Abstract
Deletions at 22q11.2 are linked to DiGeorge or velocardiofacial syndrome (VCFS), whose hallmarks include heart, limb, and craniofacial anomalies, as well as learning disabilities and increased incidence of schizophrenia. To assess the potential contribution of 22q11 genes to cognitive and psychiatric phenotypes, we determined the CNS expression of 32 mouse orthologs of 22q11 genes, primarily in the 1.5-Mb minimal critical region consistently deleted in VCFS. None are uniquely expressed in the developing or adult mouse brain. Instead, 27 are localized in the embryonic forebrain as well as aortic arches, branchial arches, and limb buds. Each continues to be expressed at apparently constant levels in the fetal, postnatal, and adult brain, except for Tbx1, ProDH2, and T10, which increase in adolescence and decline in maturity. At least six 22q11 proteins are seen primarily in subsets of neurons, including some in forebrain regions thought to be altered in schizophrenia. Thus, 22q11 deletion may disrupt expression of multiple genes during development and maturation of neurons and circuits compromised by cognitive and psychiatric disorders associated with VCFS.
There is a high incidence of schizophrenia in patients with velocardiofacial syndrome (VCFS), which is caused by heterozygous deletion at chromosome 22q11 (1). In addition, these patients have reduced cortical gray matter (2) similar to schizophrenics. It is unknown whether specific 22q11.2 genes (3, 4) mediate the neurocognitive and neuropathological aspects of VCFS. Analysis of the VCFS cardiovascular phenotype was facilitated by embryonic expression studies that suggested Tbx1 as a candidate (5, 6), a possibility that was subsequently verified genetically in mice (7, 8). Accordingly, analysis of 22q11 gene expression in the CNS might identify candidate genes that may compromise brain development or function.
Currently, there is very little data on 22q11 gene expression in the brain beyond COMT, a catecholamine catabolic enzyme independently associated with schizophrenia (9). Although COMT remains an attractive candidate, additional 22q11 genes could influence neural development or function and thus contribute to psychiatric aspects of VCFS. 22q11 genes might influence forebrain as well as heart, face, and limb development through participation in a shared developmental mechanism [nonaxial mesenchymal/epithelial (M/E) induction (10, 11)] that guides morphogenesis at each location. Furthermore, 22q11 genes might modulate later events in neural development thought to be compromised in schizophrenia, including neurogenesis, migration, process outgrowth, and synapse formation (12, 13).
To establish which 22q11 genes are the strongest candidates for the effects of VCFS on brain development or function, we undertook a comprehensive expression profile of 22q11 orthologs in the mouse, complemented by a selective analysis in the human. We focused on orthologs of genes in the proximal 1.5 megabase (Mb) portion of the 3-Mb typically deleted region because this 1.5-Mb portion includes the previously defined “minimal DiGeorge critical region” (14) thought to be crucial for VCFS phenotypes. Our results do not suggest a limited number of candidates; instead, a large subset of 22q11 genes, if dysregulated by heterozygous deletion, might compromise developmental mechanisms or vulnerable neuronal populations and thereby contribute to behavioral disorders in VCFS.
Methods
Genomic Analysis and Mapping. We confirmed and augmented previous maps of proximal 22q11.2 and the syntenic region of mouse chromosome 16 (8, 15, 16). We validated the GenBank-derived cDNA sequence for each human 22q11 gene (17) against the human EST cDNA database, as well as genomic sequences, to identify exons and map orthologous genes. In addition, we used genomic sequences to search mouse and human EST cDNA databases for previously unidentified transcripts.
PCR Analysis. PCR primers for 22q11 genes and orthologs (Table 1, which is published as supporting information on the PNAS web site) were optimized for a standardized PCR condition (60°C annealing temperature, 50-sec extension time). Primers span introns where possible and yield amplicons of ≈500 bp with modest GC content (50–60%). Amplicons were verified by sequencing.
Semiquantitative RT-PCR Analysis. We used outbred mice (ICR, Harlan Breeders, Indianapolis) that are free of developmental or brain anomalies common among inbred strains. Tissues were dissociated in TRIzol (Invitrogen), DNase-treated to remove residual genomic DNA (DNAfree, Ambion, Austin, TX), and reverse transcribed (ImPromptII, Promega) by using random hexamer primers (Invitrogen). Control reactions used an equal amount of RNA without reverse transcriptase (RT) enzyme. cDNA samples were standardized by competitive PCR (18), and equivalent dilutions of each cDNA sample were established that amplify β-actin to similar levels. Each PCR analysis was repeated two to five times from independent tissue samples, with equivalent results. Two genes (GPIBB and GSCL) not represented in these cDNAs were validated with EST or bacterial artificial chromosome DNA samples. Human cDNAs were obtained from BD Biosciences Clontech. Adult brain cDNAs reflect RNA pooled from donors between 12 and 75 years of age; fetal cDNAs were prepared from the pooled RNA of 12 fetuses aged 19–23 weeks.
