Abstract
Conversion of discoidal phospholipid (PL)-rich high density lipoprotein (HDL) to spheroidal cholesteryl ester-rich HDL is a central step in reverse cholesterol transport. A detailed understanding of this process and the atheroprotective role of apolipoprotein A-I (apoA-I) requires knowledge of the structure and dynamics of these various particles. This study, combining computation with experimentation, illuminates structural features of apoA-I allowing it to incorporate varying amounts of PL. Molecular dynamics simulated annealing of PL-rich HDL models containing unesterified cholesterol results in double belt structures with the same general saddle-shaped conformation of both our previous molecular dynamics simulations at 310 K and the x-ray structure of lipid-free apoA-I. Conversion from a discoidal to a saddle-shaped particle involves loss of helicity and formation of loops in opposing antiparallel parts of the double belt. During surface expansion caused by the temperature-jump step, the curved palmitoyloleoylphosphatidylcholine bilayer surfaces approach planarity. Relaxation back into saddle-shaped structures after cool down and equilibration further supports the saddle-shaped particle model. Our kinetic analyses of reconstituted particles demonstrate that PL-rich particles exist in discrete sizes corresponding to local energetic minima. Agreement of experimental and computational determinations of particle size/shape and apoA-I helicity provide additional support for the saddle-shaped particle model. Truncation experiments combined with simulations suggest that the N-terminal proline-rich domain of apoA-I influences the stability of PL-rich HDL particles. We propose that apoA-I incorporates increasing PL in the form of minimal surface bilayers through the incremental unwinding of an initially twisted saddle-shaped apoA-I double belt structure.
Keywords: Lipoprotein/Apolipoproteins, Lipoprotein/HDL, Lipoprotein/Structure, Computer Modeling, Molecular Dynamics, Discoidal HDL, Apolipoprotein A-I
Introduction
High density lipoprotein (HDL)4 represents a heterogeneous population of particles with apolipoprotein A-I (apoA-I) as the major protein (1). HDL biogenesis proceeds with the formation of phospholipid (PL)-rich HDL particles by addition of cell membrane-derived PL and unesterified cholesterol (UC). At this time, HDL is an underexplored and important new target for pharmacological therapy of coronary artery disease (2). Whether HDL plays a direct role in coronary artery disease prevention (e.g. removal of cholesterol from clogged arteries) or an indirect role (e.g. acts as a platform for the clustering of protective molecules, such as anti-inflammatory or antioxidant proteins), knowledge of HDL structure and dynamics is highly desirable.
However, because HDL, a supramolecular assembly of lipid and protein, is a nanoscale soft form of condensed matter easily deformable by thermal fluctuations, direct experimental methods for studying HDL structure-function have had only limited success. Adding to the problem of determining HDL structure, and partly the result of the soft matter nature of HDL, the conformation of apoA-I is highly elastic. It exists in different states as follows: lipid-free, PL-poor, and PL-rich or cholesterol ester-rich lipoproteins of different sizes. Because it appears to have a broad conformational space, a more complete understanding of HDL structure and dynamics will require liberal use of computer simulations cross-checked by experimental testing through approaches such as site-directed mutagenesis.
In one of the more significant advances in understanding the role of the amphipathic helix in lipoprotein structure since discovery of the amphipathic helix motif (3), our laboratory derived a detailed molecular double belt model for discoidal HDL (4, 5). The general features of this model have been confirmed by several laboratories using a variety of physical chemical methods (6–12). More recently, we showed that discoidal HDL complexes made from dimyristoylphosphatidylcholine (DMPC) containing two molecules of apoA-I (R2) form five discoidal particles (R2-1 to R2-5) with discrete sizes (5). Others have also shown that the nature of phospholipids affects the distribution of differently sized particles (13).
Recognizing the profound constraints imposed on the conformations of lipid-associated apolipoproteins by lipid (14), we turned to molecular dynamics (MD) simulations to aid in solving lipoprotein structure/dynamics (14–17). However, MD simulations have limitations. Multiple long simulations are required to increase the confidence that equilibrium has been achieved and that energy barriers have been overcome.
One approach for overcoming kinetic trapping is MD-simulated annealing (MDSA) (18–21). This procedure, when used to refine x-ray crystallography and NMR structures, after placing constraints on the initial protein structure in implicit solvent, applies T-jumps (to 500–1000 K or so), followed by slow cooling to physiological temperatures. Because we wanted to overcome energy barriers during the simulations, we applied MDSA without constraints to our particles in explicit solvent as an approach to bypass kinetically trapped intermediates that might exist in our previous simulations. We hypothesized that the lipid in the HDL assemblies would place its own restraints on apoA-I (14). As there is no standard protocol for unconstrained MDSA, we developed optimal conditions for unconstrained MDSA (degree and length of T-jump and rate of cooling) through trial and error. This approach provides exploration of conformational space but is not a robust way to explore longer simulation times.
In this study, we use MDSA to explore the conformational space of PL-rich HDL particles that differ in two ways from those of our previous MD simulations; they contain full-length (FL) apoA-I and UC. Through a combination of computation and experimentation, we show that apoA-I is able to encompass a continuously variable number of lipid molecules by the incremental twisting or untwisting of a saddle-shaped apoA-I double belt structure that creates minimal surface patches of lipid bilayer.
EXPERIMENTAL PROCEDURES
Computational Studies
PL-rich Particles
The particle containing 160 POPC, 24 UC, and 2 apoA-I molecules (160:24:2) was created by generating an all-atom particle containing 2 FL-apoA-I molecules in a 100% α11:3 conformation wrapped in an antiparallel double belt around a POPC bilayer of 252 lipid molecules (5). The first 32 residues of the N terminus of apoA-I (helices G0, G1, and G2) were rotated to orient their hydrophobic faces toward the surface of the POPC acyl chains. The resulting particle was subjected to random removal of 92 POPC molecules, followed by random insertion of 24 UC molecules into the space vacated by the deleted POPC to generate a particle with a POPC:UC:apoA-I molar ratio of 160:24:2. To create the 100:15:2 and 50:8:2 particles, a series of removals each followed by 5-ns simulations at 310 K were performed. Starting with the 160:24:2 particle described above, 30 central POPC were removed (15 from each leaflet), resulting in a 130:24:2 particle. This was repeated, resulting in a 100:24:2 particle. 29 central POPC were next removed, resulting in a 71:24:2 particle. Next, 14 central POPC and 2 UC were removed, resulting in a 57:22:2 particle. Finally, 7 POPC and 14 UC were removed, resulting in the 50:8:2 particle. The 100:15:2 particle was obtained from removing 9 UC from the 100:24:2 particle. POPC were removed centrally because our various particle reduction methods, described in Catte et al. (16), led to similarly shaped particles, as well as the fact that we planned to perform MDSA.
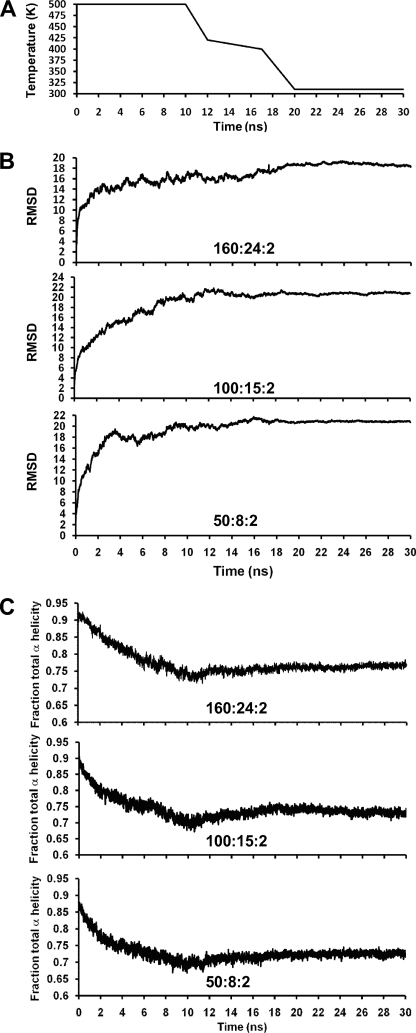
The initial particles were simulated at 310 K and 1 bar for 10 ns and then subjected four times to the following MDSA protocol (all at 1 bar): heated from 310 to 500 K in 20 ps, simulated at 500 K for 10 ns, cooled from 500 to 420 K in 2 ns, cooled from 420 to 400 K in 5 ns, cooled from 400 to 310 K in 3 ns, and finally simulated at 310 K for 10 ns, giving a total duration of 30 ns (Fig. 1A). For simulations at a fixed temperature, velocity reassignments occurred every 1 ns to prevent the “flying ice cube” effect (22).
FIGURE 1.
Changes in particles during the 30 ns of the MDSA protocol. A, plot of MDSA protocol (temperature in K versus time in nanoseconds). B, average protein r.m.s.d. calculated for the simulations of the three sets, 160:24:2, 100:15:2, and 50:8:2, as a function of time of MDSA. C, average total helicity calculated for the simulations of the three sets, 160:24:2, 100:15:2, and 50:8:2, as a function of time of MDSA.
All-atom simulations were performed using NAMD (23) as described in Catte et al. (16). Each system was ionized and charge-neutralized with NaCl to 0.15 m with the autoionize plug-in of Visual Molecular Dynamics (VMD) (24). The TIP3P water model was used (25). The CHARMM 22 (26, 27) and 27 (28, 29) force fields were used for protein and lipid molecules, respectively.
Root Mean Square Deviations
The r.m.s.d. of protein α-carbon atoms were calculated for the 160:24:2 particles over the entire duration of the four 30-ns MDSA simulations. The r.m.s.d. were calculated with respect to the 160:24:2 particles after they had been initially simulated at 310 K for 10 ns.
Lipid-Protein Salt Bridges
The distances of POPC P1-oxygen atoms to nitrogen atoms on basic protein residues were measured for selected POPC molecules over the entire duration of certain 30-ns MDSA simulations. Because the “saltbr” command of VMD only works for salt bridges between protein residues, a simple VMD script was written to measure these distances.
