Abstract
The Bcl-2 family regulates induction of apoptosis at the mitochondria. Essential to this regulation are the interactions between Bcl-2 family members, which are mediated by Bcl-2 homology (BH) domains. Vaccinia virus F1L is a unique inhibitor of apoptosis that lacks significant sequence similarity with the Bcl-2 family and does not contain obvious BH domains. Despite this, F1L inhibits cytochrome c release from mitochondria by preventing Bak and Bax activation. Although F1L constitutively interacts with Bak to prevent Bak activation, the precise mechanism of this interaction remains elusive. We have identified highly divergent BH domains in F1L that were verified by the recent crystal structure of F1L (Kvansakul, M., Yang, H., Fairlie, W. D., Czabotar, P. E., Fischer, S. F., Perugini, M. A., Huang, D. C., and Colman, P. M. (2008) Cell Death Differ. 15, 1564–1571). Here we show that F1L required these BH domains to interact with ectopically expressed and endogenous Bak. The interaction between F1L and Bak was conserved across species, and both F1L and the cellular antiapoptotic protein Mcl-1 required the Bak BH3 domain for interaction. Moreover, F1L replaced Mcl-1 during infection, as the Bak·Mcl-1 complex was disrupted during vaccinia virus infection. In contrast to UV irradiation, vaccinia virus infection did not result in rapid degradation of Mcl-1, consistent with our observation that vaccinia virus did not initiate a DNA damage response. Additionally, Mcl-1 expression prevented Bak activation and apoptosis during infection with a proapoptotic vaccinia virus devoid of F1L. Our data suggest that F1L replaces the antiapoptotic activity of Mcl-1 during vaccinia virus infection by interacting with Bak using highly divergent BH domains.
Keywords: Apoptosis, Cytochromes/Cytochrome c, Protein/Protein-Protein Interactions, Signal Transduction, Viruses/DNA, Viruses/Pox
Introduction
Apoptosis is an evolutionarily conserved process involved in tissue homeostasis and removal of unwanted, damaged, and virally infected cells (1). Mitochondria play a pivotal role in the execution of apoptosis through the release of apoptogenic molecules, such as cytochrome c (2). Mitochondrial integrity is governed by the Bcl-2 family of proteins (3, 4). Members of this family are united by the presence of one or more Bcl-2 homology (BH)3 domains, which participate in homo- and heterotypic interactions to ultimately determine the fate of a cell (3–5). Anti-apoptotic family members, such as Bcl-2, Bcl-xL, and Mcl-1, function to maintain mitochondrial integrity by inhibiting the proapoptotic molecules Bak and Bax (3, 5). The BH3-only proteins, which function upstream of Bak and Bax to sense various apoptotic insults, bear little resemblance to the rest of the family with the exception of a BH3 domain, which is crucial for their proapoptotic activity (6). Once activated, Bak and Bax oligomerize in the outer mitochondrial membrane, leading to disruption of the mitochondrial membrane potential and release of cytochrome c (7–9). Cells deficient in Bak and Bax are refractory to apoptosis, emphasizing their crucial role in regulating the mitochondrial checkpoint (10, 11).
Apoptosis serves as a potent innate barrier to viral infection, and as such, many viruses have evolved strategies to interfere with apoptotic induction at the mitochondria (12–15). For example, many DNA viruses encode Bcl-2 homologues that function to inhibit apoptosis by interfering directly with Bak and Bax activation (14, 15). Poxviruses are large DNA viruses that also encode proteins that disarm the apoptotic cascade (16). With the exception of avipoxviruses, most members of the poxvirus family lack obvious Bcl-2 homologues (17–19). We identified F1L, which lacks apparent sequence similarity to members of the Bcl-2 family, as a unique antiapoptotic protein in vaccinia virus, the prototypical member of the Orthopoxvirus genus (20–22). F1L localizes to the outer mitochondrial membrane and inhibits cytochrome c release in response to a wide variety of apoptotic stimuli (20, 23–25). Moreover, a recombinant vaccinia virus lacking the F1L open reading frame induces apoptosis upon infection, highlighting the crucial antiapoptotic activity of F1L during virus infection (20, 26).
Despite lacking obvious sequence homology to Bcl-2 family members, F1L inhibits both Bak and Bax activation by binding to Bak and the BH3-only protein Bim, a set of interactions that closely parallels the binding activity of the cellular antiapoptotic protein Mcl-1 (20, 25, 27–29). F1L interacts constitutively with Bak to prevent Bak activation (20, 23). Surprisingly, however, binding assays only reveal a strong interaction between F1L and the BH3 peptide of Bim (26, 30). Although F1L functions similarly to cellular antiapoptotic Bcl-2 members, such as Mcl-1, the mechanism behind its activity is unclear as F1L does not possess evident BH domains. However, analysis of conserved biophysical properties in a multiple sequence alignment of known Bcl-2 family members revealed patterns that we matched to the F1L sequence and, thus, predicted the presence of putative BH domains. Moreover, the recent crystal structure of F1L from vaccinia virus-modified strain Ankara confirmed that despite a lack of apparent sequence homology to Bcl-2 proteins, F1L adopts a Bcl-2-like fold (30).
Here we show that in contrast to the weak affinity of F1L for the Bak BH3 peptide in binding assays (26, 30), F1L was able to interact with Bak from a wide range of cell lines. Moreover, the interaction between F1L and Bak relied on the divergent BH domains of F1L and the BH3 domain of Bak. Upon infection, F1L appeared to replace the role of Mcl-1 in regulating Bak, as Mcl-1 was displaced from Bak during vaccinia virus infection. Infection with vaccinia virus, however, did not lead to the rapid degradation of Mcl-1, which is commonly observed after UV treatment (27, 29, 31). In addition, the expression of Mcl-1 inhibited Bak activation and apoptosis during infection with a recombinant vaccinia virus devoid of F1L, highlighting the similar functions of F1L and Mcl-1 in regulating Bak and inhibiting apoptosis.
EXPERIMENTAL PROCEDURES
Cells and Viruses
HeLa, HEK 293T, chicken leghorn male hepatoma, Jurkat, and HuTK−-143B cells were obtained from the ATCC and cultured as previously described (17, 20). Jurkat cells overexpressing Bcl-2 were generated and maintained as previously described (32), and Bak- and Bax-deficient Jurkat cells were a gift from Dr. H. Rabinowich (University of Pittsburgh School of Medicine, Pittsburgh, PA) (33). Bak-deficient baby mouse kidney cells were generously provided by Dr. E. White (Rutgers University, Piscataway, NJ) (34) and were cultured in Dulbecco's modified Eagle's medium, 10% fetal bovine serum, 2 mm l-glutamine, 50 units/ml penicillin, and 50 μg/ml streptomycin. Vaccinia virus strain Copenhagen (VV) and vaccinia virus strain Copenhagen expressing EGFP (VVEGFP) were kindly provided by Dr. G. McFadden (University of Florida, Gainesville, FL). Vaccinia virus strain Copenhagen expressing FLAG-F1L, VVFLAG-F1L, was generated as described elsewhere (35), whereas vaccinia virus strain Copenhagen deficient in F1L, VVΔF1L, was generated by insertion of EGFP into the F1L open reading frame (20). All viruses were propagated in baby green monkey kidney cells as previously described (36).
Generation of Recombinant Viruses
Recombinant VVΔF1L expressing FLAG-F1L(V104A) (VVΔF1L-FLAG-F1L(V104A)), FLAG-F1L(G144F) (VVΔF1L-FLAG-F1L(G144F)), or FLAG-Mcl-1 (VVΔF1L-FLAG-Mcl-1) were created by homologous recombination into the thymidine kinase (TK) locus of VVΔF1L (37). In brief, 1 × 106 baby green monkey kidney cells were transfected with 5 μg of pSC66-FLAG-F1L(V104A), pSC66-F1L(G144F), or pSC66-FLAG-Mcl-1 using Lipofectin (Invitrogen) and simultaneously infected with VVΔF1L at a multiplicity of infection (m.o.i.) of 0.05. Recombinant viruses were selected by growth on HuTK−-143B cells in the presence of 5-bromodeoxyuridine (Sigma) and plaque-purified using 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (Rose Scientific Ltd.) to visualize recombinant, β-galactosidase-positive viruses.
F1L BH Domain Prediction
F1L BH domains were identified by correlating F1L sequence features, in particular hydrophobic spacing, to structural constraints of known Bcl-2 protein structures together with conservation patterns in a multiple sequence alignment. Sequences used were vaccinia virus Copenhagen F1L sequence (AAA48014), Bcl-xL (NP_612815), Bcl-2 (1GJH_A), Mcl-1 (NP_068779), Bax (NP_620116), Bak (2IMS_A), myxoma virus M11L (NP_051725), monkeypox virus C7L (NP_536460), yaba monkey tumor virus 16L (NP_938273), lumpy skin disease virus 017 (NP_150451), sheep poxvirus 014 (NP_659590), rabbit fibroma virus 011L (NP_051900), and swinepox virus 012 (NP_570172). Multiple alignments were performed using ClustalW (38).
