Abstract
We have developed technologies for creating saturating libraries of sequence-defined transposon insertion mutants in which each strain is maintained. Phenotypic analysis of such libraries should provide a virtually complete identification of nonessential genes required for any process for which a suitable screen can be devised. The approach was applied to Pseudomonas aeruginosa, an opportunistic pathogen with a 6.3-Mbp genome. The library that was generated consists of 30,100 sequence-defined mutants, corresponding to an average of five insertions per gene. About 12% of the predicted genes of this organism lacked insertions; many of these genes are likely to be essential for growth on rich media. Based on statistical analyses and bioinformatic comparison to known essential genes in E. coli, we estimate that the actual number of essential genes is 300-400. Screening the collection for strains defective in two defined multigenic processes (twitching motility and prototrophic growth) identified mutants corresponding to nearly all genes expected from earlier studies. Thus, phenotypic analysis of the collection may produce essentially complete lists of genes required for diverse biological activities. The transposons used to generate the mutant collection have added features that should facilitate downstream studies of gene expression, protein localization, epistasis, and chromosome engineering.
Keywords: PAO1, MPAO1, mutagenesis, ISphoA/hah, ISlacZ/hah
Whole-genome sequences provide the foundation for the creation of relatively complete collections of strains carrying defined mutations in individual genes. Such libraries should facilitate the comprehensive identification of genes required for a wide range of biological processes. A nearly complete library of single-gene deletions of Saccharomyces cerevisiae has been assembled by an international consortium using a PCR-based mutagenesis approach (1). Other projects, also following a strategy of gene-by-gene disruption, are underway for Escherichia coli (E. coli genome project, www.genome.wisc.edu/functional/tnmutagenesis.htm), and have recently been completed for Bacillus subtilis (2).
An alternative strategy for generating mutant libraries consists of ”random” whole-genome transposon-insertion mutagenesis followed by sequence-based identification of insertion sites. The approach is cost-effective and applicable to a wide variety of microbes (3, 4). Studies with yeast, in which a collection of mutants corresponding to about one-third of the genes were represented, have illustrated that the generation of large, arrayed collections of insertion mutants is feasible (5). Other studies with bacteria have analyzed large numbers of transposon insertion mutants to identify genes essential for growth, although the mutants were analyzed within populations rather than being archived in a format allowing additional phenotypes to be examined (6-8). In this report, we describe the generation and initial phenotypic analysis of a near-saturation library of transposon insertion mutants of the opportunistic pathogen Pseudomonas aeruginosa by using technologies that should be applicable to many other bacterial species. P. aeruginosa is a bacterial pathogen that causes a variety of opportunistic infections, including pulmonary infections in cystic fibrosis patients. The mutant collection that was generated provides ≈5-fold coverage of predicted genes, corresponding to multiple insertion alleles in most nonessential genes.
Materials and Methods
Transposon Mutagenesis and Colony Selection. Transposon insertions in the PAO1 chromosome were generated by mating P. aeruginosa PAO1 obtained from B. Iglewski (Department of Microbiology, University of Rochester Medical Center, Rochester, NY) (referred to as MPAO1) with E. coli strain SM10pir/ pCM639 (ISphoA/hah insertions) or SM10pir/pIT2 (ISlacZ/hah insertions). Mutagenized cells were selected by plating on LB agar containing tetracycline (60 μg/ml), chloramphenicol (10 μg/ml) for counterselection against the donor strain, and either 5-bromo-4-chloro-3-indolyl phosphate (XP) (40 μg/ml) for detection of active phoA fusions or 5-bromo-4-chloro-3-indolyl-β-d-galactoside (X-gal) (40 μg/ml) for detection of active lacZ fusions. After incubation for 2-3 days at room temperature, tetracycline-resistant colonies were picked by using a Qpix (Genetix, Hampshire, U.K.) colony-picking robot programmed to select white or blue colonies (both colors were picked and mapped). Colonies were arrayed into 384-well plates, each well containing 90 μl of freezing medium (10 g/liter tryptone/5 g/liter yeast extract/10 g/liter NaCl/6.3 g/liter K2HPO4/1.8 g/liter KH2PO4/0.5 g/liter sodium citrate/0.9 g/liter (NH4)2SO4/4.4% glycerol) supplemented with 20 g/ml tetracycline. Plates were incubated for 18 h at 37°C, then frozen and stored at -80°C.
