Abstract
Rem, Rem2, Rad, and Gem/Kir (RGK) represent a distinct GTPase family with largely unknown physiological functions. We report here that both Rem and Rad bind directly to Ca2+ channel β-subunits (CaVβ) in vivo. No calcium currents are recorded from human embryonic kidney 293 cells coexpressing the L type Ca2+ channel subunits CaV1.2, CaVβ2a, and Rem or Rad, but CaV1.2 and CaVβ2a transfected cells elicit Ca2+ channel currents in the absence of these small G proteins. Importantly, CaV3 (T type) Ca2+ channels, which do not require accessory subunits for ionic current expression, are not inhibited by expression of Rem. Rem is expressed in primary skeletal myoblasts and, when overexpressed in C2C12 myoblasts, wild-type Rem inhibits L type Ca2+ channel activity. Deletion analysis demonstrates a critical role for the Rem C terminus in both regulation of functional Ca2+ channel expression and β-subunit association. These results suggest that all members of the RGK GTPase family, via direct interaction with auxiliary β-subunits, serve as regulators of L type Ca2+ channel activity. Thus, the RGK GTPase family may provide a mechanism for achieving cross talk between Ras-related GTPases and electrical signaling pathways.
Rem, Rem2, Rad, and Gem/Kir (RGK) are members of a Ras-related GTPase subfamily (RGK family), with many unique characteristics that distinguish them from other members of the Ras superfamily (1–5). The common structure for all RGK proteins consists of a conserved Ras-related core domain, a series of nonconservative amino acid substitutions within regions known to be involved in guanine nucleotide binding and hydrolysis, a non-CAAX-containing C-terminal extension, and large N-terminal extensions relative to other Ras family proteins. The conservation of structural features within the RGK proteins suggests shared mechanisms of regulation and the control of common cellular signal transduction networks. However, RGK subfamily members differ from each other and from other Ras-related proteins in their putative effector (G2) domains, suggesting that they interact with distinct regulatory and effector proteins, and each exhibits distinct tissue-specific expression patterns (1–5). Thus, despite their conserved structural and biochemical properties, functional evidence to suggest a unified mechanism of action has been limited.
In this study, we investigated the ability of Rem and Rad to regulate Ca2+ channel activity. Our results demonstrate that, although both proteins have distinct effector interaction domains, they each bind directly to CaVβ subunits and inhibit detectable ionic current expression from the native cardiac L type, but not from T type, Ca2+ channels when coexpressed in human embryonic kidney (HEK)293 cells. The Rem protein is expressed in striated muscle cells, and we demonstrate that Rem inhibits L type Ca2+ channel activity in differentiated C2C12 myotubes. Deletion analysis demonstrates that the C terminus of Rem plays an important role in CaVβ subunit association and is necessary for regulation of channel activity. Taken together, these studies indicate that Rem functions as a potent regulator of L type Ca2+ channel function in muscle and suggest that a conserved physiological role for the RGK GTPase gene family is the control of Ca2+ channel activity via modulation of CaVβ subunit function.
Experimental Procedures
RNase Protection Assays and Cell Culture. C2C12DS and C2C12(E) mouse muscle myoblast cell lines were from Robert Krauss (Mount Sinai School of Medicine, New York) and were cultured and induced to differentiate as described (6). Primary mouse muscle cultures and MM14 cells were provided by Phillip Bonner (Department of Biology, University of Kentucky, Lexington). The culture of HEK293 cell lines was done as described (7). RNase protection assays were performed on 20 μg of total RNA from the indicated cell lines as described (1).
Immunoblotting Rem. A rabbit polyclonal antibody (5313) was generated against the Rem6–17 peptide (QQEAKTTLRRRA) (8) and affinity purified against the same peptide. The affinity-purified antibody was used at 1 μg/ml for all experiments. The horseradish peroxidase-conjugated goat anti-rabbit secondary antibody (Zymed) was used at a 1:20,000 dilution. Super Signal (Pierce) was used as the enhanced chemiluminescence reagent.
Electrophysiological Studies. HEK293 cells were transiently transfected with plasmids 12–48 h before recordings as described (9). Transfected cells were identified by the expression of GFP. The whole-cell configuration of the patch–clamp technique was used to measure CaV1 or CaV3 ionic current. Patch electrodes with resistances of 1–2 MΩ contained: 110 mM K-glucanate, 40 mM CsCl, 1 mM MgCl2, 5 mM Mg-ATP, and 10 mM Hepes, pH 7.35. The bath solution consisted of: 140 mM NaCl or CsCl, 2.5 mM BaCl2 or CaCl2, 1 mM MgCl2, 5 mM glucose, and 5 mM Hepes, pH 7.4. Signals were amplified with an Axopatch 200B amplifier and 333kHz A/D system (Axon Instruments, Union City, CA). Data were analyzed with CLAMPFIT 8 (Axon) and ORIGIN statistical software (OriginLab, Northampton, MA). There was no difference in the Rem inhibition of Ca2+ channel current expression for either Ca2+ or Ba2+ as the charge carrier. All recordings were performed at room temperature (20–22°C).
