Abstract
Insertion of lysine during protein synthesis depends on the enzyme lysyl-tRNA synthetase (LysRS), which exists in two unrelated forms, LysRS1 and LysRS2. LysRS1 has been found in most archaea and some bacteria, and LysRS2 has been found in eukarya, most bacteria, and a few archaea, but the two proteins are almost never found together in a single organism. Comparison of structures of LysRS1 and LysRS2 complexed with lysine suggested significant differences in their potential to bind lysine analogues with backbone replacements. One such naturally occurring compound, the metabolic intermediate S-(2-aminoethyl)-l-cysteine, is a bactericidal agent incorporated during protein synthesis via LysRS2. In vitro tests showed that S-(2-aminoethyl)-l-cysteine is a poor substrate for LysRS1, and that it inhibits LysRS1 200-fold less effectively than it inhibits LysRS2. In vivo inhibition by S-(2-aminoethyl)-l-cysteine was investigated by replacing the endogenous LysRS2 of Bacillus subtilis with LysRS1 from the Lyme disease pathogen Borrelia burgdorferi. B. subtilis strains producing LysRS1 alone were relatively insensitive to growth inhibition by S-(2-aminoethyl)-l-cysteine, whereas a WT strain or merodiploid strains producing both LysRS1 and LysRS2 showed significant growth inhibition under the same conditions. These growth effects arising from differences in amino acid recognition could contribute to the distribution of LysRS1 and LysRS2 in different organisms. More broadly, these data demonstrate how diversity of the aminoacyl-tRNA synthetases prevents infiltration of the genetic code by noncanonical amino acids, thereby providing a natural reservoir of potential antibiotic resistance.
The aminoacyl-tRNA synthetases (aaRSs) are a ubiquitous family of essential enzymes responsible for producing aminoacyl-tRNAs for ribosomal protein synthesis (1). Each aaRS is able to charge with great specificity a particular set of tRNAs with their corresponding amino acids, effectively dictating the genetic code. Extensive structural and functional studies have shown that the 20 canonical aaRSs are divided into two unrelated groups, class I and class II, each with 10 members and characterized by the presence of distinct active site structural motifs (2-4). The placement of a synthetase of particular substrate specificity into one class or the other has been almost completely conserved throughout the living kingdom, indicating that the aaRSs evolved early (5). To date, only one widespread exception to this class separation has been noted, namely the lysyl-tRNA synthetases (LysRSs) that are found both as class I (LysRS1) and class II (LysRS2) proteins (6, 7), although no corresponding division among lysine-specific tRNAs was detected (8). Comparative genomic analyses have shown that LysRS2 is encoded in all sequenced eukaryotic genomes, most bacteria, and only a few archaea, whereas LysRS1 is found in the vast majority of sequenced archaea and a scattering of bacteria (9). Of relevance to this study is the finding that Bacillus subtilis encodes a single copy of LysRS2. LysRS1 and LysRS2 are almost never found together, with organisms generally containing one or the other, presumably as a result of both selective retention and horizontal gene transfer (9). The only well documented example of the coexistence of LysRS1 and LysRS2 is in the Methanosarcineae, where they function together to aminoacylate the specialized tRNAPyl suppressor species (10).
The use of structurally unrelated but functionally analogous forms of LysRS by different organisms raises the question of whether one form of the protein offers a selective advantage over another form under certain conditions. Previous investigations identified recognition of divergent tRNALys sequences by LysRS1 and LysRS2, but it is unclear whether these differences would be sufficient by themselves to explain the selection of one form of the protein over another (11-13). Structural studies of LysRS1 and LysRS2 complexed with l-lysine indicate that the two forms of the protein might also differ significantly in their ability to bind lysine analogues. In particular, the lysine-binding site of LysRS1 is more compact than that of LysRS2, indicating that it may be less able to accommodate lysine analogues with backbone substitutions (14). One such compound of particular interest is the metabolic intermediate S-(2-aminoethyl)-l-cysteine (thialysine), a naturally occurring proteinogenic lysine analogue (15-19). Thialysine is a sufficiently good mimic of lysine that it can effectively prevent cellular growth by incorporation into protein via LysRS2 (20-22). The potential reduction in thialysine incorporation offered by the use of LysRS1 suggests that it might provide an effective means to restrict the analogue's detrimental effects. The ability of a synthetase to prevent addition of a noncanonical amino acid to the genetic code was previously observed for valyl-tRNA synthetase, whose editing activity excludes aminobutyrate (23). We set out to determine whether the existence of two nonorthologous forms of LysRS might be explained by the need to prevent addition of noncanonical amino acids to the genetic code. Broader implications of our study relating to mechanisms of resistance against aaRS-targeted therapeutic agents are also discussed.
Materials and Methods
Bacterial Strains and Growth Conditions. B. subtilis strain 168 was used in this study. Cells were routinely grown aerobically in LB media or Spizizen's minimal media at 37°C (24). B. subtilis (25) and Escherichia coli TG1 (26) were transformed as described. Strains and plasmids used in this study are described in Table 2, which is published as supporting information on the PNAS web site, and oligonucleotides are described in Table 3, which is published as supporting information on the PNAS web site. Xylose was added to cultures as indicated at 1% wt/vol. All new constructs were sequenced in E. coli before introduction into B. subtilis, and stocks were used regularly to prevent suppressor mutations arising.