Transposon Insertion Location. Transposon insertion locations were determined by a two-stage semidegenerate PCR and sequencing protocol (Supporting Methods and Table 4, which are published as supporting information on the PNAS web site). In the first round of PCR, a specific primer for the transposon sequence is paired with a semidegenerate primer with a defined tail. A 0.5-μl aliquot of thawed glycerol stock was added directly to the PCR reagents as template for the first round. In the second round of PCR, a nested transposon primer is paired with a primer targeted to the tail portion of the semidegenerate primer, and 0.5 μl of PCR product from the first round was used as template. PCR products from the second round were cleaned up by using exonuclease I and shrimp alkaline phosphatase (United States Biochemical), and used as sequencing templates. Sequencing was performed by using Big Dye Terminator version 3.0 (Amersham Pharmacia) and reactions were analyzed with ABI 3700 autosequencers (Applied Biosystems).
Automated sequence analysis was accomplished by using a PERL script that compiled assessment of phred quality scores (9), crossmatch (using the Smith-Waterman algorithm) to the PAO1 sequence, and data retrieval from the PAO1 annotation table.
Quality Control. Confirmation of the positions of a randomly selected subset of transposon insertions was determined by PCR. Custom primers to the PAO1 genome were designed oriented toward the 5′ end of the transposon. Gel electrophoresis of PCR products was used to determine whether the PCR produced a major product of the expected size. This method was able to confirm the presence of the mapped strain in >95% of wells, including some wells that were identified as containing multiple strains. These ”mixed” wells were more prevalent in replica plates than in the original source plates, and in all cases the mapped strain was retrievable from mixed wells.
Statistical Analysis. To assess the statistical properties of the observed set of insertion sites, we used a neutral-candidate model in which every gene is assumed nonessential and equally likely to be hit, and a neutral-base pair model, in which every base pair is assumed equally likely to define an insertion site. The neutral-candidate model was rejected because variation in gene size had a large effect on the number of times a gene was hit. In both models, the number of times an ORF is hit follows a multinomial distribution with parameters n, p1,..., pk, where n is the number of transposon insertions, pj is the probability of landing in the jth ORF, and k is the number of ORFs. pj was estimated as the length of the ORF divided by the total length of the genome. The bias corrected estimate of the number of essential genes is 377 with a standard deviation of 77.3. See Supporting Methods for a detailed description of the statistical methods used.
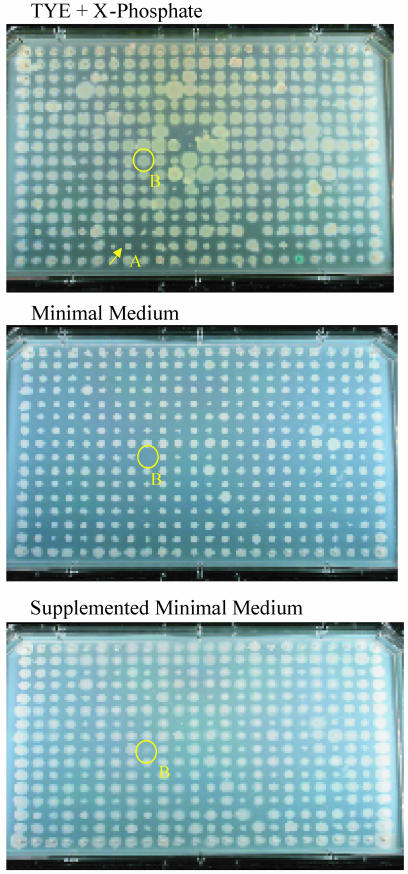
Mutant Phenotype Characterization. Twitching motility and prototrophic growth phenotypes were scored by replica printing the entire collection onto LB agar containing tetracycline and chromogenic indicator (identical to the original selection medium), Mops minimal agar, Mops minimal agar supplemented with amino acids, vitamins, purines and pyrimidines, and Pseudomonas isolation agar (PIA) (see Fig. 7). Strains were replicated by using a 384-pin plastic replicator (Genetix) or a metal replicator. The replicas were incubated for 2 days (37°C) and photographed by using high-resolution color digital imaging. Twitching motility was assessed by examining surface colony morphology from the images of the LB and supplemented minimal agar replicas; auxotrophs were scored by a comparison of growth on minimal and supplemented minimal agar. For both phenotypes, two independent blind scorings were carried out by different individuals. All potential mutants identified were included in the analysis.
Fig. 7.
