarlo 2008). By contrast, Adrbk1 was higher in females than in males supporting the hypothesis that disruption of the Adrbk1/PI3K complex preserves β-adrenergic receptor levels and signaling ability by altering the intracellular trafficking of β-adrenergic receptors (Perrino and Rockman 2007).
Interestingly, both Ank2 and Ank3 were more highly expressed in female than in male ventricles, which may be consistent with the sex differences in the type 4 long QT and Brugada syndromes (Mohler and Wehrens 2007). Because hearts of Atp1a1+/− mice are hypocontractile, while those of Atp1a2+/− are hypercontractile (Dostanic et al. 2004), we assume that the overexpression of both with very close ratios (2.17 and 2.14) in females with respect to males maintains the balance of normal contractility.
Since Adra1d, Cxadr, and Kcnq1 exhibited no significantly differential expression in the four types of samples, it is possible that these genes are part of a general HRD pathway active in both atria and ventricles, regardless of the sex. Polymorphisms of Adra1d have been associated with the response to β-blockers in dilated cardiomyopathy (Nonen et al. 2008) but no significant sex difference has been reported. Cxadr, a tight junction protein of the ID, has recently been implicated in cardiac remodeling and electrical conductance between atria and ventricle (Fischer et al. 2009; Lisewski et al. 2008). Humans heterozygous for gene variants in KCNQ1 may develop type 1 long QT syndrome, arrhythmias, and sudden cardiac death (Mohler and Wehrens 2007).
Validation of differential expression
In previous studies (e.g., Iacobas et al. 2005a; Fan et al. 2005), we have validated our microarray method by qRT-PCR. Although in this study we have limited the analysis to only 66 selected HRD genes, our GSE17324 deposited genome-wide expression array results can be compared with data of other groups. Supplementary Table 1 presents such comparisons with data obtained through Agilent cDNA microarray (Isensee et al. 2008) and Affymetrix (Witt et al. 2008). Considering the technical differences between our platform (Duke oligonucleotide array) and the other two, the similarity of the results is quite remarkable: male/female ratios of all 61 genes but neutral sphingomyelinase activation associated factor (Nsmaf) that we had quantified out of the 115 reported by Isensee et al. to be differentially expressed had the same orientation in both studies. Also, all nine genes that analyzed in common to the 10 X and Y-linked genes reported by Witt et al. showed similar bias. Moreover, our atria/ventricles ratios of Gja1, Gja5, and Gjc1 are similar to the protein level ratios of the encoded connexins reported by Manias et al. (2008; Figs. 2 and 4) for 3-week-old wild-type C57Bl/6j mice. Moreover, Tribulová et al. (2005) reported that the protein level of Cx43 was significantly higher in female than in male rat heart.
Strength of transcript abundance control
The similar transcription control of Pkp2, Gjc1, and Kcnq1 in all types of samples suggests that these genes may play important roles in both atria and ventricles, without sex differences. Remarkably, Gja1 (GES=97) in MV and Gjc1 in FV (99) and FA (89) are among the most stably expressed genes, in contrast to Pkp2, a key component of the desmosomes, that was on average the most variably expressed gene. Recent reports demonstrated that mutations in Pkp2 account for the highest proportion of hereditary cases of arrhythmogenic right ventricular cardiomyopathy, with a yield of mutational screening in a range of up to ~70% in some studies (Dalal et al. 2006; Syrris et al. 2006; Van Tintelen et al. 2006, 2007; Joshi-Mukherjee et al. 2008). Possibly, this loose control of Pkp2 expression is critical for the adaptation of the ID to the variability of the mechanical stress.
HRD gene web
The analysis of expression coordination identified pairs of genes whose expression levels are tied to each other. However, gene expression synergism, antagonism, or independence likely depends on local conditions, reflecting the plasticity of gene regulatory networks controlling not only heart rhythm but also other functional pathways with intertwined members.