Plasmid Construction
F1L-(84–226) and F1L-(109–226) were generated using the forward primer, 5′-GAATTCTCAGACAGTTGCCCACAAAGAC-3′ and 5′-GAATTCTCAATAGGGACATGAATATCAT-3′, respectively, each containing an EcoRI restriction site, and the reverse primer, 5′-GGATCCTTATCCTATCATGTATTT-3′, containing a BamHI restriction site. Residues 144–156 of F1L were deleted to generate F1LΔΒΗ1 with overlapping PCR using the F1L forward primer, 5′-GAATTCTCATGTTGTCGATGTTTATG-3′, containing an EcoRI restriction site, and the internal reverse primer, 5′-ACATCGTCTACCCAACCCCTTGTTATCCATTA-3′, to generate the N-terminal fragment. The internal forward primer, 5′-GGTAGACGATGTATGAATCC-3′, and F1L reverse primer,5′-GGATCCTTA TCCTATCATGTATTT-3′, containing a BamHI restriction site were used to generate the C-terminal fragment. The resulting PCR fragments were amplified together using the F1L forward and F1L reverse primers. F1LΔBH3 lacking residues 93–108 was generated in a similar manner to F1LΔBH1, with the exception of the internal reverse primer, 5′-CATGTCCCTATTTGATCTAGTCTTTGTGGGCA-3′, and the internal forward primer, 5′-AATAGGGACATGAATATCAT-3′. FLAG-Mcl-1 was amplified from pcDNA3-FLAG-Mcl-1 (provided by Dr. Gordon Shore, McGill University, Montreal, QC) using the forward primer, 5′-GTCGACATGGACTACAAAGACGATGACGATAAA-3′, containing a SalI restriction site, and the reverse primer, 5′-GGATCCCTATCTTATTAGATATGCCAA-3′, containing a BamHI restriction site. pcDNA3-FLAG-Bak, received from Dr. Gordon Shore (McGill University), was used to create a Bak construct lacking residues 72–87 composing the BH3 domain, FLAG-BakΔBH3, using the FLAG-Bak forward primer, 5′-AAGCTTATGGACTACAAAGACGATGACGACAAGATGGCTTCGGGGCAAGGC-3′, with a HindIII restriction site and the internal reverse primer, 5′-TGAGTCATAGCGCATGGTGCTGCTAGGTTGCAG-3′, to generate the N-terminal fragment. The C-terminal segment was amplified using the forward primer, 5′-CGCTATGACTCAGAGTTCCAG-3′, and the Bak reverse primer, 5′-GAATTCTCATGATTTGAAGAATCTTCG-3′, with an EcoRI restriction site. The resulting PCR products were combined and amplified using the FLAG-Bak forward primer and the Bak reverse primer. All PCR products were cloned into pGemT (Promega). F1L-(84–226), F1L-(109–226), F1LΔBH1, and F1LΔBH3 were subcloned into pEGFP-C3 (Clontech). FLAG-Mcl-1 was subcloned into pSC66, which places the gene under the control of a poxviral promoter (37), whereas FLAG-BakΔBH3 was subcloned into pcDNA3. pSC66-FLAG-F1L (22) was used as a template to make point mutations using the forward primer, 5′-CAACATATATTGTGATAAAGCAAGTAATGATTATAATAGGG-3′, and the reverse primer, 5′-CCCTATTATAATCATTACTTGCTTTATCACAATATATGTTC-3′, for pSC66-FLAG-F1L(V104A) and the forward primer, 5′-GGATAACAAGGGGTTGTTTGTAAGATTGGCG-3′, and reverse primer, 5′-CGCCAATCTTACAAACAACCCCTTGTTATCC-3′, for pSC66-FLAG-F1L(G144F) using QuikChange site-directed mutagenesis kit (Stratagene). pEGFP-F1L and pEGFP-Bcl-xL were generated as previously described (22, 35). pCR3.1-Mcl-1 was provided by Dr. H. Rabinowich (University of Pittsburgh School of Medicine, Pittsburgh, PA) (39), and pcDNA3-HA-Bak was provided by Dr. Grant McFadden (University of Florida, Gainesville, FL) (40). Wild-type Bak and Bak point mutations BakmtBH1 (W125A, G126E, R127A), BakmtBH2 (G175E, G176E, W177A), and BakmtBH3 (L78A, D83A) were kind gifts from Dr. D. George (University of Pennsylvania School of Medicine, Philadelphia, PA) (41).
Immunoprecipitations
To examine the interaction between F1L and endogenous Bak during virus infection, 1.4 × 107 HeLa, HEK 293T, mouse embryonic fibroblast, Jurkat, or leghorn male hepatoma cells were infected with VVFLAG-F1L or VVΔF1L at an m.o.i. of 5 for 12 h. Cells were lysed in CHAPS (Sigma) lysis buffer containing 2% (w/v) CHAPS, 150 mm NaCl, 50 mm Tris, pH 8.0, and EDTA-free protease inhibitor (Roche Diagnostics). After lysis, soluble fractions were immunoprecipitated with rabbit anti-Bak NT antibody (Upstate Biotech), and immune complexes were isolated with protein A-Sepharose (GE Healthcare). To examine regions of F1L that interact with Bak, 1 × 106 HEK 293T cells were transfected with pcDNA-HA-Bak along with pEGFP-C3 (Clontech), pEGFP-F1L, pEGFP-F1L-(84–226), pEGFP-F1L-(109–226), pEGFP-F1LΔBH1, or pEGFP-F1LΔBH3 with Lipofectamine 2000 (Invitrogen). Cells were lysed in 2% CHAPS lysis buffer and immunoprecipitated with goat anti-GFP antibody (Dr. Luc Berthiaume, University of Alberta, Edmonton, Alberta) and protein G-Sepharose (GE Healthcare). Point mutations within the BH domains of F1L were first assessed for Bak binding by infecting 7 × 106 HEK 293T cells with VV at an m.o.i. of 5 and transfecting with pSC66, pSC66-FLAG-F1L, pSC66-FLAG-F1L(V104A), or pSC66-FLAG-F1L(G144F) for 14 h. Immune complexes were isolated with mouse anti-FLAG M2 (Sigma) and protein G-Sepharose after lysis in 2% CHAPS lysis buffer. The interaction between Bak and the F1L point mutants was examined by infecting 7 × 106 HEK 293T cells with VVΔF1L, VVFLAG-F1L, VVΔF1L-FLAG-F1L(V104A), or VVΔF1L-FLAG-F1L(G144F). Twelve hours post-infection, cells were lysed in 2% CHAPS lysis buffer followed by immunoprecipitation with mouse anti-FLAG M2. To determine the region of Bak responsible for binding Mcl-1 and F1L, 1 × 106 HEK 293T cells were transfected with pEGFP-C3, pEGFP-F1L, pCR3.1-Mcl-1, pcDNA3-FLAG-Bak, or pcDNA3-FLAG-BakΔBH3 with Lipofectamine 2000 for 18 h. Cells were lysed in 2% CHAPS lysis buffer and immunoprecipitated with rabbit anti-Mcl-1 antibody (Assay Designs) and protein A-Sepharose or goat anti-GFP antibody and protein G-Sepharose. The effect of Bak point mutations on interaction with F1L was assessed by transfecting 1 × 106 Bak-deficient baby mouse kidney cells with pEGFP-C3, pEGFP-F1L, pEGFP-Bcl-xL, and wild-type Bak, BakmtBH1, BakmtBH2, or BakmtBH3. After 18 h cells were lysed in 2% CHAPS lysis buffer, and goat anti-GFP antibody and protein G-Sepharose were used to immunoprecipitate complexes. The interaction between endogenous Bak and Mcl-1 was assessed by infecting 1.4 × 107 HEK 293T cells with VV, VVFLAG-F1L, or VVΔF1L at an m.o.i. of 10 for 3, 6, or 12 h. Cells were lysed in 2% CHAPS lysis buffer and immunoprecipitated with rabbit anti-Mcl-1 antibody and protein A-Sepharose. Alternatively, 1.4 × 107 HeLa cells were treated with 2 μm staurosporine (STS) (Sigma) before immunoprecipitation with rabbit anti-Mcl-1 antibody.
Confocal Microscopy
For localization studies, 5 × 105 HeLa cells were seeded onto 18-mm coverslips (Fisher). HeLa cells were transfected with 2 μg of pEGFP, pEGFP-F1L, or pCR3.1-Mcl-1 for 18 h using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Cells were fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS) and permeabilized with 1% Nonidet P-40 in PBS. Localization of Mcl-1 was detected with rabbit anti-Mcl-1 antibody (1:100) (Assay Designs) and Alexa546 goat anti-rabbit antibody (1:400) (Invitrogen) at 543 nm, and GFP fluorescence was detected with the 488-nm laser. Fixed cells were analyzed with Zeiss LSM510 laser scanning confocal microscope.
Mitochondrial Isolation
HeLa cells (7 × 106) were infected with wild-type vaccinia virus, VVFLAG-F1L, or VVΔF1L at an m.o.i. of 10 for 24 h. Alternatively, 1.4 × 107 HeLa cells were treated with 200 mJ/cm2 UV-C and harvested up to 6 h post-treatment. Mitochondria were isolated as previously described (25). Cells were washed and resuspended in 1 ml of hypotonic lysis buffer containing 250 mm sucrose, 20 mm HEPES, pH 7.5, 10 mm KCl, 1.5 mm MgCl2, 1 mm EDTA, and 1 mm EGTA. Cells were incubated on ice for 30 min and passed through a 22-gauge needle. Crude membranes were pelleted at 750 × g for 10 min. The pellet was resuspended in 1 ml of hypotonic lysis buffer and passed through a 22-gauge needle. After centrifugation at 750 × g for 10 min, soluble fractions containing mitochondria were pelleted at 10,000 × g for 15 min at 4 °C. Isolated mitochondria were lysed at 4 °C for 1 h in 2% CHAPS lysis buffer, and protein concentration in supernatant and mitochondrial fractions was determined by standard BCA assay (Pierce).
Time Courses
To monitor the stability of Mcl-1 protein levels, HeLa cells (1 × 106) were infected at an m.o.i. of 10 with VV, VVΔF1L, or VVFLAG-F1L. Total cell lysates were collected at various time points up to 24 h post-infection. To determine Mcl-1 stability after UV treatment, 1 × 106 HeLa cells were treated with 200 mJ/cm2 UV-C, and whole cell lysates were collected. HeLa cells were also pretreated with 10 μm MG132 (Sigma-Aldrich) 1 h before UV-C treatment to prevent the degradation of Mcl-1. To determine whether vaccinia virus induced a DNA damage response, 1 × 106 HeLa cells were mock-infected or infected with VV, VVΔF1L, or VVFLAG-F1L at an m.o.i. of 10 for 6 h. Cells were then mock-treated or treated with 100 μm etoposide or 200 mJ/cm2 UV-C to trigger a DNA damage response, and whole cell lysates were harvested up to 6 h post-treatment. To detect poly(ADP-ribose) polymerase (PARP) cleavage, 1 × 106 Jurkat cells were infected with VVEGFP, VVΔF1L, or VVΔF1L-FLAG-Mcl-1 at an m.o.i. of 10. Whole cell lysates were collected at 5, 10, and 15 h post-infection and lysed in SDS-PAGE sample buffer containing 8 m urea.
Cytochrome c Release
Jurkat cells (2 × 106) were infected with VVEGFP, VVΔF1L, or VVΔF1L-FLAG-Mcl-1 at an m.o.i. of 10. At 4, 8, or 12 h post-infection, cells were permeabilized with lysis buffer containing 75 mm NaCl, 1 mm NaH2PO4, 8 mm Na2HPO4, 250 mm sucrose, and 190 μg digitonin/ml (Sigma) for 10 min on ice as previously described (21). Mitochondria-containing pellets and cytosolic supernatant fractions were separated by centrifugation at 10,000 × g for 5 min, and mitochondrial pellet fractions were resuspended in Triton X-100 lysis buffer containing 25 mm Tris, pH 8.0, and 0.1% Triton X-100 (Fisher). Cytosolic and mitochondrial fractions were then subject to SDS-PAGE.
