Abstract
One mechanism of prostate tumors for escape from androgen ablation therapies is mutation of the androgen receptor (AR). We investigated the unique properties of the AR L701H mutant, which is strongly stimulated by cortisol, by a systematic structure-function analysis. Most amino acid substitutions at position 701 did not affect AR activation by 5α-dihydrotestosterone. Further analysis of the AR Leu701 variants showed that AR L701M and AR L701Q, like AR L701H, had changed ligand responsiveness. AR L701M was strongly activated by progesterone but not by cortisol, whereas the opposite was observed for AR L701Q and AR L701H. Next, we analyzed a panel of structurally related steroids to study which of the OH groups at positions 11β, 17α, and 21, which discriminate cortisol from progesterone, underlie the differential responses to both hormones. The results showed that the 17α-OH group was essential for activation of AR L701H and AR L701Q, whereas its absence was important for activation of AR L701M. Modeling indicated a conserved H-bonding network involving the steroidal 17α-OH group, His701 or Gln701, and the backbone of Ser778. This network is absent in Leu701 and in other mutants. A hydrophobic leucine or methionine at position 701 is unfavorable for the 17α-OH group. Our results indicate that the specific amino acid residue at position 701, its interaction with the backbone of Ser778, and the steroidal 17α-hydroxyl group of the ligand are all important for the distinct transcriptional responses to progesterone and cortisol of AR mutants, including the prostate cancer mutant L701H.
Keywords: Mutant, Nuclear Receptors, Peptide Interactions, Steroid, Steroid Receptor, Androgen Receptor, Antiandrogens, Peptide, Prostate Cancer
Introduction
The androgen receptor (AR)2 is a ligand-dependent transcription factor that is activated by the androgens testosterone and 5α-dihydrotestosterone (DHT). The androgen-AR axis is essential for normal male development and plays a pivotal role in maintaining the functions of male-specific organs, including the prostate (1). A disturbed androgen-AR axis has been implicated in a number of malignancies, including androgen insensitivity syndrome (AIS) and prostate cancer (2, 3). AIS is caused by AR inactivation and is characterized by defective masculinization of 46,XY individuals. It ranges from mild undervirilization (mild AIS) to partial (PAIS) or even complete (CAIS) female phenotypic outcomes. An active AR pathway is involved in prostate cancer. Initially, prostate cancer growth is dependent on androgens. Therefore, treatment of metastasized tumors aims at inhibiting the AR pathway by suppressing testicular androgen production by luteinizing hormone-releasing hormone analogs or by blocking AR activity using antiandrogens. Despite an initial response, tumors eventually regain the ability to grow, leading to an endocrine therapy-resistant stage of the disease. Although tumor growth is androgen-independent at this stage, the AR pathway still appears to be active (4, 5). Several mechanisms that may underlie therapy failure have been proposed, including AR amplification and AR mutations (6–9).
Like other nuclear receptors, the AR contains separate functional domains: an N-terminal transactivation domain, a central DNA-binding domain, and a C-terminal ligand-binding domain (LBD) (10). The LBD is composed of 12 α-helices, of which amino acid residues in helices 3, 5, 7, and 11 form the ligand-binding pocket (11, 12). Upon binding of an agonistic ligand, the LBD undergoes major structural rearrangements to obtain an active conformation. Helix 12 closes the ligand-binding pocket and becomes part of the coactivator-binding groove. This groove then serves as a high affinity docking site for short amphipathic α-helical FXXLF-like sequences present in cofactors (13–15). In response to binding of an antagonist, helix 12 adopts a different orientation, preventing the AR LBD from obtaining the active conformation necessary to induce transcription (16).
The incidence of AR mutations is low in primary prostate tumors but increases in advanced disease during endocrine therapy (6, 7, 17). The majority of AR mutations collocate at several regions in the LBD mapping to amino acid residues 670–678, 701–730, and 874–919 (18). The first AR mutation reported was a threonine-to-alanine substitution at position 877 that was identified in the LNCaP prostate cancer cell line (19). Later, it was found that Thr877 serves as an AR mutational hot spot in recurrent prostate tumors (20). Crystallographic analysis and functional studies of the AR T877A mutant demonstrated that this amino acid substitution alters the size, shape, and properties of the ligand-binding pocket, allowing several non-natural ligands and even antiandrogens to bind and activate the receptor (12, 21–23). It seems that AR T877A drives tumor growth through aberrant activation by the antiandrogen used for treatment.
A second AR mutational hot spot identified in prostate cancers is residue 701. The AR mutation substituting Leu for His at position 701 (L701H) has been reported in hormone-refractory prostate cancer patients (24, 25). The same mutation, in combination with T877A, is also present in the AR of MDA-PCa cell lines, which were originally derived from a bone metastasis of an orchiectomized prostate cancer patient (26, 27). AR L701H and AR L701H/T877A are somewhat less sensitive to androgens but are highly responsive to the glucocorticoids cortisol and cortisone, which circulate at concentrations high enough to activate both mutant receptors (28, 29). Thus, androgen-independent growth of prostate tumors containing the L701H mutation differs from that of prostate tumors containing the T877A mutation by being driven by endogenously circulating ligands.
Previously, the T877A substitution has been studied in detail (23). In this study, we performed a systematic structure-function analysis of AR residue 701 to obtain further insight in the ligand responsiveness of AR L701H. Screening revealed that, in addition to AR L701H, also AR L701M and AR L701Q had a changed but differential ligand specificity. Functional studies with a panel of structurally related steroids showed that the presence of a hydroxyl group at position 17α is critical for activation of AR L701H and AR L701Q but not AR L701M. Modeling of the various mutations in the AR LBD structure revealed that a unique H-bonding network involving His701 or Gln701, the steroidal 17α-OH group, and the backbone oxygen of Ser778 plays an important role in the cortisol response. The structure-function analysis described in this study includes the clarification of the properties of the AR L701H mutant in prostate cancer.