Cellular Expression of Selected 22q11 Orthologs. Monoclonal antibodies (COMT, RanBP1, and Ufd1L, Transduction Laboratories, Lexington, KY) and goat polyclonal antiserum (CDC45L, Santa Cruz Biotechnology) were obtained commercially. Rabbit antisera against murine HIRA and CDCrel-1 were obtained by immunization with a 15 C-terminal amino acid synthetic peptide (Washington Biotechnology, Baltimore) and affinity-purified with the immunizing peptide. All antibodies were tested for specificity by Western blot; rabbit antisera were also tested by peptide preabsorption (data not shown). Immunolocalization was performed on mouse embryos or brain sections as described (19). All animals were handled in accordance with protocols approved by the University of North Carolina Institutional Animal Care and Use Committee.
Results
We identified, mapped, and verified sequences of 32 mouse orthologs of 22q11 genes, including 25 in the proximal 1.5-Mb portion consistently deleted in VCFS (Fig. 1a), to assemble a comprehensive PCR-based expression array (Table 1). We considered the membrane-bound isoform of COMT to be a separate transcriptional unit because its transcription start site and first exon are distinct. Primers for each remaining gene detect sequences predicted to be common to all isoforms. We identified two genes not included in previous maps, the receptor for the NoGo protein (20) and zDHHC8, a 3-kb transcript containing a DHHC-zinc finger domain. Five additional genes (DGCR6, ProDH2, BCR, E2F6, and GGT) are found within the human LCRs (15) that are thought to mediate both the typical 3-Mb deletion and the smaller 1.5-Mb deletion. Evaluation of human genes in the LCRs is complicated by the presence of multiple copies; however, we identified a single copy of each in the mouse. Two genes (CRKL and PCQAP) in the distal portion of the region typically deleted in VCFS were analyzed, because previous work suggests that they are VCFS candidates (21, 22).
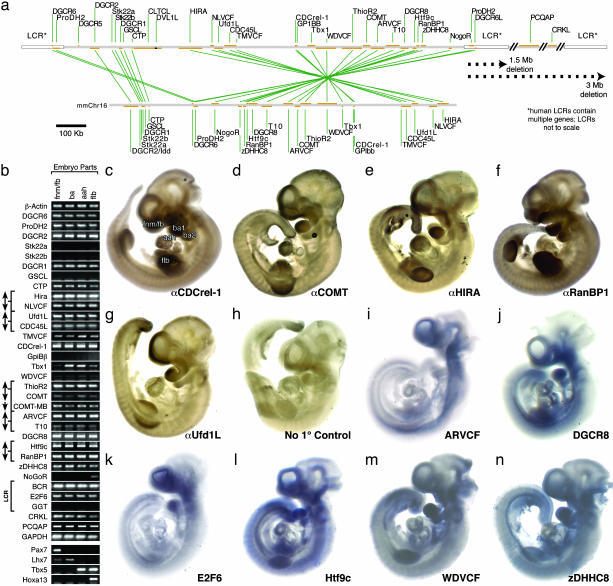
Fig. 1.
(a) The top shows a scale map of the proximal 1.5-Mb portion of chromosome 22q11 consistently deleted in DiGeorge syndrome (containing 27 of 32 transcripts examined) showing transcription units (orange bars) and low-copy repeats (LCRs, white boxes). The bottom shows that a majority of mouse orthologs are in a segment of mouse chromosome 16. (b) PCR array of 22q11.2 orthologs at sites of nonaxial M/E interaction: the frontonasal mass/forebrain (FNM/FB), branchial arches (BA), aortic arches and heart (AAH), and forelimb buds (FLB). Expression is presented in chromosomal order, centromeric (top) to telomeric (bottom). Brackets and arrows represent genes that share bidirectional promoters. The LCR bracket indicates genes found in human LCRs (not on chromosome 16 in mouse). β-actin and GAPDH are positive controls. (c–h). Whole embryonic day (E) 10/E10.5 mouse embryo immunolocalization of five 22q11 proteins shows correspondence between message and protein localization for this subset of orthologs. (i–n) Whole E10/E10.5 mouse embryo in situ hybridization of seven 22q11 orthologs shows focal expression at nonaxial inductive sites.
