Abstract
Transforming growth factor-β (Tgf-β) signaling is crucial for regulating craniofacial development. Loss of Tgf-β signaling results in defects in cranial neural crest cells (CNCC), but the mechanism by which Tgf-β signaling regulates bone formation in CNCC-derived osteogenic cells remains largely unknown. In this study, we discovered that Tgf-β regulates the basal transcriptional regulatory machinery to control intramembranous bone development. Specifically, basal transcription factor Taf4b is down-regulated in the CNCC-derived intramembranous bone in Tgfbr2fl/fl;Wnt1-Cre mice. Tgf-β specifically induces Taf4b expression. Moreover, small interfering RNA knockdown of Taf4b results in decreased cell proliferation and altered osteogenic differentiation in primary mouse embryonic maxillary mesenchymal cells, as seen in Tgfbr2 mutant cells. In addition, we show that Taf1 is decreased at the osteogenic initiation stage in the maxilla of Tgfbr2 mutant mice. Furthermore, small interfering RNA knockdown of Taf4b and Taf1 together in primary mouse embryonic maxillary mesenchymal cells results in up-regulated osteogenic initiator Runx2 expression, with decreased cell proliferation and altered osteogenic differentiation. Our results indicate a critical function of Tgf-β-mediated basal transcriptional factors in regulating osteogenic cell proliferation and differentiation in CNCC-derived osteoprogenitor cells during intramembranous bone formation.
Keywords: Cell, Development Differentiation/Organ, Growth Factors, Organisms/Mouse, Tissue/Organ Systems/Bone, Transcription/Development
Introduction
Craniofacial skeletal elements are mainly formed by intramembranous ossification through a mechanism that remains relatively uncharacterized. The majority of osteoblasts and chondrocytes in the craniofacial region are derived from cranial neural crest cells (CNCC),2 which produce the facial skeleton (1, 2). Tgf-β signaling plays a crucial role in craniofacial development, and loss of Tgf-β signaling in CNCC results in craniofacial skeletal malformations (3, 4).
Tgf-β transmits signals through a membrane receptor serine/threonine kinase complex that phosphorylates Smad2 and Smad3, and activated Smads form transcriptional complexes with Smad4 and translocate into the nucleus (5). These Tgf-β signaling complexes contain other transcription factors and target a variety of genes in an embryonic stage-dependent and cell type-specific manner, but the factors involved in this transcriptional regulatory machinery have yet to be identified. During development, the expression of many genes is associated with changes accompanied by dynamic restructuring of chromatin (6, 7). Recent studies demonstrate that basal transcriptional factors have cell- and promoter-specific functions during embryogenesis (8–12).
RNA polymerase II requires the assembly of a multiprotein complex around the transcriptional start site (13). The general transcriptional factor IID (TFIID) is a large multiprotein transcriptional factor, consisting of the TATA-binding protein and a set of 13–14 TATA-binding protein-associated factors (TAFs), that is responsible for specific binding to the TATA element found in many polymerase II promoters and also demonstrates a coactivator function during transcriptional initiation (14). TAFs are able to regulate gene transcription at multiple steps, with functions in promoter recognition, selective binding to core promoter elements, as well as direct interactions with transcriptional activators (15–17). Mutation and loss of TAFs in yeast and mammalian cells lead to cell cycle arrest and gene-specific transcriptional effects (16). The function of TAFs in gene regulation during embryogenesis has yet to be determined. Here, we show that the interaction between Tgf-β signaling and TAFs has a crucial role in regulating CNCC-derived osteogenesis during craniofacial morphogenesis.
EXPERIMENTAL PROCEDURES
Animals
Mating Tgfbr2fl/+;Wnt1-Cre with Tgfbr2fl/fl mice generated Tgfbr2fl/fl;Wnt1-Cre conditional null alleles that were genotyped using PCR primers as described previously (4).
Whole-mount Skeletal Staining
The three-dimensional architecture of the skeleton was examined using a modified whole-mount Alcian blue-Alizarin Red S staining protocol as described previously (3).
Histological Examination
Hematoxylin and eosin staining and bromodeoxyuridine staining were performed as described previously (4, 18–20). Immunohistochemical staining was performed as described previously (18). Antibody used for immunohistochemistry was anti-Taf1 rabbit polyclonal antibodies (Abcam).
Immunological Analysis
Western blots were performed as described previously (21–23). Antibodies used for Western blotting were as follows: rabbit polyclonal antibodies against cyclin D1, cyclin D2, cyclin D3, cyclin A, cyclin E, JNK, and phospho-JNK (Cell Signaling Technology); FoxO4, FoxO3a, and Taf1 (Abcam); osteopontin, osteocalcin, and osteonectin (Santa Cruz Biotechnology); and mouse monoclonal antibody against GAPDH (Chemicon).
RNA Preparation and Quantitative RT-PCR
Total RNA was isolated from mouse embryonic maxilla dissected at the indicated developmental stage or from primary MEMM cells as described previously (24). First-strand cDNA was synthesized from 1 μg of total RNA using an oligo(dT)20 primer and SuperScript III reverse transcriptase (Invitrogen), and quantitative PCR was performed in triplicate by SYBR Green (Bio-Rad) in an iCycler (Bio-Rad). A melting curve was obtained for each PCR product after each run to confirm that the SYBR Green signal corresponded to a unique and specific amplicon. The relative abundance of each transcript was calculated based on PCR efficiency and cycle number at which the fluorescence crosses a threshold for the GAPDH internal reference and the gene tested using iCycler iQ optical system software (Bio-Rad). PCR primers are available upon request.
In Situ Hybridization
To generate the probe for in situ hybridization of mouse Taf4b, DNA encoding Taf4b was amplified from E13.5 mouse maxilla cDNA by PCR. The PCR fragments were cloned into the pDrive cloning vector (Qiagen). All recombinant plasmids were verified by sequencing. In situ hybridization was performed as described previously (18). Several negative controls (e.g. sense probe and no probe) were run in parallel with the experimental reaction. Details of the experimental procedures are available upon request.