EXPERIMENTAL PROCEDURES
Hormones
DHT, progesterone, cortisol, and 11-desoxycorticosterone were purchased from Steraloids (Wilton, NH); 17α-hydroxyprogesterone, 11β-hydroxyprogesterone, 11-desoxycortisol, and 21-desoxycortisol were from Sigma; and hydroxyflutamide was from Schering (Bloomfield, NJ). Corticosterone was kindly provided by Dr. Albert Brinkmann (Erasmus Medical Center), and bicalutamide (Casodex) was a gift from AstraZeneca (Macclesfield, United Kingdom).
Plasmids
All AR L701X mutants were generated using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) in the mammalian AR cDNA expression vector pSVAR0 (30) according to the manufacturer's protocol. The primers used were 5′-CTTTGCAGCCTTGNNNTCTAGCCTCAATG-3′ (with the bases encoding AR residue 701 indicated in boldface) and its complementary sequence. AR L701H/T877A was generated by QuikChange mutagenesis of AR Leu701 in the pSVARL vector, which expresses the AR T877A mutant (19), using primer 5′-CTTTGCAGCCTTGCACTCTAGCCTCAATG-3′ (with the bases encoding mutated AR residue 701 indicated in boldface and the base substitution underlined) and its complementary sequence. All mutations were verified by sequence analysis, and Western blotting was performed to analyze size and expression of the AR mutants.
Construction of the mammalian expression plasmid encoding the Gal4 DNA-binding domain-AR FXXLF peptide and the (ARE)2TATA-Luc reporter ((PRE)2-E1b-Luc, where PRE is progesterone response element) has been described previously (31, 32). The (UAS)4TATA-Luc (where UAS is upstream activating sequence) reporter construct was kindly provided by Magda Meester.
Mammalian Cell Culture, Transient Transfections, and Luciferase Assay
Hep3B cells were maintained in α-minimal essential medium (BioWhittaker, Verviers, Belgium) supplemented with 5% fetal calf serum and antibiotics. For transient transfection assays, Hep3B cells were plated at a density of 5 × 104 cells/well of a 24-well plate and allowed to grow for 24 h. Four hours prior to transfection, the medium was replaced with α-minimal essential medium supplemented with 5% charcoal-stripped fetal calf serum, antibiotics, and hormone or vehicle. Twenty-four hours after addition of transfection mixtures (described below), cells were lysed, and luciferase activities were measured as described previously (31).
Transcriptional activation assays were performed using FuGENE 6 (Roche Diagnostics, Mannheim, Germany), 50 ng of AR expression construct, and 100 ng of (ARE)2TATA-Luc reporter construct per well. In mammalian one-hybrid assays, FuGENE 6 mixtures contained 50 ng of Gal4 DNA-binding domain-peptide, 50 ng of AR expression construct, and 150 ng of (UAS)4TATA-Luc reporter per well.
Western Blot Analysis
For Western blot analysis, Hep3B cells were transfected with 50 ng of AR expression construct as described above. Twenty-four hours after transfection, cells were lysed in 100 μl of Laemmli buffer (50 mm Tris, 10 mm dithiothreitol, 10% glycerol, 2% SDS, and 0.001% bromphenol blue). Lysates were boiled and subjected to electrophoresis on a 10% SDS-polyacrylamide gel, after which proteins were transferred to a nitrocellulose membrane. Blots were incubated with monoclonal antibodies directed against the AR N-terminal domain (F39.4.1), followed by incubation with horseradish peroxidase-conjugated goat anti-mouse antibody (Dako, Glostrup, Denmark). Proteins were visualized using SuperSignal West Pico chemiluminescent blotting substrate (Pierce), followed by exposure to x-ray film.
Modeling
Residue 701 was mutated systematically in a proprietary wild-type AR structure bound to DHT and the publicly available AR L701H structure (33). Each mutation has been minimized in YASARA using the YAMBER2 force field (34) and visually inspected.
RESULTS AND DISCUSSION
Most AR Leu701 Mutants Are Responsive to DHT
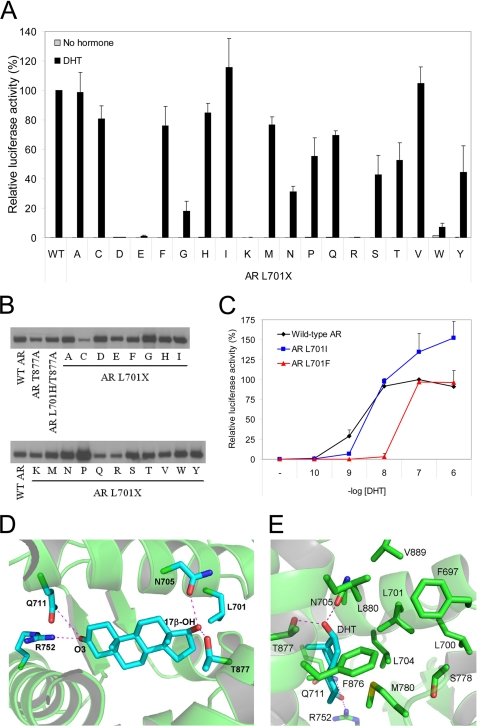
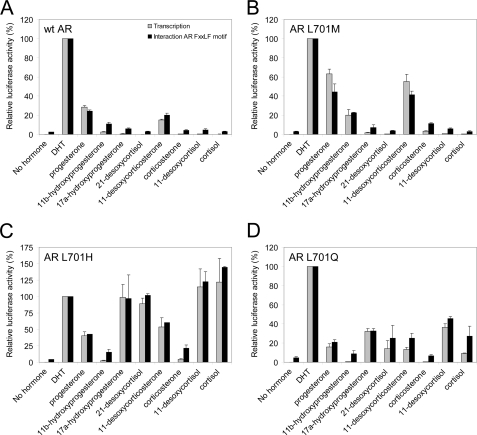
We performed a systematic structure-function analysis of AR residue 701 to obtain further insight in the effect of the L701H mutation on AR ligand responsiveness. AR Leu701 was substituted with any other amino acid residue, and the resultant AR mutant was tested for DHT (100 nm) response in transiently cotransfected Hep3B cells. With the (ARE)2TATA-Luc as reporter, most AR mutants, including AR L701H, were strongly activated by DHT (Fig. 1A). Responses were somewhat less if a polar residue (Asn, Gln, Ser, Thr, or Tyr) was present at AR position 701. Substitution of the Leu with Gly or Trp strongly reduced activation by DHT, whereas substitution with a charged residue (Asp, Glu, Arg, or Lys) completely abrogated AR transcriptional activity. Similar results were obtained if murine mammary tumor virus-Luc was used as the reporter construct (data not shown). Western blot analysis demonstrated that all AR Leu701 mutants were expressed at levels comparable with wild-type AR levels (Fig. 1B).