Annular Distribution Functions (ADF)
We first described this function, similar but not identical to a radial distribution function, in Jones et al. (14). For each trajectory frame in the last 20% of the MDSA simulations of the 160:24:2 particles, distances were measured from the nearest protein atom to each POPC or UC (using center of mass or a representative atom such as phosphorus), and the distribution of these distances was averaged and plotted. A simple VMD script was written to measure and average these distances. For circles, analysis shows that the distribution should be linear in distance from the annulus.
Fraction α-Helicity
The fraction of all protein residues in the three different sets of particles was measured every 20 ps over the course of all 30-ns trajectories of the MDSA simulations by using VMD implementation of the secondary structure determination program STRIDE (30).
The fraction of the time that each protein residue was α-helical was calculated over the last 20% of the 30-ns trajectories of each set of MDSA simulations and averaged. This was measured every 20 ps by once again using STRIDE.
Experimental Studies
Purification of Human Plasma ApoA-I and Recombinant N-terminal Truncated ApoA-I
Human plasma apoA-I was isolated from the HDL fraction of fresh human plasma and purified according to the procedure described previously (31). To produce the N-terminally truncated apoA-I, human Δ43, Δ33, Δ21, and Δ10 apoA-I cDNA in plasmid pGEMEX were expressed in Escherichia coli BL21/DE3 cells, and the recombinant protein was purified as described by Rogers et al. (32). Briefly, the expression of apoA-I was monitored by Western blotting analysis of the bacterial lysate with anti-human apoA-I antibody. The bacterial lysate containing mutant apoA-I was loaded onto a preparative reversed-phase HPLC (C4) column, and proteins were eluted and separated by a gradient of acetonitrile with 0.1% trifluoroacetic acid. Preparative HPLC fractions containing apoA-I (identified by immuno-dot blots) were then subjected to purity and identity analyses. The purity of expressed proteins was examined by analytical HPLC (C18) and SDS-PAGE. The identity of the proteins was confirmed by mass spectrometry and N-terminal amino acid sequencing. Purified proteins were lyophilized and stored at −20 or −80 °C.
Preparation of Wild Type and Mutant ApoA-I Solutions
Lyophilized protein was solubilized in 6 m guanidine hydrochloride. The solubilized protein solution was loaded onto a desalting column (Econo-Pac 10DG Disposable Chromatography Columns, Bio-Rad), and the protein was eluted by phosphate-buffered saline buffer (0.02 m phosphate, 0.15 m NaCl, pH 7.4). An extinction coefficient of 1.13 ml/(mg-cm) and 0.991 ml/(mg-cm) at 280 nm was used for determining the concentration of full-length apoA-I and N-terminal truncated apoA-I, respectively, in 6 m guanidine hydrochloride (32).
Preparation and Analysis of Reconstituted (r)HDL Assemblies
DMPC and POPC were purchased from Avanti Polar Lipids, Inc. (Alabaster, AL), and used without further purification. Reconstituted HDL assemblies were prepared by co-incubation of multilamellar vesicles of lipids with apoA-I or by the sodium cholate dialysis procedure as described previously (5, 33). The incubation temperature was set above the transition temperature for each lipid, 25 °C for DMPC and 4 °C for POPC. The particles formed were analyzed initially by nondenaturing gradient gel electrophoresis (NDGGE) on a 4–20% gradient Tris-glycine acrylamide gel (Invitrogen). High molecular weight markers (catalog no. 17-0445-01, Amersham Biosciences) with known Stokes diameter (Sd) were used as standards as follows: thyroglobulin (170 Å), ferritin (122 Å), catalase (104 Å), lactate dehydrogenase (82 Å), and albumin (71 Å). The gels were stained with the colloidal blue staining kit (Invitrogen). With reference to protein molecular markers, the Stokes diameters of differently sized particles in the complexes were calculated based on density scans of the gels by LabWorks image acquisition and analysis software (UVP Inc., Upland, CA). Following the NDGGE analysis, the well defined particles were subjected to other physicochemical analyses as described below.
Circular Dichroism Spectroscopy
The CD spectroscopy was conducted as described previously (see supplemental Materials and Methods) (32, 34).
Proton NMR Spectroscopy
Proton NMR spectroscopic analyses of samples were conducted as described previously (35). Briefly, all proton NMR spectra were measured with Bruker Avance 600 NMR spectrometer at 37 °C. The NMR data were transferred to a Silicon Graphics IRIS Indigo workstation and processed using the program FELIX (version 2004) (Accelrys Software Inc., San Diego). Reconstituted HDL assemblies were dialyzed against D2O buffer before being subjected to NMR analysis.
Electron Microscopy
The transmission electron microscopic analysis of the particles was carried out as described previously (see supplemental Materials and Methods) (16).
RESULTS
Computational Results
Structure
We used MDSA and the particle reduction methodology as described under “Experimental Procedures,” as one approach to determine the structure of R2 PL-rich HDL particles. In particular, we sought to show that FL-apoA-I forms saddle-shaped conformations on the edges of PL-rich discs stoichiometrically equivalent to R2-2 (160:24:2), R2-1 (100:15:2), and R2-0 (50:8:2) that are very similar to that seen in our initial MD simulation study of truncated apoA-I (14–16).
Changes in average r.m.s.d. of the protein of the resulting 160:24:2, 100:15:2, and 50:8:2 particles were plotted over the full 30-ns protocol of the simulations, and the results are shown in Fig. 1B. Note that the average r.m.s.d. of the protein decreases slightly or is flat over the last 6 ns of the protocol, suggesting structural convergence for all three particles.
Changes in average helicity of the protein of the resulting in 160:24:2, 100:15:2, and 50:8:2 particles were plotted over the full 30-ns protocol of the simulations, and the results are shown in Fig. 1C. For each particle, there is a decrease in helicity during the 10-ns T-jump at 500 K, followed by a slight increase in helicity during the 10 ns of cool down, and a flattening out of helicity during the 10 ns of equilibration at 310 K.
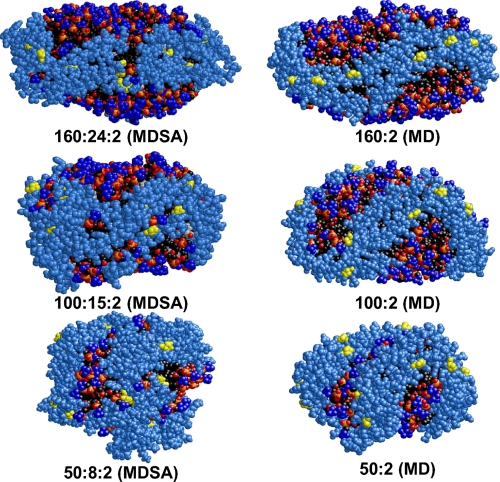
Fig. 2 compares space-filling images of one example of each of the final structures of the MDSA simulations of the three differently sized PL-rich particles with that of the final structures of our 310 K MD simulations of 160:2, 100:2, and 50:2 particles formed from truncated apoA-I (14–16). Details of the structures of the corresponding particle pairs differ somewhat. They differ particularly in their terminal (overlap) domains, the side displayed in Fig. 2; these domains of FL-apoA-I are more disordered in the three MDSA-simulated particles than in the particles containing truncated apoA-I.
FIGURE 2.
Space-filling models of representative final structures from each of the three sets subjected to the MDSA protocol compared with the initial MD simulation study of truncated apoA-I subjected to short (10 ns) MD simulations at 310 K (16). Left column, MDSA. Right column, short 310 K MD simulations. The structures are viewed from the terminal (overlap) domain side of the particles. Protein, sky blue except proline (yellow). POPC, phosphorus atoms (gold), phosphate oxygen atoms (red), choline nitrogens and methyls (blue), and acyl chains (black). UC, CPK (where CPK is Corey, Pauling, and Koltun coloring).
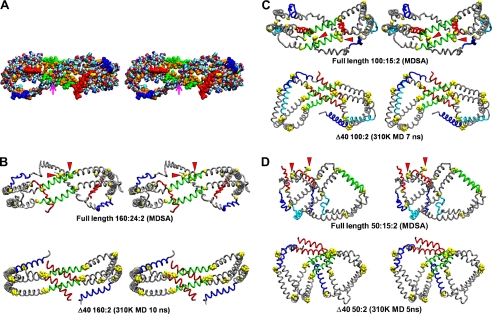
A program was written that calculates the average atomic coordinates from any set of structures aligned by VMD and that program was used to create an average structure from sets of the aligned final frames of the MDSA simulations of the 160:24:2, 100:15:2, and 50:8:2 particles. To illustrate the sort of results achievable with this program, an average all-atom structure for 160:24:2 is shown in Fig. 3A. This model shows, among other things, that essentially all of the hydrophobic residues (Fig. 3A, in gold) of the apoA-I double belt face the lipid of the disc. It also shows the tail to tail association of the pair of proline-rich N-terminal G0 domains (Fig. 3A, magenta arrow). The C-terminal helix 10 domains (Fig. 3A, red) are widely separated, and the one on the right is unassociated with the lipid of the disc.
FIGURE 3.
Cross-eyed stereo images of average final structures of apoA-I double belt in the three MDSA sets created from calculated average coordinates compared with the initial MD simulation study of particles containing truncated apoA-I subjected to short (10 ns) MD simulations at 310 K (16). A, average space-filling all-atom final structure of MDSA simulations of the 160:24:2 particle set. The particle is viewed from the overlap domain. Helix 5 (green), helix 10 (red), helix 1 (blue), leucines (gold), and remainder of protein (CPK, where CPK is Corey, Pauling, and Koltun coloring) are shown. The point of contact of the N-terminal proline-rich domains is indicated by a magenta arrow. B–D, average final structures of MDSA simulations of each of the three sets, 160:24:2, 100:15:2, and 50:8:2 (upper rows), are compared with the final structure (lower rows) of a comparable particle with truncated apoA-I subjected to short (10 ns) MD simulations at 310 K (16). The apoA-I double belts are viewed from the helix 5 domain. The protein is represented as a Cα backbone model except proline (space-filling yellow), helix 5 (green), helix 10 (red), helix 1 (blue), helix 8 (cyan), and remainder of protein (gray). The N-terminal G0 (proline-rich) domains are indicated by red arrowheads.