22q11 Genes at Sites of M/E Interaction. We first asked whether 22q11 orthologs are expressed in the developing forebrain as well as the heart, face, and limbs at midgestation [embryonic day (E) 10.5] when M/E interactions influence patterning and morphogenesis at each site (23). Previous observations indicate that at least nine murine orthologs (CDC45L, CDCrel-1, CRKL, DGCR2, DGCR8, Hira, NLVCF, RanBP1, and Ufd1L) are expressed at these sites (17). We confirmed these and evaluated the remaining 23 genes in cDNAs from microdissected FNM/FB, first and second BA, AAH, and FLB (Fig. 1b) from E10.5 mouse embryos. The identity of each cDNA sample was validated by four differentially expressed genes: Pax7 (FNM/FB), Lhx 7 (BA), Tbx5 (BA and AAH), and HoxA13 (FLB). 27 of 32 murine 22q11 orthologs are expressed at all four sites at E10.5 (Fig. 1b). Although our PCR analysis is not intended to be quantitative, we note that most transcripts are detected at comparatively high levels at all four sites except Tbx1, which, as reported in ref. 6, is enhanced in the heart and branchial arches, and NoGoR, which is enhanced in the FLB.
To evaluate whether expression is limited to inductive sites, we localized a subset of orthologs in whole mouse embryos by using antibody or nucleic acid probes. We localized five proteins [COMT, Ufd1l, CDCRel-1, RanBP1, and HIRA (Fig. 1 c–g)] by using specific antibodies that recognize bands of appropriate molecular weight by Western blot. We also selected six genes that had not been previously localized for in situ analysis at E10.5–E11: ARCVF, DGCR8, E2F6, Htf9C, WDVCF, and zDHHC8 (Fig. 1 i–n). Each was expressed in the FNM/FB, BA, AAH, and FLB. CDCrel-1, COMT, E2F6, HIRA, RanBP1, Ufd1l, and zDHHC8 are primarily limited to the four nonaxial sites, whereas Htf9C, ARVCF, WDVCF, and DGCR8 are also seen in ventral midline structures.
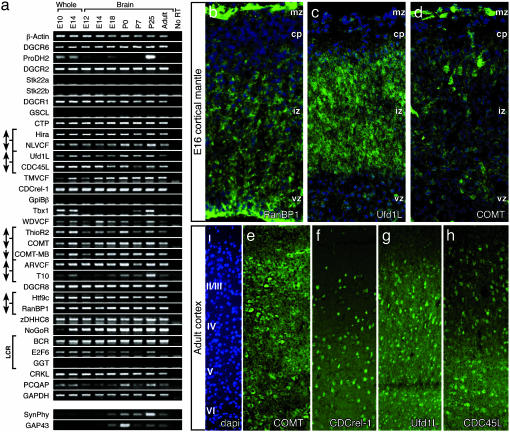
22q11 Orthologs Are Expressed in Fetal and Adolescent Brain. We next asked whether 22q11 orthologs continue to be expressed during brain development. Of 27 genes expressed in the forebrain and other sites of M/E interaction, 24 remain in the fetal, postnatal, and adult brain (Fig. 2a). In the fetal brain, the protein products of at least three genes are localized in zones where proliferating, migrating, or differentiating neurons are found in the developing neocortex. RanBP1 is seen in the ventricular zone (Fig. 2b), where proliferating precursors are found, and in the intermediate zone, where migrating neuroblasts are concentrated. Ufd1L is restricted to the intermediate zone (Fig. 2c). COMT is mostly confined to the marginal zone and cortical plate coincident with postmitotic, postmigratory neurons (Fig. 2d). In the fetal and postnatal brain, three other orthologs found at inductive sites (Tbx1, ProDH2, and T10) appear to be temporally modulated. They are not readily detectable at E12, but are robustly expressed at postnatal day (P) 25 (Fig. 2a). This is most apparent for Tbx1, which is not detected until P7, increases at P25, and returns to barely detectable levels in the adult brain. These changes coincide with that of Synaptophysin (SynPhy) (24), a protein associated with synapse formation and stabilization, and GAP43 (25), a protein associated with axon outgrowth (Fig. 2a). The five remaining transcripts were not found in the brain at any age, although three (GGT, Stk22a, and Stk22b) are found in other adult tissues (see below).
Fig. 2.
