Abstract
Pollen fecundity is crucial to crop productivity and also to biodiversity in general. Pollen development is supported by the tapetum, a metabolically active sporophytic nurse layer that devotes itself to this process. The tapetum in cereals and a vast majority of other plants is of the nonamoeboid type. Unable to reach out to microspores, it secretes nutrients into the anther locule where the microspores reside and develop. Orbicules (Ubisch bodies), studied in various plants since their discovery ≈140 years ago, are a hallmark of the secretory tapetum. Their significance to tapetal or pollen development has not been established. We have identified in wheat and rice an anther-specific single-copy gene (per haploid genome equivalent) whose suppression in rice by RNA interference nearly eliminated the seed set. The flowers in the transgenics were normal for female functions, but the pollen collapsed and became less viable. Further characterization of the gene product, named RAFTIN, in wheat has shown that it is present in pro-orbicule bodies and it is accumulated in Ubisch bodies. Furthermore, it is targeted to microspore exine. Although the carboxyl portion of RAFTINs shares short, dispersed amino acid sequences (BURP domain) in common with a variety of proteins of disparate biological contexts, the occurrence RAFTIN per se is limited to cereals; neither the Arabidopsis genome nor the vast collection of ESTs suggests any obvious dicot homologs. Furthermore, our results show that RAFTIN is essential for the late phase of pollen development in cereals.
Rice and wheat are the staple food for much of the global population. Pollen fertility is one of the limiting and poorly understood factors that affect grain yield in these highly inbreeding species. Pollen production is adversely affected when the temperature is too warm or too cold. The molecular aspects of pollen development in these cereals are still sketchy. Much of the current information is from other plant systems (1–8). Pollen grains originate from the innermost layer of the anther, and their development is sustained by the tapetum, a metabolically active sporophytic cell layer that surrounds the sporogenous cells. The sporophytic support continues until the tapetum disintegrates (4, 6, 7, 9–12), and it is essential for pollen development as shown from male sterility caused by precocious ablation of the tapetum (10, 11). There are two major types of tapetum, and their taxonomic distribution is almost mutually exclusive (13, 14). The secretory tapetal cell layer is more prevalent and is considered primitive in contrast to the amoeboid type that reaches out to the microspores in the anther locule for presumed direct delivery of the tapetal contents. Wheat and rice have secretory tapetum (15). The presence of spheroid structures of ≈1 μm is a hallmark of secretory tapetum in a vast majority of plants. Discovered by Rosanoff in 1865, these have also been called orbicules or Ubisch bodies (15). Despite their long history, a definitive function for Ubisch bodies has not been established. Some consider them to be no more than a by-product of tapetal metabolism, whereas others have suggested functions such as transport of sporopollenin, a complex of fatty acid derivatives and phenylpropanoids that form an extremely inert biopolymer in the exine to resist physical, biological and chemical attacks (15–17). They arise as pro-orbicules in the tapetum and are extruded to the locular side where they acquire a sporopollenin coat and remain attached to the peritapetal wall (17).
Ubisch bodies have not been isolated, likely because of their small size and physical association to the tapetal cell wall, and their biochemical characterization has therefore been hindered. They are absent in Cruciferae members such as Brassica spp. and Arabidopsis, where the biochemistry and genetics of tapetal contributions to pollen development have been investigated in depth (18–21). Thus, there is very little molecular insight into the function of Ubisch bodies. Here, we demonstrate that they carry a sporophytic protein that is targeted to the microspore exine and that it is essential for pollen development.
Materials and Methods
Plant Materials. Hexaploid spring wheat (Triticum aestivum L. cv. AC Karma, AABBDD), tetraploid wheat (Triticum turgidum L. cv. Sceptre, AABB), two diploid wheat species (Triticum urartu, ssp. nigrum, AA; Triticum tauschii L. china, DD) (22) and rice (Oryza sativa L. japonica var. Nipponbare; a gift of T. Sasaki, National Institute of Agrobiological Resources, Ibaraki, Japan) were used.