Fig. 3, B–D, shows the average Cα backbone structures for the 160:24:2, 100:15:2, and 50:8:2 simulations, respectively. Comparing the average MDSA simulated structures (Fig. 3, B–D, upper figures) with that of the 310 K simulations of the corresponding PL particles made from truncated apoA-I (lower figures), it can been seen that FL-apoA-I forms saddle-shaped conformations on the edges of PL-rich discs (containing UC) that are similar to those seen in our previous MD simulation study of PL-rich HDL particles containing truncated apoA-I (not containing UC) subjected to short (5–10 ns) MD simulations at 310 K (14–16). Despite the differences in terminal overlap domains, necessitated by the presence or absence of residues 1–40, the general saddle shape of the annular apoA-I double belts is similar between each corresponding particle pair. Significantly, all of the resulting 160:24:2, 100:15:2, and 50:8:2 double belt structures, when viewed from the helix 5 side, formed left-handed figure eight saddle shapes with pseudo-D2 symmetry, suggesting that saddle-shaped structures are not kinetically trapped collapsed intermediates and that MDSA is sampling a valid region of the conformational space.
There is one additional point to note about Fig. 3. The protein perimeters of the FL double belts for all three structures are closed at the terminal (overlap) domain by interaction of the N termini (the proline-rich N termini generally seem to interact, see Fig. 3, red arrowheads). However, the helix 10 C termini (Fig. 3, red), although closer in the 50:8:2 particles, are spatially separated and completely or partially detached from the lipid. The N-terminal segment of the double belt, residues 1–40, present only in the MDSA-simulated particles, is more condensed and folded upon itself than is the remainder of the apoA-I double belt structure. Conversely, the structure of the terminal (overlap) domains in the particles containing truncated apoA-I differ little from that of their remaining double belt regions.
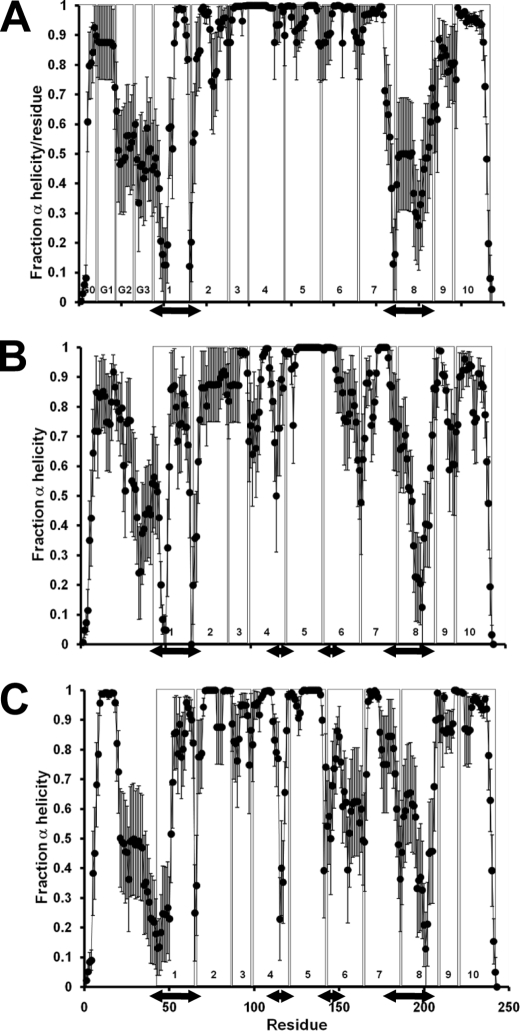
Per Residue Helicity
Fig. 4, A–C, plots the average helicity of each residue over the last 20% of the MDSA simulations of the 160:24:2, 100:15:2, and 50:8:2 particles, respectively. For 160:24:2 (Fig. 4A), there is significant loss of helicity at the juncture of helices G2-G3 with helix 1 (double-headed arrow on left) and in helix 8 (double-headed arrow on right). From Fig. 3, it can be seen that these regions are on adjacent parts of the double belt and form loops (blue and cyan, respectively) that induce saddle shapes to the belt structures. In conversion of 160:24:2 particles to 100:15:2 and then to 50:8:2 particles, the percent helicity of the loop region does not change significantly (Fig. 4, peripheral double-headed arrows); what changes is an increase in degree of bend as the angles of the loops become more acute (see Fig. 3, C and D). There is, however, a decrease in the 100:15:2 and 50:8:2 helicity as the double belt structure becomes more folded at the helix 4-helix 5 and helix 5-helix 6 junctures (central double-headed arrows in Fig. 3, B and C).
FIGURE 4.
Average local (per residue) changes in fractional α-helicity (mean ± 1 S.E.) during the last 20% of the MDSA protocols for the three sets, 160:24:2, 100:15:2, and 50:8:2. The vertical boxes in each figure denote the positions of each of the 10 helical and N-terminal G* repeats. The double-headed arrows indicate the position of the major (occurs in all three sets) and minor (occurs only in the two smaller sets) helical breaks, lateral and central pair, respectively. A, 160:24:2 set. B, 100:15:2 set. C, 50:8:2 set.
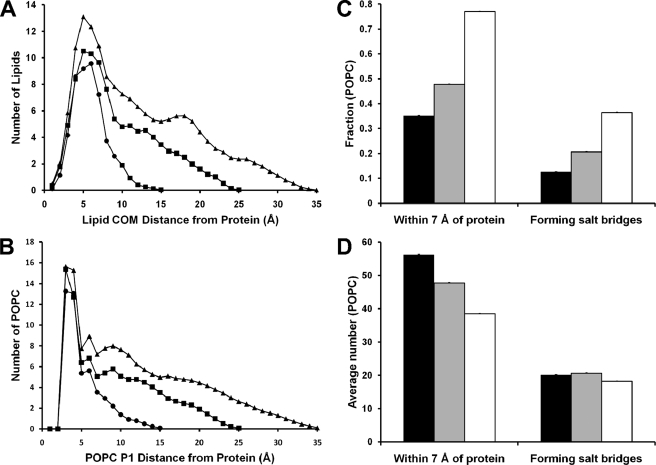
Annular POPC
Our previous MD simulations have shown that the properties of lipid adjacent to the protein belt (annular lipid) in PL-rich particles produced from truncated apoA-I are different from that of the central bulk lipid (14, 16). ADF of distances of POPC from the nearest protein atom were measured over the last 20% of the MDSA simulations, and the results were averaged over each set of 160:24:2, 100:15:2, and 50:8:2 PL-rich particles. Fig. 5A shows that in all three sets of particles, POPC forms a pronounced annular distribution shell centered ∼5 Å from the protein. A plot of the ADF of POPC phosphorus atoms from the nearest protein atom in Fig. 5B shows a sharp annular distribution shell centered ∼3 Å from the protein, suggesting that a significant fraction of the annular POPC in each particle forms salt bridges between their phosphorus atoms and basic residues on the right (R)-interfacial edge of the apoA-I double belt (4, 36).
FIGURE 5.
Properties of annular POPC for the three PL-rich HDL particle sets. A, plots of the average ADF of distances of the POPC center of mass from the nearest protein atom measured over the last 20% of the MDSA simulations for the three sets, 160:24:2 (▲), 100:15:2 (■), and 50:8:2 (●). B, plots of the average ADF of POPC phosphorus atoms from the nearest protein atom measured over the last 20% of the MDSA simulations for the three sets, 160:24:2 (▲), 100:15:2 (■), and 50:8:2 (●). C, bar plots of the average fraction (±1 S.E.) of POPC within 7 Å of protein or forming salt bridges with the protein during the last 20% of the MDSA simulations of the three sets, 160:24:2 (solid bars), 100:15:2 (gray bars), and 50:8:2 (open bars). D, bar plots of the average number (±1 S.E.) of POPC within 7 Å of protein or forming salt bridges with the protein during the last 20% of the MDSA simulations of the three sets, 160:24:2 (solid bars), 100:15:2 (gray bars), and 50:8:2 (open bars).
The estimated width at half-height of the annular POPC distribution shells in Fig. 5A were used to define annular POPC as having a center of mass ≤7 Å from the nearest protein atom. Fig. 5C shows plots for each set of PL-rich particles of the average fraction of total POPC molecules that are annular and the average fraction of annular POPC that form salt bridges. As the lipid contents of the particles decrease, an increasing fraction of total POPC becomes annular and forms salt bridges, increasing from 0.35 and 0.13 for the 160:24:2 particles to 0.77 and 0.36 for the 50:8:2 particles, respectively. However, the average total number of annular POPC progressively decreases with decreasing size, from 56 to 39, whereas the average total number of annular POPC forming salt bridges is constant at 18–21 (Fig. 5D). These results suggest that formation of the annular shell of POPC in PL-rich HDL is partially stabilized by formation of salt bridges between the phosphorus atoms of POPC and a fixed number of basic protein residues on the R-interfacial edge of the apoA-I amphipathic helices (4, 36). The total number of Arg and Lys residues in wheel position 7 on R-interfacial edges of each apoA-I double belt is 32, suggesting that not all form salt bridges with POPC at any one time.
Experimental Tests of Computer Models
Reconstitution of R2 Particles
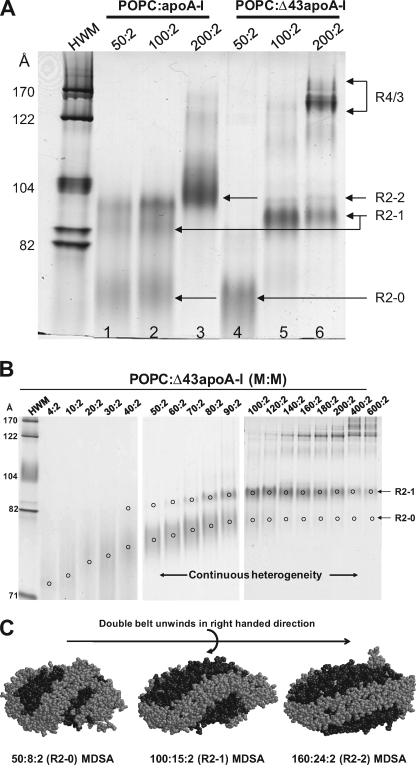
To compare the characteristics of the R2 family of POPC particles produced by MDSA simulations with the properties of in synthetico reconstituted particles, we developed protocols to produce relatively homogeneous preparations of R2-2, R2-1, and R2-0 rHDL particles. Although distinct bands could be separated and viewed by NDGGE analysis, very little material could be recovered from gel slices. Attempts to separate the particles within the R2 family by gel filtration and gradient centrifugation were also not successful. However, with the optimization of POPC to apoA-I ratios and the use of Δ43apoA-I (5), we were able to prepare relatively pure preparations of R2-2 and R2-1 and R2-0 particles (supplemental Fig. S1).