Organ Culture of Maxilla and Tgf-β Bead Implantation
Affi-Gel blue beads (Bio-Rad) were used for delivery of Tgf-β2. The beads were washed in phosphate-buffered saline and then incubated for 1 h at room temperature in 10 μg/ml Tgf-β2 (R & D Systems). Control beads were incubated with 0.1% bovine serum albumin. Tgf-β2- or bovine serum albumin-containing beads were placed adjacent to the maxilla.
Primary Cultured Cells Derived from Mouse Embryonic Maxillary Mesenchyme
Primary MEMM cells were obtained from 13.5-day-old embryos (E13.5). Briefly, maxilla was dissected at E13.5 and trypsinized for 30 min at 37 °C in a CO2 incubator. After pipetting thoroughly, cells were cultured in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum supplemented with penicillin, streptomycin, l-glutamate, sodium pyruvate, and nonessential amino acids. Proliferation of primary MEMM cells was measured using a cell counting kit 8 (Dojindo Molecular Technologies, Gaithersburg, MD). Primary MEMM cells (5 × 103 cells per well) were seeded into 96-well plates and incubated at 37 °C in a CO2 incubator for up to 72 h. Following this incubation period, sodium 2-(4-iodophenyl)-3-(4-nitrophenyl)-5-(2,4-disulfophenyl)-2H-tetrazolium was added to the culture medium to label the proliferating cells, and incubation was continued for an additional 1 h at 37 °C. The amount of reduced tetrazolium was determined by measuring the absorbance at 450 nm in a microplate reader. Osteogenic differentiation was promoted by culture in monolayers after initial seeding of cells at 1.5 × 104 cells/cm2 in complete medium supplemented with 10 mm β-glycerophosphate, 0.1 μm dexamethasone, and 0.05 mm ascorbic acid (Sigma) for 2 weeks. Alkaline phosphatase activity was measured as described previously (25).
Small Interfering RNA Transfection (siRNA)
MEMM cells (2 × 106 cells) were plated in a 6-well cell culture plate until the cells reached 60–80% confluence. siRNA duplex and reagents were purchased from Invitrogen and Santa Cruz Biotechnology, respectively. siRNA mixture in transfection medium was incubated with cells for 6 h at 37 °C in a CO2 incubator, and then 5 × 103 cells were cultured for 2 weeks in regular or osteogenic differentiation medium, including siRNA transfection mixture. Specifically, siRNA was added every 3 days into the cell culture medium throughout the 2 weeks of culture.
Statistical Analysis
Two-tailed Student's t test was applied for statistical analysis. For all graphs, data are represented as mean ± S.D. A p value of less than 0.05 was considered statistically significant.
RESULTS
Loss of Tgfbr2 in Cranial Neural Crest Cells Results in Decreased Maxilla Size in Vivo
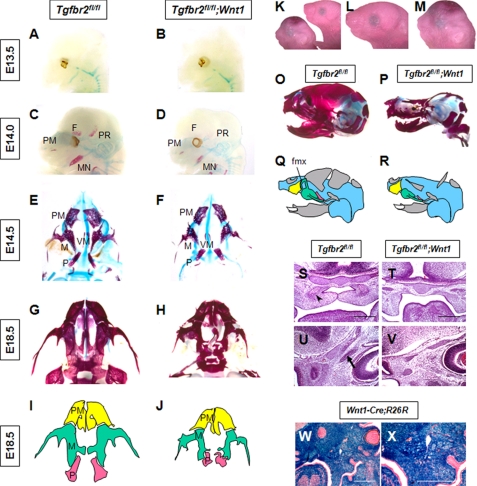
CNCC-derived osteogenic cells contribute to craniofacial bone formation that is developed through intramembranous bone ossification (1, 2). However, most facial skeletal bones are not ideal models for the analysis of intramembranous ossification. For instance, mandibular bone includes regions of both endochondral and intramembranous ossifications, and analysis of the skull region is complicated by its proximity to the dura mater. In this study, we analyzed the role of Tgf-β receptor type II in the maxilla to investigate intramembranous ossification derived from CNCC in the absence of other ossification processes or inductive tissues. The maxillary region is composed of six primordia as follows: pairs of premaxilla, maxilla, and palatine bones, which are all derived from CNCC. The size of the maxilla and palatine bones in newborn Tgfbr2fl/fl;Wnt1-Cre mice was smaller than those of Tgfbr2fl/fl control mice (Fig. 1, A–R, W, and X). Palatal and frontal processes of maxillary bone were defective in Tgfbr2fl/fl;Wnt1-Cre mice at E14.5 (Fig. 1, S–V). Thus, loss of Tgf-β signaling appears to affect intramembranous ossification.
FIGURE 1.
Development of the maxilla in Tgfbr2fl/fl;Wnt1-Cre mice. A–H, whole-mount skeletal staining with Alcian blue-Alizarin Red S. Maxilla structures of Tgfbr2fl/fl (E and G) and Tgfbr2fl/fl;Wnt1-Cre (F and H) mice are shown. The maxillary region is composed of six primordia; pairs of premaxilla, maxilla, and palatine bones, which are all derived from CNCC. The size of the maxilla and palatine bones in newborn Tgfbr2fl/fl;Wnt1-Cre mice were smaller than those of Tgfbr2fl/fl control mice. F, frontal bone; PM, premaxilla bone; M, maxilla bone; P, palatine bone; PR, parietal bone; MN, mandible; VM, vomer. I and J, schematic drawings in I and J are derived from images G and H, respectively. Note that the size of the maxilla bone is decreased in Tgfbr2fl/fl;Wnt1-Cre mice at E14.5. Premaxilla are highlighted in yellow, maxilla in green, and palatine bone in red. K–M, morphology of Tgfbr2fl/fl (K right, L) and Tgfbr2fl/fl;Wnt1-Cre (K left, M) mice. L and M are higher magnifications of K. O and P, whole-mount skeletal staining with Alcian blue-Alizarin Red S of Tgfbr2fl/fl (O) and Tgfbr2fl/fl;Wnt1-Cre (P) newborn mice. Q and R, schematic drawings in Q and R are derived from images O and P, respectively. fmx, frontal process of maxilla bone. S–V, hematoxylin and eosin staining of sections from the maxilla of Tgfbr2fl/fl (S and U) and Tgfbr2fl/fl;Wnt1-Cre (T and V) mice. Arrowhead indicates the palatal process of the maxilla bone. Arrow indicates the frontal process of maxilla bone. Scale bar, 200 μm. W and X, LacZ staining of Wnt1-Cre mice carrying the R26R reporter gene. Scale bar, 300 μm.