Cytosolic Calcium Imaging. Replication-deficient adenoviruses expressing GFP as a marker protein and Rem were constructed as described (10). C2C12(A) cells were infected with Ad-Ctr or Ad-Rem adenoviruses, placed in differentiation media (DM) for 6 days to induce robust Ca2+ channel expression (11–13), and then loaded with 1 μM fura 2 acetoxymethyl ester (fura 2-AM) with 1:1 dilution of Pluronic F127 (Molecular Probes) for 5 min at 37°C in a 5% CO2 humidified incubator. Deesterification and removal of unincorporated fura 2-AM was accomplished by rinsing cells for 20–30 min in imaging bath solution containing 140 mM NaCl, 4 mM KCl, 1 mM MgCl2, 1 mM CaCl2, 1 mM NaHPO4, 5 mM dextrose, and 10 mM Hepes, pH 7.4. All fura 2 imaging experiments were performed at 37°C. The coverslips were mounted on a temperature-controlled chamber and placed on the stage of a Zeiss inverted epifluorescence microscope. The cells were excited with light of 340 and 380 nm wavelengths, and the emission wavelength, ≈510 nm, was collected and analyzed by using Axon Instruments IMAGING WORKBENCH. The images obtained at 340 and 380 nm were divided pixel by pixel, and the ratio data were reported. Background F340/380 was determined in Ca-free (EGTA) bath solutions with permeabilized cells.
C2C12 Ca2+ transients were elicited with a rapid (<3-s) bulk flow of equimolar replacement of NaCl with 40 mM KCl. The resulting transients were monitored at a sample frequency of 2–4 Hz. Before fura 2 imaging, GFP fluorescence (excitation 488 nm) was recorded at 510 nm. The GFP image for a given field was first saved, and then 340- and 380-nm fluorescence was measured. Cells infected with control virus exhibited variable luminosities of 488-nm signal but had indistinguishable Ca2+ transients from myotubes in the same field that displayed no 488-nm signal. Therefore, Rem effects were assessed by performing statistical tests on adenoviral infected cells in a given field by sorting cells into two groups: either visible GFP signal (488 nm) or no visible GFP signal. To measure total Ca2+ flux in response to a KCl-induced depolarization, transients were integrated over the first 50 s of 40 mM KCl exposure. Within a given field, separate integrals were averaged for either GFP-positive or -negative cells. A paired t test was performed to compare the total Ca2+ GFP-positive to nondetectable GFP signals. Fields were preselected to contain ≈50% infected myotubes.
Rem-CaVβ Subunit Interactions. Hemagglutinin (HA)-Rem pCDNA, HA-Rem1–282, HA-Rem1–265, or HA-Rad pCDNA and either Flag-CaVβ2a pCMV-T7F2, Flag-CaVβ1b, Flag-CaVβ4a, or pCMV-T7F2 were cotransfected into HEK293 cells by the calcium phosphate method (8). Forty-eight hours posttransfection, the cells were washed with PBS, placed into 1 ml of Verseen (GIBCO), harvested, pelleted, and then suspended in ice-cold immunoprecipitation (IP) buffer [20 mM Tris, pH 7.5/250 mM NaCl/1% TX-100/0.5 mM DTT/1× protease inhibitor mixture (Calbiochem)/10 mM MgCl2/10 μM GTPγS]. The cells were lysed, subjected to centrifugation, and 1 mg of the supernatant incubated in a 500-μl reaction containing 10 μl of packed Protein G Sepharose (Pharmacia) and 4 μg of anti-Flag M2 monoclonal antibody (Sigma) for 3 h with gentle rotation at 4°C. The beads were pelleted and 5 μl of the supernatant saved for analysis. The beads were then washed three times with 1 ml of IP buffer. The supernatant and bound fractions were resolved on SDS/PAGE, transferred to nitrocellulose, and subjected to immunoblot analysis. HA-Rem and HA-Rad were detected by immunoblotting as described (7), except that the biotinylated HA antibody was used at 1 μg/ml, and bound protein was detected with streptavidin–horseradish peroxidase (Pierce) at a 1:40,000 dilution. The blot was subsequently probed for Flag-CaVβ2a by incubating the membranes with anti-Flag M2 monoclonal antibody (1 μg/ml) to confirm the efficiency of immunoprecipitation.