FIGURE 1.
Activation of AR Leu701 mutants by DHT. A, transcriptional responses of wild-type (WT) AR and AR Leu701 mutants to DHT (100 nm). The single-letter amino acid code of the Leu701 substitution is indicated on the x axis. Hep3B cells were transiently cotransfected with AR expression constructs and the (ARE)2TATA-Luc reporter. Transcriptional activation of wild-type AR by DHT was set to 100%. Error bars represent mean relative luciferase activities of three independent experiments performed in duplicate (±S.E.). B, Western blot analysis showing protein expression of the AR Leu701 mutants. C, dose-response curves of wild-type AR (black), AR L701I (blue), and AR L701F (red) with DHT. Hep3B cells were transiently transfected as described above and incubated for 24 h with the indicated concentrations of DHT. The response of wild-type AR to 100 nm DHT was set at 100%. Data represent the mean relative luciferase activities of three independent experiments performed in duplicate (±S.E.). D and E, structural representations of the wild-type AR LBD in complex with DHT showing the relative position of Leu701 in the ligand-binding pocket. The steroidal 3-keto and 17β-OH groups are indicated. Dotted red lines represent hydrogen bonds.
The AR L701I and AR L701F mutants have previously been identified in individuals with PAIS and CAIS, respectively (35, 36). Despite normal responses of both AR mutants at high DHT concentrations (100 nm) (Fig. 1A), the activities of AR L701F and AR L701I were substantially lower than those of wild-type AR at 1 nm DHT (Fig. 1C). These reduced DHT responses may account for the CAIS and PAIS phenotypes found in individuals carrying the AR L701F and AR L701I mutations.
Analysis of the x-ray structure of wild-type AR in complex with DHT showed that the 3-keto group of DHT is hydrogen-bonded to Gln711 and Arg752 (Fig. 1D) (12). The 17β-OH group is stabilized via hydrogen bonds to Asn705 and Thr877. Leu701 is located near the steroidal D-ring and has hydrophobic contacts with C17. The side chain of Leu701 is buried in a predominantly hydrophobic pocket consisting of Phe697, Leu700, Leu704, Ser778, Met780, Phe876, Leu880, and Val889 (Fig. 1E). We modeled the replacement of Leu701 with each amino acid and evaluated the structure. These analyses showed that, in addition to the wild-type leucine, also other aliphatic residues (Val, Ile, Met, and Ala) were tolerated sterically and formed varying degrees of favorable van der Waals interactions with the surrounding hydrophobic pocket. As expected, Ala701 and Gly701 formed less hydrophobic contacts in the receptor than the other aliphatic residues, potentially resulting in a less stable protein. This may be the cause of the strongly reduced DHT response of AR L701G. Although polar residues were sterically tolerated at position 701, their lower DHT responses are most likely due to their presence in an unfavorable hydrophobic pocket. Large residues such as Trp, Phe, and Tyr could not be tolerated in the Leu701 hydrophobic pocket without causing significant clashes with the protein structure. The presence of these large residues required structural modification of the receptor, which likely explains the strongly reduced DHT responses and the CAIS phenotype observed in the individual carrying the AR L701F mutation. The lack of a charged residue in the environment surrounding Leu701 prohibited mutation to a charged residue. Without a charged partner, this would result in the destabilization of the receptor.
AR L701H, AR L701M, and AR L701Q Display Modified Ligand Responsiveness
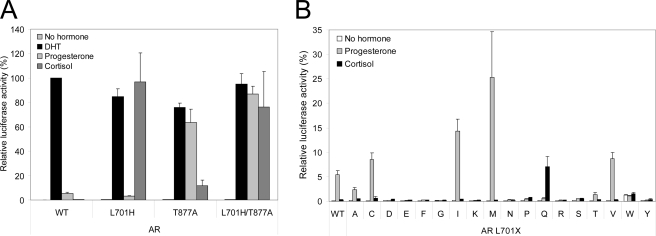
We next studied the transcriptional activities of the AR Leu701 mutants induced by progesterone and cortisol. In control experiments, we first determined the transcriptional responses of wild-type AR, the AR L701H and AR T877A single mutants, and the AR L701H/T877A double mutant to both steroids using the (ARE)2TATA-Luc reporter. In Hep3B cells, all receptor mutants responded similarly as described previously for CV-1 cells and a murine mammary tumor virus-Luc reporter (Fig. 2A) (29). Whereas wild-type AR was activated only by DHT, AR L701H was additionally activated by cortisol but not by progesterone. The opposite was observed for AR T877A. AR L701H/T877A displayed combined characteristics of both single mutants by responding strongly to both progesterone and cortisol.
FIGURE 2.