Decreased Cell Proliferation and Altered Osteogenic Differentiation in the Maxilla of Tgfbr2fl/fl;Wnt1-Cre Mice
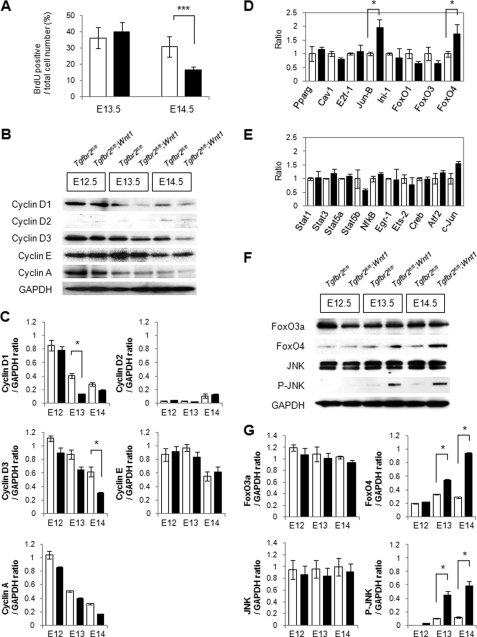
To investigate the cellular mechanism of decreased maxilla size in Tgfbr2fl/fl;Wnt1-Cre mice, we analyzed the rate of cellular proliferation and apoptosis relative to littermate wild type maxilla. In comparison with wild type control maxilla, we detected a decreased rate of cell proliferation in Tgfbr2fl/fl;Wnt1-Cre maxilla at E14.5, but apoptosis was unaffected (Fig. 2A; supplemental Fig. S1, A–D and F–K). Next, we analyzed the distribution of cells throughout the cell cycle using Tgfbr2fl/fl (control) and Tgfbr2fl/fl;Wnt1-Cre maxilla by fluorescence-activated cell sorting analyses after propidium iodide staining. We detected no significant changes in the proportion of cells at each stage of the cell cycle in Tgfbr2fl/fl and Tgfbr2fl/fl;Wnt1-Cre maxilla from E12.5 to E14.5 (supplemental Fig. 1E). D-type cyclins (cyclins D1, D2, and D3) are encoded by distinct genes that are induced in a cell lineage-specific manner (26). We found that cyclin D1 expression was reduced at E13.5 and E14.5, and cyclin D3 expression was reduced at E14.5 in Tgfbr2fl/fl;Wnt1-Cre maxilla relative to Tgfbr2fl/fl mice (Fig. 2, B and C). In contrast, there were no significant changes in the expression levels of other cyclins (Fig. 2, B and C). Gene expression of D-type cyclins is regulated by a wide array of transcriptional factors, including transactivators such as STAT proteins, NF-κB, Egr-1, Ets-2, cAMP-response element-binding protein, and c-Jun and suppressors such as peroxisome proliferator-activated receptor-γ, caveolin-1, E2F-1, Jun-B, INI1/hSNF5, and the FoxO family (26, 27). We examined the gene expression of cyclin D regulators using quantitative RT-PCR in control and Tgfbr2fl/fl;Wnt1-Cre maxilla at E13.5 (Fig. 2, D and E). FoxO4 and Jun-B were up-regulated 2-fold in Tgfbr2fl/fl;Wnt1-Cre maxilla at E13.5, but there were no significant changes in other regulators of type D cyclins. FoxO4 protein was up-regulated at E13.5 and E14.5, a time course that correlates with the reduction of cyclin D1 protein (Fig. 2, B, F, and G).
FIGURE 2.
Loss of Tgfbr2 in CNCC results in decreased type D cyclin-dependent cell proliferation during intramembranous ossification. A, ratio of bromodeoxyuridine (BrdU)-labeled nuclei in the maxilla of Tgfbr2fl/fl (white bars) and Tgfbr2fl/fl;Wnt1-Cre (black bars) mice at E13.5 and E14.5. Data are mean ± S.D. values of five mice in each group. ***, p < 0.001. B, immunoblotting analysis of Tgfbr2fl/fl and Tgfbr2fl/fl;Wnt1-Cre maxilla at E12.5, E13.5, and E14.5. Data shown are representative of three separate experiments. C, plot shows the ratios between cyclin D1, cyclin D2, cyclin D3, cyclin E, and cyclin A versus GAPDH based on quantitative densitometry of immunoblotting data in B; *, p < 0.05. Tgfbr2fl/fl, white bars; Tgfbr2fl/fl;Wnt1-Cre, black bars. D and E, quantitative RT-PCR analyses of cyclin D regulators from E13.5 maxilla of Tgfbr2fl/fl (open columns) and Tgfbr2fl/fl;Wnt1-Cre (closed columns) mice. *, p < 0.05. F, immunoblotting analysis of FoxO family members and activated JNK in the maxilla of Tgfbr2fl/fl and Tgfbr2fl/fl;Wnt1-Cre maxilla at E12.5, E13.5, and E14.5. Data shown are representative of three separate experiments. G, plot shows the ratios between FoxO3a, FoxO4, JNK, and phosphorylated JNK versus GAPDH after quantitative densitometry of immunoblotting data in F; *, p < 0.05. Tgfbr2fl/fl, white bars; Tgfbr2fl/fl;Wnt1-Cre, black bars.