Plasmid and Strain Construction. Diagrams of plasmid and strain constructions are presented in Figs. 6-9, which are published as supporting information on the PNAS web site. To construct strain B. subtilis BCJ118.49 (Fig. 6), a B. subtilis chromosomal fragment containing the 3′ part of yacF and the yacF-lysS intergenic region (with an optimized ribosome binding site, generated by using the primer pair 7F/5 and 7R/2) was ligated to a fragment containing the Borrelia burgdorferi lysK structural gene (generated by using the primer pair BbF and BbR), and the ligation mix was used as a template to assemble both fragments (using the primer pair 7F/5 and BbR). This fragment was cloned into pBCJ102 to give pBCJ103.7, which was then integrated into the B. subtilis chromosome by a Campbell-type event to give strain BCJ108.3, thereby placing the lysK gene under the control of the endogenous lysS expression signals. The lysS gene down-stream of lysK was then excised by gene replacement using linearized pBCJ113.5 to give strain BCJ118.49. To construct B. subtilis strain BCJ143.3 (Fig. 7), the Staphylococcus aureus strain N315 lysS gene was amplified by using primer pair 12F and 12R, and the resultant fragment was cloned into NdeI-digested pBCJ103.7, thereby replacing the lysK structural gene to give pBCJ121.65, which was then integrated into the chromosome by a Campbell-type event to give strain BCJ142.1. The endogenous lysS gene was excised from the chromosome by using linearized pBCJ113.15, giving strain BCJ143.3. To construct B. subtilis strain BCJ140.1 (Fig. 8) that places lysK on the chromosome under xylose-inducible expression at the amyE locus, the ClaI/ NotI fragment from pBCJ103.7 was inserted into the BamHI site of pX to give pBCJ134.1, which was then linearized and integrated into the chromosome by a double cross-over event, yielding strain BCJ140.1. To construct B. subtilis strain BCJ157.1 (Fig. 9) that places lysK on the chromosome under PrpsD expression control, the B. subtilis rpsD promoter and terminator regions were amplified by using primer pairs RpsF/1-RpsR/1 and RpsF/2-RpsR/2.2, respectively and ligated, and the fragment was cloned into pDIA5304, yielding plasmid pBCJ164.3. The lysK gene was excised from pBCJ103.7 on an NdeI fragment and inserted into the NdeI site of pBCJ164.3, giving plasmid pBCJ122.3. This PrpsD-lysK expression cassette was then excised on a BamHI fragment and cloned into BamHI-digested pDG268, giving plasmid pBCJ137.1, which was then linearized with ScaI and integrated into the B. subtilis chromosome by a double cross-over at the amyE locus, giving strain BCJ141.1. The endogenous lysS gene of BCJ141.1 was excised by gene replacement using linearized pBCJ144.3 to give strain BCJ157.1. The tRNALys1 gene from B. burgdorferi was expressed in B. subtilis by using the expression system described (27). The tRNALys1 gene was amplified from B. burgdorferi DNA by using the primer pair 14F-14R and cloned into pGEMtRNQ to give plasmid pBCJ125.1. A SalI/HinDIII fragment from pBCJ125.1 was isolated and cloned into pBCJ202 (pDG148Δkan) to give plasmid pBCJ203, which was then established as a replicating plasmid in B. subtilis.
Urea Acid Gel Electrophoresis. Exponential growth-phase cells were harvested, and the cell pellet was resuspended in 0.3 ml of 0.3 M sodium acetate, 10 mM EDTA, pH 4.5, mixed with 0.3 ml of glass beads, and snap-frozen in an ethanol/dry ice bath. Phenol/chloroform (0.3 ml., pH 4.7) was added to each tube, the cells were disrupted by vortexing (4 × 30-s pulses) and centrifuged, the aqueous layer was collected, and this extraction was repeated. The RNA was then ethanol-precipitated, the pellet was resuspended in 60 μl of 0.3 M sodium acetate (pH 4.5) and reprecipitated by ethanol, and the pellet was air-dried on ice. The RNA was resuspended in 50 μl of 10 mM sodium acetate and quantitated spectrophometrically. Either 2 μg (B. subtilis probed) or 20 μg (B. burgdorferi probed) of total RNA was fractionated on a 14% polyacrylamide gel (61 mm × 82-mm gel) with 0.3 M sodium acetate and 7 M urea as gel buffer and 0.3 M sodium acetate buffer, pH 5.0 as running buffer by using the Bio-Rad Mini Protean 3 apparatus. Gels were run at 50 V for 24hat4°C, and the electrophoresis buffer was changed every 7 h to maintain an acid pH. The separated RNA samples were electroblotted onto positively charged 0.45 μM Biodyne B nylon membranes (Pall) in buffer (10 mM Tris-acetate, pH 7.8/5 mM sodium acetate/0.5 mM EDTA) at 44 V for 2 h at 4°C and crosslinked to the membrane by UV. Oligonucleotide probes complementary to nucleotides 26-51 of B. subtilis tRNALys (oligonucleotide 24) and nucleotides 26-51 of B. burgdorferi tRNALys1 (oligonucleotide 23) were labeled with ≈30- and 2-bp tails, respectively by using the DIG Oligonucleotide Tailing Kit (Roche Diagnostics). Filters were prehybridized, hybridized, and washed according to the manufacturer's instructions, except that salmon sperm DNA was added to the prehybridization buffer. Deacylation of tRNA samples was effected by incubation for 30 min at 70°C after addition of an equal volume of 0.1 M Tris, pH 9.5 and 0.1 M NaCl.