AR L701H, AR L701M, and AR L701Q display modified ligand specificity. A, transcriptional activation of wild-type (WT) AR, the AR L701H and AR T877A single mutants, and the AR L701H/T877A double mutant by DHT (100 nm), progesterone (100 nm), and cortisol (1 μm). Activation of wild-type AR by DHT was set to 100%. Error bars represent mean relative luciferase activities of three independent experiments performed in duplicate (±S.E.). Protein expression of these AR mutants is shown in Fig. 1B. B, screening of AR Leu701 mutants for responses to progesterone (100 nm) and cortisol (1 μm). Error bars represent mean relative luciferase activities of three independent experiments performed in duplicate (±S.E.). Transcriptional activation of wild-type AR by DHT was set to 100%.
Although the majority of AR Leu701 mutants were activated by DHT (Fig. 1A), they showed weak or no responses to progesterone and cortisol (Fig. 2B). However, apart from AR L701H, three mutants had a modified but distinct ligand responsiveness. The activities of AR L701I and AR L701M were clearly induced upon incubation with progesterone but not with cortisol, whereas AR L701Q, like AR L701H, was induced by cortisol and not by progesterone (Fig. 2B). Similar results were obtained using a murine mammary tumor virus-Luc reporter construct (data not shown). Because AR L701M showed the strongest responses to progesterone, this mutant was selected for further examination.
Differential Responsiveness of AR L701M, AR L701Q, and AR L701H Is Determined by the 17α-OH Group
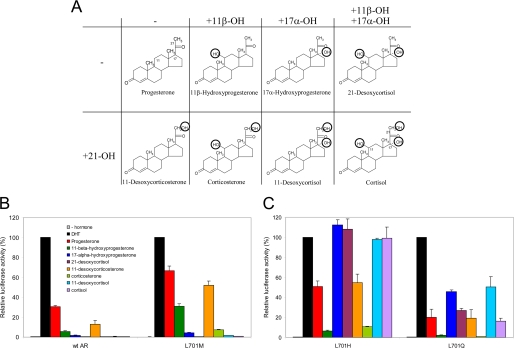
Progesterone and cortisol are structurally highly similar (Fig. 3A). Cortisol differs from progesterone by the presence of hydroxyl groups at positions 11β, 17α, and 21. To study which of these hydroxyl groups differentially affects transcriptional activation of AR L701M, AR L701H, and AR L701Q, we tested a panel of steroids intermediary between progesterone and cortisol with varying combinations of hydroxyl groups at positions 11β, 17α, and 21 (Fig. 3A).
FIGURE 3.
Differential responsiveness of AR L701M, AR L701Q, and AR L701H is determined by the 17α-OH group. A, chemical structures of steroids used in our panel. Cortisol (lower right) differs from progesterone (upper left) by the presence of hydroxyl groups at positions 11β, 17α, and 21 (indicated in the structures). All other steroids are structurally intermediary between progesterone and cortisol and differ by the positions of the hydroxyl groups. The steroids presented in the lower row differ from the steroids in the upper row by the presence of a hydroxyl group at position 21. B and C, transcriptional responses of wild-type (wt) AR, AR L701M, AR L701H, and AR L701Q to the panel of structurally related steroids. Hep3B cells were transiently transfected with expression vectors encoding the different ARs together with the (ARE)2TATA-Luc reporter plasmid. Cells were incubated for 24 h with 100 nm DHT or with the other steroids at 1 μm. For each receptor, the transcriptional activity in response to DHT was set to 100%. The responses to the other steroids are relative to their respective DHT response. Error bars represent the mean relative luciferase activities of three independent experiments performed in duplicate (±S.E.).
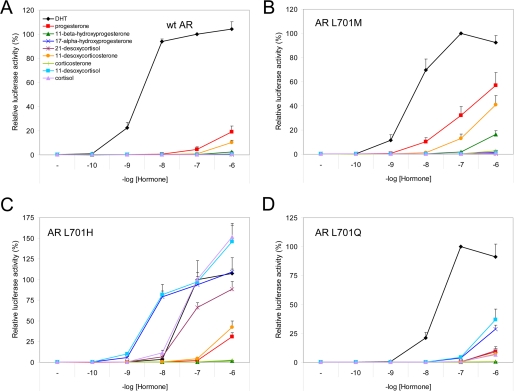
Our results demonstrate that wild-type AR and AR L701M on one hand and AR L701H and AR L701Q on the other display two different transcriptional activation profiles (Figs. 3, B and C, and 4), which are characterized by differential responses to steroids containing a hydroxyl group at position 17α (17α-hydroxyprogesterone, 21-desoxycortisol, 11-desoxycortisol, and cortisol). AR L701M and wild-type AR, which contain a hydrophobic residue at position 701, did not respond at all to these steroids (Figs. 3B and 4, A and B). In contrast, AR L701H and AR L701Q, which harbor a polar residue at position 701, were stimulated by steroids containing the 17-OH group (Figs. 3C and 4, C and D).
FIGURE 4.
Dose-response curves of wild-type AR (A), AR L701M (B), AR L701H (C), and AR L701Q (D) with the panel of steroids. Hep3B cells were transiently transfected with expression vectors encoding the different AR mutants together with the (ARE)2TATA-Luc reporter plasmid. Cells were incubated for 24 h with the indicated concentrations of hormone. For each receptor, the transcriptional activity in response to 100 nm DHT was set to 100%. Data represent the mean of three independent experiments performed in duplicate (±S.E.). wt, wild-type.
All steroids containing a hydroxyl group at position 11β (11β-hydroxyprogesterone, corticosterone, 21-desoxycortisol, and cortisol) were less capable of activating the AR mutants than steroids without this hydroxyl group (progesterone, 11-desoxycorticosterone, 17α-hydroxyprogesterone, and 11- desoxycortisol, respectively) (Figs. 3, B and C, and 4). This demonstrates that the 11β-OH group is unfavorable for AR activation. Steroids without the 21-OH group (progesterone, 11β-hydroxyprogesterone, 17α-hydroxyprogesterone, and 21-desoxycortisol) or with the 21-OH group (11-desoxycorticosterone, corticosterone, 11-desoxycortisol, and cortisol) were equally capable of activating AR L701H and AR L701Q, suggesting that the 21-OH group is not relevant for specificity (Figs. 3C and 4, C and D). The transcriptional activities of AR L701M and wild-type AR were slightly affected if the 21-OH group was present (Figs. 3B and 4, A and B).