To determine the activity of the JNK, we analyzed the phosphorylation of JNK by immunoblotting. JNK activity was up-regulated at E13.5 and E14.5 in Tgfbr2fl/fl;Wnt1-Cre maxilla compared with Tgfbr2fl/fl (Fig. 2F). These data indicate that loss of Tgf-β signaling results in up-regulated FoxO4 expression and JNK activity, followed by decreased cyclin D expression.
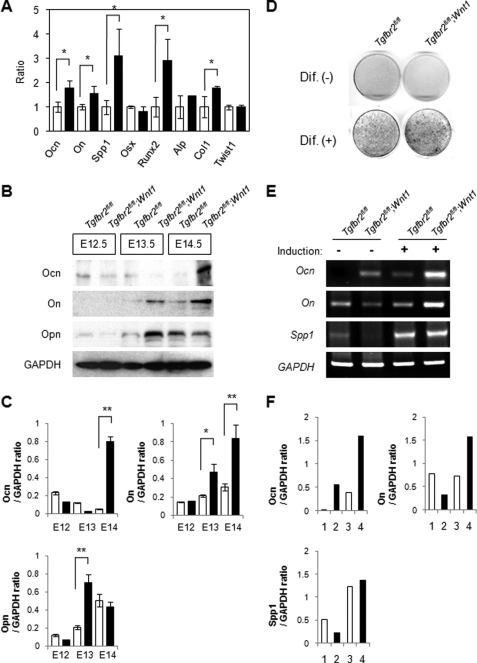
Previous studies indicated that Tgf-β signaling regulates osteogenic differentiation during bone formation (28, 29). To investigate the effect of decreased proliferation activity on cell fate determination, we compared the expression of genes involved in osteogenic differentiation in Tgfbr2fl/fl (control) and Tgfbr2fl/fl;Wnt1-Cre maxilla at E13.5 by quantitative RT-PCR. Osteopontin/Spp1 and Runx2 were up-regulated 3-fold in Tgfbr2fl/fl;Wnt1-Cre maxilla at E13.5 (Fig. 3A). Gene expression of osteocalcin, osteonectin, and type I collagen were also up-regulated 1.5-fold in Tgfbr2fl/fl;Wnt1-Cre maxilla at E13.5 (Fig. 3A). To confirm the altered osteogenic differentiation in Tgfbr2fl/fl;Wnt1-Cre maxilla, we analyzed the expression of proteins involved in osteogenic differentiation in Tgfbr2fl/fl and Tgfbr2fl/fl;Wnt1-Cre maxilla by immunoblotting. Expression of osteocalcin, osteopontin, and osteonectin was up-regulated in Tgfbr2fl/fl;Wnt1-Cre maxilla at E13.5 and E14.5 (Fig. 3, B and C). Furthermore, osteogenic differentiation was up-regulated following osteogenic induction of primary MEMM cells from Tgfbr2fl/fl;Wnt1-Cre mice compared with Tgfbr2fl/fl mice (Fig. 3D). Gene expression of osteopontin/Spp1, osteocalcin, and osteonectin was induced in Tgfbr2fl/fl MEMM cells after osteogenic induction, and these gene expressions were elevated in Tgfbr2fl/fl;Wnt1-Cre MEMM cells (Fig. 3, E and F). Thus, the reduced proliferation activity in CNCC-derived osteoprogenitor cells is followed by altered osteogenic differentiation in Tgfbr2fl/fl;Wnt1-Cre maxilla, resulting in the ossification of a reduced maxilla bone primordium at E14.5 (supplemental Fig. S4A).
FIGURE 3.
Loss of Tgfbr2 in CNCC results in altered osteogenic differentiation during intramembranous ossification. A, quantitative RT-PCR analyses of indicated genes in Tgfbr2fl/fl (open columns) and Tgfbr2fl/fl;Wnt1-Cre (closed columns) mice at E13.5. Ocn, osteocalcin; On, osteonectin; Spp1, osteopontin; Osx, Osterix; Alp, alkaline phosphatase; ColI, type I collagen. *, p < 0.05. B, immunological analysis of osteocalcin (Ocn), osteonectin (On), and osteopontin (Opn) in Tgfbr2fl/fl and Wnt1-Cre;Tgfbr2fl/fl maxilla at E12.5, E13.5, and E14.5. Data shown are representative of three separate experiments. C, plot shows the ratios between osteocalcin (Ocn), osteonectin (On), and osteopontin (Opn) versus GAPDH based on quantitative densitometry of immunoblotting data in B; *, p < 0.05. Tgfbr2fl/fl, white bars; Tgfbr2fl/fl;Wnt1-Cre, black bars. D, osteogenic differentiation of Tgfbr2fl/fl and Tgfbr2fl/fl;Wnt1-Cre primary MEMM cells cultured for 14 days in osteogenic induction medium. Alkaline phosphatase (ALP) staining of Tgfbr2fl/fl and Tgfbr2fl/fl;Wnt1-Cre MEMM cells cultured without (Dif. −) or with (Dif. +) osteogenic inducer for 2 weeks. E, mRNA expression of indicated genes after no osteogenic induction (−) or induction (+) in Tgfbr2fl/fl and Tgfbr2fl/fl;Wnt1-Cre MEMM cells. Data shown are representative of three separate experiments. F, graph shows quantitative densitometry analysis of gel electrophoresis data in E. Data shown are representative of three separate experiments. Lane 1, Tgfbr2fl/fl MEMM cells without osteogenic induction; lane 2, Wnt1-Cre;Tgfbr2fl/fl MEMM cells without osteogenic induction; lane 3, Tgfbr2fl/fl MEMM cells with osteogenic induction; lane 4, Wnt1-Cre;Tgfbr2fl/fl MEMM cells with osteogenic induction.