Immunoblot Analysis. Cell lysates of exponential growth-phase cultures were prepared and separated on 10% SDS/PAGE gels according to the method of Laemmli (28). Protein quantification was performed by using the Bio-Rad protein assay according to the manufacturer's instructions. LysRS1 from B. burgdorferi was detected by Western blotting using a primary mouse antibody (supplied by M. Theisen, State Serum Institute, Copenhagen). Chemiluminescent detection was carried out by using the Roche DIG Luminescent Detection Kit according to the manufacturer's instructions.
Antibiotic Disk Assay. Sixty milligrams of thialysine was dried onto an antimicrobial susceptibility test disk (Oxoid, Hampshire, U.K.) and placed on LB agar. Cooled molten LB agar was inoculated with 200 μl of an overnight culture of appropriate strains and poured onto the LB agar surface to just cover the test disk. Zones of clearance and inhibition were measured after overnight incubation at 37°C.
Cell-Free Protein Extracts. B. subtilis strains were grown overnight in LB at 37°C and harvested by centrifugation at 5,000 g for 10 min at 4°C. The cells were resuspended in 1 vol of 100 mM Tris·HCl (pH 7.5), 20 mM MgCl2, 1 mM EDTA, and a protease inhibitor mixture (Roche Molecular Biochemicals). Proteins were extracted by sonication of the cells at 4°C. S30 extract was prepared by centrifugation at 30,000 × g for 30 min at 4°C.
Unfractionated tRNA from B. subtilis. S30 extract was treated with phenol (vol/vol) under agitation for 2 h at 4°C. The aqueous phase was then extracted twice with phenol (vol/vol) and once with chloroform (vol/vol) before addition of 1 M NaCl. Large nucleic acids molecules were precipitated with 20% isopropanol at 4°C for 4 h. Smaller RNA and tRNA were precipitated by adjustment of the concentration of isopropanol to 60%, and the pellet was washed with 70% ethanol, dried, and resuspended in water.
LysRS-Specific Activity. The standard aminoacylation mixture (50-300 μl) contained 100 mM Na-Hepes (pH 7.2), 30 mM KCl, 2 mM ATP, 10 mM MgCl2, 10 μM l-[14C]-Lys (450 cpm/pmol), 3.8 or 5.9 mg/ml unfractionated tRNA, and 0.6-2.5 mg/ml S30 protein crude extract from B. subtilis. If necessary, S30 extract was diluted in 100 mM Hepes-Na (pH 7.2), 0.1 mg/ml BSA, 5 mM 2-mercaptoethanol, 0.1 mM EDTA, and 50% glycerol. Reactions were conducted at 37°C, and the [14C]-Lys-tRNA synthesized after 1-30 min was determined in 15-μl aliquots as described (29).
Aminoacylation Assays. The E. coli lysS-encoded LysRS2 was produced as an intein fusion protein and subsequently purified to electrophoretic homogeneity (S. Ataide and M.I., unpublished results). B. burgdorferi LysRS1 and transcript corresponding to B. burgdorferi tRNALys1 were prepared as described (11). Aminoacylation conditions for LysRS1 and LysRS2 were as before (13), except that thialysine (Sigma) was added during determination of Kis. The direct attachment of thialysine to in vitro-transcribed tRNA Lys-1 was monitored by 32P labeling of the tRNA using E. coli CCA-adding enzyme (30), followed by aminoacylation and product visualization as described (31).
Results
Expression of LysRS1 from B. burgdorferi in B. subtilis. The lysS gene in the B. subtilis genome encodes a class II-type LysRS (LysRS2). The lysS gene is essential, as demonstrated by our inability to inactivate it by insertional mutagenesis, and by the finding that when its expression is placed under the control of the Pspac promoter, growth of the strain becomes isopropyl β-d-thiogalactoside dependent (data not shown). We sought to establish whether the LysRS1 [encoded by the lysK gene (BB0659)] from B. burgdorferi could complement the endogenous LysRS2 of B. subtilis. The heterologous lysK gene was expressed in B. subtilis by using promoters of varying strengths. In strain BCJ118.49 it was expressed from the endogenous lysS expression signals, in strain BCJ140.1 it was expressed by using the xylose-inducible Pxyl promoter, and in strain BCJ157.1 it was expressed by using the rpsD promoter. The production of LysRS1 is shown in Fig. 1. A single band migrating with the expected Mr and comigrating with a band of similar Mr in the B. burgdorferi lysate was observed in strain BCJ118.49, and this band was absent from B. subtilis strain 168. It was found that the endogenous lysS gene could be deleted only from strain BCJ157.1, showing that the class I LysRS1 from B. burgdorferi can complement the endogenous LysRS activity but only when expressed from a significantly stronger promoter. Under such conditions, cellular LysRS-specific activity is broadly comparable to that of the parental strain (Table 1). As a control for this nonorthologous gene replacement, the lysS gene from Staphylococcus aureus was expressed in B. subtilis by using the endogenous lysS expression signals (strain BCJ143.3, constructed by using the strategy used for strain BCJ118.49). In this case the endogenous lysS gene could be deleted, showing that the orthologous replacement of the B. subtilis LysRS2 with a LysRS2 from S. aureus can be achieved when the heterologous LysRS is expressed by using the normal endogenous lysS expression signals, despite the lower specific activity of the foreign protein (Table 1). These data show that LysRS2 from B. subtilis can be replaced both orthologously and nonorthologously with LysRSs from S. aureus and B. burgdorferi, respectively.