Structural Analysis and Modeling Reveal That a Conserved Hydrogen-bonding Network Can Explain the Cortisol Response of AR L701H and AR L701Q
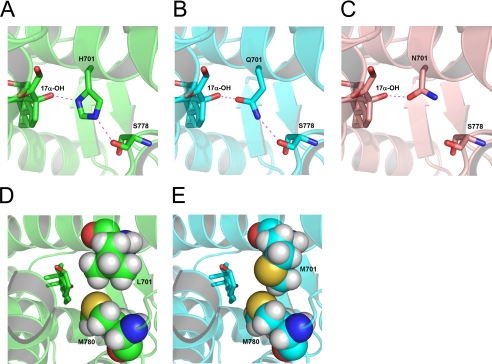
The crystal structure of wild-type AR bound to DHT showed that Leu701 has hydrophobic contacts with C17 of the steroid. The structure of the AR L701H/T877A double mutant has been elucidated in complex with 9α-fluorocortisol (33). This structure revealed an H-bond between the steroidal 17α-OH group and the polar His701. The histidine residue was also well positioned to make an H-bond to the backbone of Ser778 (Fig. 5A). On the basis of this structure, we modeled Gln and Met at position 701. Modeling revealed that Gln701 is able to make an H-bond to a 17α-OH group of the ligand in a manner similar to His701 (Fig. 5B). In addition, Gln701 also replicates the H-bond with the backbone of Ser778 that is seen in the AR L701H structure (Fig. 5B). None of the other amino acid substitutions at 701 were able to reproduce this H-bonding network. The other amide-containing side chain amino acid, Asn (Fig. 5C), and the small polar residues Thr, Ser, and Cys are sterically tolerated and were able to make the H-bond with the steroid. However, unlike His701 or Gln701, these residues are unable to interact with Ser778. The polar interaction of Asn, Thr, and Ser with the steroid's 17α-OH group should prevent the burying of this hydrophilic group into a hydrophobic part of the pocket. Despite this, the AR L701N, AR L701T, and AR L701S mutations did not show strong activation by cortisol as seen for AR L701H and AR L701Q (Fig. 2B). We therefore conclude that the additional interaction with Ser778 that is unique for AR L701H and AR L701Q must have an important role in activating the AR, perhaps by further stabilizing the protein. Although both a histidine and a glutamine residue at position 701 were able to contribute to the same H-bonding network, AR L701Q displayed lower activities than AR L701H (Figs. 3C and 4, C and D). This might be explained by potential differences in the angles, distances, and electrostatics of the polar interactions, which underlie the relative strength of a hydrogen-bonding network.
FIGURE 5.
Conserved hydrogen-bonding network around position 701 defines the cortisol response. A, the crystal structure of the AR L701H mutant (33) shows an H-bonding network between the 17α-OH group of 9α-fluorocortisol and the Nδ1 nitrogen of His701 and between the Nϵ2 nitrogen of His701 and the backbone carbonyl of Ser778. B, the same H-bonding network is maintained in the AR L701Q structure with the 17α-OH group interacting with the Oϵ1 oxygen of Gln701 and the Nϵ2 nitrogen of Gln701 forming an H-bond to the backbone of Ser778. C, contrary to this, in the AR L701N structure, an interaction between the mutated residue and the steroid's 17α-OH group is possible, but the interaction with Ser778 is missing. D and E, modeling shows that Met701 moderately improved packing with Met780, including an electrostatically favorable sulfur-sulfur contact (E), compared with the interaction between wild-type Leu701 and Met780 (D).
Contrary to this, modeling showed that the 17α-OH group in both the wild-type and L701M mutant structures is unfavorably buried in a hydrophobic pocket, explaining the lack of activity upon incubation with steroids containing the 17α-OH group (Figs. 3B and 4B). Met701 actually appears to fill the pocket marginally better than Leu701 and results in an electrostatically favorable sulfur-sulfur contact between Met701 and Met780 (Fig. 5, D and E). This improved packing and sulfur-sulfur contact could have a positive effect on protein stability, which may explain the increased activation of the AR L701M mutant compared with the wild-type receptor.
The molecular basis for progesterone activation of AR L701M and AR L701H, but not AR L701Q, is somewhat speculative. As shown in Fig. 5 (D and E), Met701 moderately improved packing compared with Leu701. Modeling suggests that also His701 better fills the pocket than Leu701 (Figs. 1D and 5A), which indicates that AR L701M and AR L701H are inherently more stable than wild-type AR. Additionally, modeling of progesterone to wild-type AR and AR L701Q appears to introduce clashes between the residues at position 701 and the 17β-group of the steroid. These clashes did not occur if progesterone was modeled to AR L701M and AR L701H. These findings, combined with the observation that progesterone does not require the receptor to adapt to the presence of a 17α-OH group, might give some indication as to why progesterone activates AR L701M and AR L701H, but not wild-type AR and AR L701Q, only at high ligand concentrations.
As shown in Fig. 3, the presence of the 11β-OH group is unfavorable for activating the AR Leu701 mutants. Modeling suggests that the most likely explanation is a clash of the steroidal 11β-OH group with Met895, which is also a Met residue in the progesterone receptor but a smaller Leu in the glucocorticoid and mineralocorticoid receptors (37). This may explain the general tolerance of 11β-OH steroids in the mineralocorticoid and glucocorticoid receptors but not in the AR and progesterone receptor.