Bak Conformational Analysis by Flow Cytometry
Jurkat cells (1 × 106), Bcl-2 expressing Jurkat cells, or Bak- and Bax-deficient Jurkat cells were infected at an m.o.i. of 10 with VVEGFP, VVΔF1L, VVΔF1L-FLAG-F1L(V104A), VVΔF1L-FLAG-F1L(G144F), or VVΔF1L-FLAG-Mcl-1 for 4 h. To induce apoptosis, cells were treated with 0.25 μm STS for 1.5 h before fixation in 0.25% paraformaldehyde. Cells were permeabilized with 500 μg/ml digitonin (Sigma) and then stained with a conformation-specific anti-Bak Ab-1 antibody (Oncogene Research Products) (7, 42) or an isotype control antibody specific for NK1.1 (PK136) (43). Phycoerythrin-conjugated anti-mouse antibody was used to counterstain cells (Jackson ImmunoResearch) before analysis by flow cytometry (FACScan; BD Biosciences) using the FL-2 channel equipped with a 585-nm filter (42 nm band pass). Data were analyzed using CellQuest software.
Immunoblotting
Cell lysates were subjected to SDS-PAGE and transferred to nitrocellulose or polyvinylidene difluoride membranes (GE Healthcare). Antibodies used for immunoblotting were as follows: rabbit anti-Bak NT (Upstate), mouse anti-Bak (BD PharMingen), mouse anti-cytochrome c (BD PharMingen), mouse anti-EGFP (Covance), mouse anti-FLAG M2 (Sigma), mouse anti-phospho-Histone H2AX (Upstate), mouse anti-Mcl-1 clone RC13 (Thermo Fisher Scientific, Inc.), rabbit anti-manganese superoxide dismutase (clone 110) (Assay Designs), mouse anti-PARP (BD PharMingen), and mouse anti-β-tubulin (ECM Bioscience). Rabbit anti-I5L serum was generated as previously described (44). Membranes were blotted with donkey anti-mouse or anti-rabbit horseradish peroxidase-conjugated antibodies (Jackson ImmunoResearch). Proteins were visualized using enhanced chemiluminescence according to the manufacturer's directions (GE Healthcare). Densitometric analysis was performed on co-immunoprecipitates using ImageQuant 1D gel analysis software (GE Healthcare).
RESULTS
The Interaction between F1L and Bak Is Conserved across Species
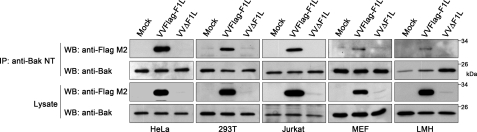
We have previously shown that F1L inhibits loss of mitochondrial membrane potential and cytochrome c release by inhibiting the proapoptotic activity of both Bak and Bax (20, 25). Although F1L inhibits both Bak and Bax, F1L interacts constitutively with only Bak, thus implicating Bak as an important binding partner of F1L (20). Despite the ability of F1L to interact with Bak by immunoprecipitation, F1L displays low micromolar affinity for the Bak BH3 peptide in binding studies (30), suggesting that other parts of the Bak sequence contribute to binding. Therefore, we further investigated the ability of F1L to bind endogenous Bak in a range of human cell lines. Cells were mock-infected or infected with VVFLAG-F1L, a recombinant vaccinia virus expressing FLAG-tagged F1L, or VVΔF1L, which is devoid of F1L. Twelve hours post-infection, cells were lysed in 2% CHAPS, a detergent that maintains the native state of Bcl-2 family members, and endogenous Bak was precipitated with an anti-Bak antibody (45, 46). Western blotting with an anti-FLAG antibody revealed that FLAG-F1L co-immunoprecipitated with Bak in HeLa, HEK 293T, and Jurkat cells, indicating that F1L was able to interact with endogenous Bak in a variety of cellular contexts (Fig. 1). Using mouse embryonic fibroblast and chicken (leghorn male) hepatoma cell lines, we also tested the binding of F1L to mouse and chicken Bak, which display 77 and 60% identity to human Bak, respectively (17, 47). In both cases F1L interacted with chicken Bak, as previously shown (17), and murine Bak (Fig. 1). Together, these data suggest that despite low affinity for the Bak BH3 peptide in binding assays (30), the constitutive interaction between F1L and Bak is highly conserved, as in addition to binding Bak in human cell lines, F1L interacts with both chicken and murine Bak.
FIGURE 1.
F1L interacts with human, murine, and chicken Bak. HeLa, HEK 293T, Jurkat, mouse embryonic fibroblast (MEF), or chicken leghorn male hepatoma (LMH) cells were mock-infected or infected at an m.o.i. of 5 with VVFLAG-F1L or VVΔF1L. Cellular lysates were immunoprecipitated (IP) with anti-Bak NT and Western-blotted (WB) with anti-FLAG M2 or anti-Bak to detect interaction. Thirty percent of the immunoprecipitates and 20% of the lysates were analyzed by Western blots.
F1L Contains Highly Divergent BH Domains
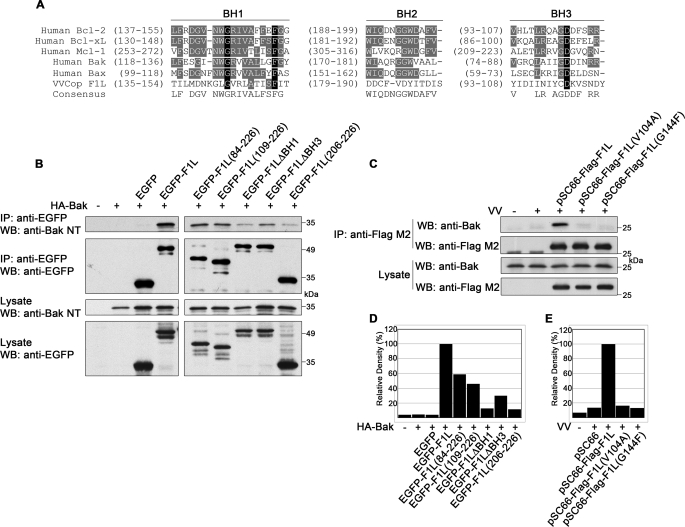
Cellular Bcl-2 proteins such as Bcl-xL and Mcl-1 interact with Bak using their BH1, BH2, and BH3 domains, which form a hydrophobic pocket to accept the Bak BH3 domain (48, 49). Although F1L lacks significant sequence identity to the highly conserved consensus sequences of Bcl-2 proteins, sequence patterns with properties compatible with the known structure of BH domains could be delineated. By correlating the F1L sequence to structural constraints of known Bcl-2 protein structures, we predicted the location of putative BH domains in F1L (Fig. 2A). Importantly, these domains were confirmed by the recent crystal structure of modified vaccinia Ankara F1L (30).
FIGURE 2.
F1L contains BH domains required for Bak interaction. A, shown is alignment of BH1, BH2, and BH3 domains from VV (Copenhagen) F1L and human Bcl-2 family members. Conserved residues are highlighted in black, and similar residues are in gray. B, HEK 293T cells were transiently transfected with pEGFP, pEGFP-F1L, pEGFP-F1L-(84–226), pEGFP-F1L-(109–226), pEGFP-F1LΔBH1, pEGFP-F1LΔBH3, or pEGFP-F1L-(206–226) along with pcDNA3-HA-Bak. Cellular lysates were immunoprecipitated (IP) with anti-EGFP, and 40% of the immunoprecipitate was Western-blotted (WB) with anti-Bak NT or anti-EGFP. Expression of EGFP constructs and Bak were confirmed by Western blotting 40% of the lysate with anti-Bak NT and anti-EGFP. C, HEK 293T cells were infected with VV at an m.o.i. of 5 and transfected with pSC66-FLAG-F1L, pSC66-FLAG-F1L(V104A), or pSC66-FLAG-F1L(G144F) followed by immunoprecipitation with anti-FLAG M2. Thirty percent of immunoprecipitates and lysates were Western-blotted with anti-Bak, and 20% of the immunoprecipitates and lysates were Western-blotted with anti-FLAG M2. D, the amount of HA-Bak co-immunoprecipitated with each EGFP-tagged protein in B (top panel) was analyzed by densitometry. E, shown is densitometric analysis of the amount of Bak co-immunoprecipitated with each FLAG-tagged protein in Fig. C, top panel.
F1L Possesses Functional BH Domains Required for Bak Interaction
To determine how the BH domains of F1L contributed to Bak binding, we generated a panel of EGFP-tagged F1L constructs, two of which contained large N-terminal deletions. EGFP-F1L-(84–226) lacks the first 83 amino acids, including the BH4 domain, whereas EGFP-F1L-(109–226) lacks the first 108 amino acids, spanning the BH3 and BH4 domains. To specifically target select BH domains, we also created EGFP-tagged F1L mutants devoid of either the BH1 or BH3 domains; EGFP-F1LΔBH1 lacks amino acids 144–156, and EGFP-F1LΔBH3 lacks amino acids 93–108. As a control, we used the transmembrane domain of F1L appended to EGFP, EGFP-F1L-(206–226), which localizes to mitochondria but fails to protect from apoptosis (24).
The generation of F1L constructs devoid of putative BH1 or BH3 domains allowed us to assess the contributions of these domains to the interaction with Bak. HEK 293T cells were transfected with EGFP-tagged wild-type or mutant F1L constructs along with HA-Bak, and interaction was assessed by immunoprecipitation with an anti-EGFP antibody. Wild-type EGFP-F1L interacted strongly with HA-Bak, whereas EGFP-F1L-(84–226), EGFP-F1L-(109–226), EGFP-F1LΔBH1, and EGFP-F1LΔBH3 displayed reduced binding (Fig. 2B). HA-Bak did not interact with EGFP as expected; however, some background binding to EGFP-F1L-(206–226) was detected. The decreased interaction was most noticeable with EGFP-F1LΔBH1 and EGFP-F1LΔBH3, which demonstrated an 8- and 3-fold decrease in Bak binding, respectively, compared with wild-type F1L (Fig. 2D). Thus, the data suggest that the BH1 and BH3 domains of F1L play an important role in Bak interaction. Similar disruptions in endogenous Bak binding during vaccinia virus infection were observed for FLAG-tagged F1L-(84–226), F1L-(109–226), and F1LΔBH1 (supplemental Fig. 1).