Fig. 1.
Quantitation of LysRS1 protein levels by immunoblot analysis. Aliquots of total cell protein were separated by SDS/PAGE, transferred to membranes, and detected by using an anti-LysRS1 polyclonal antibody. The amounts of total cell protein loaded were as follows: B. subtilis strain 168, 20 μg; strain BCJ118.49, 20 μg; strain BCJ140.1 without xylose inducer, 20 μg; strain BCJ140.1 with xylose inducer, 1 μg; and strain BCJ157.1, 0.125 and 0.25 μg. The amount of B. burgdorferi total cell lysate loaded was 3 μg. Note that the level of LysRS1 present in this bacterium is significantly higher than that found in strain BCJ118.49 or strain BCJ140.1 (induced).
Table 1. Lysyl-tRNA synthesis-specific activity in B. subtilis cell-free extracts.
| B. subtilis strain | Lysyl-tRNA synthesis-specific activity, pmol of lysine attached/min per mg of total protein |
|---|---|
| 143.3 | 91 ± 5 |
| 157.1 | 272 ± 20 |
| 168 | 484 ± 25 |
Characterization of Strains BCJ143.3 and BCJ157.1 and Comparison with B. subtilis Strain 168. To investigate the effects of heterologous LysRS on the growth of strains BCJ143.3 (containing LysRS2 from S. aureus) and BCJ157.1 (containing LysRS1 from B. burgdorferi), generation times were established for growth in LB and minimal medium. Reduced tRNALys charging during growth on minimal medium would not be expected to affect lysine biosynthesis, which is directly regulated by lysine itself (32). The generation times of WT B. subtilis strain 168 grown in LB and minimal medium are 16 and 110 min, respectively. For strain BCJ143.3 grown in LB the generation time increases to 22 min (37% increase over B. subtilis strain 168) and increases to 131 min during growth in minimal medium (20% increase). Interestingly, for strain BCJ157.1 grown in LB medium, the generation time is 27 min (65% longer than that for B. subtilis strain 168), whereas the generation time for the same strain grown in minimal medium is 93.7 min, slightly lower than that observed for B. subtilis strain 168 grown under these conditions. These data show that orthologous replacement of LysRS2 of B. subtilis with LysRS2 from S. aureus adversely affects the growth rate, with a more pronounced effect observed when cells are growing in rich medium. However, when a nonorthologous replacement of LysRS2 of B. subtilis with LysRS1 from B. burgdorferi is made, the generation time of cells grown in rich medium is adversely affected, whereas cell growth is marginally enhanced when cells are grown in minimal medium.
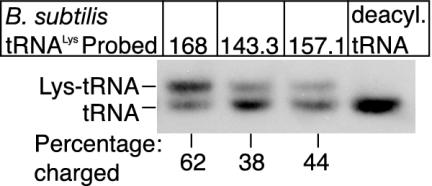
Charging of tRNALys in Vivo by Class I and Class II LysRSs. To directly demonstrate in vivo charging of B. subtilis tRNALys by LysRS2 from S. aureus and LysRS1 from B. burgdorferi, RNA was prepared from exponentially growing cultures of strains BCJ143.3 and BCJ157.1 and separated by gel electrophoresis under conditions that distinguish the acylated and nonacylated tRNA species. The RNA was then transferred to a membrane and probed with oligonucleotides specific for B. subtilis tRNALys and B. burgdorferi tRNALys1. A representative electropherogram (Fig. 2) shows only two hybridizing bands in each strain. The lower band is nonacylated tRNALys, determined by loading RNA samples that had been treated under conditions that deacylate tRNA (Fig. 2, deacyl.tRNA), whereas the upper band is acylated tRNALys. It is evident that the LysRS2 from S. aureus and LysRS1 from B. burgdorferi both can charge tRNALys from B. subtilis. However, the level of tRNALys charging differs between the strains. Approximately 62% of tRNALys is acylated in exponentially growing B. subtilis strain 168 cells. However, the level of charged tRNALys in strain BCJ143.3 is reduced to ≈38%, whereas the level of B. subtilis tRNALys charged by B. burgdorferi LysRS1 is slightly higher at ≈44%. These lower levels of charged tRNALys in strains BCJ143.3 and BCJ157.1 are consistent with the increased generation times observed for both strains grown under these conditions.
Fig. 2.
Determination of the B. subtilis tRNALys charging level by using Northern analysis. Two micrograms of total RNA from each of the strains B. subtilis 168, BCJ143.3, and BCJ157.1 grown on rich (LB) media, was separated by electrophoresis, transferred to membrane, and probed with a B. subtilis-specific tRNALys probe as detailed in Materials and Methods. Two micrograms of an RNA sample similarly prepared but treated under deacylation conditions was also loaded (deacyl.tRNA). The percentage charging for each tRNA sample is shown under each lane.