We found that not only were the selected HRD genes differently interlinked in atria and ventricles but their web also displayed considerable sex differences. Among the connexins, Gja1 has on average the largest percentage of coordinately expressed gene partners (23% compared to 11% for Gja4 and Gja5 and 4% for Gjc1). This finding emphasizes our earlier message that Cx43 may represent an important “node”, which permits the integration of multiple intracellular signaling cascades (Kardami et al. 2007). The phenotype of the neonatal Cx43 null mice includes profound developmental abnormalities resulting from impaired migration of neural crest derivatives during embryogenesis, as well as impaired activation patterns in late embryogenesis and arrhythmia (Reaume et al. 1995; Lo et al. 1997). However, the differences among the connexins are less substantial when considering the more comprehensive criterion of the average connectivity: Gja1 (50±10%), Gja4 (36±15%), Gja5 (37±10%), and Gjc1 (30±9%). Of note is also the high connectivity of Lamp2 (51 ±3%), whose mutations cause Danon disease characterized by prominent hypertrophic cardiomyopathy, myopathy, and Wolff–Parkinson–White syndrome (Maron et al. 2009). The synergistic expression of Cxadr with Gja5 in male atria and the antagonistic one with Gja1 in female atria complements the findings of other authors (e.g., Fischer et al. 2009; Lim et al. 2008; Lisewski et al. 2008) with regard to the mechanistic bases of Cxadr-dependent arrhythmia owing to Cxadr interactions with Cx43 and Cx40.
HRD gene ranking
One major finding is that the hierarchy of the HRD genes differs between atria and ventricles and is sex dependent. Remarkably, while in MV Gja1 is a very prominent gene, in FV, Gjc1 is the most prominent, indicating the importance of Cx43 and Cx45, respectively, in the regulation of ventricular rhythm in the two sexes. Pkp2 and Lmna were ranked among the least prominent within the HRD selection owing to their high expression variability. However, this high variability suggests that they may be ideally suited to adapt the heart rate to the environmental constraints, although certain mutations in these genes may cause deadly arrhythmias (e.g., Herren et al. 2009; Mounkes et al. 2005). A particularly interesting result was the remarkable prominence of Vezt that mediates recruitment of myosin VIIa to the adherens junctions of epithelial cells in a (not completely understood) process that involves the C-terminal domain of alpha-catenin (Sousa et al. 2004).
Prominence analysis further highlights the importance of the different junction types of the IDs, which are targets in multiple types of cardiomyopathy (reviewed in Spray et al. 2001; Wit and Janse 2001; Perriard et al. 2003; Dhein 2006; Schulz et al. 2007; Saffitz et al. 2007). However, while each junction type displays unique functions, we and others have shown previously that these structures frequently present intertwined relationships, and the expression of their component genes is coordinately regulated (reviewed in Derangeon et al. 2009). In this study, we provide additional information demonstrating that these interrelationships are chamber and sex specific, the knowledge of which are likely to be necessary for designing both future experimental and therapeutic strategies.
Conclusion
Our results indicate that HRD genes are expressed, controlled, and networked in different ways to generate, maintain, and modulate the normal contraction rate of each heart chamber, with major differences between the two sexes. Interestingly, none of the selected HRD genes exhibiting sex differences is located on chromosome X or Y, suggesting a complex pattern of gonad–hormone-dependent interaction with the heart rhythm. However, since the present study was conducted in mice, extrapolation of its results to humans should be taken with caution.
Supplementary Material
Acknowledgments
The research was supported by the Award Number R01HL092001 (DAI) from the National Heart, Lung, and Blood Institute (NHLBI). The content is solely the responsibility of the authors and does not necessarily represent the NHLBI official views.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s10142-009-0137-8) contains supplementary material, which is available to authorized users.