To further elucidate the importance of the BH1 and BH3 domains of F1L in binding Bak, we generated point mutations in the BH1 and BH3 domains of F1L. Based on the crystal structure of F1L, we generated FLAG-F1L(G144F), which is mutated in the BH1 domain, and FLAG-F1L(V104A), which is mutated in the BH3 domain. The corresponding mutations (G140F and V100A) in modified vaccinia Ankara F1L completely disrupted binding to the Bim BH3 peptide in binding assays (30). The contribution of the BH1 and BH3 domains in binding endogenous Bak during vaccinia virus infection was assessed by placing these point mutants under control of a poxviral promoter, therefore, limiting their expression to cells infected with vaccinia virus. As shown in Fig. 2C, wild-type FLAG-F1L pulled down endogenous Bak in HEK 293T cells, whereas Bak binding to FLAG-F1L(V104A) and FLAG-F1L(G144F) was diminished more than 6-fold compared with wild-type FLAG-F1L during infection (Fig. 2E). Thus, despite a lack of obvious sequence identity to cellular Bcl-2 proteins, F1L contains divergent BH domains that are critical for interacting with Bak. These results were not affected by the presence of viral products used to transform HEK 293T cells, as the results were reproducible in NIH/3T3 cells (supplemental Fig. 2).
The BH1 Domain of F1L Is Critical for Its Antiapoptotic Activity during Vaccinia Virus Infection
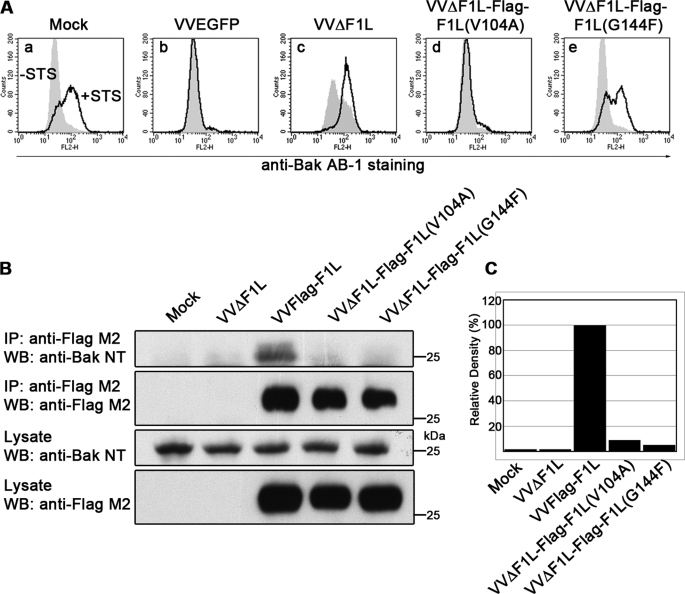
To assess the contribution of Val-104 and Gly-144 to the antiapoptotic activity of F1L, we generated recombinant vaccinia viruses expressing FLAG-tagged F1L constructs bearing the V104A or G144F mutations in the absence of endogenous F1L. Virus-induced apoptosis in infected Jurkat cells was assayed by monitoring Bak activation using a conformation-specific antibody, anti-Bak Ab-1, that detects the N-terminal exposure of Bak after its activation during apoptosis (7, 42). Additionally, virus-infected cells were treated with STS to determine the effect of the mutations on the ability of F1L to protect against an external apoptotic stimulus. Unlike mock-infected cells treated with STS, Bak was not activated in cells infected with wild-type vaccinia virus expressing EGFP, VVEGFP, in the presence or absence of STS, indicating that wild-type vaccinia virus prevented Bak activation (Figs. 3A, a and b). In contrast, Bak activation occurred in VVΔF1L-infected cells and was even more pronounced during STS treatment (Fig. 3Ac). Interestingly, the V104A mutation of F1L did not affect the antiapoptotic capacity of the protein, as VVΔF1L-FLAG-F1L(V104A) prevented virus-induced and STS-induced Bak activation (Fig. 3Ad). In contrast, the G144F mutation did not affect the ability of VVΔF1L-FLAG-F1L(G144F) to prevent virus-induced apoptosis, but this mutation dramatically rendered the virus sensitive to STS-induced apoptosis (Fig. 3Ae).
FIGURE 3.
The BH1 domain of F1L is critical for its antiapoptotic ability. A, Jurkat cells were mock-infected or infected with VVEGFP, VVΔF1L, VVΔF1L-FLAG-F1L(V104A), or VVΔF1L-FLAG-F1L(G144F) at an m.o.i. of 10 for 4 h. Cells were then treated with 250 nm STS for 1.5 h to induce apoptosis. Bak N-terminal exposure was monitored by staining cells with the conformation-specific anti-Bak AB-1 antibody. Shaded histograms, untreated cells; open histograms, STS-treated cells. B, HEK 293T cells were mock-infected or infected with VVΔF1L, VVFLAG-F1L, VVΔF1L-FLAG-F1L(V104A), or VVΔF1L-FLAG-F1L(G144F) at an m.o.i. of 5 for 12 h. FLAG-tagged constructs were immunoprecipitated with anti-FLAG M2, and both immunoprecipitates (IP) and lysates were Western-blotted (WB) with anti-FLAG M2 (20% of samples) and anti-Bak NT (30% of samples). C, quantification of the amount of endogenous Bak co-immunoprecipitated with each FLAG-tagged construct in Fig. 3B, top panel, is shown.
To verify the requirement of both Val-104 and Gly-144 for the interaction between F1L and Bak observed in the infection-transfection system of Fig. 2C, we infected HEK 293T cells with the newly generated recombinant viruses and assessed Bak binding by immunoprecipitation. Wild-type FLAG-F1L strongly interacted with Bak, whereas both FLAG-F1L(V104A) and FLAG-F1L(G144F) immunoprecipitated 90% less Bak than wild-type FLAG-F1L (Fig. 3, B and C). Together, these data suggest that both F1L constructs bearing the V104A and G144F mutations disrupt binding to Bak, but only Gly-144 plays a critical role in the antiapoptotic activity of F1L against STS.
F1L Interacts with the BH3 Domain of Bak
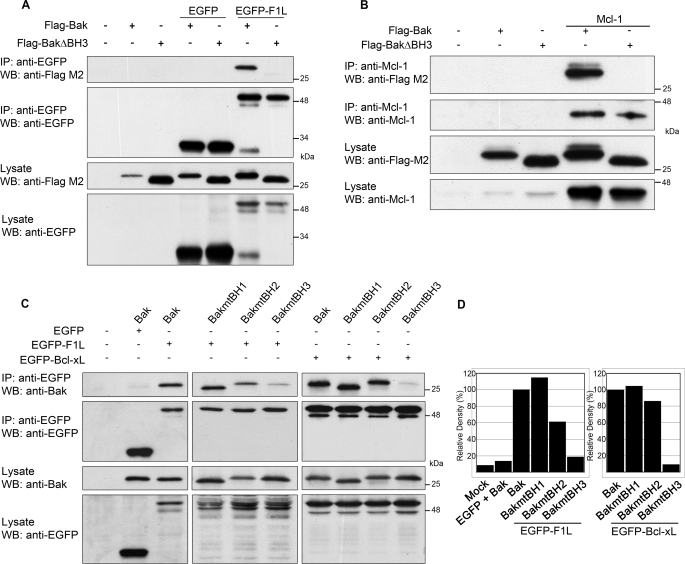
To further dissect the interaction between F1L and Bak, we generated a FLAG-tagged Bak construct lacking the BH3 domain, FLAG-BakΔBH3, by deleting amino acids 72–87 (50). HEK 293T cells were co-transfected with either FLAG-Bak or FLAG-BakΔBH3 along with pEGFP or pEGFP-F1L. Immunoprecipitation with an anti-EGFP antibody indicated that EGFP-F1L interacted strongly with FLAG-Bak, whereas the interaction between FLAG-BakΔBH3 and EGFP-F1L was undetectable (Fig. 4A). Neither FLAG-Bak nor FLAG-BakΔBH3 co-immunoprecipitated with EGFP, confirming the specificity of the interaction. As a control, Mcl-1 was co-transfected with FLAG-Bak or FLAG-BakΔBH3 and co-immunoprecipitated with an anti-Mcl-1 antibody. Unlike FLAG-Bak, which interacted with Mcl-1, deletion of the Bak BH3 domain completely abrogated binding to Mcl-1 (Fig. 4B).
FIGURE 4.
F1L interacts with the BH3 domain of Bak. A, HEK 293T cells were transiently transfected with pEGFP or pEGFP-F1L and pcDNA3-FLAG-Bak or pcDNA3-FLAG-BakΔBH3 and immunoprecipitated (IP) with anti-EGFP. Forty percent of immunoprecipitates and lysates were Western-blotted (WB) with anti-FLAG M2 and anti-EGFP. B, HEK 293T cells transiently transfected with pcDNA3-FLAG-Bak or pcDNA3-FLAG-BakΔBH3 with or without pCR3.1-Mcl-1 were immunoprecipitated with anti-Mcl-1, and 40% of immunoprecipitates and lysates were Western-blotted with anti-Bak and anti-Mcl-1. C, Bak−/− baby mouse kidney cells were transiently transfected with pEGFP, pEGFP-F1L, or pEGFP-Bcl-xL and either pcDNA3-Bak, pcDNA3-BakmtBH1, pcDNA3-BakmtBH2, or pcDNA3-BakmtBH3. Cellular lysates were immunoprecipitated with anti-EGFP, and 40% of immunoprecipitates and lysates were Western-blotted with anti-Bak and anti-EGFP. D, shown is a densitometric analysis of the amount of wild-type or Bak mutants that co-immunoprecipitated with EGFP-F1L (left) or EGFP-Bcl-xL (right) in C, top panel.