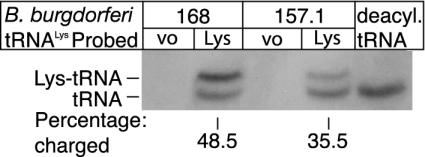
To investigate in vivo charging of B. burgdorferi tRNALys1 by LysRS2 from B. subtilis and LysRS1 from B. burgdorferi, strains were constructed in which the tRNALys1 gene was expressed by using the system developed by Henkin and colleagues (27). RNA was prepared from cells containing the vector only (168 and 157.1) and with the vector expressing tRNALys1 (168 and 157.1), and the level of charged tRNALys1 was determined as described. Control RNA preparations (containing vector only) show no hybridizing bands (Fig. 3). Two bands are observed in RNA samples prepared from cells containing the vector expressing tRNALys1, showing that both LysRS2 from B. subtilis and LysRS1 can charge tRNALys1 from B. burgdorferi. That the upper band is acylated-tRNALys1 is confirmed by its disappearance when samples are treated under deacylating conditions (Fig. 3, deacyl.tRNA). Repeated experiments show that the level of charging of tRNALys1 by the heterologous LysRS2 from B. subtilis is consistently higher (48.5%) than that observed in cells carrying the homologous LysRS1 from B. burgdorferi (35.5%). These studies show that tRNALys species from B. subtilis and B. burgdorferi both can be acylated in vivo by LysRS1 from B. burgdorferi and LysRS2 from B. subtilis, respectively, with relative aminoacylation levels broadly correlated with LysRS-specific activity (Table 1).
Fig. 3.
Determination of the level of B. burgdorferi tRNALys charging by Northern analysis. Twenty micrograms of total RNA from each of the strains B. subtilis with vector only (168 VO), B. subtilis expressing tRNALys1 (168 Lys), BCJ157.1 with vector only (157.1 VO), and BCJ157.1 expressing tRNALys1 (157.1 Lys) was separated by electrophoresis, transferred to membrane, and probed with a B. burgdorferi tRNALys1-specific probe as detailed in Materials and Methods. Twenty micrograms of an RNA sample was treated under deacylation conditions before loading (deacyl.tRNA). The percentage charging of tRNALys1 is shown under each lane.
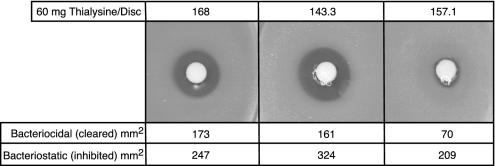
Inhibition of Class I and Class II LysRSs in Vivo. The effects of three LysRS inhibitors (lysine hydroxamate, cadaverine, and thialysine) on growth of B. subtilis strains 168, BCJ143.3, and BCJ157.1 were tested by using disk assays. No differences in the sensitivities of the three strains to lysine hydroxamate and cadaverine were observed (data not shown). In contrast, the strains were differentially sensitive to thialysine (Fig. 4). For B. subtilis strains 168 and BCJ143.3, there is a zone of clearing surrounding the drug-containing disk, indicating that thialysine levels are bactericidal in this area. However, a significantly smaller zone of clearing surrounds the disk in the case of BCJ157.1. The areas of the zones in which thialysine is bactericidal are approximately the same for strains 168 and BCJ143.3 (173 and 161 mm2, respectively), whereas the bactericidal zone for strain BCJ157.1 is only ≈70 mm2. Surrounding each of the bactericidal zones is a second zone where the level of thialysine is bacteriostatic. The areas of these zones are 247 mm2 (strain 168), 374 mm2 (strain BCJ143.3), and 209 mm2 (strain BCJ157.1). These data show that cells harboring LysRS1 and LysRS2 are equally sensitive to lysine hydroxamate and cadaverine, whereas LysRS1 from B. burgdorferi confers a high level of resistance to thialysine.
Fig. 4.
Analysis of growth inhibition by thialysine by using disk assay. Sixty milligrams of thialysine was loaded onto each disk, which was placed in the center of an agar Petri dish that had been lawned with the appropriate bacterial strain. The area of the bactericidal zone is shown at the bottom.
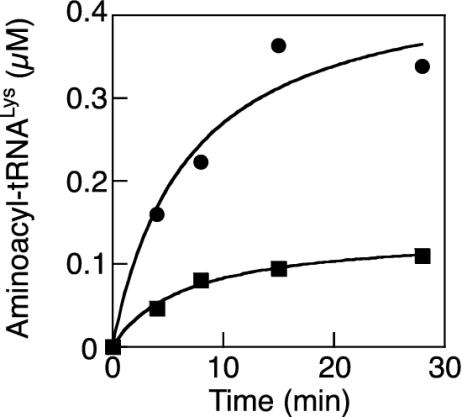
Thialysine Prefrentially Inhibits LysRS2 in Vitro. The ability of thialysine to inhibit in vitro aminoacylation was investigated for E. coli LysRS2 (lysS) and B. burgdorferi LysRS1. Thialysine acted as a competitive inhibitor of both LysRS1 and LysRS2, lowering the KM but not the kcat for both enzymes during steady-state aminoacylation (Fig. 10, which is published as supporting information on the PNAS web site). The Kis for thialysine were calculated to be 1.3 ± 0.08 mM for LysRS1 and 6.6 ± 0.5 μM for LysRS2. These results indicated that thialysine is a considerably more potent inhibitor of LysRS2 than it is of LysRS1. Previous studies (15) showed that thialysine not only inhibits but is also a substrate for the aminoacylation of tRNALys by LysRS2. To determine whether thialysine is also a substrate for LysRS1, 3′ 32P-labeled B. burgdorferi tRNALys1 was used for aminoacylation under standard conditions (Fig. 5). These experiments showed that thialysine is a substrate for aminoacylation by LysRS1. Taken together, these findings are consistent with the above in vivo observation that LysRS1 offers a more effective barrier than LysRS2 to the inhibitory effects of thialysine.