Replica plating and phenotyping. One 384-well plate of phoA transposon-containing strains was replica plated onto three conditions: rich medium plus indicator (Top), minimal medium (Middle), and minimal medium plus supplemented nutrients (Bottom). (Top) Colony A (arrow) represents a twitch- phenotype; thus, the mutant is deficient in the gene pilY1, a type-4 fimbrial biogenesis protein. Colony B (circled) is an auxotrophic mutant (showing growth on LB and supplemented, but not minimal, media) and carries an argG (argininosuccinate synthetase) insertion.
Results
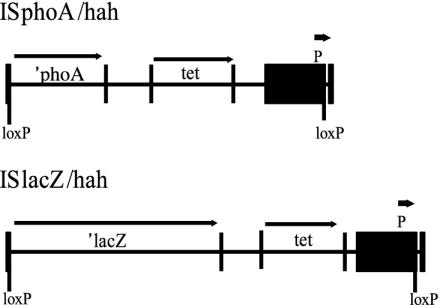
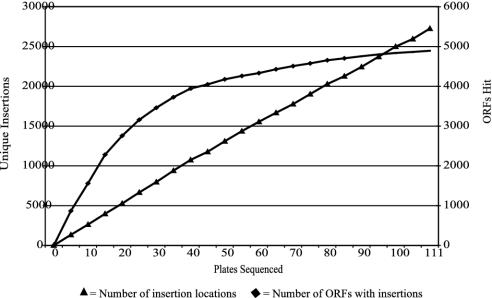
Mutant Production. A genome-wide random-insertion library was generated for the MPAO1 isolate of P. aeruginosa strain PAO1. Two different transposon Tn5 IS50L derivatives, ISphoA/hah (10) and ISlacZ/hah, were used to generate mutant strains (Fig. 1). Insertion of either transposon confers tetracycline resistance and leads to a blue colony phenotype on indicator medium when they are positioned in-frame in appropriately expressed genes. Tetracycline-resistant strains were arrayed in 384-well plates and assessed for transposon-insertion position and phenotype. Insertion locations were mapped by using PCR amplification and sequencing of the 5′ transposon boundary (10). A total of 42,240 mutant strains (110 plates of 384 individual mutants) were mapped, corresponding to 45,409 attempted sequencing reads, and 36,154 matches to the PAO1 genome, for an average success rate of 80% (Table 1). Elimination of exact-duplicate-insertion locations left 30,100 unique insertions, split evenly between the two transposons (15,063 ISphoA/hah insertions, 15,037 ISlacZ/hah insertions). Of the unique insertion locations, 27,263 were within predicted ORFs, corresponding to the 89% of the genome comprised of coding sequence (11). The distribution of hits among ORFs did not conform well to a Poisson distribution, but near-saturation was nonetheless achieved (Fig. 2). As expected, more hits occurred in larger ORFs (Fig. 3), and there was a larger-than-expected zero class. The average number of hits per ORF was 5.05, and was 5.75 among ORFs hit at least once.
Fig. 1.
Transposons used for insertion mutagenesis. Transposons ISphoA/ hah (4.83 kbp) and ISlacZ/hah (6.16 kbp) are derived from the IS50L element of transposon Tn5 and generate alkaline phosphatase (′phoA) or β-galactosidase (′lacZ) translational gene fusions if appropriately inserted in a target gene. An outward-facing neomycin phosphotransferase promoter is expected to reduce polar effects on downstream gene expression for appropriately oriented insertions. Cre-mediated recombination excises sequences situated between the loxP sites in each transposon, leaving a 63-codon insertion that encodes an influenza-hemagglutinin epitope and a hexahistidine metal-affinity purification tag (together referred to as ”hah,” see ref. 10). ′phoA, alkaline phosphatase gene; ′lacZ, β-galactosidase gene; tet, tetracycline resistance determinant; loxP, Cre recognition sequence; P, neomycin phosphotransferase promoter.
Table 1. Summary of the results of the transposon library.
| Data set | N |
|---|---|
| Mutants arrayed | 42,240 |
| Mapped insertion locations | 36,154 |
| Identical insertions | 4,423 |
| Unique insertion locations | 30,100 |
| Insertions inside ORFs | 27,263 |
| Insertions between ORFs | 2,837 |
| ORFs hit internally | 4,892 |
| ORFs never hit internally | 678 |
| Average hits per ORF | 5.05 |
| In-frame insertion locations | 4,823 |
| Blue colony in-frame insertions | 2,546 |
| ORFs with in-frame insertions | 2,582 |
| Mutants scored for colony phenotype | 42,240 |
| Twitchless mutants | 709 |
| ORFs with twitchless mutants | 360 |
| Auxotroph mutants | 813 |
| ORFs with auxotroph mutants | 546 |
Fig. 2.