Contributor Information
D. A. Iacobas, Email: dumitru-andrei.iacobas@einstein.yu.edu.
S. Iacobas, Email: sanda.iacobas@einstein.yu.edu.
N. Thomas, Email: nthomas@einstein.yu.edu.
D. C. Spray, Email: david-conover.spray@einstein.yu.edu.
References
- Adams W. The normal duration of the electrocardiographic ventricular complex. J Clin Invest. 1936;15:335–342. doi: 10.1172/JCI100784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert CM, McGovern BA, Newell JB, Ruskin JN. Sex differences in cardiac arrest survivors. Circulation. 1996;15:1170–1176. doi: 10.1161/01.cir.93.6.1170. [DOI] [PubMed] [Google Scholar]
- AVID. The antiarrhythmics versus implantable defibrillators (AVID) investigators. A comparison of antiarrhythmic drug therapy with implantable defibrillators in patients resuscitated from near-fatal ventricular arrhythmias. N Engl J Med. 1997;337:1576–1583. doi: 10.1056/NEJM199711273372202. [DOI] [PubMed] [Google Scholar]
- Bazett H. An analysis of the time-relations of electrocardiograms. Heart. 1920;7:353–370. [Google Scholar]
- Benjamin EJ, Wolf PA, D’Agostino RB, Silbershatz H, Kannel WB, Levy D. Impact of atrial fibrillation on the risk of death: the Framington Heart Study. Circulation. 1998;98(10):946–952. doi: 10.1161/01.cir.98.10.946. [DOI] [PubMed] [Google Scholar]
- Boden MJ, Kennaway DJ. Circadian rhythms and reproduction. Reproduction. 2006;132(3):379–392. doi: 10.1530/rep.1.00614. Review. [DOI] [PubMed] [Google Scholar]
- Boles MK, Wilkinson BM, Maxwell A, Lai L, Mills AA, Nishijima I, Salinger AP, Moskowitz I, Hirschi KK, Liu B, Bradley A, Justice MJ. A mouse chromosome 4 balancer ENU-mutagenesis screen isolates eleven lethal lines. BMC Genet. 2009;10:12. doi: 10.1186/1471-2156-10-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buxton AE, Hafley GE, Lehmann MH, Gold M, O’Toole M, Tang A, Coromilas J, Hook B, Stamato NJ, Lee KL. Prediction of sustained ventricular tachycardia inducible by programmed stimulation in patients with coronary artery disease. Utility of clinical variables. Circulation. 1999a;99(14):1843–1850. doi: 10.1161/01.cir.99.14.1843. [DOI] [PubMed] [Google Scholar]
- Buxton AE, Lee KL, Fisher JD, Josephson ME, Prystowsky EN, Hafley G. A randomized study of the prevention of sudden death in patients with coronary artery disease. Multi-center unsustained tachycardia trial investigators. N Engl J Med. 1999b;341(25):1882–90. doi: 10.1056/NEJM199912163412503. Erratum in: N Engl J Med (2000)342(17):1300. [DOI] [PubMed] [Google Scholar]
- Chugh SS, Whitesel S, Turner M, Roberts CT, Jr, Nagalla SR. Genetic basis for chamber-specific ventricular phenotypes in the rat infarct model. Cardiovasc Res. 2003;57(2):477–485. doi: 10.1016/s0008-6363(02)00703-4. [DOI] [PubMed] [Google Scholar]
- Dalal D, James C, Devanagondi R, Tichnell C, Tucker A, Prakasa K, Spevak PJ, Bluemke DA, Abraham T, Russell SD, Calkins H, Judge DP. Penetrance of mutations in plakophilin-2 among families with arrhythmogenic right ventricular dysplasia/cardiomyopathy. J Am Coll Cardiol. 2006;48(7):1416–1424. doi: 10.1016/j.jacc.2006.06.045. [DOI] [PubMed] [Google Scholar]
- Danik SB, Rosner G, Lader J, Gutstein DE, Fishman GI, Morley GE. Electrical remodeling contributes to complex tachyar-rhythmias in connexin43-deficient mouse hearts. FASEB J. 2008;22(4):1204–1212. doi: 10.1096/fj.07-8974com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derangeon M, Spray DC, Bourmeyster N, Sarrouilhe D, Herve JC. Reciprocal influence of connexins and apical junction proteins on their expressions and functions. Biochim Biophys Acta. 2009;1788(4):768–778. doi: 10.1016/j.bbamem.2008.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhein S. Cardiac ischemia and uncoupling: gap junctions in ischemia and infarction. Adv Cardiol. 2006;42:198–212. doi: 10.1159/000092570. Review. [DOI] [PubMed] [Google Scholar]