These studies were extended using three additional Bak mutants that contain point mutations in the BH1, BH2, or BH3 domains. Wild-type Bak, BakmtBH1 (W125A, G126E, R127A), BakmtBH2 (G175E, G176E, W177A), and BakmtBH3 (L78A, D83A) were cotransfected with either pEGFP, pEGFP-F1L, or pEGFP-Bcl-xL in Bak-deficient baby mouse kidney cells (41). Lysis in 2% CHAPS and immunoprecipitation with anti-EGFP revealed that EGFP-F1L coprecipitated both wild-type Bak and BakmtBH1 equally (Fig. 4C). However, in contrast to wild-type Bak, the interaction between F1L and BakmtBH2 was reduced by 40% (Fig. 4, C and D). Additionally, the interaction between EGFP-F1L and BakmtBH3 was reduced by 80% compared with wild-type Bak, indicating that Leu-78 and Asp-83 in the BH3 domain of Bak play a critical role in binding F1L (Figs. 4, C and D). Similarly, Bcl-xL displayed binding to wild-type Bak, BakmtBH1, and BakmtBH2 (Figs. 4, C and D). In contrast, binding to BakmtBH3 was reduced to 10% compared with the interaction between Bcl-xL and wild-type Bak (Figs. 4, C and D). Thus, similar to cellular Bcl-2 proteins, such as Bcl-xL and Mcl-1, F1L bound the BH3 domain of Bak, and deletion or point mutations in this domain abrogated the interaction between F1L and Bak.
Vaccinia Virus Infection Disrupts the Bak·Mcl-1 Complex
In healthy cells, Bak is complexed with Mcl-1 until an apoptotic stimulus causes dissociation of Mcl-1, resulting in Bak activation (27, 29, 41). Given that F1L constitutively binds Bak during infection, we sought to determine the fate of the Bak·Mcl-1 association during vaccinia virus infection. To this end, HEK 293T cells were mock-infected or infected with either wild-type VV that endogenously expresses F1L, VVFLAG-F1L, or VVΔF1L. After lysis, endogenous Mcl-1 was immunoprecipitated. Blotting for endogenous Bak revealed a strong interaction between Mcl-1 and Bak in mock-infected cells (Fig. 5A). However, the Bak·Mcl-1 complex was disrupted in cells infected with VV and VVFLAG-F1L. Infection with the proapoptotic virus, VVΔF1L, also disrupted the Bak·Mcl-1 complex, likely due to apoptosis induced in the absence of F1L, as previously reported (20). To examine the kinetics of the dissociation between Mcl-1 and Bak, we performed immunoprecipitations at various times over a 12-h period in mock-infected cells and cells infected with VV or VVΔF1L. As expected, the Bak·Mcl-1 complex remained stable over 12 h in mock-infected cells (Fig. 5B). However, in vaccinia virus-infected cells, the interaction between Mcl-1 and Bak was diminished by 6 h post-infection. The same trend of dissociation, with slightly faster kinetics, was observed in cells infected with the proapoptotic virus, VVΔF1L (20, 23). A similar disruption of the Bak·Mcl-1 complex was also detected after treatment of HeLa cells with STS (Fig. 5C).
FIGURE 5.
Vaccinia virus infection disrupts the Bak·Mcl-1 complex. A, HEK 293T cells were mock-infected or infected with VV, VVFLAG-F1L, or VVΔF1L at an m.o.i. of 10 for 12 h before immunoprecipitation (IP) with anti-Mcl-1. Thirty percent of the immunoprecipitates and lysates were Western-blotted (WB) with anti-Bak and anti-Mcl-1. B, HEK 293T cells were infected with VV or VVΔF1L for 3, 6, or 12 h before immunoprecipitation and Western blotting as in A. C, HeLa cells were treated with 2 μm STS for 1, 2, or 3 h to induce apoptosis before immunoprecipitation as in A. Fifty percent of the immunoprecipitates and lysates were Western-blotted with anti-Bak, and 30% were Western-blotted with anti-Mcl-1.
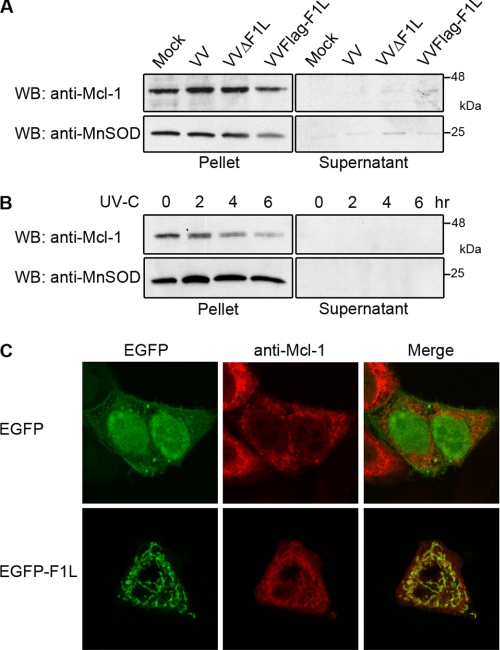
Mcl-1 Is Not Rapidly Degraded during Vaccinia Virus Infection
During treatment with certain apoptotic stimuli such as UV irradiation, Mcl-1 dissociates from Bak, and the unbound Mcl-1 is ubiquitinated and degraded in a proteasome-dependent manner (27, 29, 31). To determine the stability of Mcl-1 during vaccinia virus infection, we mock-infected HeLa cells or infected with VV, VVΔF1L, or VVFLAG-F1L at an m.o.i. of 10 and harvested cell lysates at various time points. In cells infected with VV, VVΔF1L, or VVFLAG-F1L, Mcl-1 remained relatively stable during the course of infection (Fig. 6A). Western blotting for β-tubulin served as a loading control, whereas infection was confirmed by the presence of the viral late protein I5L (44). In contrast to vaccinia virus infection, HeLa cells treated with UV-C demonstrated a dramatic reduction in Mcl-1 protein levels (Fig. 6B). However, pretreatment with the proteasome inhibitor MG132 before UV exposure completely prevented the degradation of Mcl-1, indicating that the 26 S proteasome is actively involved in UV-induced Mcl-1 loss (Fig. 6B) (29, 31). Thus, despite being displaced from Bak during vaccinia virus infection, Mcl-1 protein levels remain relatively stable, in contrast to the quick loss of Mcl-1 after treatment with UV. In agreement with the above results, we found that vaccinia virus did not induce a DNA damage response, which likely accounts for the absence of rapid Mcl-1 degradation (Fig. 6C). Using an antibody specific for phosphorylated histone H2AX as a marker for DNA double-strand breaks (51, 52), we found no evidence of a DNA damage response in mock-infected or VV-, VVFLAG-F1L-, or VVΔF1L-infected cells up to 12 h post-infection (Fig. 6C). However, treatment with etoposide and UV-C resulted in DNA lesions as demonstrated by phosphorylation of histone H2AX (Fig. 6C). This was also noted in mock-infected and vaccinia virus-infected cells treated with etoposide or UV-light, demonstrating that vaccinia virus does not induce a DNA damage response, and infection with vaccinia virus is unable to prevent the DNA response after treatment with DNA damaging agents.
FIGURE 6.
Mcl-1 is not rapidly degraded during vaccinia virus infection. A, HeLa cells were mock-infected or infected at an m.o.i. of 10 with VV, VVΔF1L, or VVFLAG-F1L. Cellular lysates were collected over 24 h and Western-blotted (WB) with anti-Mcl-1 (20% of sample), anti-I5L (5% of sample), and anti-β-tubulin (20% of sample). B, HeLa cells were treated with UV light in the absence or presence of 10 μm MG132 to inhibit the proteasome. Cellular lysates were harvested at 2, 4, or 6 h post-UV treatment, and 20% of whole cell lysates were Western-blotted with anti-Mcl-1 and anti-Bak. C, mock-infected HeLa cells or HeLa cells infected with VV, VVFLAG-F1L, or VVΔF1L at an m.o.i. of 10 for 6 h were mock-treated or treated with 100 μm etoposide or 200 mJ/cm2 UV-C. Whole cell lysates were harvested at 0, 2, 4, or 6 h post-etoposide or UV-C treatment, and 20% of these samples were subject to Western blotting with an antibody specific for phosphorylated histone H2AX to determine the induction of a DNA damage response.
Mcl-1 Remains at the Mitochondria during Vaccinia Virus Infection
Our observations indicated that the dissociation of Mcl-1 from Bak during vaccinia virus infection was not followed by rapid degradation of Mcl-1. To further investigate the effect of vaccinia virus infection on Mcl-1, we assessed the presence of Mcl-1 at the mitochondria. HeLa cells were either mock-infected or infected with vaccinia virus before mitochondrial fractionation. To detect Mcl-1 levels at the mitochondria, isolated supernatant and mitochondrial fractions were blotted for Mcl-1. We verified the purity of the supernatant and mitochondrial fractions by Western blotting for the mitochondrial-resident protein, manganese superoxide dismutase. Upon infection with VV, VVΔF1L, or VVFLAG-F1L, Mcl-1 remained at the mitochondria, similar to mock-infected cells (Fig. 7A). Thus, disruption of the Bak·Mcl-1 complex does not result in a change in Mcl-1 localization. In contrast, treatment of HeLa cells with UV-C before mitochondrial isolation resulted in loss of Mcl-1 from mitochondria (Fig. 7B). Although Mcl-1 was observed at the mitochondria 2 h post-UV treatment, the presence of Mcl-1 was reduced at 4 and 6 h, consistent with the degradation observed in whole cell lysates (Figs. 6B and 7B). Given that both F1L and Mcl-1 localize to mitochondria and interact with Bak, we sought to determine the effect of F1L expression on the localization of Mcl-1 in the absence of infection. HeLa cells were transiently transfected with pCR3.1-Mcl-1 and pEGFP-F1L or pEGFP. No co-localization was detected between EGFP and Mcl-1; however, clear co-localization was detected between Mcl-1 and EGFP-F1L, indicating that Mcl-1 remained at the mitochondria even in the presence of overexpressed F1L (Fig. 7C). Therefore, upon Mcl-1 displacement from Bak during infection, Mcl-1 remained stable and maintained its mitochondrial localization, in contrast to the loss of Mcl-1 after UV-C treatment. This suggests that Mcl-1 may have a role during vaccinia virus infection aside from inhibiting Bak.
FIGURE 7.
Mcl-1 remains at the mitochondria during VV infection. A, HeLa cells were mock-infected or infected with VV, VVΔF1L, or VVFLAG-F1L at an m.o.i. of 10 for 24 h, and mitochondria were isolated. Ten micrograms of supernatant and mitochondrial fractions were Western-blotted (WB) with anti-Mcl-1 and anti-MnSOD. B, HeLa cells were treated with UV-light for 2, 4, or 6 h before mitochondrial isolation and Western blotting as in A. C, HeLa cells were transfected with pEGFP or pEGFP-F1L and pCR3.1-Mcl-1 for 16 h before fixing and staining with a polyclonal anti-Mcl-1 antibody to visualize the localization of Mcl-1.