Saturation transposon mutagenesis. A total of 110 384-well plates of transposon-containing strains of P. aeruginosa were analyzed for transposon insertion location. Insertions were mapped to 27,263 locations within ORFs with another 2,837 between ORFs. Of the 5,570 ORFs in the P. aeruginosa genome, 4,892 were hit at least once by a transposon insertion. The number of unique insertion locations increased linearly with new strains, whereas the number of ORFs hit approached a plateau.
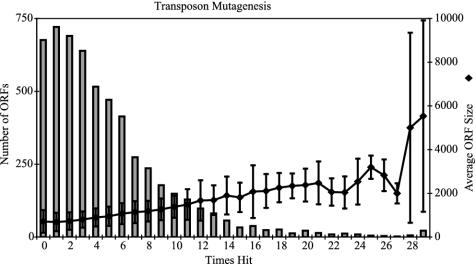
Fig. 3.
Distribution of transposon hits among ORFs. The number of ORFs for which a transposon insertion wasn't recovered was 678, and 721 were hit only once. The number of times an ORF was hit increases with ORF size. Error bars are one standard deviation in each direction.
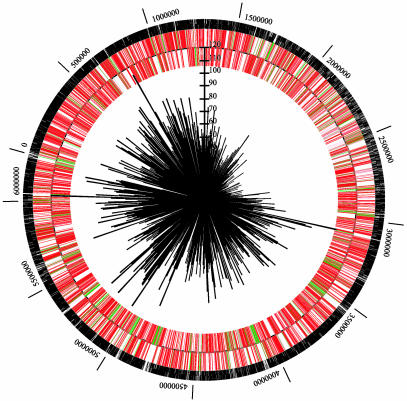
Candidate-Essential Genes. Transposon insertions were not recovered in 678 ORFs. Genes may have been missed either by chance, because of sequence-specific insertion rates, or because mutations are lethal. The number of genes missed is a small fraction of the total (12%); thus, it is likely that many of these genes are essential for growth on a rich medium. Genes without insertions, designated candidate-essential genes appear to be distributed randomly throughout the PAO1 genome (Fig. 4). Transposon insertion density is lowest in the area between coordinates 1.5 Mbp and 3 Mbp; the cause for this is unknown.
Fig. 4.
Distribution of transposon insertions and candidate-essential genes. The circular 6.2-Mbp P. aeruginosa genome was hit in 30,100 locations with individual transposon insertions. The black circular line represents the genome sequence with the origin of replication at coordinate zero. Bars outside the line represent genes transcribed clockwise, whereas those inside are transcribed counterclockwise. Red bars represent ORFs that contain transposon insertions, and green bars represent ORFs not hit (candidate-essential genes). Black marks on the outside of the circle represent transposon insertions. The sunburst pattern represents the number of insertions per 10,000 bp, with the scale extending from the center.
Several models were used to analyze the transposon insertion results. A neutral-candidate ORF model (i.e., each gene may be hit with equal probability) (12), was rejected after initial trials showed ORF size has an effect on the number of insertions. When a neutral-base pair model (each base pair position in the genome is hit with equal probability and the likelihood of each gene being hit is proportional to its size) is used, the expected number of missed ORFs is 307 with a standard deviation of 15.33 (see Materials and Methods). When the data from Fig. 2 are fitted to this model, it is predicted that a maximum of 5,206 ORFs may be hit, leaving 364 essential genes. From these analyses, we conclude that it is likely the actual number of essential genes in P. aeruginosa is between 300 and 400.
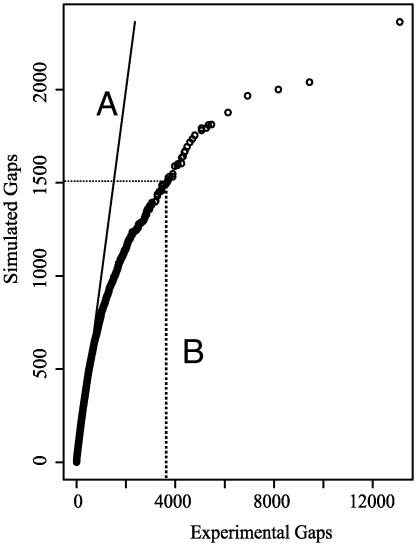
To investigate whether the transposon-insertion positions approximated a random distribution, observed gaps between insertions were compared with simulated gaps from a random positioning of an equal number of insertions in the genome (Fig. 5). From this analysis, it is clear that the deviation from a random distribution is significant, and is caused by numerous larger-than-expected gaps between transposon insertions. This result matches the observation that large gaps between insertions contain candidate-essential gene ”clusters.”