Results
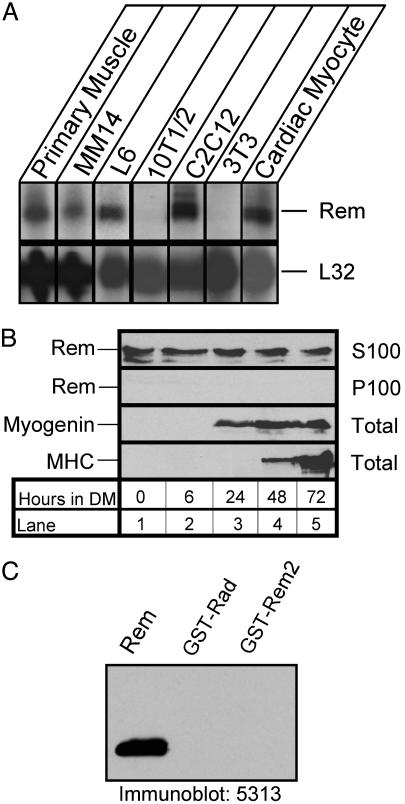
Rem Expression During C2C12 Myoblast Differentiation. Rem mRNA is abundantly expressed in mouse cardiac and skeletal muscle (1). To identify a cellular system in which to examine the contribution of Rem to muscle function, RNase protection analysis was used to assess Rem expression (1). Total RNA was isolated from the myoblast cell lines C2C12, L6, and MM14, from primary mouse skeletal muscle and cardiac myocytes, and from 10T1/2 and 3T3 fibroblast cell lines, and used for RNase protection analysis. Rem mRNA was detected in each of the myoblast cell lines and in primary mouse skeletal and cardiac myocytes examined but not in fibroblast cell lines (Fig. 1A). To confirm that C2C12 cells expressed Rem protein, we performed immunoblots with a polyclonal antipeptide antibody directed against amino acids 6–17 of Rem. Fig. 1B shows that this antibody reacted with a protein that migrated on SDS gels with an apparent molecular weight of 42,000 and bound strongly to recombinant Rem but did not bind detectably to other RGK family proteins (Fig. 1C). Antibody binding to this 42-kDa protein was abolished in the presence of the synthetic peptide used to make the antibody, and transfection of HEK293 cells with a recombinant Rem expression plasmid produced a 42-kDa protein by immunoblot analysis (data not shown).
Fig. 1.
Muscle expression of Rem. (A) Total RNA (20 μg) from the indicated primary muscle cell cultures and cell lines was hybridized with radiolabeled antisense Rem and L32 riboprobes and then subjected to RNase protection analysis. (B) C2C12DS myoblasts were cultured and differentiated into myotubes by replacing the growth media with DM; cell lysates were harvested at the indicated time, fractionated into a 100,000 × g soluble (S100) and particulate (P100) fractions, and subjected to immunoblot analysis. Rem expression was analyzed in both fractions with the 5313 antibody; myogenin expression was analyzed with the F5D monoclonal antibody, and myosin heavy chain (MHC) expression was analyzed with the MF-20 antibody, as indicated on the left. (C) Immunoblot analysis using affinity-purified anti-Rem (5313) antibody.
C2C12 cells have been used extensively as an in vitro model of skeletal muscle differentiation (14). Under the conditions used in this study, myotubes began to form 2 days after transfer to DM, and the majority of cell nuclei were present in large multinucleate myotubes after 4 days in DM (see Fig. 6, which is published as supporting information on the PNAS web site). C2C12 cells expressed Rem protein regardless of cell density, and Rem expression remained constant throughout the differentiation process (Fig. 1B). To investigate the potential mechanistic importance of Rem function during myoblast differentiation, we generated replication-defective adenovirus engineered to express either wild-type Rem or a control virus containing the Rem cDNA cloned in the reverse orientation. These adenoviruses express the GFP gene under the control of a separate promoter, allowing the direct observation of the efficiency of infection by following GFP expression (10, 15). Rem overexpressing cells did not differ from control cells or uninfected cells in gross morphology or in their ability to differentiate into mature myotubes, suggesting that Rem does not control myogenic differentiation (Fig. 6).
Rem Inhibits L Type Ca2+ Channel Function in C2C12 Cells. Voltagegated Ca2+ channels play essential roles in many cellular functions, including excitation–contraction coupling in muscle cells (16). Recent studies suggest that the Rem-related protein Gem regulates Ca2+ channel function (17). To determine whether Rem acts similarly, C2C12 cells were infected with Rem or control adenovirus and induced to differentiate by exposure to DM for 6 days (Fig. 2C). To assess the role of voltage-gated Ca2+ channels in triggering Ca2+ entry, we activated channels by KCl-induced depolarization. Addition of 40 mM KCl elicits a Ca2+ transient in all C2C12 myotubes tested (Fig. 2). However, myotubes overexpressing exogenous Rem had significantly reduced transients compared with uninfected myotubes (Fig. 2 A; P = 0.003, n = 39 Rem-infected vs. 33 noninfected cells in five fields). The mean difference of the response between GFP-positive and nondetectable-GFP cells (five fields) after Rem adenovirus exposure was 14.2 ± 3.2, whereas control virus-infected C2C12 cells showed no significant difference in the Ca2+ transients to uninfected cells (mean difference was –1 ± 0.6) (Fig. 2B).
Fig. 2.