Cloning, Sequence Analysis, and Nucleic Acid Hybridization. The 5′ sequence of an anther-specific cDNA (A71) identified from an anther cDNA library of hexaploid wheat (22) was found not to show any resemblance to the Arabidopsis genome in the nucleotide or deduced amino acid sequence (www.arabidopsis.org; blast). A 346-bp amplicon from the 5′ portion, PCR-amplified with primers OL3044 and OL3045, was32P-labeled and used to probe ≈500,000 Uni-ZAP XR clones of the anther cDNA library (all primers are listed in Table 1, which is published as supporting information on the PNAS web site). The inserts from all 26 positive clones were sequenced and found to be only of two polymorphic types. The longest clone of each class, confirmed by 5′ RACE as full-length cDNAs, is referred to as taRAFTIN1a and taRAFTIN1b, respectively. BLAST analysis of the entire GenBank records identified a rice ortholog (GenBank accession no. AP000364). The predicted ORF (osRAFTIN1) was RT-PCR amplified with primers OL4382 and OL4383 and directionally cloned into the KpnI–XbaI sites of plasmid pBluescript SK (Stratagene). The genomic counterparts of the wheat and rice RAFTIN cDNAs, obtained by PCR, were cloned into T/A vector (Invitrogen) and sequenced. The oligonucleotide synthesis, DNA sequencing and sequences analyses were carried out as described (22). Genomic DNA isolation and Southern blot analysis were performed as described (22). The entire ORFs of taRAFTIN1a and osRAFTIN1 retrieved by PCR were used as probes for hybridization of genomic DNA. The upstream genomic region of the taRAFTIN1a and taRAFTIN1b ORFs were isolated by using a Universal GenomeWalker kit (Clontech). Two nested primers, OL3070 and OL3071, were used for the first and second PCR. The resulting two fragments of 1.7 kb for taRAFTIN1a and 2.1 kb for taRAFTIN1b were cloned into a T/A vector (Invitrogen). A 1,458-bp segment upstream of the osRAFTIN1 ORF was retrieved by using primers OL3079 and OL3080.
Plant Vectors and Genetic Transformation. Rice transformation was as described (23) with plasmid constructs shown in Fig. 7, which is published as supporting information on the PNAS web site.
RT-PCR Analysis of RAFTIN1 Gene Expression. First-strand cDNA was generated in a 20-μl reaction containing 5 μg of total RNA isolated from appropriate wheat/rice tissues, 0.5 μg of oligo(dT)18 and 20 units of SUPERSCRIPT II RNase H- Reverse Transcriptase (Invitrogen). A total of 150 ng of RNA-derived cDNA were used for a 100-μl PCR in the presence of 10 units of TaqDNA polymerase (Amersham Pharmacia). Primers OL3044 and OL3073 were used for wheat taRAFTIN1a to produce an 875-bp amplicon, and primers OL3148 and OL3815 were used for a 441-bp amplicon from rice osRAFTIN1. Primers OL4556 and OL4557 were used for generating 562-bp control for a housekeeping gene, GAPDH (GenBank accession no. U31676). All PCRs were carried out with a Techne Genius thermocycler (Duxford, Cambridge, U.K.): 35 cycles of 94°C, 30 sec; 56°C, 30 sec; and 72°C, 1 min; finally a 10-min extension at 72°C. Five microliters of the reaction was used for agarose gel analysis.
Protein Methods. taRAFTIN1a ORF was amplified with primers OL3174 and OL3175 and cloned in-frame into the BamH1–EcoRI sites of plasmid pTrxFus (Invitrogen) to yield plasmid pAMWthio-A71. The fusion protein produced in Escherichia coli strain GI724 (Invitrogen) was purified and used for immunizing rabbits as described (22). Antiserum IgG was initially purified (24) and further purified with Affi-Gel 10 (Bio-Rad) following the suppliers' instructions. Plant protein extraction and Western blot analysis were performed essentially as described (25).
Microscopy. For transmission electron microscopy, samples were fixed in 3% glutaraldehyde in 0.025 M phosphate buffer (pH 6.8) overnight at 4°C and postfixed in 1% osmium tetroxide on ice for 8 h. After dehydration in a graded ethanol series, the samples were embedded in acrylic resin (London Resin, Reading, Berkshire, U.K.). Ultra-thin sections (50–70 nm) were made by using a Reichert Jung Ultracut E microtome (Leica, Vienna, Austria), and double-stained with 2% (wt/vol) uranyl acetate and 2.6% (wt/vol) lead citrate. The section was viewed and photographed with a Philips CM-10 transmission electron microscope (Philips Electron Optics). In situ RNA hybridization and immunoblotting were as described (22). Scanning electron microscopy was as described (26). For immunogold labeling, wheat anthers were fixed with 1.5% glutaraldehyde in 0.025 M phosphate buffer (PB, pH 6.8) for 1 h, then with 3% glutaraldehyde in PB for 3 h, followed by a rinse with PB at 4°C overnight and dehydration in a graded ethanol series. The anthers were infiltrated with LR-white resin (London Resin) and polymerized by UV light. Microtome sections were mounted on a grid (Canemco, Quebec) and incubated with blocking solution containing 1% BSA in PBS buffer (10.14 mM Na2HPO4/1.76 mM KH2PO4, pH 7.4/136.9 mM NaCl/2.69 mM KCl) for 30 min, followed with 1 h of incubation with the Affi-Gel column-purified antibody in the blocking solution. After washing with PBS buffer, the sections were treated with colloidal gold-conjugated anti-rabbit IgG developed in goat (1:75; EMGAR15; British BioCell International, Cardiff, Wales) for 1 h. After washing with PBS and rinsing with distilled water, the grids were stained with 2% uranyl acetate for 20 min, washed with distilled water for 1 h, and incubated with 0.3% lead citrate for 10 min. After a rinse with distilled water, the sections were viewed and photographed with a Philips 410 LS electron microscope (Philips Electron Optics). All of the above steps were performed at room temperature unless otherwise stated.