- Donnelly-Roberts DL, Namovic MT, Han P, Jarvis MF. Mammalian P2X7 receptor pharmacology: comparison of recombinant mouse, rat and human P2X7 receptors. Br J Pharmacol. 2009;157(7):1203–1214. doi: 10.1111/j.1476-5381.2009.00233.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dostanic I, Schultz Jel J, Lorenz JN, Lingrel JB. The alpha 1 isoform of Na, K-ATPase regulates cardiac contractility and functionally interacts and co-localizes with the Na/Ca exchanger in heart. J Biol Chem. 2004;279(52):54053–54061. doi: 10.1074/jbc.M410737200. [DOI] [PubMed] [Google Scholar]
- Drici MD, Knollmann BC, Wang WX, Woosley RL. Cardiac actions of erythromycin: influence of female sex. JAMA. 1998;280(20):1774–1776. doi: 10.1001/jama.280.20.1774. [DOI] [PubMed] [Google Scholar]
- Duffy HS, Fort AG, Spray DC. Cardiac connexins: genes to nexus. Adv Cardiol. 2006;42:1–17. doi: 10.1159/000092550. [DOI] [PubMed] [Google Scholar]
- Fan C, Iacobas DA, Zhou D, Chen Q, Gavrialov O, Haddad GG. Gene expression and phenotypic characterization of mouse heart after chronic constant and intermittent hypoxia. Physiol Genomics. 2005;22:292–307. doi: 10.1152/physiolgenomics.00217.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer R, Poller W, Schultheiss HP, Gotthardt M. CAR-diology-a virus receptor in the healthy and diseased heart. J Mol Med. 2009;87(9):879–884. doi: 10.1007/s00109-009-0489-5. [DOI] [PubMed] [Google Scholar]
- Fishman GI. Gap junction remodeling and ventricular arrhythmias. Heart Rhythm. 2005;2(8):887–889. doi: 10.1016/j.hrthm.2005.05.001. [DOI] [PubMed] [Google Scholar]
- Gilbert JS, Nijland MJ. Sex differences in the developmental origins of hypertension and cardiorenal disease. Am J Physiol Regul Integr Comp Physiol. 2008;295(6):R1941–R1952. doi: 10.1152/ajpregu.90724.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Györke S. Molecular basis of catecholaminergic polymorphic ventricular tachycardia. Heart Rhythm. 2009;6(1):123–129. doi: 10.1016/j.hrthm.2008.09.013. [DOI] [PubMed] [Google Scholar]
- Herren T, Gerber PA, Duru F. Arrhythmogenic right ventricular cardiomyopathy/dysplasia: a not so rare “disease of the desmosome” with multiple clinical presentations. Clin Res Cardiol. 2009;98(3):141–158. doi: 10.1007/s00392-009-0751-4. [DOI] [PubMed] [Google Scholar]
- Hervé JC, Derangeon M, Théveniau-Ruissy M, Miquerol L, Sarrouilhe D, Gros D. Connexins and junctional channels. Roles in the spreading of cardiac electrical excitation and heart development. Pathol Biol (Paris) 2008;56(5):334–341. doi: 10.1016/j.patbio.2008.05.009. [DOI] [PubMed] [Google Scholar]
- Harris AL. Connexin specificity of second messenger permeation: real numbers at last. J Gen Physiol. 2008;131(4):287–292. doi: 10.1085/jgp.200809998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacobas DA, Fan C, Iacobas S, Haddad GG. Integrated transcriptomic response to cardiac chronic hypoxia: translation regulators and response to stress in cell survival. Funct Integr Genomics. 2008a;8(3):265–275. doi: 10.1007/s10142-008-0082-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacobas DA, Iacobas S, Li WE, Zoidl G, Dermietzel R, Spray DC. Genes controlling multiple functional pathways are transcriptionally regulated in connexin43 null mouse heart. Physiol Genomics. 2005a;20:211–223. doi: 10.1152/physiolgenomics.00229.2003. [DOI] [PubMed] [Google Scholar]