Rem inhibits depolarization-induced Ca2+ transients in C2C12 myotubes. Representative Ca2+ transients recorded from multiple myotubes in a given field. (A) C2C12 myotubes infected with Rem-GFP. The Ca2+ transient was elicited by 40 mM K induced depolarization. The black line represents average response of cells that did not express GFP fluorescence; the green line is transient recorded from myotubes from the same field expressing the GFP signal. (Bars = SE.) (B) C2C12 myotubes infected with control adenovirus expressing unfused GFP. (C) Cell lysates from adenoviral infected C2C12 cells were incubated with anti-Rem antibody for immunoblot analysis.
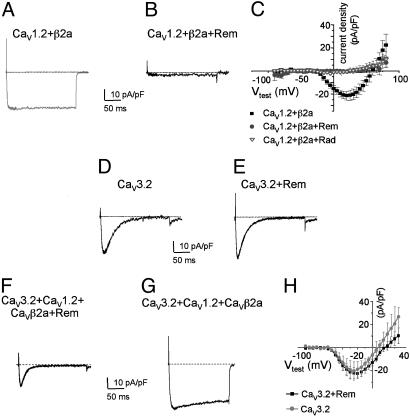
Rem Inhibits L Type, but Not T Type, Ca2+ Channel Function. Myoblasts express two classes of voltage-gated Ca2+ channel: in the early stages of differentiation, T type (CaV3.2) channels are essential for fusion (18), and during differentiation, L type channel expression becomes prominent (12, 19). The experiments of Fig. 2 suggested that Rem expression resulted in the attenuation of L type Ca2+ current, because slow depolarization would be expected to voltage inactivate T type channels (20). Therefore, we next tested the ability of Rem to prevent current expression of heterologously expressed L type (CaV1) or T type (CaV3) Ca2+ channels.
As seen in Fig. 3A, HEK293 cells transiently cotransfected with CaV1.2 and CaVβ2a express >20 pA/pF of peak inward current. In contrast, coexpression of wild-type Rem with CaV1.2 and CaVβ2a results in a complete absence of detectable ionic current expression (Fig. 3B). ICa,L was undetectable for all potentials when coexpressed with Rem (Fig. 3C). These results are consistent with the ability of the Gem GTPase to prevent L type Ca2+ current expression (17). Next, we tested the effect of Rem coexpression on CaV3.2 function, the T type channel expressed by skeletal myoblasts. To control for any nonspecific effects of Rem on current expression, we coexpressed Rem with the T type Ca2+ channels, CaV3.1 or CaV3.2, with or without the auxiliary CaVβ subunit, even though CaV3.2 channels do not require accessory subunits for ionic current expression (21, 22). CaVβ subunit coexpression had no effect on CaV3 channel current expression (data not shown). In contrast to L type ionic current expression, Rem did not inhibit expression of currents through the CaV3 family of channels [average peak current densities for CaV3.2 alone, -22.2 ± 4.4 pA/pF (n = 8); CaV3.2 and Rem, -20.1 ± 8.0 (n = 8)] (Fig. 3 D and E; similar results were seen for CaV3.1). Furthermore, Rem had no effect on CaV3 functional properties (Fig. 3E). Finally, we tested the ability of Rem to selectively regulate L type Ca2+ currents in cells coexpressing CaV1.2, CaVβ2a, and CaV3.2. As expected, coexpression of functional CaV1.2 and CaV3.2 channels showed a complex kinetic pattern (Fig. 3G), but expression of Rem in these cells resulted in ionic current similar to CaV3.2 alone (Fig. 3 D and F).
Fig. 3.
Rem and Rad prevent de novo expression of CaV1.2 ionic current but not T type Ca2+ currents. (A) Representative Ba2+ current elicited by a voltage step from –80 to 10 mV in HEK293 cells transiently transfected with expression vectors encoding CaV1.2, β2a, and GFP. (Bars = 10 pA/pF and 50 ms.) (B) HEK293 cells cotransfected with CaV1.2, β2a, and GFP-tagged wild-type Rem or Rad (same conditions as A). No current is detectable. (C) Current–voltage relationships for cells transfected with CaV1.2, β2a, and unfused GFP (filled squares) or CaV1.2, β2a, and GFP-Rem (filled circles) or Rad (open triangles). Average and SE from six to eight experiments for each group are shown. (D and E) Representative control CaV3.2 ionic current and CaV3.2 ionic current coexpressed with GFP-Rem. Also, coexpression of β2a with CaV3.1 or 3.2 or coexpression of CaVβ2a, GFP-Rem, and CaV3.1 or 3.2 had no effect on T type Ca2+ channel current expression (data not shown for CaV3.1). (F and G) Coexpression of CaV3.2, CaV1.2, β2a display a complex kinetic pattern that resolves to an ionic current that on Rem expression is indistinguishable from the CaV3.2 expressed alone. (H) The current–voltage curve shows that CaV3.2 current amplitude expression is not altered by Rem coexpression. Average and SE from six to eight experiments for each group are shown.