Transcriptional Activation of AR Leu701 Mutants Corresponds with AR FXXLF Peptide Interaction
Different coactivator groove-interacting peptides display distinct binding modes (31). For example, LXXLL peptides are shifted in the AR groove toward Lys720 compared with FXXLF peptides (38–41). In addition, AR T877A bound to cyproterone acetate (CPA) strongly interacts with LXXLL motifs but less with FXXLF motifs, whereas the opposite is observed if it is bound to hydroxyflutamide (42, 43). We therefore determined the interaction capacities of distinct peptides containing FXXLF, FXXFF, FXXMF, and LXXLL sequences with the different AR Leu701 mutants in the presence of steroids from our panel. Peptide interactions were studied in mammalian one-hybrid assays as described previously (31).
As shown in Fig. 6, for AR L701M, AR L701H, and AR L701Q, the relative interaction of the AR FXXLF peptide correlated with the relative transcriptional activation capacity. Similar results were obtained for AR T877A and AR L701H/T877A (data not shown). Although we found that AR T877A preferred binding of LXXLL motifs in the presence of CPA and FXXLF-like motifs in the presence of hydroxyflutamide as demonstrated previously (42, 43), we did not observe a similar effect for the AR Leu701 mutants (supplemental Fig. 1). Also, the interaction patterns of FXXLF-like motifs with the AR Leu701 mutants in the presence of progesterone and cortisol were similar to those with DHT and corresponded to the capabilities of these steroids to induce transcription (supplemental Fig. 2, A–D). In line with these data, the AIS mutants AR L701I and AR L701F showed an altered binding preference for neither FXXLF nor LXXLL peptides (supplemental Fig. 2E). The relative peptide interaction capacities of both mutant receptors were comparable with the transcriptional activities at different DHT concentrations (compare Fig. 1C and supplemental Fig. 2E). Together, these results suggest that, unlike AR T877A, amino acid residue 701 in the AR does not directly or indirectly influence the conformation of the coactivator groove.
FIGURE 6.
Transcriptional activities of wild-type AR and selected AR mutants correspond with AR FXXLF peptide interaction capacities. Using the panel of structurally related natural steroids, transcriptional activation (gray bars) of wild-type (wt) AR (A), AR L701M (B), AR L701H (C), and AR L701Q (D) was compared with the capacity to bind the AR FXXLF peptide (black bars). Transcriptional activation was determined as described in the legend to Fig. 1. Mammalian one-hybrid assays were carried out to determine peptide interactions. Hep3B cells were transiently transfected with expression constructs encoding the peptide fused to the Gal4 DNA-binding domain, which served as bait for the different AR constructs. In the case of interaction between the peptide and the AR, the AR N-terminal domain transactivates the luciferase reporter. Cells were incubated for 24 h with 100 nm DHT or with the other steroids at 1 μm. For each receptor, both the transcriptional activity and the AR FXXLF peptide interaction in response to DHT were set to 100%. The responses and interactions in the presence of the other steroids are relative to their respective DHT response. Error bars represent the mean relative luciferase activities of two independent experiments performed in duplicate (±S.D.).
AR Leu701 Mutants Hardly Respond to Antiandrogens
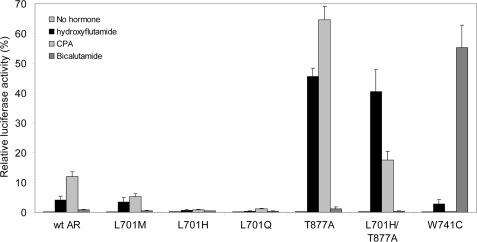
Many AR mutations found in prostate cancer, including T877A and W741C, result in antiandrogen-responsive receptors, leading to failure of antiandrogen treatment of metastasized prostate cancer (23, 44). As shown previously (23), AR T877A is strongly activated by hydroxyflutamide and CPA but not by bicalutamide (Fig. 7). Vice versa, AR W741C is strongly activated by bicalutamide but not by hydroxyflutamide (44) and CPA (Fig. 7). It was shown previously that AR L701H is not responsive to hydroxyflutamide (29).
FIGURE 7.
Transcriptional activities of wild-type AR and selected AR mutants with antiandrogens. Hep3B cells were transiently transfected with expression constructs encoding wild-type (wt) AR or the indicated mutant receptors. Cells were treated for 24 h with hydroxyflutamide (10 μm), CPA (1 μm), or bicalutamide (10 μm). Responses to the different antiandrogens are relative to the DHT (100 nm) response, which was set to 100% for each receptor (not shown). AR T877A and AR L701H/T877A served as controls for hydroxyflutamide and CPA, whereas AR W741C served as a control for activation by bicalutamide (44). Error bars represent the mean relative luciferase activities of three independent experiments performed in duplicate (±S.E.).
Here, we extended these observations by investigating the effects of the antiandrogens hydroxyflutamide, bicalutamide, and CPA on the transcriptional activities of AR L701H and the other AR Leu701 mutants. Fig. 7 shows that, similar to the responses to the panel of steroids (Fig. 3), AR L701H and AR L701Q on one hand and wild-type AR and AR L701M on the other displayed different responses to the antiandrogens. AR L701H and AR L701Q were activated neither by hydroxyflutamide nor by bicalutamide or CPA. AR L701M showed weak agonistic responses to hydroxyflutamide and CPA, which were comparable with wild-type AR. Both receptors could not be activated by bicalutamide.
The Leu-to-His substitution is the only mutation found at position 701 in prostate cancer patients (24, 25, 27). AR L701M and AR L701Q mutations have never been found, possibly because two base substitutions are needed to mutate the Leu codon into a codon for Met or Gln. The lack of activation of the AR L701H mutant by antiandrogens strongly suggests that this AR mutant does not drive prostate tumor growth upon binding of an antiandrogen used for treatment. This provides an additional clue for the conclusion that, in these cases, tumor growth is dependent on endogenously circulating ligands such as cortisol. This finding indicates a different mechanism of tumor growth than observed for the AR T877A and AR W741C mutants, which are dependent on antiandrogens for their transcriptional activity.