Fig. 5.
Quantile-quantile plot of transposon-insertion gap size distribution. The 30,100-transposon insertion locations in the 6.2-Mbp P. aeruginosa genome were compared with an equal number of random insertions in a simulated 6.2-Mbp genome. The x axis represents the observed size distribution of gaps (the distance between adjacent insertions) and the y axis represents the simulated distribution of gaps. Each point plots the same quantile for both distributions. Line A represents a 1:1 relationship, where the points would lie if the observed and the randomly generated data sets were identical. The size distribution of the observed gaps is significantly larger than that of the random data set. For example, an equal proportion of gaps fell below 3,800 bp in the observed data set as fell below 1,500 bp in the random data set (represented by line B).
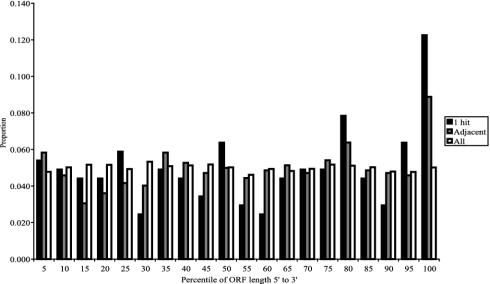
In addition to the 678 ORFs never hit, 721 ORFs were hit only once. We expect this class of ORFs to contain some essential genes whose functions were not fully disrupted by the insertion (a ”wounded” phenotype). Analysis of these single-hit ORFs showed that the hits were distributed approximately evenly throughout the ORFs, with a small bias toward the extreme 3′ of the gene. Of the 721 ORFs that were hit only once, 204 are also adjacent to candidate-essential genes. These ORFs were more highly biased toward insertions in their extreme 3′ end (Fig. 6). Hence, it is likely that some of the single-hit ORFs are essential and, more rarely, that ORFs were hit more than once (see below).
Fig. 6.
Hit distribution relative to position within ORF. The proportion of transposon insertions according to their relative position within ORFs is represented in a histogram. Hits are nearly evenly distributed (e.g., 5% of hits occur in the first 5% of ORFs) when all insertions are considered. For ORFs that were hit only once (dark gray), particularly those adjacent to ORFs never hit, the proportion of hits is highly skewed toward the 3′ end of the gene.
The deduced list of candidate-essential genes was compared with the overall PAO1 gene complement for functional representation. PAO1 genes have been grouped into 25 functional classes, with an additional class for unknown hypothetical genes (11). When the proportion of candidate-essential genes falling into each class was compared with the whole genome, several categories were highly over- or underrepresented (Table 2). ORFs found most commonly in the list of candidate-essential genes included translation machinery and cell-division control genes (overrepresented by 3.5-4 times), whereas underrepresented categories included chemotaxis and two-component regulatory systems.
Table 2. Comparison of the proportion of genes (sorted by function) represented in the list of candidate essentials, versus their proportions in the P. aeruginosa genome.
| Primary function | No. of genes | No. not hit | Relative representation among candidate essentials |
|---|---|---|---|
| Translation, posttranslational modification, degradation | 149 | 75 | 4.14 |
| Cell division | 26 | 11 | 3.48 |
| Cell wall/lipopolysaccharide/capsule | 86 | 30 | 2.87 |
| Biosynthesis of cofactors, prosthetic groups, and carriers | 132 | 44 | 2.74 |
| Transcription, RNA processing, and degradation | 45 | 13 | 2.37 |
| Fatty acid and phospholipid metabolism | 57 | 15 | 2.16 |
| Nucleotide biosynthesis and metabolism | 60 | 14 | 1.92 |
| Energy metabolism | 170 | 35 | 1.69 |
| Protein secretion/export apparatus | 84 | 16 | 1.56 |
| DNA replication, recombination, modification, and repair | 81 | 15 | 1.52 |
| Chaperones and heat shock proteins | 52 | 8 | 1.26 |
| Adaptation and protection | 66 | 9 | 1.12 |
| Related to phage, transposon, or plasmid | 62 | 8 | 1.06 |
| Hypothetical, unclassified, unknown | 2,381 | 261 | 0.90 |
| Central intermediary metabolism | 65 | 7 | 0.88 |
| Membrane proteins | 43 | 4 | 0.76 |
| Amino acid biosynthesis and metabolism | 151 | 13 | 0.71 |
| Secreted factors (toxins, enzymes, alginate) | 60 | 5 | 0.68 |
| Carbon compound metabolism | 134 | 11 | 0.67 |
| Transcriptional regulators | 403 | 27 | 0.55 |
| Putative enzymes | 457 | 25 | 0.45 |
| Antibiotic resistance and susceptibility | 19 | 1 | 0.43 |
| Transport of small molecules | 559 | 26 | 0.38 |
| Motility and attachment | 67 | 2 | 0.25 |
| Two-component regulatory systems | 116 | 3 | 0.21 |
| Chemotaxis | 45 | 0 | 0.00 |
| Total | 5,570 | 678 |