Fig. 5.
Aminoacylation of tRNALys1 with thialysine by LysRS1. Aminoacylation reactions were performed as described by using either 3 mM thialysine (▪) or 3 mM lysine (•), in the presence of 2 μM tRNALys1 and 100 nM B. burgdorferi LysRS1.
Discussion
Functional Genomics in B. subtilis. Complete genome sequences have given unprecedented insight into the physiology and inter-relationships among bacterial genera. Although >112 complete bacterial sequences are publicly available, many are refractory to genetic analysis for reasons including (i) a lack of a transformation system, (ii) a dearth of appropriate genetic tools, (iii) the inability to propagate the bacterium under laboratory conditions, or (iv) propagation requiring specialist facilities because of growth fastidiousness or pathogenicity. In this study, we have demonstrated that gene replacement and complementation in B. subtilis is a viable strategy to characterize genes from heterologous bacteria. We chose to illustrate this strategy by using genes encoding LysRSs from S. aureus (lysS) and B. burgdorferi (lysK), because they represent orthologous and nonorthologous replacements of the endogenous B. subtilis lysS, respectively. The LysRS2 from the closely related Gram-positive pathogen S. aureus complemented the endogenous LysRS2 from B. subtilis when expressed by using the normal B. subtilis expression signals as expected, because the LysRS2 proteins and cognate tRNA sequences are very similar between these bacteria. Despite these similarities a retarded growth phenotype and lowered steady-state level of lysyl-tRNALys during growth in rich medium was observed, showing that LysRS2 from S. aureus is not optimized for the internal B. subtilis milieu. Complementation by the nonorthologous LysRS1 was achieved only when it was expressed from a stronger promoter than found for the gene encoding the endogenous LysRS2 protein. A strain expressing LysRS1 at such levels had a growth-deficient phenotype and lowered levels of charged tRNALys during propagation at fast growth rates. Significantly, our data show that the tRNALys species from B. subtilis and B. burgdorferi both can be charged by the heterologous LysRSs, showing that LysRS1 and LysRS2 can perform similar functions in vivo. Therefore, B. subtilis strain BCJ157.1 can now be used to functionally characterize the B. burgdorferi LysRS1 in vivo with regard to substrate recognition and to identify potential inhibitors of activity. The recent report of 271 essential genes in B. subtilis, and their high conservation among bacteria, means that their orthologues from industrially important or pathogenic bacteria can be transferred into this genetically amenable bacterium for characterization (33).
LysRS1 Presents a Barrier to Addition of a Nonprotein Amino Acid to the Genetic Code. LysRS1 and LysRS2 are analogous proteins in vivo, as clearly demonstrated by the finding that either one can support the growth of B. subtilis under normal conditions. This functional equivalence does not readily equate with the very different phylogenetic distributions of LysRS1 and LysRS2, which suggests there might be some underlying difference between the two proteins. Earlier attempts to address this question focused on evolutionary, structural and functional comparisons of how LysRS1 and LysRS2 recognize tRNALys (8, 9, 14). Differences were found in the ability of LysRS1 and LysRS2 to recognize tRNALys acceptor stem variants, although it is doubtful this would favor one form of the enzyme over the other in vivo (11, 12). Our in vivo findings indicate that LysRS1 and LysRS2 can recognize both B. subtilis and B. burgdorferi tRNALys with similar efficiencies under normal growth conditions. In contrast, LysRS1 and LysRS2 differ significantly in their ability to recognize thialysine and attach it to tRNALys, with the class II enzyme >200 times more sensitive to inhibition by the lysine analogue in vitro. This difference in sensitivity is sufficient to provide a strong selective advantage in vivo to LysRS1-carrying cells, which are able to grow under conditions that prevent growth of LysRS2-producing cells. The growth advantage offered by LysRS1 is realized only in the absence of LysRS2, as shown by the fact that the merodiploid (which contains lysK and lysS) and the parental strain grow similarly on thialysine (B.C.J. and K.M.D., unpublished results).
The Role of Amino Acid Recognition During Evolution of the Two Forms of LysRS. The finding that LysRS1 only confers resistance to a proteinogenic lysine analogue when alone offers a possible explanation for the unusual evolutionary distribution of the enzyme, although no extensive information is currently available on thialysine concentrations in different niches. Although both LysRS1 and LysRS2 are thought to have been present in the common ancestor, LysRS1 appears to have been retained and LysRS2 lost in the archaea, α-proteobacteria, and spirochetes (9). The almost universal absence of LysRS2 from these organisms suggests that the ability of LysRS1 to prevent incorporation of certain nonprotein amino acids, such as thialysine, into protein could have exerted a selective pressure during the evolution of lysyl-tRNA synthesis, leading to the loss of LysRS2. The converse may have occurred in most other lineages where instead LysRS2 was retained and LysRS1 lost, suggesting the existence at some point in evolution of another opposing selective pressure.