Because Rad is also expressed in muscle, the effects of wild-type Rad on heterologously expressed L type Ca2+ channels was examined. Coexpression of Rad with CaV1.2 and CaVβ2a results in a complete inhibition of detectable ionic current expression (Fig. 3C). Taken together, these data strongly support the notion that both Rem and Rad serve as regulators of L type Ca2+ channel activity.
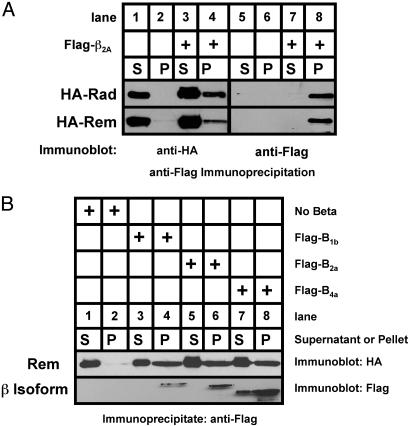
Rem and Rad Bind to CaVβ Subunits. Gem has been reported to associate with CaVβ subunits (17). To determine whether Rem and Rad interact with Ca2+ channel β-subunits in vivo, HEK293 cells were transiently transfected with expression vectors encoding Flag-tagged CaVβ2a alone or cotransfected with vectors expressing HA-tagged Rem or Rad. Flag-tagged CaVβ2a was then isolated by immunoprecipitation and bound HA-tagged proteins visualized by SDS gel electrophoresis and immunoblotting. As seen in Fig. 4A (compare lanes 2 and 4), HA-tagged Rem and Rad were found in the pelleted fraction in a Flag-CaVβ2a-dependent manner. HA-tagged Rem was also found to interact with CaVβ1b and CaVβ4a, indicating that Rem can directly interact with a variety of β-subunit isoforms (Fig. 4B top, lanes 4 and 8). Thus, both Rem and Rad proteins demonstrate direct interaction with CaVβ subunits.
Fig. 4.
In vivo interaction of Rem and Rad with CaVβ2a.(A) HEK293 cells were cotransfected with expression vectors encoding HA-tagged Rem or Rad with or without a vector encoding Flag-tagged CaVβ2a. Cell extracts were prepared, 1 mg of the supernatant was subjected to immunoprecipitation with anti-FLAG antibody in a 500-μl reaction volume, and the entire bound fraction or 5 μl of the unbound fraction was analyzed by immunoblotting with anti-HA to detect Rem/Rad (left) and anti-Flag to detect β2a (right). (B) Coimmunoprecipitation analysis demonstrates association of Rem and both β1b and β4a.
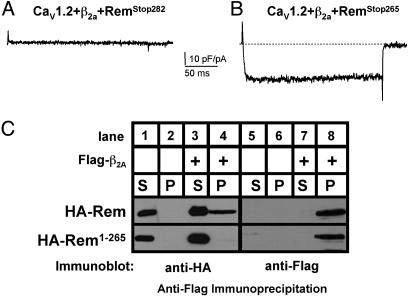
Rem C-Terminal Domain Is Crucial for Regulation of Ca2+ Channel Function. Although Rem, Rad, and Gem each directly associates with β subunits, they have distinct effector domains, which is significant because the effector domain of Ras-related GTPases plays a central role in defining cellular effector protein interactions and is often perfectly conserved within Ras subfamilies (1, 2). However, RGK GTPases all contain an extended and highly conserved polybasic C-terminal domain, suggesting a possible role for this domain in their function (2, 7). To determine the importance of the C terminus to Rem-mediated regulation of Ca2+ channel function, a series of Rem C-terminal deletion mutants were generated and examined for their ability to regulate Ca2+ channel activity. As seen in Fig. 5, coexpression of CaV1.2, β2a, and a short C-terminal deletion (RemStop282) resulted in a complete absence of Ca2+ channel function, whereas coexpression of a larger deletion (RemStop265) resulted in expression of current densities that were the same as expressing the empty GFP control vector [mean current density RemStop282: 0 current (n = 8); RemStop265: -26.2 ± 11.1 pA/pF (n = 5)]. Identical results were seen when RadStop276, containing an equivalent C-terminal deletion to the conserved C-terminal domain of Rad, was coexpressed with CaV1.2 and CaVβ2a (data not shown). Thus, the C-terminal 32 amino acids in Rem and Rad contain residues that play a critical role in Rem- and Rad-mediated inhibition of Ca2+ channel function.
Fig. 5.