(a) PCR array showing expression of 22q11 orthologs in whole E10 and E14 mouse embryos and in the brain (anterior from the medulla) from E12 through adulthood. A no-RT lane is included as a negative control for sample contamination, and β-actin and GAPDH were used as positive controls. (b–d) Immunolocalization (green) of three 22q11 proteins in the developing neocortex at E16 shows expression in distinct domains of the developing cortex (cytoarchitecture shown with blue nuclear counterstain). vz, ventricular zone; iz, intermediate zone; cp, cortical plate. (e–h) Immunolocalization (green) of four 22q11 proteins in the adult neocortex suggests subtle variations in laminar frequency of labeled cells. Layer identity, demonstrated by blue nuclear counterstain, is shown at left.
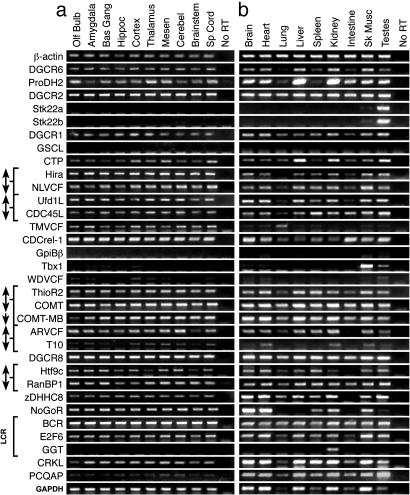
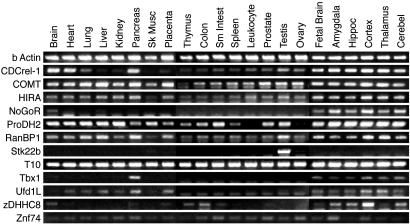
22q11 Gene Expression in the Adult Brain. We next asked whether 22q11 gene expression varies in distinct brain regions. Twenty-seven genes are expressed in the adult brain; however, ProDH2, TMVCF, WDVCF, T10, and Tbx1 are more difficult to detect. Nevertheless, all 27 are expressed in all major forebrain subdivisions: the neocortex, hippocampus, basal ganglia, amygdala/basal forebrain, and olfactory bulb, as well as all other CNS regions (Fig. 3a). No ortholog is limited to the brain; most are expressed in other adult tissues (Fig. 3b). Only three genes are restricted: the metabolic enzyme GGT in kidney, and two small putative serine kinases (Stk22a and Stk22b) primarily in testes. Thus, no 22q11 orthologs are uniquely expressed in the developing or mature CNS, nor are any orthologs limited to specific CNS regions.
Fig. 3.
(a) PCR array of microdissected adult forebrain regions and other CNS domains shows expression of the 22q11 orthologs. (b) A PCR array of several organs shows widespread expression of most 22q11 orthologs outside the CNS.
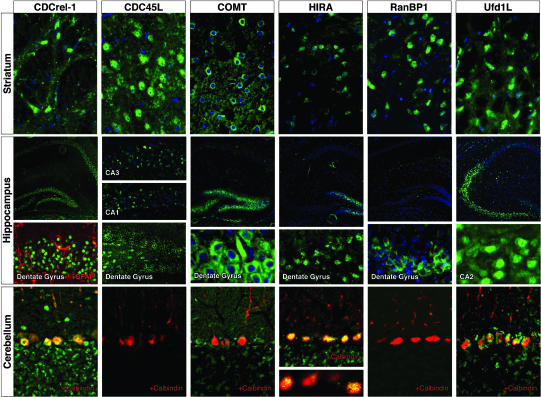
To begin to distinguish whether 22q11 proteins are expressed in neurons or in nonneural cells, we immunolocalized CDCrel-1, CDC45L, COMT, HIRA, RanBP1, and Ufd1L in the mature brain (Fig. 4). This subset represents a broad range of functional classes and is widely distributed in the proximal 1.5-Mb portion (17). All are expressed primarily in neurons; we saw no labeling in astroglia or oligodendroglia (data not shown). CDCrel-1, CDC45L, COMT, and Ufd1l are expressed by subsets of neurons in multiple neocortical layers (Fig. 2c). There is no laminar specificity; however, the frequency of CDCrel-1-labeled cells is apparently reduced in layers 2 and 3, and that for CDC45L is diminished in layer 4. RanBP1 and HIRA are expressed lightly throughout the cortex; however, labeled cells are more frequent in layer 6 for both (data not shown). All six proteins are also expressed in subsets of basal ganglia neurons, with no apparent specificity for any striatal compartment or cell class. In contrast, hippocampal expression is not uniform: Ufd1L is primarily found in CA2 and 3, COMT and HIRA are found in CA4 and the dentate gyrus, and RanBP1 is found in small clusters in the hilus of the dentate gyrus (Fig. 4). In contrast, CDCrel-1 and CDC45L are found in pyramidal cells throughout all hippocampal fields. There is also neuronal expression in the olfactory bulb, thalamus, and cerebellum (data not shown and Fig. 4). A substantial number of neurons in the neocortex, basal ganglia, and hippocampus express more than one 22q11 protein (Fig. 6, which is published as supporting information on the PNAS web site); however, 22q11 protein-expressing neuronal populations do not overlap completely. 22q11 proteins coincide in neurons in other brain regions, including the cerebellum, where CDCrel-1, HIRA, and Ufd1L are expressed in calbindin-positive Purkinje cells (Fig. 4, bottom row). We cannot rule out the possibility that other 22q11 proteins are expressed primarily in glial or cerebral vascular cells; nevertheless, there is significant neuronal-restricted expression of at least these six proteins throughout the brain, including in regions potentially compromised in behavioral and psychiatric disorders associated with VCFS.