Limiting the Addition of Nonprotein Amino Acids to the Genetic Code Provides a Natural Reservoir of Antibiotic Resistance. Although the existence of two LysRSs is unique in that the two enzymes are unrelated, the existence of evolutionarily diverged duplicated aaRS homologues is widespread with at least nine examples known to date in bacteria alone (5). In some cases aaRS duplicates with different tRNA substrate specificities are found in the same organism, presumably to provide versatility in the choice of certain aminoacyl-tRNA synthesis pathways (34, 35). Both closely related (36) and highly diverged homologues have also been described that differ in their amino acid recognition properties (37, 38). For example, several bacteria encode two distantly related forms of isoleucyl-tRNA synthetase, IleRS1 and IleRS2, the second of which confers resistance to pseudomonic acid A, a competitive inhibitor of isoleucine binding (39, 40). Pseudomonic acid A is a naturally occurring antibiotic, suggesting that the acquisition of IleRS2 would provide a selective advantage for certain organisms under the appropriate conditions. Two methionyl-tRNA synthetases, MetRS1 and MetRS2, have also been described, the second of which confers resistance to a spectrum of synthetic methionine competitors (41). When taken together with the example of LysRS, these various instances demonstrate that extensive aaRS duplication exists as a means to protect against the potentially detrimental effects on protein synthesis of amino acid competitors. It has been suggested that, at least for IleRS, MetRS, and tryptophanyl-tRNA synthetase, duplication and divergence resulted from a necessity for antibiotic resistance, even though no such natural antibiotic has yet been found for MetRS. A more general scenario, suggested both by our findings and the extremely wide distribution of IleRS2 beyond pseudomonic acid A-containing environments, is that the diverged aaRS activities originally arose from a need to prevent addition of nonproteinogenic amino acids to the genetic code. Resistance to antibiotics that compete for cognate amino acid binding sites would be an inherent property of such synthetases, thereby providing a large natural reservoir of potential antibiotic resistance alleles.
Supplementary Material
Acknowledgments
We thank S. Ataide (Ohio State University) for the gift of E. coli LysRS2 (lysS) and critical reading of the manuscript, M. Theisen for providing antibodies, and T. Foster (Trinity College) for the gift of S. aureus N315. This work was supported by Fifth Framework Program of the European Commission Grant QLG-CT-99-00660 (to K.M.D. and M.I.) and National Institute of General Medical Sciences Grant 65183 (to M.I.).
Abbreviations: aaRS, aminoacyl-tRNA synthetase; LysRS, lysyl-tRNA synthetase; IleRS, isoleucyl-tRNA synthetase.
References
- 1.Ibba, M. & Söll, D. (2000) Annu. Rev. Biochem. 69, 617-650. [DOI] [PubMed] [Google Scholar]
- 2.Cusack, S., Berthet-Colominas, C., Hartlein, M., Nassar, N. & Leberman, R. (1990) Nature 347, 249-255. [DOI] [PubMed] [Google Scholar]
- 3.Eriani, G., Delarue, M., Poch, O., Gangloff, J. & Moras, D. (1990) Nature 347, 203-206. [DOI] [PubMed] [Google Scholar]
- 4.Ribas De Pouplana, L. & Schimmel, P. (2001) Cell 104, 191-193. [DOI] [PubMed] [Google Scholar]
- 5.Woese, C. R., Olsen, G. J., Ibba, M. & Söll, D. (2000) Microbiol. Mol. Biol. Rev. 64, 202-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ibba, M., Morgan, S., Curnow, A. W., Pridmore, D. R., Vothknecht, U. C., Gardner, W., Lin, W., Woese, C. R. & Söll, D. (1997) Science 278, 1119-1122. [DOI] [PubMed] [Google Scholar]
- 7.Ibba, M., Bono, J. L., Rosa, P. A. & Söll, D. (1997) Proc. Natl. Acad. Sci. USA 94, 14383-14388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ribas De Pouplana, L., Turner, R. J., Steer, B. A. & Schimmel, P. (1998) Proc. Natl. Acad. Sci. USA 95, 11295-11300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ambrogelly, A., Korencic, D. & Ibba, M. (2002) J. Bacteriol. 184, 4594-4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Polycarpo, C., Ambrogelly, A., Ruan, B., Tumbula-Hansen, D., Ataide, S. F., Ishitani, R., Yokoyama, S., Nureki, O., Ibba, M. & Söll, D. (2003) Mol. Cell 12, 287-294. [DOI] [PubMed] [Google Scholar]