Results
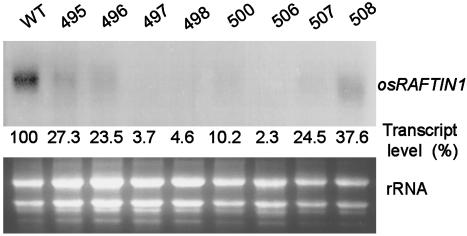
RAFTIN Gene of Wheat and Its Paralogs and the Rice Ortholog Are Highly Anther-Specific in Expression, and RAFTIN Is Essential for Male Fertility. The expression pattern of RAFTIN1 in wheat and rice was studied in detail (Fig. 1). The RAFTIN1 transcripts of wheat and rice (≈1.3 kb) were evident in young inflorescence but not in root, stem, leaf tissues, or emasculated inflorescence. RTPCR analyses showed that the negative samples here were indeed devoid of RAFTIN mRNA. Even after 35 cycles no amplicon was generated in these, whereas it was found from the 22nd cycle onward in the anther samples. Thus, these experiments established anther-specific expression of RAFTIN1 in both wheat and rice. Given the spatial specificity in rice and the absence of an ortholog in the Arabidopsis genome, we generated RAFTIN-defective phenocopy in rice to determine whether the gene had a discernible function. Intron hairpin (ihp) RNA-induced gene silencing strategy (27) was used. A combinatorial series of eight constructs (Fig. 7) that had rice or wheat ihp RAFTIN1 sequences were made and introduced into rice by particle bombardment. Fifty-four transgenic lines with these constructs and nine lines with a selection marker gene (as a control) were obtained. RNA isolated from the anthers (at the vacuolated microspore stage) of a randomly chosen representative line for each construct was used for Northern blot analysis to evaluate the silencing efficacy (Fig. 2). Of the eight representative ihp transgenic lines, the ones made with pAMW497, pAMW498, and pAMW506 showed osRAFTIN1 transcript levels of <5% relative to the control (WT), whereas the others had 10–37.6% (Fig. 2). All of the transgenic lines including those analyzed above were grown in a greenhouse, and all of them had a similar vegetative growth phenotype (Fig. 8 A and B, which is published as supporting information on the PNAS web site). Tillering and leafing ability, leaf size, internode elongation, and overall plant sizes were similar, and panicle initiation in all of the lines occurred ≈90–100 days after transplanting. All of the lines had similar floral morphology, but the spikelet of the osRAFTIN1-silenced lines did not open and anthesis did not occur at maturity even over an extended time (6 weeks) after panicle emergence. No seeds or only a few seeds per plant (<1 seed per panicle) were produced in the osRAFTIN1-silenced lines, in contrast to >10 seeds per panicle in the control lines (Fig. 8 C–H). The palea in the osRAFTIN1-silenced lines remained green, whereas that in the control lines turned yellow (Fig. 8 E and F). The lack of seed set was found to be due to male sterility; the osRAFTIN1-silenced lines crossed with pollen from a control line produced seeds, showing that female fertility was unimpaired (Fig. 8I).
Fig. 1.