Overall Sequence Conservation of Pseudomonas Candidate-Essential Genes. It is generally observed that significant overlap exists among sets of essential genes in genomes of Gram-negative bacteria. To examine the overlap between sets of essential genes in genomes of PAO1 and E. coli, ORF translations from these two genomes were compared by using mass BLASTP analyses. Candidate-essential and candidate-nonessential genes from this work were compared with the list of known essential and nonessential E. coli genes in the PEC database (www.shigen.nig.ac.jp/ecoli/pec/index.jsp) (Table 3). A total of 215 ORFs from PAO1 have a strict ortholog in the list of known essential genes from E. coli. A majority of these orthologues (133 ORFs) are on our list of candidate-essential genes.
Table 3. Homology of PAO1 and E. coli candidate-essential genes.
| Gene class query | Gene class subject | Strict orthologues |
|---|---|---|
| PAO1 ORFs (5,570) | E. coli known essential genes | 215 |
| PAO1 candidate-nonessential genes (4,892) | E. coli known essential genes | 82 |
| PAO1 candidate-essential genes (678) | E. coli known essential genes | 133 |
| E. coli known essential genes (232) | PAO1 candidate-essential genes | 135 |
Comparison of predicted translations of PAO1 ORFs to E. coli. Strict orthologues were defined as the top BLAST hit that had a match length of at least 75% of the length of the query sequence.
The median percent position (5′ to 3′) of insertions in PAO1 ORFs only hit once was 49.1%, but was 86.7% for those that had a match to an E. coli known-essential gene. This observation suggests that we did recover mutations in some essential genes, but that the positions of these insertions were strongly biased toward the extreme 3′ end of the genes. The fraction of apparently wounded genes (i.e., genes have one or more hits, but none in the first 90% of their ORFs, a total of 109 ORFs) from PAO1 that have homology to known essential genes was similar to the fraction in our set of candidate-essential genes (21% or 23 ORFs). We consider these ORFs additional candidate-essential genes. These results match the mathematical modeling prediction that approximately half of our candidate-essential genes are truly nonessential.
Mutant Phenotypes. To examine the use of the strain collection for mutant identification, we screened for loss of surface (”twitching”) motility and inability to grow on minimal medium (auxotrophy) (Fig. 7). These ”reference” phenotypes were chosen because their genetic bases are well characterized; hence, we could measure our recovery rate of mutants relative to previous mutational analyses. Twitching motility, which is a pilus-based process dependent on 34 identified genes, leads to the production of colonies with a distinctive lacy edge (13). In the screen for twitching defects, we identified mutations in or immediately upstream to all of the previously identified genes except pilZ, (Table 5, which is published as supporting information on the PNAS web site). However, an insertion near the 3′ end of a functionally unrelated gene immediately upstream of pilZ did cause a nontwitching phenotype, perhaps because of a polar effect on pilZ expression. These results confirm excellent correlation between phenotype and genotype. However, numerous insertions in the known twitching motility genes were not detected (Table 5). In the 33 genes known to affect surface motility for which we had at least one transposon mutation, 12.5-100% of the insertions caused a twitch- phenotype. The majority of ”missed” mutants were in wells containing a mixture of (genetically stable) twitch+ and twitch- cells. The occurrence of mixed populations in wells was observed to increase through rounds of replica plating, although quality control experiments were able to identify the sequenced strain in most cases. Even low-level contamination with twitch+ cells is expected to obscure the sharp colony edge that defines the twitch- phenotype. For one gene in which mutants were detected inefficiently (pilS), further analysis revealed that those undetected alleles we examined had ”leaky” phenotypes. Insertions in four genes (chpB-chpE) implicated in surface motility (14) were not associated with significant motility defects. Overall, in the mutant collection, twitchless-phenotype-producing hits occurred in 366 ORFs, 80 of which were confirmed by a second hit. An additional 16 produced a phenotype in their only hit. Of the 80 ”confirmed-twitch-” ORFs, 26 were among the previously known ORFs. An additional 31 of these ORFs had been functionally annotated as ”mobility and attachment” genes, and another 23 had no functional annotation.