Effects of Rem C-terminal mutants on Ca2+ channel current expression. Representative Ba2+ current elicited by a voltage step from –80 to 10 mV in HEK293 cells transiently transfected with expression vectors encoding CaV1.2, β2a, and either GFP-RemStop282 (A) or GFP-RemStop265 (B). (Bars = 10 pA/pF and 50 ms.) (C) HEK293 cells transiently cotransfected with expression vectors encoding HA-tagged Rem or HA-RemStop265 and either Flag-CaVβ2a or empty Flag vector. Whole-cell extracts were immunoprecipitated with anti-FLAG antisera and analyzed by immunoblotting as described in Fig. 4. Data are representative of a typical experiment performed three independent times.
We next asked whether the Rem C-terminal domain was necessary for β-subunit interaction. Although Flag-tagged CaVβ2a interacted with wild-type Rem, the deletion of residues 265–282 within the C-terminal domain greatly reduced CaVβ2a binding (Fig. 5C, compare lanes 4 and 8). These results suggest that the C-terminal domain is necessary for Rem-mediated regulation of L type Ca2+ channel activity, in part by contributing to CaVβ2a binding.
Discussion
The most important finding in this study is that two members of the RGK GTPase family, Rem and Rad, have a common cellular function. Both GTPases block L type Ca2+ channel current expression via interactions with auxiliary CaVβ subunits. Coupled with recent findings for a third member of this family, Kir/Gem (17), we now propose that regulation of Ca2+ channel expression via CaVβ interaction is a general function of all RGK GTPases. Moreover, the regulation of Ca2+ channel density is central to the regulation of skeletal muscle function. Because Rem and Rad are both expressed in skeletal muscle myotubes, we suggest that these GTPases represent a previously unrecognized regulatory mechanism for regulating muscle cell Ca2+ signaling.
Although Rem and Rad operate via a common mechanism, their expression is temporally distinct. Striated muscle cells, including skeletal muscle myoblasts, myotubes and cardiac myocytes express Rem, and Rem expression is unchanged during myogenic differentiation (Fig. 1). This profile contrasts with Rad, which is not expressed in proliferating myoblasts (23) or Gem, which is not expressed in muscle cells (3, 4). To directly examine the involvement of Rem in myoblast differentiation, we infected C2C12 cells with recombinant adenovirus and assessed the effects of wild-type Rem expression on C2C12 differentiation. C2C12 cells overexpressing Rem undergo differentiation and continue to express muscle-specific marker proteins that are a hallmark of normal adult muscle (see Fig. 6), demonstrating that the process of myogenic differentiation is not overtly affected by Rem expression.
Using a calcium-sensitive dye and fluorescence microscopy to monitor changes in the cytosolic free Ca2+ concentration in differentiated C2C12 cells, we demonstrate that Rem-expressing C2C12 myotubes have significantly reduced Ca2+ transients compared with uninfected myotubes or GFP-expressing control virus-infected cells (Fig. 2). Taken together with Rem blockade of Ca2+ current expression, these data indicate that Rem regulates myoblast Ca2+ channel function, which raises an interesting point. As with mature skeletal muscle, the differentiated C2C12 intracellular store Ca2+ release can be triggered by depolarization via electrical stimulation but without appreciable transplasma-membrane Ca2+ influx (e.g., ref. 23). In that same study, 40 mM KCl induced transients that were inhibited by 10 μM nifedipine, but only after a 30-min incubation. This result is consistent with the contribution of long-term Ca2+ entry via L type Ca2+ channels. Also, recent reports of fusing myoblasts showed that window current via T, and presumably L, type channels may contribute to cytosolic Ca2+ (18). In other words, voltage-gated Ca2+ channels permit a steady-state influx of Ca2+ to the cytosol. More to the point of the present study, our data concur that the L type channels' contribution may be setting tonic Ca2+ levels in the cytosol.
Recent studies suggest that RGK proteins regulate cytoskeleton dynamics (24–26). Because reorganization of the cytoskeleton might interfere with Ca2+ channel function, it is important to distinguish between selective Rem-dependent effects on Ca2+ channels and a general blockade of protein sorting. The absence of any obvious effects on the actin cytoskeleton after adenovirus-mediated expression of wild-type Rem in C2C12 myoblasts, as monitored by immunostaining with Texas red phalloidin (data not shown), or on T type Ca2+ channel current expression (Fig. 3E), which are synthesized in the endoplasmic reticulum and must undergo proper intracellular trafficking to be functionally expressed at the plasma membrane, is consistent in both cases with the specific inhibition of CaV1 function by Rem.