CD Spectroscopy of R2 Particles
Samples of R2-2, R2-1, and R2-0 POPC and R2-2 and R2-1 DMPC assemblies and lipid-free apoA-I/Δ43apoA-I were subjected to CD spectroscopic analysis. The results are shown in supplemental Fig. S2A.The percent α-helicity of the particles decreased with a decrease in particle size as follows: R2-2, 84 ± 3%; R2-1, 73 ± 3%; and R2-0, 60 ± 2%.
These values can be compared with the average percent helicity calculated from Fig. 1C for the corresponding MDSA-simulated particles 160:24:2, 100:15:2, and 50:8:2, respectively. Published experimental values for percent helix of POPC-containing R2-2 particles, 72 and 80% (37, 38), give a mean percent helicity of 76% and, in combination with our value of 84%, a mean percent helicity of 79%; both values are close to our MDSA value of 77%. Our CD-determined value of 73% for R2-1, apparently the only percent helicity reported for this particle in the literature, is identical to our MDSA-determined value of 73%. The percent α-helicity of our previous 310 K simulations of PL particles was clearly much more problematic because the values were greater than 90% for all three particles (16).
There is, however, a significant discrepancy in the percent helicity of simulated and reconstituted R2-0 particles of 72 versus 60%, respectively. Possible reasons for this discrepancy will be addressed under the “Discussion.”
Negative Stain Electron Microscopy of R2 Particles
The reconstituted R2-2, R2-1, and R2-0 POPC assemblies were subjected to negative stain EM analysis. Both the POPC and DMPC R2-2 rHDL particles appeared as typical discs stacked in rouleaux, whereas the POPC/DMPC R2-1 and POPC R2-0 particles had an ellipsoidal appearance (supplemental Fig. S2B), suggesting that the R2-1 and R2-0 particles do not form planar disc shapes. Finally, the R2-2 and R2-1 particles containing POPC or DMPC were similar in size and shape.
1H NMR Spectroscopic Analysis of R2 Particles
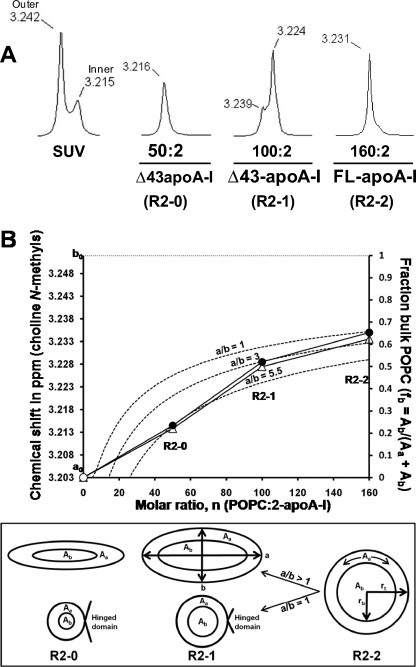
In a previous 1H NMR spectroscopic study of small unilamellar vesicles (39), we showed that, as the vesicle size decreased, the split between the phosphatidylcholine N-methyl resonances, corresponding to lipids residing in the nonexchangeable outer and inner monolayers, became more pronounced (see the small unilamellar vesicle spectra in Fig. 6A). We showed that the increasing split is due to the dramatic upfield shift of the inner monolayer choline resonance due to increased headgroup packing with decreasing vesicle size. In a second 1H NMR spectroscopic study, we showed that the chemical shift of choline N-methyl proton resonances contains information about the chemical environment of PL in complexes with apoA-I; choline N-methyl proton resonances of annular PL are shifted upfield compared with central or bulk PL (35). Because the rate of lipid exchange between the annular and central lipid is more rapid than the 1H NMR relaxation time (35), as the particle size increases, the time averaged resonance forms a single peak that shifts downfield, reflecting the increasing ratio of bulk to annular lipid.
FIGURE 6.
Analysis of the 1H NMR spectra of the choline N-methyl region of PL-rich HDL particles reconstituted with POPC to apoA-I molar ratios of 160:2, 100:2, and 50:2 representing R2-2, R2-1, and R2-0 particles. A, 1H NMR spectra of the choline N-methyl region of small unilamellar vesicle (SUV) formed from POPC and PL-rich HDL particles reconstituted with POPC to apoA-I molar ratios of 160:2, 100:2, and 50:2 representing R2-2, R2-1, and R2-0 particles. The chemical shifts are shown above each resonance peak. B, plot of chemical shifts versus fits provided by several models. ●, average of two 1H NMR chemical shifts for reconstituted R2-2, R2-1, and R2-0 from two independent experiments plotted against their molar ratios, n. ○, chemical shift of pure annular POPC, ao, calculated as described under “Results.” ···, chemical shift of pure bulk POPC, bo, calculated as described under “Results.” Δ, chemical shifts calculated as described under “Results” by using fractions of annular and bulk POPC derived from the MDSA models (Fig. 5C). - - -, chemical shifts calculated by assuming simple circular/elliptical planar lipid surface geometry. To develop this model for a circle (ellipse with axial ratio a/b = 1), we made the following assumptions (lower panel): (i) the particles shrink in size via a hinged domain that detaches from the disc perimeter (5,35); (ii) the POPC of all particles form circular planar discs with radius rt; (iii) annular POPC forms a 7-Å wide band (from Fig. 5A) with area Aa around the bulk POPC having area Ab and radius rb; (iv) therefore, rt − rb = 7 Å; (v) total POPC area = Aa + Ab = n/2 × 65 Å2, where n = molar ratio of each particle (POPC·2-apoA-I) and 65 Å2 is the surface area per POPC; (vi) fraction bulk lipid (fb) = Ab/(Aa + Ab). It follows that fb = (1 − 2.17/(n)1/2)2. A general equation for elliptical models with an axial ratio of a/b is as follows: fb = (1 − 2.17/((a/b)1/2·(n)1/2))·(1 − 2.17·(a/b)1/2/(n)1/2).
To study the environment of POPC in differently sized particles, three reconstituted PL-rich particles with POPC to apoA-I molar ratios of 160:2, 100:2, and 50:2 representing R2-2, R2-1, and R2-0 particles (supplemental Fig. S1) were subjected to 1H NMR spectroscopic analysis, and the results were compared with the chemical shifts of small unilamellar vesicles (Fig. 6A). The results show the presence of a progressive upfield chemical shift of the choline N-methyl resonance from the largest particle (R2-2) with a 160:2 molar ratio to the smallest (R2-0) with a 50:2 molar ratio (Fig. 6A), indicating that smaller particles contain a larger fraction of their POPC in their annular shell than the larger particles. The minor peak in the spectrum for R2-1 particles with a 100:2 molar ratio indicates the existence of a residual amount of larger R2-2 particles in the preparation (see supplemental Fig. S1).
A plot of the average of two measurements of chemical shifts of the 1H NMR spectra of the three reconstituted particles versus the POPC to apoA-I molar ratio (n) from two independent experiments are shown in Fig. 6B as closed circles. Making the assumption that the chemical shifts of the three particles are linear functions of the relative proportions of their annular to bulk POPC, three equations can be written: R2-2, 3.235 ppm = 0.35ao + 0.65bo; R2-1, 3.229 ppm = 0.48ao + 0.52bo; and R2-0, 3.2145 ppm = 0.77ao + 0.23bo, where ao and bo are the chemical shifts in parts/million for pure annular and bulk POPC, respectively, and the constants are the fractions of annular (fa) and bulk (fb) POPC calculated for the 160:24:2, 100:15:2, and 50:8:2 MDSA-simulated particles (Fig. 5C), and the chemical shifts are averages of two separate 1H NMR spectra for each particle. An approximate solution of this set of overdetermined simultaneous linear equations gives ao = 3.203 ppm and bo = 3.252 ppm (plotted in Fig. 6B as an open circle and a horizontal dashed line, respectively). Thus, if a particle has only annular POPC, its chemical shift would equal ao = 3.203 ppm, and if it has only bulk POPC, its chemical shift would equal bo = 3.252 ppm; if the particle contains a mixture of annular and bulk POPC, its chemical shift will vary as a linear function of these values.
We then plotted the values for fb (fraction bulk POPC) that we measured from the three MDSA-simulated particles (Fig. 5C, y axis) against n and mapped these fb to the chemical shifts (Fig. 6B, left-hand y axis) by linear transformation (open triangles). The resulting calculated chemical shifts closely fit the measured chemical shifts for the three reconstituted particles.
As we did previously (35), we then derived a model based upon simple circular/elliptical geometry for calculating fb as a function of the molar ratios of the particles, n. The equations used to develop this model are described in the legend to Fig. 6B. Substituting bo and ao as the upper and lower limits, respectively, for fb, this equation was used to plot chemical shift curves for a circle (an ellipse with an axial ratio a/b = 1) and for two ellipses with axial ratios a/b = 3 and 5.5.
The circular model (dashed curve a/b = 1 in Fig. 6B) clearly does not fit the chemical shift data for the two smaller particles. However, it does fit the chemical shift data for R2-2, the only one of the three MDSA models that approximates a circular planar disc.
The elliptical models (dashed curves a/b = 3 and a/b = 5.5 in Fig. 6B) give different results. Curve a/b = 3 fits the chemical shift of R2-1 but not R2-0, whereas curve a/b = 5.5 fits the chemical shift of R2-0 but not R2-1. This shows that the lipid surface geometries of all three particles approximate ellipses with axial ratios that increase with decreasing particle size or lipid content. The lipid surface geometries of the MDSA structures (see Fig. 2) clearly display the same general trend toward ellipses with increasing axial ratios.
Kinetics of R2 Particles
We demonstrated previously that pre-formed DMPC·apoA-I assemblies can take up additional bulk DMPC to form larger particles (5); this reaction is reversible (supplemental Fig. S3A). Furthermore, regardless of the method of preparation of pre-formed particles, the addition of apoA-I drove the formation of smaller particles in a temperature-dependent manner (supplemental Fig. S3B). These findings indicate that there is a relatively low energy barrier for exchange of components between the discontinuously variable DMPC assemblies.