Intergenic transposon insertions at 39 unique locations produced a twitch- phenotype. Of those, seven were adjacent to genes that produced a twitch- phenotype when hit internally, and another five were in two large intergenic spaces. The observation that these two large intergenic spaces produced twitch- phenotypes each time they were hit suggests that genetic information essential for twitching exists there, albeit in sequences that provide no immediate clues to function.
Insertions resulting in auxotrophic phenotypes were distributed among 546 ORFs. Of those, 110 ORFs that had two or more unique hits that resulted in the same phenotype were considered ”confirmed auxotrophs.” There were also 21 ORFs in which the only hit produced an auxotrophic phenotype. ”Auxotroph” ORFs were 10- and 6-fold overrepresented in amino acid biosynthesis and metabolism and nucleotide biosynthesis and metabolism genes, respectively. Auxotrophic phenotypes resulted from 58 intergenic hits, all but one of which were adjacent to ORFs that had produced an auxotrophic phenotype at least once.
For the arginine, histidine, isoleucine-valine, leucine, and tryptophan biosynthetic pathways, we compared our results with previous data (Table 6, which is published as supporting information on the PNAS web site). Of the 36 genes known to be required for these five pathways, mutations in all except one (hisE, the shortest of the 36 ORFs) were represented in our collection. Mutants with insertions in 30 of the genes were detected as auxotrophs. In the other five cases, the existence of redundant genes appears to explain our failure to find insertions with auxotrophic phenotypes (Table 6). For three histidinebiosynthetic genes, the recovery of auxotrophic insertions in only one of two paralogues implies that these genes (hisC1, hisF1, and hisH1) are primarily responsible for biosynthesis.
Discussion
The genome sequence and associated annotation information for the PAO1 strain of P. aeruginosa have been used to facilitate high-throughput generation of a comprehensive mutant library. By using random transposon-insertion mutagenesis, nearly 90% of the ORFs in the PAO1 genome have been disrupted at least once. This mutant collection is both a significant resource for future research and a source of immediate functional insight into the genome of P. aeruginosa.
Several large mutant collections have provided significant data with regard to mutant phenotypes and essential genes. Our transposon-insertion library is distinct in providing virtually complete coverage of the genome with a set of strains archived in a format that facilitates mutant retrieval and phenotypic analysis. Archived collections of deletion mutants (S. cerevisiae) and inactivating-insertion mutants (B. subtilis) have been created by using gene-by-gene knockout strategies that required the participation of large consortia of laboratories (2). More cost-effective high-throughput technologies, developed here for the analysis of P. aeruginosa, should be directly applicable to a wide variety of bacterial species.
Development of therapeutic agents may be directed by a comprehensive understanding of all of the gene products individually required for survival of P. aeruginosa. By generating a near-saturation-mutant collection, we have arrived empirically at a list of 678 candidate-essential genes. Many of the candidate-essential genes are expected (i.e., they code for nonredundant machinery central to cell survival), whereas others are new, including the 263 previously unclassified ORFs that are on the candidate-essential gene list. Our statistical and bioinformatic analyses predict that approximately half of the genes on our list are truly essential.
To test the suitability of the P. aeruginosa strain collection for direct phenotypic screening, we identified mutants defective in two previously studied traits (twitching motility and prototrophic growth). In both cases, we identified nearly all genes expected from the earlier studies. Several previously undescribed genes apparently required for twitching motility were also identified. These initial tests show that phenotypic screens of the entire collection are feasible and may produce essentially complete lists of candidate genes implicated in biological processes of interest. Furthermore, the existence of multiple alleles for most genes in our mutant collection meant that low-level contamination did not undermine the phenotypic analysis.
The potential utility of the library extends beyond screens for new mutants. Because the strains in the collection may be readily retrieved, the effects of mutations in any gene can be studied by a ”reverse-genetic” strategy. When a gene attracts interest on the basis of bioinformatic or functional genomic analysis, appropriate phenotype tests can be immediately pursued. In addition, the properties of the transposons used to generate the mutant set should facilitate downstream manipulations. Deletion of transposon sequences by site-specific (cre-loxP) recombination eliminates the tetracycline-resistance determinant and facilitates constructing double mutants by using other insertions in the collection. Such double mutants may be useful for epistasis analysis or in engineering genomes with deletions between two insertion sites (15). In-frame insertions of the transposons generate reporter-gene fusions that may be used to study expression. Such fusions may also be readily converted into derivatives carrying internal epitope/affinity purification tags for analysis of unfused polypeptides (10).