Mcl-1 Compensates for the Loss of F1L during Vaccinia Virus Infection
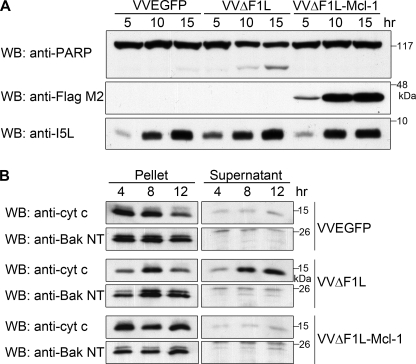
The ability of F1L to bind both Bak and the BH3-only protein Bim and inhibit apoptosis resembles the activity of Mcl-1 (28, 29, 39). This parallel function suggests that F1L may replace the antiapoptotic role of Mcl-1 during infection by interacting with Bak and Bim and inhibiting cytochrome c release. To test whether Mcl-1 could compensate for the loss of F1L during VVΔF1L infection, we generated VVΔF1L-FLAG-Mcl-1, a recombinant F1L-deficient virus expressing FLAG-tagged Mcl-1, and determined whether VVΔF1L-FLAG-Mcl-1 could prevent virus-induced apoptosis. Jurkat cells were analyzed for PARP cleavage after infection with VVΔF1L-FLAG-Mcl-1, the parental virus VVΔF1L, or wild-type vaccinia expressing EGFP, VVEGFP. As expected, infection with VVEGFP failed to induce apoptosis, indicated by the presence of full-length PARP up to 15 h post-infection; however, infection with VVΔF1L resulted in a loss of full-length PARP at 10 and 15 h post-infection and an increase in the amount of cleaved PARP (Fig. 8A). In contrast to VVΔF1L, infection with VVΔF1L-FLAG-Mcl-1 did not induce PARP cleavage, demonstrating that Mcl-1 protected from virus-induced apoptosis in the absence of F1L. The expression of FLAG-Mcl-1 was detected by blotting with anti-FLAG antibody, and infection was detected using an antibody to the late viral protein I5L (Fig. 8A) (44).
FIGURE 8.
Mcl-1 prevents apoptosis induced by VVΔF1L. A, Jurkat cells were infected at an m.o.i. of 10 with VVEGFP, VVΔF1L, or VVΔF1L-Mcl-1. At 5, 10, and 15 h post-infection, cell lysates were harvested and Western-blotted (WB) with anti-PARP (20% of sample), anti-FLAG M2 (10% of sample), and anti-I5L (5% of sample). B, Jurkat cells were infected with VVEGFP, VVΔF1L, or VVΔF1L-Mcl-1 at an m.o.i. of 10 for 4, 8, or 12 h. Mitochondria-containing pellets and cytosolic supernatants were separated after lysis with digitonin. Pellet fractions were subsequently resuspended in 0.1% Triton lysis, and 20% of pellet fractions and 50% of supernatant fractions were Western-blotted with anti-cytochrome c (cyt c) or anti-Bak NT.
To investigate events upstream of PARP cleavage, we determined whether Mcl-1 could replace the function of F1L by monitoring cytochrome c release. Jurkat cells infected with VVEGFP, VVΔF1L, or VVΔF1L-FLAG-Mcl-1 were separated into cytosolic fractions and mitochondria-containing fractions. In VVEGFP-infected cells, cytochrome c release was inhibited (Fig. 8B). In contrast, cytochrome c was predominantly found in cytosolic fractions after 12 h of infection with VVΔF1L, signifying loss of mitochondrial integrity. Notably, VVΔF1L-FLAG-Mcl-1 protected against the release of cytochrome c. As a control, pellet and supernatant fractions were blotted for Bak, which served as a mitochondrial marker (Fig. 8B). Together, the data suggest that the presence of Mcl-1 preserved mitochondrial integrity and downstream apoptosis in the absence of F1L during vaccinia virus infection.
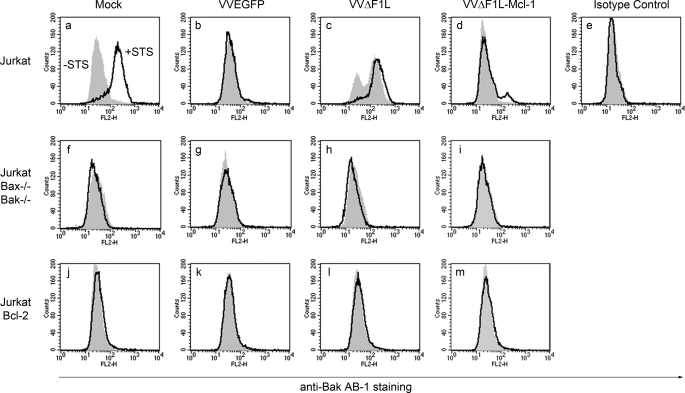
To establish whether VVΔF1L-FLAG-Mcl-1 could prevent VVΔF1L-induced Bak activation, we infected Jurkat cells at an m.o.i. of 10 with VVEGFP, VVΔF1L, or VVΔF1L-FLAG-Mcl-1 for 4 h. Infected cells were also treated with STS to determine whether VVΔF1L-FLAG-Mcl-1 could prevent Bak activation in response to an external stimulus in addition to virus-induced apoptosis. Once again, Bak activation was monitored using a conformation specific antibody, anti-Bak Ab-1. As shown in Fig. 9a, mock-infected Jurkat cells treated with STS demonstrated an increase in anti-Bak Ab-1 fluorescence, indicative of Bak activation and exposure of its N terminus. The measurement of Bak N-terminal exposure was specific because an isotype control antibody did not display an increase in fluorescence after STS treatment (Fig. 9e). Jurkat cells infected with VVEGFP did not undergo Bak activation, and these cells were protected from STS-induced Bak activation (Fig. 9b). However, VVΔF1L-infected cells demonstrated an increase in Bak Ab-1 fluorescence as expected, as this virus induces apoptosis upon infection (Fig. 9c) (17, 20). Bak activation was even more pronounced after STS treatment in VVΔF1L-infected cells, highlighting the inability of this proapoptotic virus to protect from external apoptotic stimuli (Fig. 9c). In contrast to VVΔF1L, the presence of Mcl-1 in VVΔF1L-FLAG-Mcl-1 was sufficient to prevent Bak activation during infection and in response to STS treatment (Fig. 9d). This suggests that the expression of Mcl-1 renders VVΔF1L protective against both virus- and STS-induced Bak activation and subsequent apoptosis. As shown previously, Bcl-2-overexpressing Jurkat cells infected with VVΔF1L and treated with STS prevented Bak activation (Fig. 9, j–m) (17, 20). The specificity of the assay was confirmed using Jurkat cells deficient in Bak and Bax (33), which displayed no increase in anti-Bak Ab-1 fluorescence after VVΔF1L infection and STS treatment (Fig. 9, f–i). Together these data indicate that although F1L prevents the activation of Bak during vaccinia virus infection, Mcl-1 can replace this activity and protect against virus- and STS-induced apoptosis during VVΔF1L infection.
FIGURE 9.
Mcl-1 inhibits Bak activation during VVΔF1L infection. Jurkat cells (a–e), Jurkat cells devoid of Bax and Bak (f–i), and Jurkat cells overexpressing Bcl-2 (j–m) were mock-infected or infected with VVEGFP, VVΔF1L, or VVΔF1L-Mcl-1 for 4 h at an m.o.i. of 10 before treatment with 250 nm STS for 1.5 h to induce apoptosis. Bak N-terminal exposure was monitored by staining cells with the conformation-specific anti-Bak AB-1 antibody or anti-NK1.1 as an isotype control antibody. Shaded histograms, untreated cells; open histograms, STS-treated cells.
DISCUSSION
Apoptosis is a powerful antiviral barrier; thus, many viruses have evolved strategies to overcome cell death to ensure viral replication and propagation (12, 53). Many DNA viruses encode viral Bcl-2 (vBcl-2) homologues that inhibit the mitochondrial checkpoint (14, 15), yet several poxviruses encode novel inhibitors of apoptosis that do not display apparent sequence similarity to members of the Bcl-2 family (54). Vaccinia virus F1L interacts with Bak and Bim to prevent mitochondrial depolarization and release of cytochrome c (20, 21, 25). However, the mechanism mediating these interactions has remained unclear, because unlike cellular and viral Bcl-2 proteins, F1L does not contain obvious BH domains.
Although F1L lacks sequence homology to the Bcl-2 family of proteins, we and others have previously identified Bak as a major interacting partner and target of F1L (20, 23). Here we show that the association between F1L and Bak is conserved in human, murine, and chicken cells (Fig. 1). Based on the conservation of hydrophobic residues important for mediating interactions between cellular Bcl-2 proteins, we identified highly divergent BH domains in F1L (Fig. 2). Importantly, these domains coincide with the proposed BH domains based on the recently published crystal structure of F1L (30). The BH1 and BH3 domains of F1L display little sequence similarity to those of cellular Bcl-2 proteins; however, these domains were essential for binding to Bak (Fig. 2). Additionally, mutating a single amino acid in either the BH1 or BH3 domain of F1L completely abrogated the interaction with endogenous Bak during vaccinia virus infection (Figs. 2 and 3). Likewise, mutations in the BH1 domain of Kaposi sarcoma-associated herpesvirus Bcl-2, adenovirus E1B-19K, African swine fever virus A179L, and Epstein-Barr virus BHRF1 eliminate the antiapoptotic activity of these vBcl-2 proteins (55–58). The ability of F1L to bind Bak in a cellular context contrasts with its low affinity for a Bak BH3 peptide in binding assays (26, 30), suggesting that the interaction may require the presence of a membrane given that these proteins are anchored in the outer mitochondrial membrane (7, 24). Alternatively, the conformation of full-length Bak may be important for binding to F1L. The discrepancy between immunoprecipitation data and in vitro binding assays using BH3 peptides has been observed previously for Bak, underscoring the importance of examining interactions between Bcl-2 family members in a cellular context (59).