The intracellular trafficking of Ca2+ channels from their site of synthesis in the endoplasmic reticulum to the cell surface is an absolute requirement for their function (16). Ca2+ channel α1 subunits contain cytoplasmic endoplasmic reticulum retention signals, which act to prevent the expression of ion channels at the cell surface (27). Coexpression of auxiliary cytoplasmic CaVβ subunits masks these retention signals and allows for expression of functional channels in the plasma membrane (28). Recent studies have suggested that binding of the Gem GTPase to β-subunits inhibits their assembly with Ca2+ channel α1 subunits, such that coexpressing Gem eliminated Ca2+ channel expression (17). In this regard, the predominant localization of Rem to the cytoplasm (Fig. 1B and ref. 2) may act to sequester Rem:CaVβ–subunit complexes from the membrane fraction and inhibit Ca2+ channel expression at the cell surface. However, coimmunoprecipitation studies suggest that only a fraction of total Rem and Rad protein is found in association with CaVβ subunits when overexpressed in HEK293 cells (Fig. 4), although it is possible that muscle-specific proteins may stabilize the complex or that yet- to-be-defined posttranslational modification(s) or GTP binding may stabilize a higher-affinity complex. In parallel, CaVβ regulation of CaVα is a dynamic process that may involve transient association of preformed channels in the surface membrane (29). This information, coupled with our finding that Rem and Rad potently inhibit L type but not heterologously expressed T type Ca2+ channels (Fig. 3 C, E–G), suggests that regulation occurs in CaVβ-dependent fashion but may not involve a blockade of functional L type Ca2+ channel trafficking. Our results also indicate an important role for the Rem and Rad C terminus in CaVβ-subunit binding (Fig. 5). The C-terminal domains of Rad and Gem contain calmodulin-binding regions, whereas Rem and Rad interact with 14–3-3 proteins in a phosphorylation-dependent manner within this domain, suggesting a possible link between these GTPases and both calcium and kinase signaling events (7, 30, 31).
The potential importance of RGK-mediated regulation of voltage-dependent Ca2+ channels is highlighted by recent genetic study by Lynch et al. (32), which has defined a link between a nonsynonymous single-nucleotide polymorphism within the Rad gene (RadQ66P) and patients with congestive heart failure. The finding that Rad efficaciously inhibits de novo expression of L type Ca2+ channels (Fig. 3C) suggests that the RadQ66P mutation may be causative for disease by virtue of its regulation of Ca2+ entry. Clearly, additional studies will be needed to clarify the upstream signals that regulate Rem/Rad activity and the structural basis of CaVβ-subunit association, the effects of Rem/Rad expression on the interaction of α1:β-subunits, and the subcellular localization of both CaVβ and α1 subunits on RGK protein expression.
Conclusion
The expression of Rem and Rad in skeletal and cardiac muscle, together with their dynamic effects on Ca2+ channel function, suggests that they play important roles in the regulation of intracellular Ca2+ signaling. In particular, we demonstrate that Rem and Rad regulate the activity of voltage-dependent Ca2+ channels in a CaVβ-subunit-dependent fashion. Our present results indicate that the RGK GTPase family may provide a mechanism for achieving cross talk between Ras-related GTPases and electrical signaling pathways. Further investigations are necessary to understand the role of Rem and Rad in regulating voltage-dependent Ca2+ channels and how RGK-mediated changes in the individual response properties of voltage-sensitive ion channels may effect excitation–contraction coupling and calcium-signaling pathways in cardiac and skeletal muscle.
Supplementary Material
Acknowledgments
The excellent technical assistance of Renee Bartlett is gratefully acknowledged. We also thank Drs. P. Bonner and R. Krauss for muscle cell lines; Dr. T. Kamp (University of Wisconsin, Madison) for the CaV1.2 and CaVβ2a plasmids; and members of the laboratory for comments on the manuscript. This work was supported in part by the American Heart Association [(to D.A.A.); J.S. is an Established Investigator of the American Heart Association]; and by National Institutes of Health Grants HL072936 (to D.A.A.), HL63416, and HL74091 (to J.S.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: HA, hemagglutinin; RGK, Rem, Rem2, Rad, and Gem/Kir Ras-related GTP-binding proteins; DM, differentiation media.