Tgf-β Signaling Regulates Gene Expression of Basal Transcriptional Factors
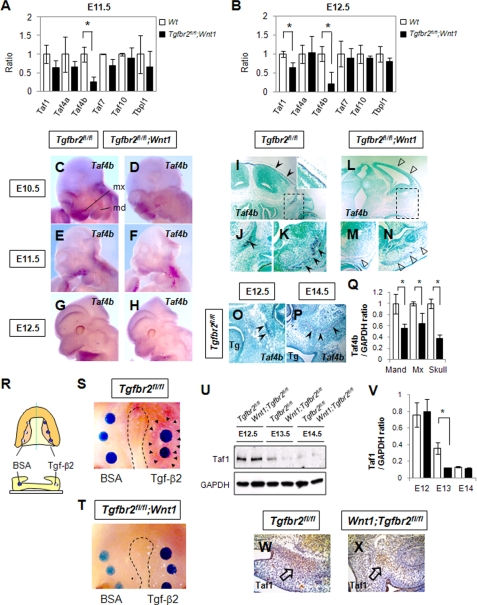
Previous studies revealed that some basal transcriptional factors are expressed in a tissue- and cell-specific manner (14, 30). Mutations of these basal transcriptional factors resulted in decreased cell proliferation (15). To explore potential osteoprogenitor cell-specific regulation of basal transcriptional factors by Tgf-β signaling, we analyzed the gene expression of basal transcriptional factors in the maxilla of Tgfbr2fl/fl and Tgfbr2fl/fl;Wnt1-Cre mice using quantitative RT-PCR. Interestingly, Taf4b was down-regulated in Tgfbr2fl/fl;Wnt1-Cre maxilla at E11.5 and E12.5 (Fig. 4, A and B). Gene expression of Taf1 was down-regulated at E12.5 but not E11.5 (Fig. 4B). Loss of Taf4 results in increased gene expression of Tgfb1, Tgfb3, and Ctgf (9), and overexpression of Taf4b results in the altered gene expression of Ctgf and Tgfb ligands (9), suggesting that the transcriptional regulation of Taf4b and its paralogue Taf4 is closely related to Tgf-β signaling. Taken together, these data suggest that the stoichiometry of basal transcriptional factors incorporated into TFIID regulates the fate of osteoprogenitor cells.
FIGURE 4.
Osteoprogenitor cell-specific expression of Taf4b in mouse embryos and reduced Taf4b expression in the maxillary process of Tgfbr2fl/fl;Wnt1-Cre mice. A and B, quantitative RT-PCR analyses of indicated genes in the maxilla of Tgfbr2fl/fl (open columns) and Tgfbr2fl/fl;Wnt1-Cre (closed columns) mice at E11.5 (A) and E12.5 (B). Wt, wild type. C–H, whole-mount in situ hybridization of Taf4b in Tgfbr2fl/fl and Tgfbr2fl/fl;Wnt1-Cre mice at E10.5, E11.5, and E12.5. Taf4b mRNA was strongly expressed in the maxilla and limb and weakly expressed in the mandible and frontal primordia. mx, maxillary process; md, mandibular process. I–P, in situ hybridization of Taf4b mRNA in sections of Tgfbr2fl/fl and Tgfbr2fl/fl;Wnt1-Cre mice at E14.5. Taf4b was strongly expressed by osteoprogenitor cells in the skull, frontal bone, mandible, and maxilla of wild-type mice, whereas the gene expression of Taf4b was significantly reduced in Tgfbr2fl/fl;Wnt1-Cre mice. Boxed areas in I and L are magnified in J and M, respectively. Wild-type maxilla of each developmental stage is shown in O and P. Arrowheads point to expression of Taf4b mRNA. Open arrowheads indicate areas negative for Taf4b expression. Tg is tongue. Q, quantitative RT-PCR analyses of Taf4b from E14.5 skull, mandible (Mand), and maxilla (Max) of Tgfbr2fl/fl (open columns) and Tgfbr2fl/fl;Wnt1-Cre (closed columns) mice. *, p < 0.05. R–T, Tgf-β2 or bovine serum albumin (BSA) bead implantation experiment in maxillas from Tgfbr2fl/fl (S) and Tgfbr2fl/fl;Wnt1-Cre (T) mice at E13.5. R is a schematic diagram of the experiment design. Arrowheads indicate the expression of Taf4b mRNA detected by whole-mount in situ hybridization. Dotted line outlines the edge of palates. U, immunoblotting analysis of Taf1 in Tgfbr2fl/fl and Tgfbr2fl/fl;Wnt1-Cre maxilla at E12.5, E13.5, and E14.5. Data shown are representative of three separate experiments. V, plot shows the ratios between Taf1 and GAPDH after quantitative densitometry of immunoblotting data in U. *, p < 0.05. Tgfbr2fl/fl (white bars) and Tgfbr2fl/fl;Wnt1-Cre (black bars). W and X, immunohistochemical staining of Taf1 in sections of Tgfbr2fl/fl (W) and Tgfbr2fl/fl;Wnt1-Cre (X) mice at E14.0. Taf1 expression was significantly reduced in Tgfbr2fl/fl;Wnt1-Cre mice. Arrows point to expression of Taf1.