Relevant to this study, POPC·apoA-I complexes were compared with DMPC·apoA-I complexes (both prepared by cholate dialysis). Unlike DMPC·apoA-I complexes, freshly prepared POPC·apoA-I complexes appeared as broad bands by NDGGE analysis (Fig. 7A). However, incubating the complexes at 4 °C for a sustained period of time (e.g. 2–4 weeks) after dialysis sharpened the bands on NDGGE to produce particles of more uniform size (Fig. 7B), similar to those produced by DMPC·apoA-I (Fig. 7C). Further studies determined that the narrowing of POPC·apoA-I particle size distribution was also temperature-dependent; increasing incubation temperature overcame the energy barriers and increased the rate of equilibration (data not shown). Therefore, each of the several distinct stepwise heterogeneous R2 PL·apoA-I rHDL particles must represent a local energy minimum.
FIGURE 7.
Effect of incubation time on kinetic and thermodynamic distribution of POPC·apoA-I assemblies. A, NDGGE analysis of freshly prepared POPC·apoA-I complexes in a 4–20% polyacrylamide gel run for 48 h. B, NDGGE analysis of the same complexes as in A after being incubated at 4 °C for 4 weeks. C, NDGGE analysis of freshly prepared DMPC·apoA-I complexes in a 4–20% polyacrylamide gel run for 48 h.
Size Distribution of R2 Particles
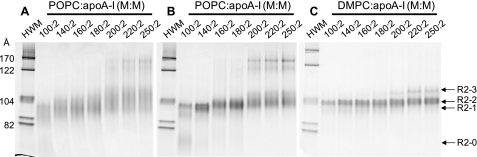
Comparison of POPC·apoA-I and DMPC·apoA-I rHDL particles with identical lipid to apoA-I ratios revealed common, as well as distinct, features of the particle size distribution of the two types of complexes (Fig. 8A). The results show that, at the ratio of 100:2 (lipid:apoA-I), R2-2 and R2-1 rHDL particles were formed by both POPC·apoA-I and DMPC·apoA-I complexes, although the R2-1 band is more pronounced in the POPC rHDL particles. Of significance, as we and others previously observed (35), in POPC·apoA-I rHDL particles, a particle smaller than R2-1, absent from equivalent DMPC complexes, with a rather broad distribution (designated R2-0, Sd = 72–78 Å) was formed. At a ratio of 160:2, the predominant assemblies are R2-2 for both POPC and DMPC complexes.
FIGURE 8.
Effects of lipids and individual N-terminal G* domains of apoA-I on the distribution of discretely sized rHDL particles. A, comparison between DMPC·apoA-I and POPC·apoA-I complexes. NDGGE analysis of complexes in a 4–20% polyacrylamide gel run for 48 h comparing DMPC (lanes 2, 4, 6, and 8) and POPC (lanes 1, 3, 5, and 7) complexes with apoA-I at different lipid:protein molar ratios. B, NDGGE analyses of DMPC particles reconstituted with apoA-I constructs containing progressively truncated N-terminal G* domains of apoA-I.
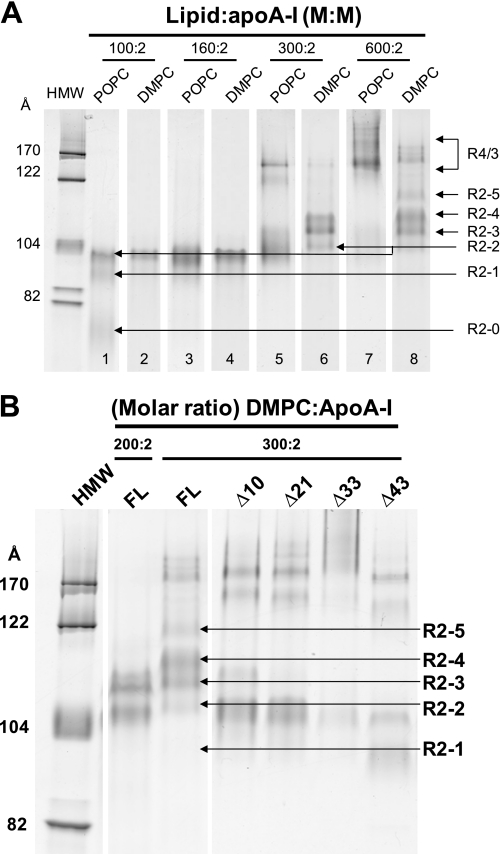
At a DMPC to apoA-I ratio of 300:2, R2-2 becomes a minor particle with a shift to the larger particles, R2-3 and R2-4. At this ratio for POPC to apoA-I, the R2-2 rHDL particle is the predominant particle; no discernable R2-4 band is seen (Fig. 8A). Assemblies with more than two molecules of apoA-I (Sd >122 Å) appear in both complexes, the bands being more pronounced in POPC. At 600:2, DMPC·apoA-I complexes form the largest R2 particle, R2-5, in addition to significant amounts of R2-4 and R2-3 particles. At 600:2, POPC·apoA-I complexes form minor amounts of R2-2 and R2-3 but no discernable R2-4 and R2-5 particles; the vast majority of the complexes formed are of the R3/4 variety containing three/four molecules of apoA-I (Sd 140–200 Å) (Fig. 8A and supplemental Fig. S4).
Therefore, although POPC·apoA-I complexes may share certain structural features with DMPC·apoA-I complexes, they also exhibit significant differences. It seems likely that the uniqueness to POPC of the R2-0 particle, as well as a relative increase in R2-1 compared with DMPC, has physiological significance.
Effects of ApoA-I Sequence on Stability of R2 Particles Larger than R2-2
We showed previously that the N-terminal G-helical domain (residues 1–43) of apoA-I is required for formation of larger sized (R2-3 to R2-5) DMPC-containing rHDL particles and affects the relative distribution of smaller sized (R2-2 and R2-1) particles. We further suggested that the G* amphipathic helices in the N-terminal 1–43 residues of apoA-I contribute to the stepwise increase in size of DMPC-containing rHDL particles larger than R2-2 (5). The MDSA-generated structural model for R2-2 formed from full-length apoA-I (Fig. 2, A and B) suggested to us the hypothesis that the proline-rich N-terminal G* domain, G0 (residues 1–10), may be involved in stabilizing R2-2 particles in a separate and distinct manner from the other G* domains, G1, G2, and G3, residues 11–21, 22–32, and 33–43, respectively.
To determine the role of each of the three G* amphipathic helices in the N terminus of apoA-I in size heterogeneity of discoidal HDL, serial deletion mutants were constructed to produce Δ10, Δ21, Δ33, and Δ43 apoA-I, missing G0, G0-G1, G0-G2, and G0-G3, respectively; the common feature is they all are missing G0. Discoidal complexes of these mutants with DMPC were prepared and compared with complexes with FL-apoA-I by NDGGE analysis (Fig. 8B).
The results show that, although FL-apoA-I produces all five discretely sized DMPC particles from R2-1 to R2-5, the N-terminal deletion mutants displayed a decreasing ability to form particles larger than R2-2 with rank order Δ43 < Δ33 < Δ21 < Δ10 apoA-I. Interestingly, although Δ10, Δ21, and Δ33 apoA-I produced R2-2 particles as the predominant product, Δ43 apoA-I produced R2-1 particles as the predominant product. These results demonstrate the following: (i) each of the three G* amphipathic helices in the N-terminal G-helical domain of apoA-I contributes to the size heterogeneity of HDL particles, and (ii) although the formation of the R2-2 particle does not require the presence of the N-terminal 43 residues, the stability of this particle is affected without the N terminus, and at least one G* amphipathic helix is required for the stability of the R2-2 particles.
Because Δ10 apoA-I, which has three intact G* helices but no G0, does not form R2-4 and R2-5 particles as does FL-apoA-I, the G0 domain of apoA-I, residues 1–10, appears to play a role in the formation and stability of particles larger than R2-2. Our MDSA simulations show that the protein perimeters of the FL double belts for all three sizes of R2 particles are closed at the terminal (overlap) domain by interactions of the two proline-rich N-terminal G0 domains with one another (Fig. 2). The results of Fig. 8B is therefore compatible with the hypothesis that the proline-rich N-terminal G0 is of major importance in stabilizing R2 particles in a separate and distinct manner from the other G* domains, G1, G2, and G3, residues 11–21, 22–32, and 33–43, respectively.
Properties of the Smallest R2 Particle, R2-0
We hypothesized that the R2-0 particle might have a physiological relevance to biogenesis of HDL. Complexes of apoA-I or Δ43apoA-I with POPC at three different lipid:protein molar ratios were prepared and subjected to NDGGE analysis (Fig. 9A). The results indicate that, in addition to formation of R2-2 and R2-1 assemblies, the full-length plasma apoA-I formed significant amounts of the small R2-0 particles at the lower lipid:protein molar ratios of 50:2 and 100:2. The Δ43apoA-I, on the other hand, formed exclusively R2-0 assemblies at a 50:2 ratio and predominantly the R2-1 particles at a 100:2 ratio, the latter similar to the Δ43apoA-I complexes made with DMPC (Fig. 8B). At a 200:2 ratio, R2-2 became the predominant R2 particle in POPC·apoA-I complexes and R2-1 remained the predominant R2 particle in POPC·Δ43apoA-I. However, in the latter case, large amounts of assemblies with more than two molecules of apoA-I (Sd 150–200 Å, R4/3) were formed.
FIGURE 9.