Conclusions
In addition to the prostate cancer mutant L701H, we detected two other mutations, L701M and L701Q, that result in an AR with modified ligand specificity. We have shown that these mutants can be subdivided on the basis of their transcriptional activation profiles and structural conformation, in which the interaction between AR residue 701, the backbone of Ser778, and the steroidal 17α-hydroxyl group plays a crucial role. The largely identical structural and functional properties of AR L701H and AR L701Q were instrumental in explaining the altered ligand specificity of the AR L701H mutant.
Supplementary Material
Acknowledgments
We thank Natasja Dits for technical assistance and Dr. Albert Brinkmann and AstraZeneca for providing corticsterone and bicalutamide, respectively.
This work was supported by Grant DDHK2001-2402 from the Dutch Cancer Society.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2 and additional references.
- AR
- androgen receptor
- DHT
- 5α-dihydrotestosterone
- AIS
- androgen insensitivity syndrome
- PAIS
- partial AIS
- CAIS
- complete AIS
- LBD
- ligand-binding domain
- ARE
- androgen response element
- Luc
- luciferase
- CPA
- cyproterone acetate.
REFERENCES
- 1.Brinkmann A. O., Blok L. J., de Ruiter P. E., Doesburg P., Steketee K., Berrevoets C. A., Trapman J. (1999) J. Steroid Biochem. Mol. Biol. 69, 307–313 [DOI] [PubMed] [Google Scholar]
- 2.Brinkmann A. O. (2001) Mol. Cell. Endocrinol. 179, 105–109 [DOI] [PubMed] [Google Scholar]
- 3.Taplin M. E. (2007) Nat. Clin. Pract. Oncol. 4, 236–244 [DOI] [PubMed] [Google Scholar]
- 4.Mostaghel E. A., Page S. T., Lin D. W., Fazli L., Coleman I. M., True L. D., Knudsen B., Hess D. L., Nelson C. C., Matsumoto A. M., Bremner W. J., Gleave M. E., Nelson P. S. (2007) Cancer Res. 67, 5033–5041 [DOI] [PubMed] [Google Scholar]
- 5.van der Kwast T. H., Schalken J., Ruizeveld de Winter J. A., van Vroonhoven C. C., Mulder E., Boersma W., Trapman J. (1991) Int. J. Cancer 48, 189–193 [DOI] [PubMed] [Google Scholar]
- 6.Taplin M. E., Bubley G. J., Ko Y. J., Small E. J., Upton M., Rajeshkumar B., Balk S. P. (1999) Cancer Res. 59, 2511–2515 [PubMed] [Google Scholar]
- 7.Taplin M. E., Bubley G. J., Shuster T. D., Frantz M. E., Spooner A. E., Ogata G. K., Keer H. N., Balk S. P. (1995) N. Engl. J. Med. 332, 1393–1398 [DOI] [PubMed] [Google Scholar]
- 8.Trapman J. (2001) Eur. J. Cancer 37, Suppl. 7, S119–S125 [DOI] [PubMed] [Google Scholar]
- 9.Visakorpi T., Hyytinen E., Koivisto P., Tanner M., Keinänen R., Palmberg C., Palotie A., Tammela T., Isola J., Kallioniemi O. P. (1995) Nat. Genet. 9, 401–406 [DOI] [PubMed] [Google Scholar]
- 10.Mangelsdorf D. J., Thummel C., Beato M., Herrlich P., Schütz G., Umesono K., Blumberg B., Kastner P., Mark M., Chambon P., Evans R. M. (1995) Cell 83, 835–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matias P. M., Donner P., Coelho R., Thomaz M., Peixoto C., Macedo S., Otto N., Joschko S., Scholz P., Wegg A., Bäsler S., Schäfer M., Egner U., Carrondo M. A. (2000) J. Biol. Chem. 275, 26164–26171 [DOI] [PubMed] [Google Scholar]
- 12.Sack J. S., Kish K. F., Wang C., Attar R. M., Kiefer S. E., An Y., Wu G. Y., Scheffler J. E., Salvati M. E., Krystek S. R., Jr., Weinmann R., Einspahr H. M. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 4904–4909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dubbink H. J., Hersmus R., Pike A. C., Molier M., Brinkmann A. O., Jenster G., Trapman J. (2006) Mol. Endocrinol. 20, 1742–1755 [DOI] [PubMed] [Google Scholar]
- 14.He B., Minges J. T., Lee L. W., Wilson E. M. (2002) J. Biol. Chem. 277, 10226–10235 [DOI] [PubMed] [Google Scholar]
- 15.van de Wijngaart D. J., van Royen M. E., Hersmus R., Pike A. C., Houtsmuller A. B., Jenster G., Trapman J., Dubbink H. J. (2006) J. Biol. Chem. 281, 19407–19416 [DOI] [PubMed] [Google Scholar]
- 16.Masiello D., Cheng S., Bubley G. J., Lu M. L., Balk S. P. (2002) J. Biol. Chem. 277, 26321–26326 [DOI] [PubMed] [Google Scholar]
- 17.Marcelli M., Ittmann M., Mariani S., Sutherland R., Nigam R., Murthy L., Zhao Y., DiConcini D., Puxeddu E., Esen A., Eastham J., Weigel N. L., Lamb D. J. (2000) Cancer Res. 60, 944–949 [PubMed] [Google Scholar]
- 18.Buchanan G., Greenberg N. M., Scher H. I., Harris J. M., Marshall V. R., Tilley W. D. (2001) Clin. Cancer Res. 7, 1273–1281 [PubMed] [Google Scholar]