Taf4b Is Specifically Expressed in Maxillary Bone Primordium
Taf4b is specifically expressed in gonad tissues in adult mice (12, 30, 31); however, the expression pattern and function of Taf4b are still unknown during embryogenesis. To examine the expression pattern of Taf4b during embryonic development, we performed whole-mount in situ hybridization (Fig. 4, C–H). Taf4b expression was prominent in the maxilla and limbs from E10.5 to E12.5, and weaker staining was detectable in the mandible and frontal bone primordia. Taf4b expression was detectable in the osteogenic primordia of wild type mice at E14.5 (Fig. 4, I–K and P), but it was significantly reduced in the bone primordia of Tgfbr2fl/fl;Wnt1-Cre mice (Fig. 4, L–N and Q). In contrast, we detected Taf1 expression throughout the craniofacial region in wild type mice and reduced expression in Tgfbr2fl/fl;Wnt1-Cre mice at E13.5 and E14.0 (Fig. 4, U–X). To investigate Tgf-β regulation of Taf4b in vivo, we implanted beads containing Tgf-β2 protein into organ cultures of maxilla derived from Tgfbr2fl/fl and Tgfbr2fl/fl;Wnt1-Cre mice (Fig. 4, R–T). At 24 h after the administration of Tgf-β2, Taf4b gene expression was up-regulated around the beads in controls but not in Tgfbr2fl/fl;Wnt1-Cre maxilla (Fig. 4, S and T). Furthermore, gene expression of Taf4b was up-regulated following osteogenic induction of primary MEMM cells from wild type mice but not in that of Tgfbr2fl/fl;Wnt1-Cre mice (supplemental Fig. S3). We conclude that Tgf-β signaling regulates the gene expression of Taf4b.
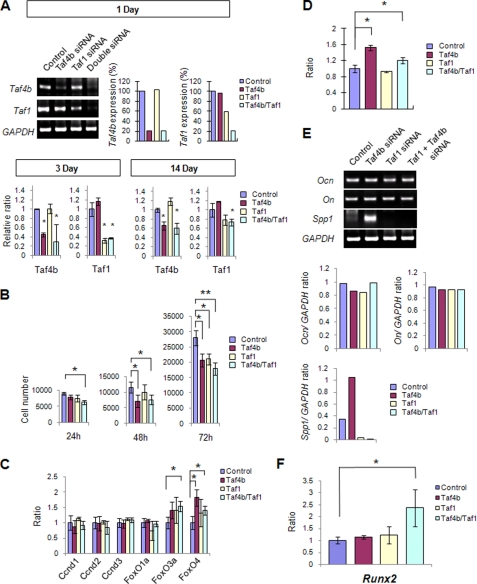
Double Knockdown of Taf4b and Taf1 Affects Gene Expression Related to Cellular Proliferation, Initiation of Bone Formation, and Osteogenic Differentiation
To test the functional significance of Taf4b and Taf1 in regulating the fate of osteogenic progenitor cells, we reduced the gene expression of Taf4b and Taf1 in primary MEMM cells derived from E13.5 maxilla using an siRNA knockdown approach (Fig. 5A). Gene expression of Taf4b and Taf1 was successfully suppressed by the siRNA treatment (Fig. 5A). We found that the simultaneous down-regulation of Taf4b and Taf1 resulted in reduced cell proliferation (Fig. 5B). Gene expression of FoxO3 and FoxO4, which are cyclin D suppressors, was significantly increased by 1.4- and 1.5-fold after siRNA treatment of Taf4b alone and a 1.8- and 1.4-fold change after siRNA treatment of Taf1 and Taf4b together, respectively (Fig. 5C). To analyze osteogenic differentiation, we cultured primary MEMM cells treated with Taf4b, Taf1, and Taf4b/Taf1 double siRNA for 2 weeks with osteogenic induction medium and then analyzed alkaline phosphatase activity. Specifically, siRNA mixture was added every 3 days into the cell culture medium throughout the 2 weeks of culture. The success of our siRNA knockdown experiments was demonstrated by quantitative gene expression analyses (Fig. 5A). We detected increased osteogenic differentiation in samples with combined down-regulation of Taf4b and Taf4b/Taf1 (Fig. 5D). To investigate osteogenic differentiation following Taf4b and Taf1 siRNA treatment, we analyzed the expression of genes related to bone formation. We found that Spp1 was specifically up-regulated after Taf4b siRNA treatment but not Taf1 siRNA treatment (Fig. 5E). These data confirm that Taf4b has unique functions in CNCC-derived osteoprogenitor cells. Interestingly, Runx2 expression was specifically up-regulated after siRNA knockdown of Taf4b/Taf1 together, although synergistic changes were not seen in other osteogenic factors, suggesting that a combination of Taf4b and Taf1 regulates the initiation of bone formation (Fig. 5F). Runx2 is required for mesenchymal cell differentiation into osteoblasts (32). Runx2 activates expression of several genes expressed by mature osteoblasts and chondrocytes (33, 34). Basal transcriptional factors may regulate Runx2 gene expression via Tgf-β signaling to promote the initiation of osteogenic differentiation. Thus, basal transcriptional factors have multifunctional physiological roles in CNCC-mediated osteoprogenitor cells that include regulation of cell proliferation, osteogenic fate determination, and differentiation, and Taf1 and Taf4b work synergistically during intramembranous bone development following regulation by Tgf-β signaling (supplemental Fig. S4B).
FIGURE 5.
Osteogenic progenitor cell proliferation and differentiation in primary MEMM cells after siRNA knockdown of Taf1 and Taf4b. A, Taf4b and Taf1 mRNA expression in primary MEMM cells isolated from wild-type maxilla after a 24-h treatment with Taf4b, Taf1, or Taf4b and Taf1 (double) siRNA. *, p < 0.05. Antisense siRNA treatment was used as control. Graph shows quantitative densitometry analysis of gel electrophoresis data. Data shown are representative of three separate experiments. Quantitative RT-PCR of Taf4b and Taf1 was performed at 3 and 14 days during siRNA treatment. B, cell proliferation was assayed by cell number after siRNA treatment of MEMM cells at 24, 48, and 72 h. Cell culture was started at 5 × 103 cells (0 h). Data are the mean values from three independent experiments. *, p < 0.05; **, p < 0.01. C, quantitative RT-PCR of indicated genes after siRNA treatment. *, p < 0.05. D, alkaline phosphatase enzyme activities measured by β-galactosidase assay following siRNA treatments and 2 weeks culture in osteogenic induction medium. Data are expressed as ratio of absorbance at 405 nm after alkaline phosphatase staining compared with control siRNA. Alkaline phosphatase enzyme activity indicates osteogenic cell differentiation. Data are the mean values from three independent experiments. *, p < 0.05. E, mRNA expression of indicated genes after siRNA treatments. Data shown are representative of three separate experiments. Graph shows quantitative densitometry analysis of gel electrophoresis data. F, quantitation of Runx2 mRNA level by real time RT-PCR after siRNA treatment. *, p < 0.05.