- Iacobas DA, Iacobas S, Spray DC. Use of cDNA arrays to explore gene expression in genetically manipulated mice and cell lines. In: Dhein S, Mohr FW, Delmar M, editors. Practical methods in cardiovascular research. Springer; New York: 2005b. pp. 907–915. [Google Scholar]
- Iacobas DA, Fan C, Iacobas S, Spray DC, Haddad GG. Transcriptomic changes in developing kidney exposed to chronic hypoxia. Biochem Biophys Res Comm. 2006;349(1):329–338. doi: 10.1016/j.bbrc.2006.08.056. [DOI] [PubMed] [Google Scholar]
- Iacobas DA, Iacobas S, Spray DC. Connexin43 and the brain transcriptome of the newborn mice. Genomics. 2007a;89:113–123. doi: 10.1016/j.ygeno.2006.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacobas DA, Iacobas S, Spray DC. Connexin-dependent transcellular transcriptomic networks in mouse brain. Prog Biophys Mol Biol. 2007b;94(1–2):168–184. doi: 10.1016/j.pbiomolbio.2007.03.015. [DOI] [PubMed] [Google Scholar]
- Iacobas DA, Iacobas S, Urban-Maldonado M, Scemes E, Spray DC. Similar transcriptomic alterations in Cx43 knock-down and knock-out astrocytes. Cell Commun Adhes. 2008b;15(1):195–206. doi: 10.1080/15419060802014222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isensee J, Witt H, Pregla R, Hetzer R, Regitz-Zagrosek V, Noppinger PR. Sexually dimorphic gene expression in the heart of mice and men. J Mol Med. 2008;86(1):61–74. doi: 10.1007/s00109-007-0240-z. Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi-Mukherjee R, Coombs W, Musa H, Oxford E, Taffet S, Delmar M. Characterization of the molecular phenotype of two arrhythmogenic right ventricular cardiomyopathy (ARVC)-related plakophilin-2 (PKP2) mutations. Heart Rhythm. 2008;5(12):1715–1723. doi: 10.1016/j.hrthm.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiński KA, Oledzka E, Białobrzewska K, Kozuch M, Musiał WJ, Winnicka MM. The effects of moderate physical exercise on cardiac hypertrophy in interleukin 6 deficient mice. Adv Med Sci. 2007;52:164–168. [PubMed] [Google Scholar]
- Kanagaratnam P, Cherian A, Stanbridge RD, Glenville B, Severs NJ, Peters NS. Relationship between connexins and atrial activation during human atrial fibrillation. J Cardiovasc Electrophysiol. 2004;15(2):206–216. doi: 10.1046/j.1540-8167.2004.03280.x. [DOI] [PubMed] [Google Scholar]
- Kannel WB, Wilson PWF, D’Agostino RB. Sudden coronary death in women. Am Heart J. 1998;136:205–212. doi: 10.1053/hj.1998.v136.90226. [DOI] [PubMed] [Google Scholar]
- Kardami E, Dang X, Iacobas DA, Nickel BE, Jeyaraman M, Srisakuldee W, Makazan J, Tanguy S, Spray DC. The role of connexins in controlling cell growth and gene expression. Prog Biophys Mol Biol. 2007;94(1–2):245–264. doi: 10.1016/j.pbiomolbio.2007.03.009. [DOI] [PubMed] [Google Scholar]
- Killeen MJ, Thomas G, Sabir IN, Grace AA, Huang CL. Mouse models of human arrhythmia syndromes. Acta Physiol (Oxf) 2008;192(4):455–469. doi: 10.1111/j.1748-1716.2007.01822.x. [DOI] [PubMed] [Google Scholar]
- Kreuzberg MM, Söhl G, Kim JS, Verselis VK, Willecke K, Bukauskas FF. Functional properties of mouse connexin30.2 expressed in the conduction system of the heart. Circ Res. 2005;96(11):1169–1177. doi: 10.1161/01.RES.0000169271.33675.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RS, Lam CW, Lai CK, Yuen YP, Chan KY, Shek CC, Chan AY, Chow CB. Carnitine-acylcarnitine translocase deficiency in three neonates presenting with rapid deterioration and cardiac arrest. Hong Kong Med J. 2007;13(1):66–68. [PubMed] [Google Scholar]
- Lehman MH, Hardy S, Archibald D, MacNeil DJ. JTc prolongation with d, I-sotalol in women versus men. Am J Cardiol. 1999;83:354–359. doi: 10.1016/s0002-9149(98)00868-6. [DOI] [PubMed] [Google Scholar]