Although both V104A and G144F mutations in F1L disrupted binding to Bak (Figs. 2 and 3), only G144F inhibited the antiapoptotic activity of F1L in response to STS (Fig. 3). Gly-144 is the only conserved residue in F1L that resembles the NWGR sequence in both cellular and viral Bcl-2 homologues (Fig. 2) (60). Mutating this glycine in Bcl-2 or Kaposi sarcoma-associated herpesvirus Bcl-2 abrogates the pro-survival activity of these proteins (56, 61); therefore, Gly-144 likely plays an equally important role in the antiapoptotic function of F1L (Fig. 3). Interestingly, the V104A mutation prevented Bak binding but had no effect on the ability of F1L to prevent virus- or STS-induced apoptosis (Figs. 2 and 3). This suggests that the antiapoptotic function of F1L does not rely solely on binding and inhibiting Bak. F1L(V104A) does not have measurable affinity for a Bim BH3 peptide in binding assays (30), yet the effect of this mutation on binding full-length Bim in a cellular context remains to be determined. Although F1L(V104A) may bind to Bim, we cannot exclude the possibility that F1L(V104A) may retain the ability to interact with other BH3-only proteins or components of the permeability transition pore to inhibit apoptosis.
With the exception of the avipoxviruses, which possess obvious Bcl-2 homologues, most members of the poxvirus family encode antiapoptotic proteins that lack sequence similarity to cellular Bcl-2 proteins. For example, myxoma virus M11L and Orf virus ORFV125 inhibit the activation of both Bak and Bax but display limited sequence homology to cellular Bcl-2 proteins (40, 62–64). Despite divergence at the sequence level, however, M11L adopts a Bcl-2-like fold highly reminiscent of Bcl-xL (65, 66). Additionally, the recent crystal structure of F1L indicates that, despite limited sequence homology, F1L also retains a Bcl-2-like fold (30). Unlike previously described vBcl-2 proteins and their cellular counterparts, however, F1L forms a domain-swapped homodimer that is unique among apoptotic inhibitors (30). It remains to be established whether the interaction with Bak requires F1L in a dimer conformation and whether this conformation has a role in the ability of F1L to inhibit apoptosis. Intriguingly, mutation in a previously reported BH-like domain of F1L abrogates Bak binding and the antiapoptotic activity of F1L (23). This is possibly due to disruption of the structure of F1L, as this region is thought to be involved in F1L dimerization (30).
Bak and Bax are the targets of many vBcl-2 proteins as they are the critical executioners of apoptosis (14, 15). Most vBcl-2 proteins bind to Bak or Bax to inhibit their activation. A notable exception is Kaposi sarcoma-associated herpesvirus Bcl-2, which adopts a Bcl-2 fold and binds Bax and Bak BH3 peptides yet does not bind full-length Bak or Bax (56, 67). In contrast, adenovirus E1B-19K interacts with Bak, Bax, and the BH3-only protein Bik (27, 55, 68–70). Our laboratory has recently demonstrated that the vBcl-2 from fowlpox virus, FPV039, interacts with Bak, Bax, and the BH3-only proteins Bim and Bik to inhibit apoptosis (17, 35). Although overall sequence conservation between vBcl-2 proteins and cellular Bcl-2 proteins is low, there is significant sequence homology in the BH domains (14, 60). All vBcl-2 proteins contain an identifiable BH1 domain and at least one other BH2 or BH3 domain (14). Although F1L is not considered a true vBcl-2 protein at the sequence level, the divergent BH1 and BH3 domains we have identified are required for interacting with Bak, indicating that F1L is a functional Bcl-2 homologue despite the absence of significant sequence homology.
In addition to identifying the BH1 and BH3 domains of F1L as essential for Bak binding, we have also confirmed that F1L required the Bak BH3 domain for interaction, in a similar manner to Mcl-1 and Bcl-xL (Fig. 4) (29, 41). The shared requirement for the Bak BH3 domain suggested that F1L may out-compete Mcl-1 for Bak binding. In agreement with this idea, Bak and Mcl-1 dissociated during vaccinia virus infection (Fig. 5). If F1L actively displaces Mcl-1, the disruption of the Bak·Mcl-1 complex observed during infection with VVΔF1L was likely due to apoptosis initiated in the absence of F1L. Alternatively, vaccinia virus infection may disrupt the Bak·Mcl-1 complex, and F1L may be required to bind and sequester Bak after its dissociation from Mcl-1. In contrast to the rapid loss of Mcl-1 after UV irradiation, Mcl-1 displacement from Bak during vaccinia virus infection did not result in Mcl-1 degradation (Fig. 6) (27, 29, 31). The stabilization of Mcl-1 was consistent with the observation that vaccinia virus infection did not induce a DNA damage response (Fig. 6). In contrast, adenovirus infection induces a DNA damage response resulting in rapid degradation of Mcl-1, but Bak is inhibited by binding the vBcl-2 protein E1B-19K (27).
Although Mcl-1 was displaced from Bak, we found that Mcl-1 was retained at the mitochondria during infection, suggesting that Mcl-1 may contribute to the inhibition of apoptosis during vaccinia virus infection (Fig. 7). Interestingly, mcl-1 is up-regulated upon infection with vaccinia virus and the apoptosis-inducing VVΔE3L, supporting a possible role for Mcl-1 during infection (71). In addition to binding Bak, Mcl-1 binds a variety of BH3-only proteins, including Bim, Puma, Noxa, tBid, and Bik (28, 29, 39, 72–74). Mcl-1 may, therefore, bind one or more of these BH3-only proteins and aid in sustaining a prosurvival environment during vaccinia virus infection. Notably, both F1L and Mcl-1 interact with Bim (25, 26, 28), but the fate of the Bim·Mcl-1 complex has not been investigated during infection. Alternatively, Mcl-1 may not be required in the presence of F1L due to the overlapping antiapoptotic activities of the two proteins. Indeed, Mcl-1 was able to replace the function of F1L during infection with a vaccinia virus devoid of F1L, as demonstrated by the inhibition of Bak activation, cytochrome c release, and PARP cleavage (Figs. 8 and 9). Together the data highlight the functional similarities between these two proteins.
Given the need for constitutive inhibition of Bak to prevent apoptosis, many poxviruses encode antiapoptotic proteins that interact with Bak (17, 20, 40). F1L binds and inhibits Bak in a manner similar to cellular Bcl-2 proteins, such as Mcl-1, despite a lack of obvious resemblance to Bcl-2 proteins at the sequence level. The Bcl-2-like fold and the presence of highly divergent BH domains classifies F1L as a novel inhibitor of apoptosis that folds and functions like a Bcl-2 family member (30). Although the sequence conservation between cellular Bcl-2 proteins is maintained throughout eukaryotes, poxviruses encode a variety of unique inhibitors of apoptosis (22, 62, 64). The divergence of F1L as well as M11L and ORFV125 from cellular Bcl-2 proteins suggests that these proteins have maintained only the minimal requirements for the inhibition of Bak and Bax and, thus, serve as useful tools to dissect the mitochondrial pathway of apoptosis.
Supplementary Material
Acknowledgments
We thank L. Banadyga and N. van Buuren for discussion and critical review of the manuscript and D. Quilty for pEGFP-F1L-(84–226) and pcDNA3-FLAG-BakΔBH3.
This work was supported by grants from the Canadian Institutes of Health Research and the Howard Hughes Medical Institute (to M. B.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
- BH
- Bcl-2 homology
- EGFP
- enhanced green fluorescent proteins
- m.o.i.
- multiplicity of infection
- STS
- staurosporine
- VV
- vaccinia virus
- CHAPS
- 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate
- HEK cells
- human embryonic kidney cells
- GFP
- green fluorescent protein
- PARP
- poly(ADP-ribose) polymerase
- HA
- hemagglutinin
- vBcl-2
- viral Bcl-2.