- 11.Ibba, M., Losey, H. C., Kawarabayasi, Y., Kikuchi, H., Bunjun, S. & Söll, D. (1999) Proc. Natl. Acad. Sci. USA 96, 418-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schimmel, P. & Ribas De Pouplana, L. (1999) Proc. Natl. Acad. Sci. USA 96, 327-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Söll, D., Becker, H. D., Plateau, P., Blanquet, S. & Ibba, M. (2000) Proc. Natl. Acad. Sci. USA 97, 14224-14228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Terada, T., Nureki, O., Ishitani, R., Ambrogelly, A., Ibba, M., Söll, D. & Yokoyama, S. (2002) Nat. Struct. Biol. 9, 257-262. [DOI] [PubMed] [Google Scholar]
- 15.Stern, R. & Mehler, A. H. (1965) Biochem. Z. 342, 400-409. [PubMed] [Google Scholar]
- 16.Di Girolamo, M., Busiello, V., Blarzino, C. & Cini, C. (1989) Biochem. Int. 19, 1195-1203. [PubMed] [Google Scholar]
- 17.Pitari, G., Maurizi, G., Flati, V., Ursini, C. L., Spera, L., Dupre, S. & Cavallini, D. (1992) Biochim. Biophys. Acta 1116, 27-33. [DOI] [PubMed] [Google Scholar]
- 18.Christner, P., Yankowski, R. L., Benditt, M. & Jimenez, S. A. (1996) Biochim. Biophys. Acta 1294, 37-47. [DOI] [PubMed] [Google Scholar]
- 19.Yu, S., Sugahara, K., Zhang, J., Ageta, T., Kodama, H., Fontana, M. & Dupre, S. (1997) J. Chromatogr. B Biomed. Sci. Appl. 698, 301-307. [DOI] [PubMed] [Google Scholar]
- 20.Hirshfield, I. N. & Zamecnik, P. C. (1972) Biochim. Biophys. Acta 259, 330-343. [PubMed] [Google Scholar]
- 21.Hirshfield, I. N., Tomford, J. W. & Zamecnik, P. C. (1972) Biochim. Biophys. Acta 259, 344-356. [DOI] [PubMed] [Google Scholar]
- 22.Di Girolamo, M., Busiello, V., Cini, C., Foppoli, C. & De Marco, C. (1982) Mol. Cell Biochem. 46, 43-48. [DOI] [PubMed] [Google Scholar]
- 23.Döring, V., Mootz, H. D., Nangle, L. A., Hendrickson, T. L., De Crécy-Lagard, V., Schimmel, P. & Marlière, P. (2001) Science 292, 501-504. [DOI] [PubMed] [Google Scholar]
- 24.Harwood C. R. & Cutting S. M. (1990) Molecular Biological Methods for Bacillus (Wiley, New York).
- 25.Anagnostopoulos, C. & Spizizen, J. (1961) J. Bacteriol. 81, 741-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sambrook, J., Fritsch, E. F. & Maniatis. T. (1989) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY).
- 27.Grundy, F. J., Collins, J. A., Rollins, S. M. & Henkin, T. M. (2000) RNA 6, 1131-1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laemmli, U. K. (1970) Nature 227, 680-685. [DOI] [PubMed] [Google Scholar]
- 29.Kern, D., Dietrich, A., Fasiolo, F., Renaud, M., Giegé, R. & Ebel, J. P. (1977) Biochimie 59, 453-462. [DOI] [PubMed] [Google Scholar]
- 30.Seth, M., Thurlow, D. L. & Hou, Y. M. (2002) Biochemistry 41, 4521-4532. [DOI] [PubMed] [Google Scholar]
- 31.Wolfson, A. D. & Uhlenbeck, O. C. (2002) Proc. Natl. Acad. Sci. USA 99, 5965-5970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grundy, F. J., Lehman, S. C. & Henkin, T. M. (2003) Proc. Natl. Acad. Sci. USA 100, 12057-12062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kobayashi, K., Ehrlich, S. D., Albertini, A., Amati, G., Andersen, K. K., Arnaud, M., Asai, K., Ashikaga, S., Aymerich, S., Bessieres, P., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 4678-4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Curnow, A. W., Tumbula, D. L., Pelaschier, J. T., Min, B. & Söll, D. (1998) Proc. Natl. Acad. Sci. USA 95, 12838-12843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Becker, H. D. & Kern, D. (1998) Proc. Natl. Acad. Sci. USA 95, 12832-12837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brevet, A., Chen, J., Leveque, F., Blanquet, S. & Plateau, P. (1995) J. Biol. Chem. 270, 14439-14444. [DOI] [PubMed] [Google Scholar]
- 37.Brown, J. R., Zhang, J. & Hodgson, J. E. (1998) Curr. Biol. 8, R365-R367. [DOI] [PubMed] [Google Scholar]
- 38.Kitabatake, M., Ali, K., Demain, A., Sakamoto, K., Yokoyama, S. & Söll, D. (2002) J. Biol. Chem. 277, 23882-23887. [DOI] [PubMed] [Google Scholar]
- 39.Brown, J. R., Gentry, D., Becker, J. A., Ingraham, K., Holmes, D. J. & Stanhope, M. J. (2003) EMBO Rep. 4, 1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yanagisawa, T. & Kawakami, M. (2003) J. Biol. Chem. 278, 25887-25894. [DOI] [PubMed] [Google Scholar]
- 41.Gentry, D. R., Ingraham, K. A., Stanhope, M. J., Rittenhouse, S., Jarvest, R. L., O'Hanlon, P. J., Brown, J. R. & Holmes, D. J. (2003) Antimicrob. Agents Chemother. 47, 1784-1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.