- 19.Veldscholte J., Ris-Stalpers C., Kuiper G. G., Jenster G., Berrevoets C., Claassen E., van Rooij H. C., Trapman J., Brinkmann A. O., Mulder E. (1990) Biochem. Biophys. Res. Commun. 173, 534–540 [DOI] [PubMed] [Google Scholar]
- 20.Gottlieb B., Beitel L. K., Wu J. H., Trifiro M. (2004) Hum. Mutat. 23, 527–533 [DOI] [PubMed] [Google Scholar]
- 21.Bohl C. E., Gao W., Miller D. D., Bell C. E., Dalton J. T. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 6201–6206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bohl C. E., Wu Z., Miller D. D., Bell C. E., Dalton J. T. (2007) J. Biol. Chem. 282, 13648–13655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Steketee K., Berrevoets C. A., Dubbink H. J., Doesburg P., Hersmus R., Brinkmann A. O., Trapman J. (2002) Eur. J. Biochem. 269, 5780–5791 [DOI] [PubMed] [Google Scholar]
- 24.Suzuki H., Sato N., Watabe Y., Masai M., Seino S., Shimazaki J. (1993) J. Steroid Biochem. Mol. Biol. 46, 759–765 [DOI] [PubMed] [Google Scholar]
- 25.Watanabe M., Ushijima T., Shiraishi T., Yatani R., Shimazaki J., Kotake T., Sugimura T., Nagao M. (1997) Jpn. J. Clin. Oncol. 27, 389–393 [DOI] [PubMed] [Google Scholar]
- 26.Navone N. M., Olive M., Ozen M., Davis R., Troncoso P., Tu S. M., Johnston D., Pollack A., Pathak S., von Eschenbach A. C., Logothetis C. J. (1997) Clin. Cancer Res. 3, 2493–2500 [PubMed] [Google Scholar]
- 27.Zhao X. Y., Boyle B., Krishnan A. V., Navone N. M., Peehl D. M., Feldman D. (1999) J. Urol. 162, 2192–2199 [DOI] [PubMed] [Google Scholar]
- 28.Krishnan A. V., Zhao X. Y., Swami S., Brive L., Peehl D. M., Ely K. R., Feldman D. (2002) Endocrinology 143, 1889–1900 [DOI] [PubMed] [Google Scholar]
- 29.Zhao X. Y., Malloy P. J., Krishnan A. V., Swami S., Navone N. M., Peehl D. M., Feldman D. (2000) Nat. Med. 6, 703–706 [DOI] [PubMed] [Google Scholar]
- 30.Brinkmann A. O., Faber P. W., van Rooij H. C., Kuiper G. G., Ris C., Klaassen P., van der Korput J. A., Voorhorst M. M., van Laar J. H., Mulder E., Trapman J. (1989) J. Steroid Biochem. 34, 307–310 [DOI] [PubMed] [Google Scholar]
- 31.Dubbink H. J., Hersmus R., Verma C. S., van der Korput H. A., Berrevoets C. A., van Tol J., Ziel-van der Made A. C., Brinkmann A. O., Pike A. C., Trapman J. (2004) Mol. Endocrinol. 18, 2132–2150 [DOI] [PubMed] [Google Scholar]
- 32.Jenster G., Spencer T. E., Burcin M. M., Tsai S. Y., Tsai M. J., O'Malley B. W. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 7879–7884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matias P. M., Carrondo M. A., Coelho R., Thomaz M., Zhao X. Y., Wegg A., Crusius K., Egner U., Donner P. (2002) J. Med. Chem. 45, 1439–1446 [DOI] [PubMed] [Google Scholar]
- 34.Krieger E., Darden T., Nabuurs S. B., Finkelstein A., Vriend G. (2004) Proteins 57, 678–683 [DOI] [PubMed] [Google Scholar]
- 35.Ahmed S. F., Cheng A., Dovey L., Hawkins J. R., Martin H., Rowland J., Shimura N., Tait A. D., Hughes I. A. (2000) J. Clin. Endocrinol. Metab. 85, 658–665 [DOI] [PubMed] [Google Scholar]
- 36.Chávez B., Vilchis F., Zenteno J. C., Larrea F., Kofman-Alfaro S. (2001) Clin. Genet. 59, 185–188 [DOI] [PubMed] [Google Scholar]
- 37.Poujol N., Wurtz J. M., Tahiri B., Lumbroso S., Nicolas J. C., Moras D., Sultan C. (2000) J. Biol. Chem. 275, 24022–24031 [DOI] [PubMed] [Google Scholar]
- 38.Askew E. B., Gampe R. T., Jr., Stanley T. B., Faggart J. L., Wilson E. M. (2007) J. Biol. Chem. 282, 25801–25816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Estébanez-Perpiñá E., Moore J. M., Mar E., Delgado-Rodrigues E., Nguyen P., Baxter J. D., Buehrer B. M., Webb P., Fletterick R. J., Guy R. K. (2005) J. Biol. Chem. 280, 8060–8068 [DOI] [PubMed] [Google Scholar]
- 40.He B., Gampe R. T., Jr., Kole A. J., Hnat A. T., Stanley T. B., An G., Stewart E. L., Kalman R. I., Minges J. T., Wilson E. M. (2004) Mol. Cell 16, 425–438 [DOI] [PubMed] [Google Scholar]
- 41.Hur E., Pfaff S. J., Payne E. S., Grøn H., Buehrer B. M., Fletterick R. J. (2004) PLoS Biol. 2, e274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brooke G. N., Parker M. G., Bevan C. L. (2008) Oncogene 27, 2941–2950 [DOI] [PubMed] [Google Scholar]
- 43.Ozers M. S., Marks B. D., Gowda K., Kupcho K. R., Ervin K. M., De Rosier T., Qadir N., Eliason H. C., Riddle S. M., Shekhani M. S. (2007) Biochemistry 46, 683–695 [DOI] [PubMed] [Google Scholar]
- 44.Hara T., Miyazaki J., Araki H., Yamaoka M., Kanzaki N., Kusaka M., Miyamoto M. (2003) Cancer Res. 63, 149–153 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.