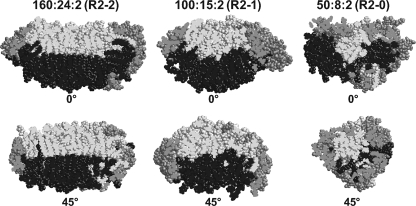
Properties of the smallest reconstituted PL-rich HDL particle, R2-0. A, NDGGE analysis of POPC·apoA-I (lanes 1–3) or Δ43apoA-I (lanes 4–6) complexes at different lipid:protein molar ratios in a 4–20% polyacrylamide gel run for 48 h. B, NDGGE analyses illustrating the dynamic range of lipid binding possessed by R2-0 PL-rich HDL particles. C, space-filling models of a representative example of the final structures of the MDSA simulations of the 160:24:2 100:15:2, and 50:8:2 particles illustrating the mechanism whereby we suggest that the apoA-I saddle-shaped double belt particle can accommodate a wide range of lipid concentrations. Protein is shown in gray and lipids in black. These models illustrate the hypothesis that PL-rich HDL is able to incorporate increasing PL by an incremental unwinding (left to right in the figure) of the twisted apoA-I double belt saddle shape of the relatively lipid-poor R2-0 particle to produce a planar discoidal R2-2 particle.
To determine the lipid binding range of R2-0 assemblies experimentally, a broad spectrum of POPC·Δ43apoA-I ratios (4:2 to 600:2) was used to prepare the complexes (Fig. 9B). The results show that the R2-0 family of particles displays a wide range of particle size varying from Stokes diameters of 72–78 Å and lipid binding that varies from a few to 50 or more POPC per particle.
Fig. 9C, showing space-filling models of the final structures of the MDSA simulations of the 160:24:2, 100:15:2, and 50:8:2 particles, illustrates the mechanism whereby we suggest that the apoA-I saddle-shaped double belt particle can accommodate a wide range of lipid concentrations. Based upon these models, we propose that PL-rich HDL is able to incorporate increasing PL by an incremental unwinding (left to right in Fig. 9C) of the apoA-I double belt structure of the relatively lipid-poor R2-0 particle to produce, ultimately, a planar discoidal particle, R2-2.
DISCUSSION
Strengths and Weaknesses of MDSA
Use of MD to examine the structure of apoA-I in PL-rich discoidal HDL particles involves simulations of periodic boundary boxes containing several hundred thousand atoms. Because of current limitations on simulation, we used a combination of all-atom MDSA to bypass kinetic energy barriers to the global minimum structures of these particles and T-jump MD simulations to scan the per-residue helix stability of truncated apoA-I in PL-rich HDL (14). The pros and cons of the use of temperatures as high as 500 K during MD simulations been discussed in detail elsewhere (14).
Experimental Tests of MDSA Models
Studies of reconstituted PL-rich HDL particles reported here provide support for certain features of the MDSA models.
(i) The general size and shape of the 160:24:2 (R2-2), 100:15:2 (R2-1), and 50:8:2 (R2-0) MDSA particles match the size and/or shape of reconstituted particles with comparable stoichiometries determined by NDGGE and negative stain EM. Although the reconstituted particles do not contain UC, their size and shape also match that of truncated apoA-I particles containing no UC (16).
Table 1 represents a comparison of the average dimensions of the 160:24:2, 100:15:2, and 50:8:2 particles simulated by MDSA with dimensions of previous 310 K MD simulations of comparable particles containing truncated apoA-I (16) and dimensions of reconstituted particles analyzed by NDGGE (5). EM measurements have been reported and discussed in our previous publication (16). Because drying induces mechanical stresses to particles subjected to negative stain EM, and because HDL is a soft form of condensed matter that is easily deformable by mechanical stresses, the reliability of negative stain EM for measuring the dimensions of these particles could be suspect.
TABLE 1.
Dimensions of MD-simulated PL-rich particles
Comparison of current MDSA simulations of particles containing FL-apoA-I with previous 310 K simulations of particles containing truncated apoA-I and reconstituted particles analyzed by NDGGE.
| Current MDSA particles | Average dimensions ± S.D. |
||
|---|---|---|---|
| x | y | z | |
| Å | |||
| 160:24:2 (R2-2) | 119.9 ± 3.7 | 106.6 ± 9.2 | 68.7 ± 4.5 |
| x:y ratio | 1.12:1 | ||
| 100:15:2 (R2-1) | 109.7 ± 8.1 | 96.3 ± 1.9 | 62.5 ± 2.8 |
| x:y ratio | 1.14:1 | ||
| 50:8:2 (R2-0) | 108.7 ± 13.3 | 73.3 ± 6.6 | 66.4 ± 3.8 |
| x:y ratio | 1.48:1 | ||
| Previous MD particles containing truncated apoA-I (16) | Dimensions |
||
|---|---|---|---|
| x | y | z | |
| Å | |||
| 160:2 (R2-2) | 112.8 | 97.7 | 59.6 |
| x:y ratio | 1.15:1 | ||
| 100:2 (R2-1) | 110.5 | 86.4 | 63.0 |
| x:y ratio | 1.28:1 | ||
| 50:2 (R2-0) | 94.7 | 68.5 | 60.5 |
| x:y ratio | 1.38:1 | ||
| NDGGE | Dimensions, Stokes diameter |
|---|---|
| Å | |
| R2-2:POPC (full length) | 102 |
| R2-2:DMPC (full length) | 105.5 ± 0.7 (5) |
| R2-2:DMPC (Δ43) | 105.5 ± 0.6 (5) |
| R2-1:POPC (full length) | 93–95 |
| R2-1:POPC (Δ43) | 92–93 |
| R2-1:DMPC (full length) | 97.9 ± 0.5 (5) |
| R2-1:DMPC (Δ43) | 98.6 ± 0.9 (5) |
| R2-0:POPC (full length) | 73 |
| R2-0:POPC (Δ43) | 72–78 |
From Table 1, the dimensions of the full-length particles are generally larger than those of the truncated particles. The x:y axis ratios, reflecting the particles intermediate oblate/prolate ellipsoidal shapes, are more comparable.
Also from Table 1, the dimensions of the x axes of both MDSA- and MD-simulated particles are significantly larger than the Stokes diameters measured by NDGGE for the corresponding reconstituted R2-2-, R2-1, and R2-0 particles; the Stokes diameters correspond more closely to the dimensions of the y axes. We hypothesize that the intermediate y axes play the dominant role in determining the NDGGE mobility of oblate/prolate ellipsoid particles like reconstituted PL-rich HDL particles; the z axes will have little or no influence, whereas the x axes, on average, will tend to orient parallel to the direction of particle motion in the electric field.
(ii) With a single exception, physical-chemical properties of reconstituted R2-2, R2-1, and R2-0 (percent helicity and chemical shifts of choline N-methyl proton resonances) are compatible with measurement of comparable properties for the 160:24:2, 100:15:2, and 50:8:2 MDSA particles. The exception is that percent helicity for the experimental R2-0 and the simulated 50:8:2 particles, 60 and 73%, respectively, are not comparable. There are four possible reasons for this discrepancy as follows. (a) The discrepancy might be the result of the use of Δ43 apoA-I for our CD studies. (b) It may be related to the presence of UC in the MDSA particle but not in the rHDL particle. (c) The discrepancy could suggest that the 50:8:2 MDSA structure is not fully equilibrated. (d) The CD results do not accurately reflect the helicity.
The measurements of total helicity as a function of time of simulation for the three different particles (Fig. 1C) show that there is a 3–4% increase in helicity with cool down between 500 and 310 K (10–20 ns); 160:24:2 increases from 74 to 77%; 100:15:2 increases from 70 to 73%, and 50:8:2 increases from 68 to 72%. The 4% increase in helicity for 50:8:2 with cool down suggests that the equilibration helicity for the particle is greater than 68%; if it were lower, one would think the percent helicity would continue to decrease. This increase in helicity represents one piece of evidence that the 60% helicity measured by CD for the R2-0 rHDL particle may be too low. Alternatively, it is possible that simulation technologies are not yet able to accurately model smaller particles that may exhibit significant regions of unfolded structure.
The two approaches used to model the 1H NMR spectroscopy chemical shifts for reconstituted PL-rich particles are good support for the MDSA-determined structures. The simple circular/elliptical lipid surface geometries used to model the chemical shift data suggest a circular discoidal structure for R2-2 and elliptical structures with increasing axial ratios for R2-1 and R2-0, respectively, compatible with the MDSA simulation results. This simple geometric approach is validated by the excellent fit achieved for all three of the reconstituted particles by using the annular/bulk POPC measurements of the MDSA simulations to model the chemical shift data. Taken together, the 1H NMR data provide strong evidence for the general correctness of the saddle-shaped structures reported here and in our previous computer simulations (14, 16).
(iii) MDSA structures provide a structural mechanism for explaining the effects of truncations of the G* domains of apoA-I on the size distribution of reconstituted R2 particles. Our MDSA simulations show that the protein perimeters of the full-length double belts for all three sized R2 particles are closed by interactions of the two proline-rich N-terminal G0 domains with one another (Fig. 3). We propose that the antiparallel proline-rich N-terminal G0 domains, through end to end association, are of major importance in stabilizing R2-2 particles in a separate and distinct manner from the other G* domains.
Structural Relevance of MDSA Models
The MDSA studies reported here show clearly that FL-apoA-I forms saddle-shaped conformations on the edges of PL-rich discs similar to those seen in our initial MD simulation study of truncated apoA-I (14–16). All of the resulting 160:24:2, 100:15:2, and 50:8:2 double belt structures form left-handed figure eight saddle shapes with pseudo-D2 symmetry, suggesting that MDSA is sampling a valid region of the conformational space.
In our previous publication of MD simulations at 310 K of PL-rich HDL particles containing truncated apoA-I without UC (16), we showed that decreasing the POPC surface area, even though the protein boundary length was unchanged, for example, during conversion from R2-2 to R2-1, produced a saddle-shaped curvature of the POPC monolayer-monolayer interface that resembled a patch of a minimal surface. Fig. 10 shows that the monolayer-monolayer interfaces of all three PL-rich HDL particles containing FL-apoA-I and UC simulated by MDSA also approximate patches of minimal surface that increase in curvature with decreasing particle size; alternating curvatures with opposite signs with a period of 90° are recognizable in the monolayer-monolayer interface (see Fig. 9 in Ref. 16 for more detailed examples).
FIGURE 10.
Rotational fit of space-filling models of a representative example of each of the final structures of the MDSA simulations of the 160:24:2, 100:15:2, and 50:8:2 particles to a patch of minimal surface. Side view of space-filling images of a cross-section of each particle rotated with a periodicity of 45° to show conversion of a curved monolayer-monolayer interface to a flat interface. This can be extrapolated to the alternating curvature of a minimal surface lipid bilayer monolayer-monolayer interface that has a periodicity of 90°. See Ref. 16 for a more complete illustration of the 90° periodicity of a minimal surface. ApoA-I molecules are shown in gray; POPC molecules of the two leaflets are shown in light gray and black, respectively.