- Li J, Patel VV, Radice GL. Dysregulation of cell adhesion proteins and cardiac arrhythmogenesis. Clin Med Res. 2006;4:42–52. doi: 10.3121/cmr.4.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim BK, Xiong D, Dorner A, Youn TJ, Yung A, Liu TI, Gu Y, Dalton ND, Wright AT, Evans SM, Chen J, Peterson KL, McCulloch AD, Yajima T, Knowlton KU. Coxsackievirus and adenovirus receptor (CAR) mediates atrioventricular-node function and connexin 45 localization in the murine heart. J Clin Invest. 2008;118:2758–2770. doi: 10.1172/JCI34777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisewski U, Shi Y, Wrackmeyer U, Fischer R, Chen C, Schirdewan A, Jüttner R, Rathjen F, Poller W, Radke MH, Gotthardt M. The tight junction protein CAR regulates cardiac conduction and cell-cell communication. J Exp Med. 2008;205(10):2369–2379. doi: 10.1084/jem.20080897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo CW, Cohen MF, Huang GY, Lazatin BO, Patel N, Sullivan R, Pauken C, Park SM. Cx43 gap junction gene expression and gap junctional communication in mouse neural crest cells. Dev Genet. 1997;20:119–132. doi: 10.1002/(SICI)1520-6408(1997)20:2<119::AID-DVG5>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- Lujan HL, Dicarlo SE. Sex differences to myocardial ischemia and beta-adrenergic receptor blockade in conscious rats. Am J Physiol Heart Circ Physiol. 2008;294(4):H1523–H1529. doi: 10.1152/ajpheart.01241.2007. [DOI] [PubMed] [Google Scholar]
- Manias JL, Plante I, Gong XQ, Shao Q, Churko J, Bai D, Laird DW. Fate of connexin43 in cardiac tissue harbouring a disease-linked connexin43 mutant. Cardiovasc Res. 2008;80(3):385–395. doi: 10.1093/cvr/cvn203. [DOI] [PubMed] [Google Scholar]
- Maron BJ, Roberts WC, Arad M, Haas TS, Spirito P, Wright GB, Almquist AK, Baffa JM, Saul JP, Ho CY, Seidman J, Seidman CE. Clinical outcome and phenotypic expression in LAMP2 cardiomyopathy. JAMA. 2009;301(12):1253–1259. doi: 10.1001/jama.2009.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCurley AT, Callard GV. Characterization of housekeeping genes in zebrafish: male–female differences and effects of tissue type, developmental stage and chemical treatment. BMC Mol Biol. 2008;9:102. doi: 10.1186/1471-2199-9-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merri M, Benhorin J, Alberti M, Locati E, Moss AJ. Electrocardiographic quantitation of ventricular repolarization. Circulation. 1989;80:1301–1308. doi: 10.1161/01.cir.80.5.1301. [DOI] [PubMed] [Google Scholar]
- Mohler PJ, Wehrens XH. Mechanisms of human arrhythmia syndromes: abnormal cardiac macromolecular interactions. Physiology (Bethesda) 2007;22:342–350. doi: 10.1152/physiol.00018.2007. Review. [DOI] [PubMed] [Google Scholar]
- Mounkes LC, Kozlov SV, Rottman JN, Stewart CL. Expression of an LMNA-N195K variant of A-type lamins results in cardiac conduction defects and death in mice. Hum Mol Genet. 2005;14(15):2167–2180. doi: 10.1093/hmg/ddi221. [DOI] [PubMed] [Google Scholar]
- Nao T, Ohkusa T, Hisamatsu Y, Inoue N, Matsumoto T, Yamada J, Shimizu A, Yoshiga Y, Yamagata T, Kobayashi S, Yano M, Hamano K, Matsuzaki M. Comparison of expression of connexin in right atrial myocardium in patients with chronic atrial fibrillation versus those in sinus rhythm. Am J Cardiol. 2003;15:678–83. doi: 10.1016/s0002-9149(02)03403-3. [DOI] [PubMed] [Google Scholar]
- Nattel S, Maguy A, Le Bouter S, Yeh YH. Arrhythmogenic ion-channel remodeling in the heart: heart failure, myocardial infarction, and atrial fibrillation. Physiol Rev. 2007;87(2):425–456. doi: 10.1152/physrev.00014.2006. [DOI] [PubMed] [Google Scholar]