DISCUSSION
We investigated CNCC-derived intramembranous bone formation and the downstream targets of the Tgf-β signaling using Tgfbr2fl/fl;Wnt1-Cre mice. The proliferation period of osteoprogenitor cells derived from Tgfbr2fl/fl;Wnt1-Cre mice is shorter than that of wild type mice. The consequence of the decreased proliferation term is the early onset of osteogenic differentiation. Thus, the decreased size of the maxilla in Tgfbr2fl/fl;Wnt1-Cre mice results from a decreased number of osteoprogenitor cells (supplemental Fig. S4A). The expression of FoxO4 and Runx2 was increased in Tgfbr2fl/fl;Wnt1-Cre maxilla at E13.5 but not at E11.5 and E12.5 (supplemental Fig. S2). Moreover, after we reduced expression of Taf4b and Taf1 in primary MEMM cells, we found similar changes in the expression of FoxO4 and Runx2, consistent with a role for these basal transcriptional factors in regulating osteogenic gene expression. Our results suggest that Tgf-β signaling regulates the expression of basal transcriptional factors in a time- and tissue-dependent manner (supplemental Fig. S4B).
A previous study indicated that Taf4b mediates Tgf-β signaling more efficiently than Taf4 (9). Taf4 is ubiquitously expressed, whereas Taf4b is expressed in a tissue- and cell type-specific manner (30, 31). Taf4 knock-out mice have premature mortality at E9.5 (9); however Taf4b knock-out mice show no visible phenotype except defects in gonad tissue (31). Taf4b-TFIID and Taf4-TFIID may utilize similar mechanisms to activate gene expression in the Tgf-β signaling cascade, but Taf4 can apparently compensate for the loss of Taf4b function and not vice versa. In this study, we found that Taf1 was down-regulated in Tgfbr2fl/fl;Wnt1-Cre maxilla at E12.5. Mutations in Taf1 result in decreased cell proliferation in vitro (35, 36). Future studies of Taf1/Taf4b double heterozygous mutant mice may demonstrate that they recapitulate the phenotype of Tgfbr2fl/fl;Wnt1-Cre mice. This finding underlines the importance of the stoichiometry of the Taf1/Taf4b subunits in regulating intramembranous ossification.
CNCC-derived mesenchymal cells progress through osteogenic proliferation and then commit to the transition from preosteoblastic progenitors to osteoblasts. Osteopontin and osteocalcin were up-regulated in Tgfbr2fl/fl;Wnt1-Cre mice at E13.5. The promoter region of Taf4b has a putative Tgf-β-response element from −293 to −284 bp, osteocalcin motif from −521 to −513 bp, and osteopontin-response element from −149 to −135 bp, consistent with our hypothesis that gene expression of Taf4b is regulated by Tgf-β signaling directly and/or indirectly. Furthermore, osteogenic inducers may also provide feedback to Taf4b transcriptional regulation. Our study demonstrates that Tgf-β-regulated Taf4b gene expression is a tightly controlled process during intramembranous maxillary bone formation.
Bone formation requires a cascade of transcriptional events to control the spatial and temporal expression of osteoblast-specific genes. Our findings show that Tgf-β signaling regulates cell proliferation and osteogenic initiation via basal transcriptional factors in osteoprogenitor cells. Tgf-β-mediated basal transcriptional factors appear to exert their functional specificity by controlling downstream target genes. Variations of the Taf(s) complex may contribute to the multifunctional role of Tgf-β signaling during embryogenesis. Thus, the interactions between Tgf-β signaling and basal transcriptional factors have a crucial function in regulating osteogenic cell proliferation and differentiation during intramembranous bone formation.
Supplementary Material
Acknowledgments
We thank H. Moses for the Tgfbr2fl/fl mice and Julie Mayo for critical reading of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants DE012711, DE014078, and U01 DE020065 from NIDCR (to Y. C.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
- CNCC
- cranial neural crest cells
- JNK
- Jun oncogene N-terminal kinase
- MEMM
- mouse embryonic maxillary mesenchymal
- TAF
- TATA-binding protein-associated factors
- Tgf-β
- transforming growth factor-β
- siRNA
- small interfering RNA
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- RT
- reverse transcription
- TFIID
- transcription factor IID
- E
- embryonic day.