Fig. 4.
Immunolocalization (green) of six 22q11 proteins in the adult mouse brain shows expression in subsets of neurons in the striatum, hippocampus, and cerebellum; a blue nuclear counterstain indicates the position of cell bodies. Double labeling with glial (second row, red) or neuronal (third row, red) markers suggests that the cells expressing these 22q11 proteins are neurons.
Murine Analysis Predicts Human 22q11 Gene Expression. To determine whether essential aspects of the mouse expression profile are retained in humans, we analyzed a subset of nine human 22q11 genes in cDNAs from fetal and adult human tissues. We selected five genes (CDCrel-1, COMT, HIRA, RanBP1, and Ufd1L) whose murine orthologs are expressed robustly at inductive sites and continue to be expressed throughout development, and whose protein products can be localized to subsets of forebrain neurons. Our limited analysis of human expression suggests that these genes are expressed widely in human tissues (Fig. 5), as they are in mouse. Two additional genes (Tbx1 and Stk22b, whose murine orthologs have restricted expression) are similarly restricted in human tissues. The two transcripts not previously analyzed in either mouse or human (zDHHC8 and NogoR) are detected in human tissues including the brain. Finally, in mouse, Znf74 does not encode a functional transcript (16); however, it is expressed in the developing and mature human brain and in nonneural tissues.
Fig. 5.
PCR expression array of selected human 22q11 genes. Human cDNA pools were obtained from BD Biosciences Clontech. cDNAs from whole brain and organ tissues were evaluated, and samples of forebrain and other CNS regions are included at the far right.
Discussion
Our expression analysis of a comprehensive set of 22q11 orthologs in the developing and adult mouse as well as a limited set of 22q11 genes in the human does not suggest a single or small number of candidate genes for schizophrenia or other cognitive disorders associated with VCFS. Instead, most 22q11 genes are expressed in the brain from early development through maturity. Accordingly, increased vulnerability to schizophrenia in VCFS may reflect consequences of heterozygous deletion of multiple 22q11 genes, some of which have distinct functions during specific developmental epochs or in specific neuronal classes in the mature brain.
27 of 32 orthologs are expressed in the forebrain during its initial patterning, as well as at other sites phenotypically compromised by 22q11.2 deletion: the limb buds, aortic arches, and branchial arches. A common cellular mechanism, M/E induction, guides morphogenesis of these sites (23, 26–28). Disruption of at least two inductive signals (retinoic acid and FGF8) phenocopies aspects of VCFS (11, 29–31). Moreover, FGF8 and another inductive signal, sonic hedgehog, modulate expression of at least one 22q11 ortholog, Tbx1 (32), and heterozygous deletion of FGF8 enhances penetrance of cardiovascular phenotypes when Tbx1 is deleted homozygously (33). Our results suggest that additional 22q11 genes may be involved in M/E induction. Thus, in addition to anomalies of the limbs, heart, and face, VCFS may subtly alter initial regional development and subsequent cell and circuit differentiation in the forebrain.