The presence of minimal surface monolayer-monolayer interfaces in our simulations of both truncated and FL-apoA-I-containing PL-rich HDL particles also suggests that MDSA is sampling a valid region of the conformational space. The recent confirmation in reconstituted particles by Miyazaki et al. (40) of our computational observations of saddle-shaped minimal surfaces in PL-rich HDL particles provides strong evidence for the general correctness of the saddle-shaped structures reported here.
The two regions of the double belt with extensive loss of helicity in Fig. 4 lie on adjacent parts of the double belt as follows: at the juncture of helices G2-G3 with helix 1 and within helix 8. These two domains form reproducible loops (Fig. 3, C and D, blue and cyan, respectively) that are responsible for conversion of the belt structures from planar rings to twisted saddle shapes. As the particle size decreases, there is an increase in degree of bend as the angle of the loops becomes more acute. The effect is much like a two-sided clam shell (see Fig. 7 in Ref. 16) but with a twist.
We have performed sequence alignment and conservation analyses on a data base of apoA-I sequences from 11 species of fish, 2 species of amphibians, 3 species of birds, and 18 species of mammals.5 There are three tyrosine residues, Tyr18, Tyr115, and Tyr195, representing three out of seven tyrosines in human apoA-I, that are completely conserved from fish to mammals out of a total of eight completely conserved residues.
Tyr18 in helix G2 has been shown by Tanaka et al. (41) to be important for assembly of HDL complexes through interactions of apoA-I with phospholipid; “….the α-helix around Tyr18 (conceals) a potential lipid-binding region in the N-terminal domain, which (is) exposed by the disruption of the helix-bundle structure.” Tyr18 may be required to maintain a certain conformation in which the initial lipid-binding sites of the protein are accessible to the lipid (42). It is therefore significant that Tyr18 resides in a helically stable region (Fig. 4).
The residues Tyr115 and Arg116 in helix 4 are conserved as a pair in all but three of the 31 species and form, after Glu191–Tyr192, the second most conserved sequence pair in apoA-I. Tyr115 is in a region that becomes much less helical as 160:24:2 converts to 100:15:2 and 50:8:2 (Fig. 4).
The residues Tyr192 and Glu191 in helix 8 form a 100% conserved sequence motif, suggesting strongly that this site in apoA-I has an important biological function. It is therefore of interest that Shao et al. (43) suggest that Tyr192 acts as a preferred atherogenic oxidation target in apoA-I. It is significant that the conserved Glu191–Tyr192 pair is located within a solvent-exposed loop on the edge of the bilayer disc, a conformation ideally suited to act as an oxidation target within HDL (Fig. 4).
Tyr166, well conserved only in mammals, has been suggested also to act as a preferred atherogenic oxidation target in apoA-I (44). Like Tyr115, Tyr166 lies in a region of apoA-I that becomes much less helical as 160:24:2 converts to 100:15:2 and 50:8:2 (Fig. 4).
In this study, we have shown that the size distribution of pre-formed DMPC or POPC·apoA-I assemblies is regulated by both kinetic and thermodynamic factors. Compared with DMPC·apoA-I assemblies, POPC·apoA-I particles have a broader size distribution when formed, but the two types of complexes become more similar with time. This represents the significantly poorer transferability of POPC versus DMPC and is most likely related to the lower critical micelle concentration of the former versus the latter (45).
The most important lesson from these results is that the sharpening of bands with time, equivalent to a narrowing of distribution of assembly size toward a much more monodisperse particle, suggests that each discontinuous heterogeneous particle (R2-1, R2-2, and R2-0) has a relatively narrow thermodynamic energy minimum. Thus, for POPC assemblies formed by cholate dialysis, the initial particles have not equilibrated kinetically and form a broad distribution of particle sizes around the most energetically favorable particle. This broad distribution decays with time or temperature with POPC transfer between particles to coalesce into particles that fall into a narrower free energy minimum.
POPC·apoA-I assemblies, however, differ quantitatively from DMPC·apoA-I assemblies in that the former do not form the larger R2-4 and R2-5 particles and form an additional smaller particle, R2-0. The R2-0 family of POPC·apoA-I particles are unique in that they have an extraordinary range in stoichiometry of lipid binding, from a few molecules of POPC to 50 or more. As already noted, we previously hypothesized that lipid-free apoA-I incorporates PL, perhaps one molecule at a time, within a highly elastic lipid pocket to produce the relatively PL-poor R2-0 particle. The proposed lipid pocket is an extremely elastic clam-like structure, with pseudo-D2 symmetry (16); each of the “jaws” back to back encompasses one of the two POPC monolayers (Fig. 9C).
We hypothesize that the distinct local energy minima of R2-0 compared with the less stable R2-1 particle is driven by the predominance of protein-lipid interactions, particularly the high percent of bound POPC that form phosphorus-nitrogen salt bridges with the protein. This model suggests that the reason DMPC does not form R2-0 particles is related to low hydrophobicity and/or bilayer thickness relative to POPC.
Wu et al. (46) recently published a model for rHDL particles with a composition similar to R2-2 that is dramatically different from previous “discoidal” rHDL models. In their model, termed a double super helix, apoA-I possesses an open helical shape that twists around a central ellipsoidal lipid phase.
The double super helix model for R2-2 is interesting. It is similar to our model for 50:8:2 (R2-0) shown in Fig. 9C in that the proposed double super helix forms a left-handed spiral as it wraps around the central lipid. The major difference between the models is that in the double super helix model the terminal domains are separated at opposite ends of the ellipsoidal structure, whereas in our model the G0 domains wrap back to self-associate (Fig. 3B).
Our 1H NMR data suggest that annular POPC accounts for 35% of total POPC in the R2-2 particle. Analysis of the double super helix model available on line shows that ∼33% of the total POPC is within 7 Å of the protein, so that in principle the double super helix model is compatible with our 1H NMR data. However, the double super helix model possesses no classic bulk bilayer lipid but rather is organized as an end-capped hexagonal I phase lipid (rod-shaped micelle). The hexagonal I phase is seen only under unique, specialized conditions. Because detergents, such as lyso-PC, form hexagonal I phases only under conditions of very low water content, it is not clear how apoA-I could stabilize such a structure in a lipid, such as POPC, that is not a detergent nor is it clear what sort of 1H NMR signal would be seen if it could. Although outside the scope of the present discussion, additional evidence against the double super helix model also includes stage-tilted cryo-electron microscopy (47) that clearly shows the presence of discs.
There are discrepancies in the literature for the Stokes diameter of catalase in NDGGE (6, 48, 49, 50–52). Our laboratory has been using 104 Å as the Stokes diameter for catalase in our previous studies (5, 35) and in this study. As most rHDL assemblies are around 100 Å, the size used for catalase is particularly important for the determination of Stokes diameters of particles. Furthermore, we have observed that the length of time for running the gradient gel affects the determination of particle size by NDGGE. Under our experimental conditions, the gel (4–20% polyacrylamide) has to be run for at least 48 h at 125 V to reach an equilibrium at which the assemblies will no longer move (5). For these reasons, and because individual rHDL particles, e.g. R2-0 (Fig. 9B), are continuously heterogeneous in size, we suggest that the nomenclature for rHDL particles would best be based upon terminology that is independent of size.
Conclusions
In this study, we show that apoA-I can assemble a range of dynamic lipoprotein particles containing a continuously variable number of lipid molecules. Through a combination of computation and experimentation, we provide evidence that apoA-I accomplishes this feat of unique flexibility by the incremental twisting or untwisting of a saddle-shaped apoA-I double belt structure that creates minimal surface patches of lipid bilayer. These results, especially the excellent fit of the simulations to the 1H NMR data, combined with the recent confirmation in reconstituted particles by Miyazaki et al. (40) of saddle-shaped minimal surfaces in reconstituted PL-rich HDL particles, provide strong evidence for the global correctness of the saddle-shaped structures reported here and in our previous computer simulations (14, 16).
Supplementary Material
Acknowledgments
We thank Drs. Vinod Mishra and Michael J. Jablonsky for their assistance in proton NMR and CD spectroscopy. We also thank the University of Alabama at Birmingham Information Technology and the Department of Mechanical Engineering for use of the IBM Blue Gene/L rack that they jointly maintain. EM analysis was performed in part through the use of the Vanderbilt University Medical Center Research EM Resource (supported by National Institutes of Health Grants DK20539, CA68485, and DK58404).
This work was supported, in whole or in part, by National Institutes of Health Grant P01 HL-34343.

The on-line version of this article (available at http://www.jbc.org) contains supplemental “Materials and Methods,” Figs. S1–S4, and additional references.
D. Bashtovyy and J. P. Segrest, unpublished data.
- HDL
- high density lipoprotein
- ADF
- annular distribution functions
- apo
- apolipoprotein
- DMPC
- dimyristoylphosphatidylcholine
- FL
- full-length
- MD
- molecular dynamics
- MDSA
- molecular dynamics simulated annealing
- NDGGE
- nondenaturing gradient gel electrophoresis
- PL
- phospholipids
- POPC
- palmitoyloleoylphosphatidylcholine
- R2 or R3 or R4
- reconstituted high density lipoprotein particles containing 2, 3, or 4 molecules of apoA-I
- rHDL
- reconstituted high density lipoproteins
- r.m.s.d.
- root mean square deviation
- UC
- unesterified cholesterol
- VMD
- visual molecular dynamics
- HPLC
- high pressure liquid chromatography.