Infections with P. aeruginosa are the leading cause of death in cystic fibrosis patients, and also lead to several other clinically important infections. The development of new therapies for these infections will be challenging because of the complex biology of P. aeruginosa. The comprehensive mutant library we have constructed will allow an accelerated genetic dissection of traits such as metabolic flexibility and inherent drug resistance that make P. aeruginosa such a tenacious pathogen.
Supplementary Material
Acknowledgments
We thank Peter Chapman, David D'Argenio, Larry Gallagher, Arnold Kas, and Doug Passey for technical assistance and advice. We also acknowledge Kenneth Stover and members of his group for sharing unpublished information about a transposon-mutagenesis project on P. aeruginosa that they initiated at Pathogenesis, Inc. (subsequently acquired by Chiron). This work is supported by the Cystic Fibrosis Foundation Grants JACOBS03F0 and MILLER00V0.
References
- 1.Giaever, G., Chu, A. M., Ni, L., Connelly, C., Riles, L., Veronneau, S., Dow, S., Lucau-Danila, A., Anderson, K., Andre, B., et al. (2002) Nature 418, 387-391. [DOI] [PubMed] [Google Scholar]
- 2.Kobayashi, K., Ehrlich, S. D., Albertini, A., Amati, G., Andersen, K. K., Arnaud, M., Asai, K., Ashikaga, S., Aymerich, S., Bessieres, P., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 4678-4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berg, C. M. & Berg, D. E. (1995) in Mobile Genetic Elements, ed. Sherratt, D. (Oxford Univ. Press, Oxford), pp. 38-68.
- 4.Kumar, A., des Etages, S. A., Coelho, P. S., Roeder, G. S. & Snyder, M. (2000) Methods Enzymol. 328, 550-574. [DOI] [PubMed] [Google Scholar]
- 5.Ross-Macdonald, P., Coelho, P. S., Roemer, T., Agarwal, S., Kumar, A., Jansen, R., Cheung, K. H., Sheehan, A., Symoniatis, D., Umansky, L., et al. (1999) Nature 402, 413-418. [DOI] [PubMed] [Google Scholar]
- 6.Gerdes, S. Y., Scholle, M. D., D'Souza, M., Bernal, A., Baev, M. V., Farrell, M., Kurnasov, O. V., Daugherty, M. D., Mseeh, F., Polanuyer, B. M., et al. (2002) J. Bacteriol. 184, 4555-4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lehoux, D. E., Sanschagrin, F. & Levesque, R. C. (2002) FEMS Microbiol. Lett. 210, 73-80. [DOI] [PubMed] [Google Scholar]
- 8.Akerley, B. J., Rubin, E. J., Novick, V. L., Amaya, K., Judson, N. & Mekalanos, J. J. (2002) Proc. Natl. Acad. Sci. USA 99, 966-971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ewing, B., Hillier, L., Wendl, M. C. & Green, P. (1998) Genome Res. 8, 175-185. [DOI] [PubMed] [Google Scholar]
- 10.Bailey, J. & Manoil, C. (2002) Nat. Biotechnol. 20, 839-842. [DOI] [PubMed] [Google Scholar]
- 11.Stover, C. K., Pham, X. Q., Erwin, A. L., Mizoguchi, S. D., Warrener, P., Hickey, M. J., Brinkman, F. S., Hufnagle, W. O., Kowalik, D. J., Lagrou, M., et al. (2000) Nature 406, 959-964. [DOI] [PubMed] [Google Scholar]
- 12.Johnson, N. L. & Kotz, S. (1977) Urn Models and Their Application: An Approach to Modern Discrete Probability Theory (Wiley, New York).
- 13.Semmler, A. B., Whitchurch, C. B. & Mattick, J. S. (1999) Microbiology 145, 2863-2873. [DOI] [PubMed] [Google Scholar]
- 14.Mattick, J. S. (2002) Annu. Rev. Microbiol. 56, 289-314. [DOI] [PubMed] [Google Scholar]
- 15.Yu, B. J., Sung, B. H., Koob, M. D., Lee, C. H., Lee, J. H., Lee, W. S., Kim, M. S. & Kim, S. C. (2002) Nat. Biotechnol. 20, 1018-1023. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.