Expression of most 22q11 orthologs continues in the fetal and adolescent brain, throughout the critical processes of neuronal proliferation, migration, and circuit differentiation. The localization of COMT, Ufd1L, and RanBP1 in proliferating, migrating, or early postmigratory cortical cells, as well as the persistent expression of at least 21 other orthologs in the fetal brain, suggests that multiple 22q11.2 genes influence neurogenesis and migration. In contrast, ProDH2, Tbx1, and T10 transcripts are expressed in the adolescent mouse brain: after most migration as well as axonal and dendritic growth, but during the refinement and stabilization of synaptic connections (34). This synaptic refinement is likely compromised in schizophrenics, based on studies of histopathology and altered gene expression in postmortem brain tissue (13, 35). Thus, heterozygous deletion of 22q11 may alter the expression of genes required for proper development and function of neuronal circuits in the CNS.
Schizophrenia is generally thought to be a multigenic disease with a distinct natural history that encompasses disrupted neural development, maturation, and function (36). Thus, it is not unexpected that our data enlarges, rather than limits, the list of potential contributors associated with the high incidence of schizophrenia in 22q11.2-deleted VCFS patients (1). Several 22q11 genes have been linked to schizophrenia independently of VCFS, including COMT (9), ProDH2 (37), Ufd1L (38), and PCQAP (22). Our data strengthen the case that each may contribute independently to altered brain function in VCFS. Furthermore, several genes implicated in developmental or behavioral deficits associated with 22q11 deletion, based on loss of function in mice, are expressed in brain regions where local disruption of gene function might compromise differentiation or maintenance of relevant brain circuits. Abnormal sensorimotor gating in COMT (39) or ProDH2 (40) null mice, and similar deficiencies in heterozygous deletions of large regions of mouse chromosome 16 (41), may reflect direct alteration of developing or mature neural circuits, rather than some general systemic or metabolic effect. Homozygous deletion of several other orthologs, including CDC45L, CRKL, Hira, Ufd1L, and Tbx1, is either lethal at early stages or results in morphogenetic defects (17). Further analysis (e.g., using conditional inactivation to circumvent early lethality) will be necessary to better understand the contribution of these genes to behavioral pathology associated with VCFS.
Our comprehensive expression analysis reinforces a conclusion suggested by multiple genetic linkages within 22q11.2: that the increased risk for schizophrenia associated with VCFS reflects the concerted action of multiple 22q11 genes. Given the regional, temporal, and cellular coincidence shown here, there may also be interactions between multiple 22q11 genes in the CNS. The deletion of 22q11.2 genes as a group (as commonly occurs in VCFS) may synergistically disrupt processes required for proper brain development or function, leading to a greatly increased risk for schizophrenia.
Note Added in Proof. A brief description of zDHHC8 was included (as KIAA1292) in Liu et al. (42).
Supplementary Material
Acknowledgments
The revision of this manuscript followed the untimely death of Dr. Patricia Goldman-Rakic, whose pioneering work on the neurobiology of cognition and schizophrenia inspired the authors. We dedicate this paper, which she generously and rigorously edited, to her memory. We thank Carl Fisher, Andrew Gassman, Tanya Hartney, Cliff Heindel, and Kenichi Tagoku for their assistance during different phases of this project. Sharon Milgram provided valuable advice on production and characterization of antisera. This work was supported by National Institutes of Health Grants HD-29178 (to A.-S.L.), MH-33127 (to J.A.L.), and HD-40127 (to T.M.M.).
Abbreviations: AAH, aortic arches and heart; BA, branchial arches; En, embryonic day n; FLB, forelimb buds; FNM/FB, frontonasal mass/forebrain; LCR, low-copy repeat; Mb, megabase; M/E, mesenchymal/epithelial; Pn, postnatal day n; RT, reverse transcriptase; VCFS, velocardiofacial syndrome.