REFERENCES
- 1.Hengartner M. O. (2000) Nature 407, 770–776 [DOI] [PubMed] [Google Scholar]
- 2.Wang X. (2001) Genes Dev. 15, 2922–2933 [PubMed] [Google Scholar]
- 3.Cory S., Adams J. M. (2002) Nat. Rev. Cancer 2, 647–656 [DOI] [PubMed] [Google Scholar]
- 4.Marsden V. S., Strasser A. (2003) Annu. Rev. Immunol. 21, 71–105 [DOI] [PubMed] [Google Scholar]
- 5.Youle R. J., Strasser A. (2008) Nat. Rev. Mol. Cell Biol. 9, 47–59 [DOI] [PubMed] [Google Scholar]
- 6.Willis S. N., Adams J. M. (2005) Curr Opin Cell Biol. 17, 617–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Griffiths G. J., Dubrez L., Morgan C. P., Jones N. A., Whitehouse J., Corfe B. M., Dive C., Hickman J. A. (1999) J. Cell Biol. 144, 903–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wei M. C., Lindsten T., Mootha V. K., Weiler S., Gross A., Ashiya M., Thompson C. B., Korsmeyer S. J. (2000) Genes Dev. 14, 2060–2071 [PMC free article] [PubMed] [Google Scholar]
- 9.Antonsson B., Montessuit S., Sanchez B., Martinou J. C. (2001) J. Biol. Chem. 276, 11615–11623 [DOI] [PubMed] [Google Scholar]
- 10.Lindsten T., Ross A. J., King A., Zong W. X., Rathmell J. C., Shiels H. A., Ulrich E., Waymire K. G., Mahar P., Frauwirth K., Chen Y., Wei M., Eng V. M., Adelman D. M., Simon M. C., Ma A., Golden J. A., Evan G., Korsmeyer S. J., MacGregor G. R., Thompson C. B. (2000) Mol. Cell 6, 1389–1399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wei M. C., Zong W. X., Cheng E. H., Lindsten T., Panoutsakopoulou V., Ross A. J., Roth K. A., MacGregor G. R., Thompson C. B., Korsmeyer S. J. (2001) Science 292, 727–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Benedict C. A., Norris P. S., Ware C. F. (2002) Nat Immunol 3, 1013–1018 [DOI] [PubMed] [Google Scholar]
- 13.Boya P., Pauleau A. L., Poncet D., Gonzalez-Polo R. A., Zamzami N., Kroemer G. (2004) Biochim. Biophys. Acta 1659, 178–189 [DOI] [PubMed] [Google Scholar]
- 14.Cuconati A., White E. (2002) Genes Dev. 16, 2465–2478 [DOI] [PubMed] [Google Scholar]
- 15.Galluzzi L., Brenner C., Morselli E., Touat Z., Kroemer G. (2008) PLoS Pathog. 4, e1000018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taylor J. M., Barry M. (2006) Virology 344, 139–150 [DOI] [PubMed] [Google Scholar]
- 17.Banadyga L., Gerig J., Stewart T., Barry M. (2007) J. Virol. 81, 11032–11045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Afonso C. L., Tulman E. R., Lu Z., Zsak L., Kutish G. F., Rock D. L. (2000) J. Virol. 74, 3815–3831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tulman E. R., Afonso C. L., Lu Z., Zsak L., Kutish G. F., Rock D. L. (2004) J. Virol. 78, 353–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wasilenko S. T., Banadyga L., Bond D., Barry M. (2005) J. Virol. 79, 14031–14043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wasilenko S. T., Meyers A. F., Vander Helm K., Barry M. (2001) J. Virol. 75, 11437–11448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wasilenko S. T., Stewart T. L., Meyers A. F., Barry M. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 14345–14350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Postigo A., Cross J. R., Downward J., Way M. (2006) Cell Death Differ 13, 1651–1662 [DOI] [PubMed] [Google Scholar]
- 24.Stewart T. L., Wasilenko S. T., Barry M. (2005) J. Virol. 79, 1084–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taylor J. M., Quilty D., Banadyga L., Barry M. (2006) J. Biol. Chem. 281, 39728–39739 [DOI] [PubMed] [Google Scholar]
- 26.Fischer S. F., Ludwig H., Holzapfel J., Kvansakul M., Chen L., Huang D. C., Sutter G., Knese M., Häcker G. (2006) Cell Death Differ 13, 109–118 [DOI] [PubMed] [Google Scholar]
- 27.Cuconati A., Mukherjee C., Perez D., White E. (2003) Genes Dev. 17, 2922–2932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Han J., Goldstein L. A., Gastman B. R., Rabinovitz A., Rabinowich H. (2005) J. Biol. Chem. 280, 16383–16392 [DOI] [PubMed] [Google Scholar]
- 29.Willis S. N., Chen L., Dewson G., Wei A., Naik E., Fletcher J. I., Adams J. M., Huang D. C. (2005) Genes Dev. 19, 1294–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kvansakul M., Yang H., Fairlie W. D., Czabotar P. E., Fischer S. F., Perugini M. A., Huang D. C., Colman P. M. (2008) Cell Death Differ. 15, 1564–1571 [DOI] [PubMed] [Google Scholar]
- 31.Nijhawan D., Fang M., Traer E., Zhong Q., Gao W., Du F., Wang X. (2003) Genes Dev. 17, 1475–1486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barry M., Heibein J. A., Pinkoski M. J., Lee S. F., Moyer R. W., Green D. R., Bleackley R. C. (2000) Mol. Cell. Biol. 20, 3781–3794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang G. Q., Wieckowski E., Goldstein L. A., Gastman B. R., Rabinovitz A., Gambotto A., Li S., Fang B., Yin X. M., Rabinowich H. (2001) J. Exp. Med. 194, 1325–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Degenhardt K., Sundararajan R., Lindsten T., Thompson C., White E. (2002) J. Biol. Chem. 277, 14127–14134 [DOI] [PubMed] [Google Scholar]
- 35.Banadyga L., Veugelers K., Campbell S., Barry M. (2009) J. Virol. 83, 7085–7098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stuart D., Graham K., Schreiber M., Macaulay C., McFadden G. (1991) J. Virol. 65, 61–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Earl P. L., Moss B., Wyatt L. S., Carroll M. W. (1998) in Current Protocols in Molecular Biology (Ausubel F. M., Brent R., Kingston R. E., Moore D. D., Seidman J. G., Smith J. A., Struhl K. eds) pp. 16.17.1–16.17.19, Wiley Interscience, New York [Google Scholar]
- 38.Thompson J. D., Higgins D. G., Gibson T. J. (1994) Nucleic Acids Res. 22, 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Han J., Goldstein L. A., Gastman B. R., Froelich C. J., Yin X. M., Rabinowich H. (2004) J. Biol. Chem. 279, 22020–22029 [DOI] [PubMed] [Google Scholar]
- 40.Wang G., Barrett J. W., Nazarian S. H., Everett H., Gao X., Bleackley C., Colwill K., Moran M. F., McFadden G. (2004) J. Virol. 78, 7097–7111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leu J. I., Dumont P., Hafey M., Murphy M. E., George D. L. (2004) Nat. Cell Biol. 6, 443–450 [DOI] [PubMed] [Google Scholar]
- 42.Griffiths G. J., Corfe B. M., Savory P., Leech S., Esposti M. D., Hickman J. A., Dive C. (2001) Oncogene 20, 7668–7676 [DOI] [PubMed] [Google Scholar]
- 43.Koo G. C., Peppard J. R. (1984) Hybridoma 3, 301–303 [DOI] [PubMed] [Google Scholar]
- 44.van Buuren N., Couturier B., Xiong Y., Barry M. (2008) J. Virol. 82, 9917–9927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hsu Y. T., Wolter K. G., Youle R. J. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 3668–3672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hsu Y. T., Youle R. J. (1997) J. Biol. Chem. 272, 13829–13834 [DOI] [PubMed] [Google Scholar]
- 47.Ulrich E., Kauffmann-Zeh A., Hueber A. O., Williamson J., Chittenden T., Ma A., Evan G. (1997) Genomics 44, 195–200 [DOI] [PubMed] [Google Scholar]
- 48.Muchmore S. W., Sattler M., Liang H., Meadows R. P., Harlan J. E., Yoon H. S., Nettesheim D., Chang B. S., Thompson C. B., Wong S. L., Ng S. L., Fesik S. W. (1996) Nature 381, 335–341 [DOI] [PubMed] [Google Scholar]
- 49.Sattler M., Liang H., Nettesheim D., Meadows R. P., Harlan J. E., Eberstadt M., Yoon H. S., Shuker S. B., Chang B. S., Minn A. J., Thompson C. B., Fesik S. W. (1997) Science 275, 983–986 [DOI] [PubMed] [Google Scholar]
- 50.Chittenden T., Flemington C., Houghton A. B., Ebb R. G., Gallo G. J., Elangovan B., Chinnadurai G., Lutz R. J. (1995) EMBO J. 14, 5589–5596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rogakou E. P., Boon C., Redon C., Bonner W. M. (1999) J. Cell Biol. 146, 905–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rogakou E. P., Pilch D. R., Orr A. H., Ivanova V. S., Bonner W. M. (1998) J. Biol. Chem. 273, 5858–5868 [DOI] [PubMed] [Google Scholar]
- 53.Roulston A., Marcellus R. C., Branton P. E. (1999) Annu. Rev. Microbiol. 53, 577–628 [DOI] [PubMed] [Google Scholar]
- 54.Barry M., Wasilenko S. T., Stewart T. L., Taylor J. M. (2004) Prog. Mol. Subcell. Biol. 36, 19–37 [DOI] [PubMed] [Google Scholar]
- 55.Han J., Sabbatini P., Perez D., Rao L., Modha D., White E. (1996) Genes Dev. 10, 461–477 [DOI] [PubMed] [Google Scholar]
- 56.Huang Q., Petros A. M., Virgin H. W., Fesik S. W., Olejniczak E. T. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 3428–3433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Khanim F., Dawson C., Meseda C. A., Dawson J., Mackett M., Young L. S. (1997) J. Gen. Virol. 78, 2987–2999 [DOI] [PubMed] [Google Scholar]
- 58.Revilla Y., Cebrián A., Baixerás E., Martínez C., Viñuela E., Salas M. L. (1997) Virology 228, 400–404 [DOI] [PubMed] [Google Scholar]
- 59.Zhai D., Jin C., Huang Z., Satterthwait A. C., Reed J. C. (2008) J. Biol. Chem. 283, 9580–9586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Petros A. M., Olejniczak E. T., Fesik S. W. (2004) Biochim. Biophys. Acta 1644, 83–94 [DOI] [PubMed] [Google Scholar]
- 61.Yin X. M., Oltvai Z. N., Korsmeyer S. J. (1994) Nature 369, 321–323 [DOI] [PubMed] [Google Scholar]
- 62.Everett H., Barry M., Lee S. F., Sun X., Graham K., Stone J., Bleackley R. C., McFadden G. (2000) J. Exp. Med. 191, 1487–1498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Su J., Wang G., Barrett J. W., Irvine T. S., Gao X., McFadden G. (2006) J. Virol. 80, 1140–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Westphal D., Ledgerwood E. C., Hibma M. H., Fleming S. B., Whelan E. M., Mercer A. A. (2007) J. Virol. 81, 7178–7188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Douglas A. E., Corbett K. D., Berger J. M., McFadden G., Handel T. M. (2007) Protein Sci. 16, 695–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kvansakul M., van Delft M. F., Lee E. F., Gulbis J. M., Fairlie W. D., Huang D. C., Colman P. M. (2007) Mol. Cell 25, 933–942 [DOI] [PubMed] [Google Scholar]
- 67.Cheng E. H., Nicholas J., Bellows D. S., Hayward G. S., Guo H. G., Reitz M. S., Hardwick J. M. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 690–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Boyd J. M., Gallo G. J., Elangovan B., Houghton A. B., Malstrom S., Avery B. J., Ebb R. G., Subramanian T., Chittenden T., Lutz R. J., et al. (1995) Oncogene 11, 1921–1928 [PubMed] [Google Scholar]
- 69.Han J., Modha D., White E. (1998) Oncogene 17, 2993–3005 [DOI] [PubMed] [Google Scholar]
- 70.Han J., Sabbatini P., White E. (1996) Mol. Cell. Biol. 16, 5857–5864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Langland J. O., Kash J. C., Carter V., Thomas M. J., Katze M. G., Jacobs B. L. (2006) J. Virol. 80, 10083–10095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Clohessy J. G., Zhuang J., de Boer J., Gil-Gómez G., Brady H. J. (2006) J. Biol. Chem. 281, 5750–5759 [DOI] [PubMed] [Google Scholar]
- 73.Mei Y., Du W., Yang Y., Wu M. (2005) Oncogene 24, 7224–7237 [DOI] [PubMed] [Google Scholar]
- 74.Shimazu T., Degenhardt K., Nur-E-Kamal A., Zhang J., Yoshida T., Zhang Y., Mathew R., White E., Inouye M. (2007) Genes Dev. 21, 929–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.