References
- 1.Finlin, B. S. & Andres, D. A. (1997) J. Biol. Chem. 272, 21982-21988. [DOI] [PubMed] [Google Scholar]
- 2.Finlin, B. S., Shao, H., Kadono-Okuda, K., Guo, N. & Andres, D. A. (2000) Biochem. J. 347, 223-231. [PMC free article] [PubMed] [Google Scholar]
- 3.Cohen, L., Mohr, R., Chen, Y. Y., Huang, M., Kato, R., Dorin, D., Tamanoi, F., Goga, A., Afar, D., Rosenberg, N., et al. (1994) Proc. Natl. Acad. Sci. USA 91, 12448-12452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maguire, J., Santoro, T., Jensen, P., Siebenlist, U., Yewdell, J. & Kelly, K. (1994) Science 265, 241-244. [DOI] [PubMed] [Google Scholar]
- 5.Reynet, C. & Kahn, C. R. (1993) Science 262, 1441-1444. [DOI] [PubMed] [Google Scholar]
- 6.Kang, J. S., Mulieri, P. J., Miller, C., Sassoon, D. A. & Krauss, R. S. (1998) J. Cell Biol. 143, 403-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Finlin, B. S. & Andres, D. A. (1999) Arch. Biochem. Biophys. 368, 401-412. [DOI] [PubMed] [Google Scholar]
- 8.Andres, D. A., Shao, H., Crick, D. C. & Finlin, B. S. (1997) Arch. Biochem. Biophys. 346, 113-124. [DOI] [PubMed] [Google Scholar]
- 9.Zhang, Y., Cribbs, L. L. & Satin, J. (2000) Am. J. Physiol. 278, H184-H193. [DOI] [PubMed] [Google Scholar]
- 10.He, T. C., Zhou, S., da Costa, L. T., Yu, J., Kinzler, K. W. & Vogelstein, B. (1998) Proc. Natl. Acad. Sci. USA 95, 2509-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McMahon, D. K., Anderson, P. A., Nassar, R., Bunting, J. B., Saba, Z., Oakeley, A. E. & Malouf, N. N. (1994) Am. J. Physiol. 266, C1795-C1802. [DOI] [PubMed] [Google Scholar]
- 12.Lorenzon, P., Giovannelli, A., Ragozzino, D., Eusebi, F. & Ruzzier, F. (1997) Eur. J. Neurosci. 9, 800-808. [DOI] [PubMed] [Google Scholar]
- 13.Varadi, G., Orlowski, J. & Schwartz, A. (1989) FEBS Lett. 250, 515-518. [DOI] [PubMed] [Google Scholar]
- 14.Blau, H. M., Chiu, C. P. & Webster, C. (1983) Cell 32, 1171-1180. [DOI] [PubMed] [Google Scholar]
- 15.Spencer, M. L., Shao, H., Tucker, H. M. & Andres, D. A. (2002) J. Biol. Chem. 277, 17605-17615. [DOI] [PubMed] [Google Scholar]
- 16.Catterall, W. A. (2000) Annu. Rev. Cell Dev. Biol. 16, 521-555. [DOI] [PubMed] [Google Scholar]
- 17.Beguin, P., Nagashima, K., Gonoi, T., Shibasaki, T., Takahashi, K., Kashima, Y., Ozaki, N., Geering, K., Iwanaga, T. & Seino, S. (2001) Nature 411, 701-706. [DOI] [PubMed] [Google Scholar]
- 18.Bijlenga, P., Liu, J. H., Espinos, E., Haenggeli, C. A., Fischer-Lougheed, J., Bader, C. R. & Bernheim, L. (2000) Proc. Natl. Acad. Sci. USA 97, 7627-7632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schmid, A., Renaud, J. F., Fosset, M., Meaux, J. P. & Lazdunski, M. (1984) J. Biol. Chem. 259, 11366-11372. [PubMed] [Google Scholar]
- 20.Satin, J. & Cribbs, L. L. (2000) Circ. Res. 86, 636-642. [DOI] [PubMed] [Google Scholar]
- 21.Lacinova, L., Klugbauer, N. & Hofmann, F. (1999) J. Physiol. 516, 639-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cribbs, L. L., Lee, J. H., Yang, J., Satin, J., Zhang, Y., Daud, A., Barclay, J., Williamson, M. P., Fox, M., Rees, M. & Perez-Reyes, E. (1998) Circ. Res. 83, 103-109. [DOI] [PubMed] [Google Scholar]
- 23.Huang, C. F., Flucher, B. E., Schmidt, M. M., Stroud, S. K. & Schmidt, J. (1994) Neuron 13, 167-177. [DOI] [PubMed] [Google Scholar]
- 24.Ward, Y., Yap, S. F., Ravichandran, V., Matsumura, F., Ito, M., Spinelli, B. & Kelly, K. (2002) J. Cell Biol. 157, 291-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Piddini, E., Schmid, J. A., de Martin, R. & Dotti, C. G. (2001) EMBO J. 20, 4076-4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pan, J. Y., Fieles, W. E., White, A. M., Egerton, M. M. & Silberstein, D. S. (2000) J. Cell Biol. 149, 1107-1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walker, D. & De Waard, M. (1998) Trends Neurosci. 21, 148-154. [DOI] [PubMed] [Google Scholar]
- 28.Bichet, D., Cornet, V., Geib, S., Carlier, E., Volsen, S., Hoshi, T., Mori, Y. & De Waard, M. (2000) Neuron 25, 177-190. [DOI] [PubMed] [Google Scholar]
- 29.Garcia, R., Carrillo, E., Rebolledo, S., Garcia, M. C. & Sanchez, J. A. (2002) J. Physiol. 545, 407-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fischer, R., Wei, Y., Anagli, J. & Berchtold, M. W. (1996) J. Biol. Chem. 271, 25067-25070. [DOI] [PubMed] [Google Scholar]
- 31.Moyers, J. S., Bilan, P. J., Zhu, J. & Kahn, C. R. (1997) J. Biol. Chem. 272, 11832-11839. [DOI] [PubMed] [Google Scholar]
- 32.Lynch, R. A., Wagoner, L., Li, S., Sparks, L., Molkentin, J. & Dorn, G. W. II (2002) Physiol. Genomics 10, 159-168. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.