References
- 1.Bassett, A. S. & Chow, E. W. (1999) Biol. Psychiatry 46, 882-891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van Amelsvoort, T., Daly, E., Robertson, D., Suckling, J., Ng, V., Critchley, H., Owen, M. J., Henry, J., Murphy, K. C. & Murphy, D. G. (2001) Br. J. Psychiatry 178, 412-419. [DOI] [PubMed] [Google Scholar]
- 3.Driscoll, D. A., Budarf, M. L. & Emanuel, B. S. (1992) Am. J. Hum. Genet. 50, 924-933. [PMC free article] [PubMed] [Google Scholar]
- 4.Carlson, C., Sirotkin, H., Pandita, R., Goldberg, R., McKie, J., Wadey, R., Patanjali, S. R., Weissman, S. M., Anyane-Yeboa, K., Warburton, D., et al. (1997) Am. J. Hum. Genet. 61, 620-629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chieffo, C., Garvey, N., Gong, W., Roe, B., Zhang, G., Silver, L., Emanuel, B. S. & Budarf, M. L. (1997) Genomics 43, 267-277. [DOI] [PubMed] [Google Scholar]
- 6.Chapman, D. L., Garvey, N., Hancock, S., Alexiou, M., Agulnik, S. I., Gibson-Brown, J. J., Cebra-Thomas, J., Bollag, R. J., Silver, L. M. & Papaioannou, V. E. (1996) Dev. Dyn. 206, 379-390. [DOI] [PubMed] [Google Scholar]
- 7.Lindsay, E. A., Vitelli, F., Su, H., Morishima, M., Huynh, T., Pramparo, T., Jurecic, V., Ogunrinu, G., Sutherland, H. F., Scambler, P. J., et al. (2001) Nature 410, 97-101. [DOI] [PubMed] [Google Scholar]
- 8.Merscher, S., Funke, B., Epstein, J. A., Heyer, J., Puech, A., Lu, M. M., Xavier, R. J., Demay, M. B., Russell, R. G., Factor, S., et al. (2001) Cell 104, 619-629. [DOI] [PubMed] [Google Scholar]
- 9.Shifman, S., Bronstein, M., Sternfeld, M., Pisante-Shalom, A., Lev-Lehman, E., Weizman, A., Reznik, I., Spivak, B., Grisaru, N., Karp, L., et al. (2002) Am. J. Hum. Genet. 71, 1296-1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maynard, T. M., Sikich, L., Lieberman, J. A. & LaMantia, A. S. (2001) Schizophr. Bull. 27, 457-476. [DOI] [PubMed] [Google Scholar]
- 11.LaMantia, A. S. (1999) Biol. Psychiatry 46, 19-30. [DOI] [PubMed] [Google Scholar]
- 12.Akbarian, S., Kim, J. J., Potkin, S. G., Hetrick, W. P., Bunney, W. E., Jr., & Jones, E. G. (1996) Arch. Gen. Psychiatry 53, 425-436. [DOI] [PubMed] [Google Scholar]
- 13.Mirnics, K., Middleton, F. A., Marquez, A., Lewis, D. A. & Levitt, P. (2000) Neuron 28, 53-67. [DOI] [PubMed] [Google Scholar]
- 14.Gong, W., Emanuel, B. S., Collins, J., Kim, D. H., Wang, Z., Chen, F., Zhang, G., Roe, B. & Budarf, M. L. (1996) Hum. Mol. Genet. 5, 789-800. [DOI] [PubMed] [Google Scholar]
- 15.Shaikh, T. H., Kurahashi, H., Saitta, S. C., O'Hare, A. M., Hu, P., Roe, B. A., Driscoll, D. A., McDonald-McGinn, D. M., Zackai, E. H., Budarf, M. L. & Emanuel, B. S. (2000) Hum. Mol. Genet. 9, 489-501. [DOI] [PubMed] [Google Scholar]
- 16.Lund, J., Chen, F., Hua, A., Roe, B., Budarf, M., Emanuel, B. S. & Reeves, R. H. (2000) Genomics 63, 374-383. [DOI] [PubMed] [Google Scholar]
- 17.Maynard, T., Haskell, G., Lieberman, J. & LaMantia, A. (2002) Int. J. Dev. Neurosci. 20, 407-419. [DOI] [PubMed] [Google Scholar]
- 18.Maynard, T. M., Haskell, G. T., Bhasin, N., Lee, J. M., Gassman, A. A., Lieberman, J. A. & LaMantia, A. S. (2002) Mech. Dev. 111, 177-180. [DOI] [PubMed] [Google Scholar]
- 19.Thompson Haskell, G., Maynard, T. M., Shatzmiller, R. A. & Lamantia, A. S. (2002) J. Comp. Neurol. 452, 228-241. [DOI] [PubMed] [Google Scholar]
- 20.Fournier, A. E., GrandPre, T. & Strittmatter, S. M. (2001) Nature 409, 341-346. [DOI] [PubMed] [Google Scholar]
- 21.Guris, D. L., Fantes, J., Tara, D., Druker, B. J. & Imamoto, A. (2001) Nat. Genet. 27, 293-298. [DOI] [PubMed] [Google Scholar]