- Nonen S, Okamoto H, Fujio Y, Takemoto Y, Yoshiyama M, Hamaguchi T, Matsui Y, Yoshikawa J, Kitabatake A, Azuma J. Polymorphisms of norepinephrine transporter and adrenergic receptor alpha1D are associated with the response to beta-blockers in dilated cardiomyopathy. Pharmacogenomics J. 2008;8(1):78–84. doi: 10.1038/sj.tpj.6500450. [DOI] [PubMed] [Google Scholar]
- O’Connell TD, Swigart PM, Rodrigo MC, Ishizaka S, Joho S, Turnbull L, Tecott LH, Baker AJ, Foster E, Grossman W, Simpson PC, et al. Alpha1-adrenergic receptors prevent a maladaptive cardiac response to pressure overload. J Clin Invest. 2006;116(4):1005–1015. doi: 10.1172/JCI22811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perriard J-C, Hirschy A, Ehler E. Dilated cardiomyopathy: a disease of the intercalated disc? Trends Cardiovascu Med. 2003;13:30–38. doi: 10.1016/s1050-1738(02)00209-8. [DOI] [PubMed] [Google Scholar]
- Perrino C, Rockman HA. Reversal of cardiac remodeling by modulation of adrenergic receptors: a new frontier in heart failure. Curr Opin Cardiol. 2007;22(5):443–449. doi: 10.1097/HCO.0b013e3282294d72. [DOI] [PubMed] [Google Scholar]
- Peters RW, Gold MR. The influence of gender on arrhythmia. Cardiol Rev. 2004;12:97–105. doi: 10.1097/01.crd.0000096416.94304.bd. [DOI] [PubMed] [Google Scholar]
- Pham TV, Rosen MR. Sex, hormones and repolarization. Cardiovasc Res. 2002;53:740–751. doi: 10.1016/s0008-6363(01)00429-1. [DOI] [PubMed] [Google Scholar]
- Pilote L, Dasgupta K, Guru V, Humphries KH, McGrath J, Norris C, Rabi D, Tremblay J, Alamian A, Barnett T, Cox J, Ghali WA, Grace S, Hamet P, Ho T, Kirkland S, Lambert M, Libersan D, O’Loughlin J, Paradis G, Petrovich M, Tagalakis V. A comprehensive view of sex-specific issues related to cardiovascular disease. CMAJ. 2007;176(6):S1–S44. doi: 10.1503/cmaj.051455. Review. Erratum in 24 176(9):1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polontchouk L, Haefliger JA, Ebelt B, Schaefer T, Stuhlmann D, Mehlhorn U, Kuhn-Regnier F, De Vivie ER, Dhein S. Effects of chronic atrial fibrillation on gap junction distribution in human and rat atria. J Am Coll Cardiol. 2001;38(3):883–891. doi: 10.1016/s0735-1097(01)01443-7. [DOI] [PubMed] [Google Scholar]
- Rautaharju PM, Zhou SH, Calhoun WS, HPBGS, Prineas R, Davignon A. Sex differences in the evolution of the electrocardiographic QT interval with age. Can J Cardiol. 1992;8(7):690–695. [PubMed] [Google Scholar]
- Reaume AG, de Sousa PA, Kulkarni S, Langille BL, Zhu D, Davies TC, Juneja SC, Kidder GM, Rossant J. Cardiac malformation in neonatal mice lacking connexin43. Science. 1995;267:1831–1834. doi: 10.1126/science.7892609. [DOI] [PubMed] [Google Scholar]
- Reinoehl J, Frankovich D, Machado C, Kawasaki R, Baga JJ, Pires LA, Steinman RT, Fromm BS, Lehmann MH. Probucol-associated tachyarrhythmic events and QT prolongation: importance of gender. Am Heart J. 1996;131(6):1184–1191. doi: 10.1016/s0002-8703(96)90095-2. [DOI] [PubMed] [Google Scholar]
- Rodriguez LM, de Chillou C, Schläpfer J, Metzger J, Baiyan X, van den Dool A, Smeets JL, Wellens HJ. Age at onset and gender of patients with different types of supraventricular tachycardias. Am J Cardiol. 1992;70(13):1213–1215. doi: 10.1016/0002-9149(92)90060-c. [DOI] [PubMed] [Google Scholar]
- Saffitz JE, Hames KY, Kanno S. Remodeling of gap junctions in ischemic and nonischemic forms of heart disease. J Membr Biol. 2007;218(1–3):65–71. doi: 10.1007/s00232-007-9031-2. [DOI] [PubMed] [Google Scholar]
- Schulz R, Boengler K, Totzeck A, Luo Y, Garcia-Dorado D, Heusch G. Connexin 43 in ischemic pre- and postconditioning. Heart Fail Rev. 2007;12(3–4):261–266. doi: 10.1007/s10741-007-9032-3. [DOI] [PubMed] [Google Scholar]