REFERENCES
- 1.Linsel-Nitschke P., Tall A. R. (2005) Nat. Rev. Drug Discov. 4, 193–205 [DOI] [PubMed] [Google Scholar]
- 2.Segrest J. P., Anantharamaiah G. M. (1994) Curr. Opin. Cardiol. 9, 404–410 [PubMed] [Google Scholar]
- 3.Segrest J. P., Jackson R. L., Morrisett J. D., Gotto A. M., Jr. (1974) FEBS Lett. 38, 247–258 [DOI] [PubMed] [Google Scholar]
- 4.Segrest J. P., Jones M. K., Klon A. E., Sheldahl C. J., Hellinger M., De Loof H., Harvey S. C. (1999) J. Biol. Chem. 274, 31755–31758 [DOI] [PubMed] [Google Scholar]
- 5.Li L., Chen J., Mishra V. K., Kurtz J. A., Cao D., Klon A. E., Harvey S. C., Anantharamaiah G. M., Segrest J. P. (2004) J. Mol. Biol. 343, 1293–1311 [DOI] [PubMed] [Google Scholar]
- 6.Li H., Lyles D. S., Thomas M. J., Pan W., Sorci-Thomas M. G. (2000) J. Biol. Chem. 275, 37048–37054 [DOI] [PubMed] [Google Scholar]
- 7.Panagotopulos S. E., Horace E. M., Maiorano J. N., Davidson W. S. (2001) J. Biol. Chem. 276, 42965–42970 [DOI] [PubMed] [Google Scholar]
- 8.Davidson W. S., Hilliard G. M. (2003) J. Biol. Chem. 278, 27199–27207 [DOI] [PubMed] [Google Scholar]
- 9.Bhat S., Sorci-Thomas M. G., Alexander E. T., Samuel M. P., Thomas M. J. (2005) J. Biol. Chem. 280, 33015–33025 [DOI] [PubMed] [Google Scholar]
- 10.Martin D. D., Budamagunta M. S., Ryan R. O., Voss J. C., Oda M. N. (2006) J. Biol. Chem. 281, 20418–20426 [DOI] [PubMed] [Google Scholar]
- 11.Bhat S., Sorci-Thomas M. G., Tuladhar R., Samuel M. P., Thomas M. J. (2007) Biochemistry 46, 7811–7821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davidson W. S., Thompson T. B. (2007) J. Biol. Chem. 282, 22249–22253 [DOI] [PubMed] [Google Scholar]
- 13.Zorich N. L., Kézdy K. E., Jonas A. (1987) Biochim. Biophys. Acta 919, 181–189 [DOI] [PubMed] [Google Scholar]
- 14.Jones M. K., Catte A., Patterson J. C., Gu F., Chen J., Li L., Segrest J. P. (2009) Biophys. J. 96, 354–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klon A. E., Segrest J. P., Harvey S. C. (2002) J. Mol. Biol. 324, 703–721 [DOI] [PubMed] [Google Scholar]
- 16.Catte A., Patterson J. C., Jones M. K., Jerome W. G., Bashtovyy D., Su Z., Gu F., Chen J., Aliste M. P., Harvey S. C., Li L., Weinstein G., Segrest J. P. (2006) Biophys. J. 90, 4345–4360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Catte A., Patterson J. C., Bashtovyy D., Jones M. K., Gu F., Li L., Rampioni A., Sengupta D., Vuorela T., Niemelä P., Karttunen M., Marrink S. J., Vattulainen I., Segrest J. P. (2008) Biophys. J. 94, 2306–2319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kerr I. D., Sankararamakrishnan R., Smart O. S., Sansom M. S. (1994) Biophys. J. 67, 1501–1515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sankararamakrishnan R., Sansom M. S. (1995) Biophys. Chem. 55, 215–230 [DOI] [PubMed] [Google Scholar]
- 20.Bassolino-Klimas D., Tejero R., Krystek S. R., Metzler W. J., Montelione G. T., Bruccoleri R. E. (1996) Protein Sci. 5, 593–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li H., Tejero R., Monleon D., Bassolino-Klimas D., Abate-Shen C., Bruccoleri R. E., Montelione G. T. (1997) Protein Sci. 6, 956–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harvey S. C., Tan R. K., Cheatham T. E. (1998) J. Comput. Chem. 19, 726–740 [Google Scholar]
- 23.Kalé L., Skeel R., Bhandarkar M., Brunner R., Gursoy A., Krawetz N., Phillips J., Shinozaki A., Varadarajan K., Schulten K. (1999) J. Comp. Phys. 151, 283–312 [Google Scholar]
- 24.Humphrey W., Dalke A., Schulten K. (1996) J. Mol. Graph. 14, 33–38, 27–38 [DOI] [PubMed] [Google Scholar]
- 25.Jorgensen W. L., Chandrasekhar J., Madura J. D., Impey R. W., Klein M. L. (1983) J. Chem. Phys. 79, 926–935 [Google Scholar]
- 26.MacKerell A. D., Jr., Bashford D., Bellot M., Dunbrack R. L., Jr., Evanseck J., Field M. J., Fischer S., Gao J., Guo H., Ha S., Joseph D., Kuchnir L., Kuczera K., Lau F. T., Mattos C., Michnick S., Ngo T., Nguyen D. T., Prodhom B., Reiher W. E., 3rd, Roux B., Schlenkrich M., Smith J., Stote R., Straub J., Watanabe M., Wiorkiewicz-Kuczera J., Yin D., Karplus M. (1998) J. Phys. Chem. B 102, 3586–3616 [DOI] [PubMed] [Google Scholar]
- 27.Brooks B. R., Bruccoleri R. E., Olafson B. D., States D. J., Swaminathan S., Karplus M. (1983) J. Comput. Chem. 4, 187–217 [Google Scholar]
- 28.Feller S. E., Pastor R. W. (1997) Pac. Symp. Biocomput. 142–150 [PubMed] [Google Scholar]
- 29.Schlenkrich M., Brinkmann J., MacKerell A. D., Jr., Karplus M. (1996) in Biological Membranes: A Molecular Perspective from Computation and Experiment (Merz K. M., Roux B. eds) pp. 31–81, Birkhauser Boston, Inc., Cambridge, MA [Google Scholar]
- 30.Frishman D., Argos P. (1995) Proteins 23, 566–579 [DOI] [PubMed] [Google Scholar]
- 31.Anantharamaiah G. M., Garber D. W. (1996) Methods Enzymol. 263, 267–282 [DOI] [PubMed] [Google Scholar]
- 32.Rogers D. P., Brouillette C. G., Engler J. A., Tendian S. W., Roberts L., Mishra V. K., Anantharamaiah G. M., Lund-Katz S., Phillips M. C., Ray M. J. (1997) Biochemistry 36, 288–300 [DOI] [PubMed] [Google Scholar]
- 33.Jonas A. (1986) Methods Enzymol. 128, 553–582 [DOI] [PubMed] [Google Scholar]
- 34.Mishra V. K., Palgunachari M. N., Datta G., Phillips M. C., Lund-Katz S., Adeyeye S. O., Segrest J. P., Anantharamaiah G. M. (1998) Biochemistry 37, 10313–10324 [DOI] [PubMed] [Google Scholar]
- 35.Brouillette C. G., Jones J. L., Ng T. C., Kercret H., Chung B. H., Segrest J. P. (1984) Biochemistry 23, 359–367 [DOI] [PubMed] [Google Scholar]
- 36.Segrest J. P., Li L., Anantharamaiah G. M., Harvey S. C., Liadaki K. N., Zannis V. (2000) Curr. Opin. Lipidol. 11, 105–115 [DOI] [PubMed] [Google Scholar]
- 37.Jonas A., Wald J. H., Toohill K. L., Krul E. S., Kézdy K. E. (1990) J. Biol. Chem. 265, 22123–22129 [PubMed] [Google Scholar]
- 38.Davidson W. S., Hazlett T., Mantulin W. W., Jonas A. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 13605–13610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brouillette C. G., Compans R. W., Brandts J. F., Segrest J. P. (1982) J. Biol. Chem. 257, 12–15 [PubMed] [Google Scholar]
- 40.Miyazaki M., Nakano M., Fukuda M., Handa T. (2009) Biochemistry 48, 7756–7763 [DOI] [PubMed] [Google Scholar]
- 41.Tanaka M., Vedhachalam C., Sakamoto T., Dhanasekaran P., Phillips M. C., Lund-Katz S., Saito H. (2006) Biochemistry 45, 4240–4247 [DOI] [PubMed] [Google Scholar]
- 42.Fang Y., Gursky O., Atkinson D. (2003) Biochemistry 42, 13260–13268 [DOI] [PubMed] [Google Scholar]
- 43.Shao B., Oda M. N., Bergt C., Fu X., Green P. S., Brot N., Oram J. F., Heinecke J. W. (2006) J. Biol. Chem. 281, 9001–9004 [DOI] [PubMed] [Google Scholar]
- 44.Wu Z., Wagner M. A., Zheng L., Parks J. S., Shy J. M., 3rd, Smith J. D., Gogonea V., Hazen S. L. (2007) Nat. Struct. Mol. Biol. 14, 861–868 [DOI] [PubMed] [Google Scholar]
- 45.Tanford C. (1980) The Hydrophobic Effect: Formation of Micelles and Biological Membranes, 2nd Ed., p. 112, John Wiley & Sons, Inc., New York [Google Scholar]
- 46.Wu Z., Gogonea V., Lee X., Wagner M. A., Li X. M., Huang Y., Undurti A., May R. P., Haertlein M., Moulin M., Gutsche I., Zaccai G., Didonato J. A., Hazen S. L. (2009) J. Biol. Chem. 284, 36605–36619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van Antwerpen R., Chen G. C., Pullinger C. R., Kane J. P., LaBelle M., Krauss R. M., Luna-Chavez C., Forte T. M., Gilkey J. C. (1997) J. Lipid Res. 38, 659–669 [PubMed] [Google Scholar]
- 48.Ji Y., Jonas A. (1995) J. Biol. Chem. 270, 11290–11297 [DOI] [PubMed] [Google Scholar]
- 49.Maiorano J. N., Jandacek R. J., Horace E. M., Davidson W. S. (2004) Biochemistry 43, 11717–11726 [DOI] [PubMed] [Google Scholar]
- 50.Duong P. T., Collins H. L., Nickel M., Lund-Katz S., Rothblat G. H., Phillips M. C. (2006) J. Lipid Res. 47, 832–843 [DOI] [PubMed] [Google Scholar]
- 51.Sorci-Thomas M. G., Curtiss L., Parks J. S., Thomas M. J., Kearns M. W. (1997) J. Biol. Chem. 272, 7278–7284 [DOI] [PubMed] [Google Scholar]
- 52.Laccotripe M., Makrides S. C., Jonas A., Zannis V. I. (1997) J. Biol. Chem. 272, 17511–17522 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.