- 22.De Luca, A., Conti, E., Grifone, N., Amati, F., Spalletta, G., Caltagirone, C., Bonaviri, G., Pasini, A., Gennarelli, M., Stefano, B., et al. (2003) Am. J. Med. Genet. 116B, 32-35. [DOI] [PubMed] [Google Scholar]
- 23.LaMantia, A. S., Bhasin, N., Rhodes, K. & Heemskerk, J. (2000) Neuron 28, 411-425. [DOI] [PubMed] [Google Scholar]
- 24.Knaus, P., Betz, H. & Rehm, H. (1986) J. Neurochem. 47, 1302-1304. [DOI] [PubMed] [Google Scholar]
- 25.Biffo, S., Verhaagen, J., Schrama, L. H., Schotman, P., Danho, W. & Margolis, F. L. (1990) Eur. J. Neurosci. 2, 487-499. [DOI] [PubMed] [Google Scholar]
- 26.Johnson, R. L. & Tabin, C. J. (1997) Cell 90, 979-990. [DOI] [PubMed] [Google Scholar]
- 27.Creazzo, T. L., Godt, R. E., Leatherbury, L., Conway, S. J. & Kirby, M. L. (1998) Annu. Rev. Physiol. 60, 267-286. [DOI] [PubMed] [Google Scholar]
- 28.Schneider, R. A., Hu, D. & Helms, J. A. (1999) Cell Tissue Res. 296, 103-109. [DOI] [PubMed] [Google Scholar]
- 29.Vermot, J., Niederreither, K., Garnier, J. M., Chambon, P. & Dolle, P. (2003) Proc. Natl. Acad. Sci. USA 100, 1763-1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frank, D. U., Fotheringham, L. K., Brewer, J. A., Muglia, L. J., Tristani-Firouzi, M., Capecchi, M. R. & Moon, A. M. (2002) Development (Cambridge, U.K.) 129, 4591-4603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goodman, A. B. (1998) Proc. Natl. Acad. Sci. USA 95, 7240-7244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garg, V., Yamagishi, C., Hu, T., Kathiriya, I. S., Yamagishi, H. & Srivastava, D. (2001) Dev. Biol. 235, 62-73. [DOI] [PubMed] [Google Scholar]
- 33.Vitelli, F., Taddei, I., Morishima, M., Meyers, E. N., Lindsay, E. A. & Baldini, A. (2002) Development (Cambridge, U.K.) 129, 4605-4611. [DOI] [PubMed] [Google Scholar]
- 34.Rakic, P., Bourgeois, J. P. & Goldman-Rakic, P. S. (1994) Prog. Brain Res. 102, 227-243. [DOI] [PubMed] [Google Scholar]
- 35.Selemon, L. D. & Goldman-Rakic, P. S. (1999) Biol. Psychiatry 45, 17-25. [DOI] [PubMed] [Google Scholar]
- 36.Freedman, R., Leonard, S., Olincy, A., Kaufmann, C. A., Malaspina, D., Cloninger, C. R., Svrakic, D., Faraone, S. V. & Tsuang, M. T. (2001) Am. J. Med. Genet. 105, 794-800. [DOI] [PubMed] [Google Scholar]
- 37.Liu, H., Heath, S. C., Sobin, C., Roos, J. L., Galke, B. L., Blundell, M. L., Lenane, M., Robertson, B., Wijsman, E. M., Rapoport, J. L., et al. (2002) Proc. Natl. Acad. Sci. USA 99, 3717-3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.De Luca, A., Pasini, A., Amati, F., Botta, A., Spalletta, G., Alimenti, S., Caccamo, F., Conti, E., Trakalo, J., Macciardi, F., et al. (2001) Am. J. Med. Genet. 105, 529-533. [DOI] [PubMed] [Google Scholar]
- 39.Gogos, J. A., Morgan, M., Luine, V., Santha, M., Ogawa, S., Pfaff, D. & Karayiorgou, M. (1998) Proc. Natl. Acad. Sci. USA 95, 9991-9996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gogos, J. A., Santha, M., Takacs, Z., Beck, K. D., Luine, V., Lucas, L. R., Nadler, J. V. & Karayiorgou, M. (1999) Nat. Genet. 21, 434-439. [DOI] [PubMed] [Google Scholar]
- 41.Paylor, R., McIlwain, K. L., McAninch, R., Nellis, A., Yuva-Paylor, L. A., Baldini, A. & Lindsay, E. A. (2001) Hum. Mol. Genet. 10, 2645-2650. [DOI] [PubMed] [Google Scholar]
- 42.Liu, H., Abecasis, G. R., Heath, S. C., Knowles, A., Demars, S., Chen, Y.-J., Roos, J. L., Rapoport, J. L., Gogos, J. A. & Karayiorgou, M. (2002) Proc. Natl. Acad. Sci. USA 99, 16859-16864. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.