- Silander K, Alanne M, Kristiansson K, Saarela O, Ripatti S, Auro K, Karvanen J, Kulathinal S, Niemelä M, Ellonen P, Vartiainen E, Jousilahti P, Saarela J, Kuulasmaa K, Evans A, Perola M, Salomaa V, Peltonen L. Gender differences in genetic risk profiles for cardiovascular disease. PLoS ONE. 2008;3(10):e3615. doi: 10.1371/journal.pone.0003615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa S, Cabanes D, El-Amraoui A, Petit C, Lecuit M, Cossart P. Unconventional myosin VIIa and vezatin, two proteins crucial for Listeria entry into epithelial cells. J Cell Sci. 2004;117(Pt 10):2121–2130. doi: 10.1242/jcs.01066. [DOI] [PubMed] [Google Scholar]
- Spray DC, Iacobas DA. Organizational principles of the connexin-related brain transcriptome. J Membr Biol. 2007;218(1–3):39–47. doi: 10.1007/s00232-007-9049-5. [DOI] [PubMed] [Google Scholar]
- Spray DC, Suadicani S, Srinivas M, Gutstein DE, Fishman GI. Handbook of physiology: section 2: the cardiovascular system, vol 1: the heart: gap junctions in the cardiovascular system. Oxford University Press; New York: 2001. Gap junctions in the cardiovascular system; pp. 169–212. [Google Scholar]
- Syrris P, Ward D, Asimaki A, Sen-Chowdhry S, Ebrahim HY, Evans A, Hitomi N, Norman M, Pantazis A, Shaw AL. Clinical expression of plakophilin-2 mutations in familial arrhythmogenic right ventricular cardiomyopathy. Circulation. 2006;113:356–364. doi: 10.1161/CIRCULATIONAHA.105.561654. [DOI] [PubMed] [Google Scholar]
- Teng GQ, Zhao X, Lees-Miller JP, Quinn FR, Li P, Rancourt DE, London B, Cross JC, Duff HJ. Homozygous missense N629D hERG (KCNH2) potassium channel mutation causes developmental defects in the right ventricle and its outflow tract and embryonic lethality. Circ Res. 2008;103(12):1483–1491. doi: 10.1161/CIRCRESAHA.108.177055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tribulová N, Dupont E, Soukup T, Okruhlicová L, Severs NJ. Sex differences in connexin-43 expression in left ventricles of aging rats. Physiol Res. 2005;54(6):705–708. [PubMed] [Google Scholar]
- van Tintelen JP, Entius MM, Bhuiyan ZA, Jongbloed R, Wiesfeld AC, Wilde AA, van der Smagt J, Boven LG, Mannens MM, van Langen IM, Hofstra RM, Otterspoor LC, Doevendans PA, Rodriguez LM, van Gelder IC, Hauer RN. Plakophilin-2 mutations are the major determinant of familial arrhythmogenic right ventricular dysplasia/cardiomyopathy. Circulation. 2006;113(13):1650–1658. doi: 10.1161/CIRCULATIONAHA.105.609719. [DOI] [PubMed] [Google Scholar]
- van Tintelen JP, Hofstra RM, Wiesfeld AC, van den Berg MP, Hauer RN, Jongbloed JD. Molecular geneticsof arrhythmogenic right ventricular cardiomyopathy: emerging horizon? Curr Opin Cardiol. 2007;22(3):185–192. doi: 10.1097/HCO.0b013e3280d942c4. [DOI] [PubMed] [Google Scholar]
- Wilhelm M, Kirste W, Kuly S, Amann K, Neuhuber W, Weyand M, Daniel WG, Garlichs C. Atrial distribution of connexin 40 and 43 in patients with intermittent, persistent, and postoperative atrial fibrillation. Heart Lung Circ. 2006;15:30–37. doi: 10.1016/j.hlc.2005.06.011. [DOI] [PubMed] [Google Scholar]
- Wit AL, Janse MJ. Reperfusion arrhythmias and sudden cardiac death: a century of progress toward an understanding of the mechanisms. Circ Res. 2001;89:741–743. [PubMed] [Google Scholar]
- Witt H, Schubert C, Jaekel J, Fliegner D, Penkalla A, Tiemann K, Stypmann J, Roepcke S, Brokat S, Mahmoodzadeh S, Brozova E, Davidson MM, Ruiz Noppinger P, Grohé C, Regitz-Zagrosek V. Sex-specific pathways in early cardiac response to pressure overload in mice. J Mol Med. 2008;86(9):1013–1024. doi: 10.1007/s00109-008-0385-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.