REFERENCES
- 1.Jiang X., Iseki S., Maxson R. E., Sucov H. M., Morriss-Kay G. M. (2002) Dev. Biol. 241, 106–116 [DOI] [PubMed] [Google Scholar]
- 2.Chai Y., Maxson R. E., Jr. (2006) Dev. Dyn. 235, 2353–2375 [DOI] [PubMed] [Google Scholar]
- 3.Chai Y., Ito Y., Han J. (2003) Crit. Rev. Oral Biol. Med. 14, 78–88 [DOI] [PubMed] [Google Scholar]
- 4.Ito Y., Yeo J. Y., Chytil A., Han J., Bringas P., Jr., Nakajima A., Shuler C. F., Moses H. L., Chai Y. (2003) Development 130, 5269–5280 [DOI] [PubMed] [Google Scholar]
- 5.Ross S., Hill C. S. (2008) Int. J. Biochem. Cell Biol. 40, 383–408 [DOI] [PubMed] [Google Scholar]
- 6.Müller C., Leutz A. (2001) Curr. Opin. Genet. Dev. 11, 167–174 [DOI] [PubMed] [Google Scholar]
- 7.de la Serna I. L., Ohkawa Y., Imbalzano A. N. (2006) Nat. Rev. Genet. 7, 461–473 [DOI] [PubMed] [Google Scholar]
- 8.Hiller M., Chen X., Pringle M. J., Suchorolski M., Sancak Y., Viswanathan S., Bolival B., Lin T. Y., Marino S., Fuller M. T. (2004) Development 131, 5297–5308 [DOI] [PubMed] [Google Scholar]
- 9.Mengus G., Fadloun A., Kobi D., Thibault C., Perletti L., Michel I., Davidson I. (2005) EMBO J. 24, 2753–2767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Metcalf C. E., Wassarman D. A. (2007) Dev. Dyn. 236, 2836–2843 [DOI] [PubMed] [Google Scholar]
- 11.Wang X., Truckses D. M., Takada S., Matsumura T., Tanese N., Jacobson R. H. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 7839–7844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu W. L., Coleman R. A., Grob P., King D. S., Florens L., Washburn M. P., Geles K. G., Yang J. L., Ramey V., Nogales E., Tjian R. (2008) Mol. Cell 29, 81–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hampsey M. (1998) Microbiol. Mol. Biol. Rev. 62, 465–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Veenstra G. J., Wolffe A. P. (2001) Trends Biochem. Sci. 26, 665–671 [DOI] [PubMed] [Google Scholar]
- 15.Davidson I., Kobi D., Fadloun A., Mengus G. (2005) Cell Cycle 4, 1486–1490 [DOI] [PubMed] [Google Scholar]
- 16.Albright S. R., Tjian R. (2000) Gene 242, 1–13 [DOI] [PubMed] [Google Scholar]
- 17.Lemon B., Tjian R. (2000) Genes Dev. 14, 2551–2569 [DOI] [PubMed] [Google Scholar]
- 18.Sasaki T., Ito Y., Bringas P., Jr., Chou S., Urata M. M., Slavkin H., Chai Y. (2006) Development 133, 371–381 [DOI] [PubMed] [Google Scholar]
- 19.Iwata J., Ezaki J., Komatsu M., Yokota S., Ueno T., Tanida I., Chiba T., Tanaka K., Kominami E. (2006) J. Biol. Chem. 281, 4035–4041 [DOI] [PubMed] [Google Scholar]
- 20.Kawakubo T., Okamoto K., Iwata J., Shin M., Okamoto Y., Yasukochi A., Nakayama K. I., Kadowaki T., Tsukuba T., Yamamoto K. (2007) Cancer Res. 67, 10869–10878 [DOI] [PubMed] [Google Scholar]
- 21.Komatsu M., Waguri S., Ueno T., Iwata J., Murata S., Tanida I., Ezaki J., Mizushima N., Ohsumi Y., Uchiyama Y., Kominami E., Tanaka K., Chiba T. (2005) J. Cell Biol. 169, 425–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Komatsu M., Waguri S., Chiba T., Murata S., Iwata J., Tanida I., Ueno T., Koike M., Uchiyama Y., Kominami E., Tanaka K. (2006) Nature 441, 880–884 [DOI] [PubMed] [Google Scholar]
- 23.Sou Y. S., Waguri S., Iwata J., Ueno T., Fujimura T., Hara T., Sawada N., Yamada A., Mizushima N., Uchiyama Y., Kominami E., Tanaka K., Komatsu M. (2008) Mol. Biol. Cell 19, 4762–4775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oka K., Oka S., Sasaki T., Ito Y., Bringas P., Jr., Nonaka K., Chai Y. (2007) Dev. Biol. 303, 391–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hatakeyama Y., Tuan R. S., Shum L. (2004) J. Cell. Biochem. 91, 1204–1217 [DOI] [PubMed] [Google Scholar]
- 26.Coqueret O. (2002) Gene 299, 35–55 [DOI] [PubMed] [Google Scholar]
- 27.Schmidt M., Fernandez de Mattos S., van der Horst A., Klompmaker R., Kops G. J., Lam E. W., Burgering B. M., Medema R. H. (2002) Mol. Cell. Biol. 22, 7842–7852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Javed A., Bae J. S., Afzal F., Gutierrez S., Pratap J., Zaidi S. K., Lou Y., van Wijnen A. J., Stein J. L., Stein G. S., Lian J. B. (2008) J. Biol. Chem. 283, 8412–8422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seo H. S., Serra R. (2009) Dev. Biol. 334, 481–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dikstein R., Zhou S., Tjian R. (1996) Cell 87, 137–146 [DOI] [PubMed] [Google Scholar]
- 31.Freiman R. N., Albright S. R., Zheng S., Sha W. C., Hammer R. E., Tjian R. (2001) Science 293, 2084–2087 [DOI] [PubMed] [Google Scholar]
- 32.Komori T. (2002) J. Cell. Biochem. 87, 1–8 [DOI] [PubMed] [Google Scholar]
- 33.Stricker S., Fundele R., Vortkamp A., Mundlos S. (2002) Dev. Biol. 245, 95–108 [DOI] [PubMed] [Google Scholar]
- 34.Yoshida C. A., Furuichi T., Fujita T., Fukuyama R., Kanatani N., Kobayashi S., Satake M., Takada K., Komori T. (2002) Nat. Genet. 32, 633–638 [DOI] [PubMed] [Google Scholar]
- 35.Maile T., Kwoczynski S., Katzenberger R. J., Wassarman D. A., Sauer F. (2004) Science 304, 1010–1014 [DOI] [PubMed] [Google Scholar]
- 36.Hilton T. L., Li Y., Dunphy E. L., Wang E. H. (2005) Mol. Cell. Biol. 25, 4321–4332 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.