Anther-specific expression of RAFTIN1 in wheat and rice. (A and C) Northern blot analysis of RAFTIN1 expression in hexaploid wheat and rice, respectively. (A) Probed with the entire taRAFTIN1a ORF. (C) Probed with the entire osRAFTIN1 ORF. The panels immediately underneath A and C are ethidium bromide-stained gels. (B and D) RT-PCR analysis of RAFTIN1 expression in wheat and rice, respectively. For RT-PCR, the panel underneath is the control RT-PCR of a housekeeping gene, GAPDH (taGAPDH for wheat, osGAPDH for rice). Primers and PCR conditions were described in Materials and Methods. Total RNA isolated from different tissues was used for RT-PCR and Northern blotting analyses. Fl w/o anther, young flower tissue with the anthers removed.
Fig. 2.
Silencing osRAFTIN1 in transgenic rice by using an intron-containing haipin RNA strategy. Northern blot analysis of osRAFTIN1 expression in transgenic rice transformed with intron-containing hairpin constructs. Percentage of osRAFTIN1 expression in different intron hairpin lines relative to that of WT (control) normalized against total RNA loaded is given. The numbers above the lanes refer to the pAMW constructs listed in Fig. 7.
Suppression of RAFTIN1 Impairs Pollen Grain Maturation. The male sterile phenotype was studied further for investigating the function of RAFTINs. Scanning electron microscopy showed that mature anthers in the sterile line were nondehiscent and 10–15% smaller in length (Fig. 9 A and B, which is published as supporting information on the PNAS web site) and that the pollen grains had collapsed (Fig. 9 C and D). There was no gross difference in the pollen surfaces (Fig. 9 E and F). In pollen germination assays, the average germination frequency of pollen in the sterile lines was 4.7%, whereas that of the control lines was 74.3%. Thus, in the sterile lines, vegetative growth and flower development in general were normal before anthesis, but pollen grain development was apparently impaired.
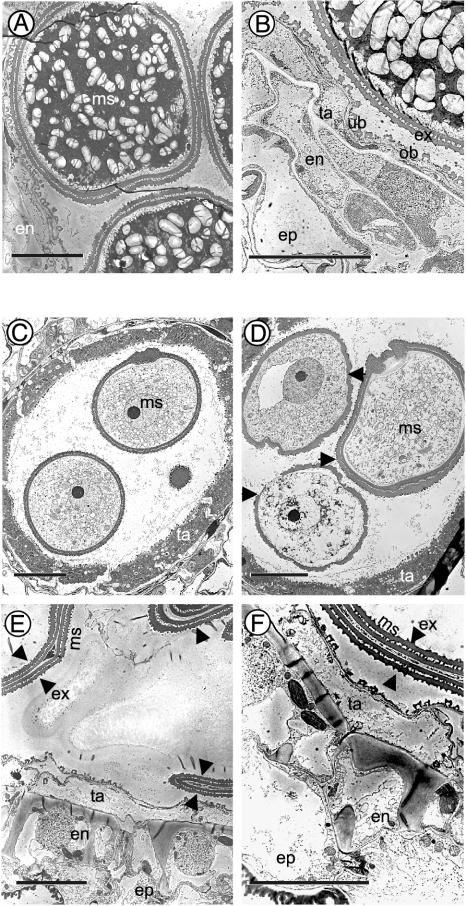
Transmission electron microscopy showed no appreciable differences between the sterile and control lines in terms of the orbicular wall and Ubisch bodies of the sporophyte or the exine of the gametophyte. Furthermore, in all these lines, microspore development was typical (28) up to the free young microspore stage. In the control line, the tapetum showed signs of degeneration at the vacuolated microspore stage and the microspore continued its rapid expansion. During the subsequent stages, the tapetum degenerated to release its metabolites for microspore development and the microspore underwent sequential mitotic divisions and developed into tri-nucleate pollen grains rich in starch granules and other cytoplasmic contents (Fig. 3 A and B). The tapetal degeneration proceeded to completion, showing few remnants. In contrast, the vacuolated microspores of the osRAFTIN1-silenced lines started to collapse and the tapetum did not initiate degeneration at the end of the vacuolated microspore stage (Fig. 3D). The collapsed microspores appeared as flat pollen grains (Fig. 3E). The tapetal degeneration was arrested apparently at the vacuolated microspore stage, leaving the partly degenerated tapetum ≈4 μm thick at the very mature stage when the endothecium wall had thickened (Fig. 3 E and F).
Fig. 3.
Transmission electron micrographs of anthers and pollen grains from osRAFTIN1-silenced rice (line 507-5) and control (empty vector alone). (A) Mature anther from the control line. (B) Enlargement of A. (C–F) Anther from line 507-5 at different developmental stages. (C) Free microspore stage. (D) Vacuolated microspore stage. (E) Mature stage. (F) Enlargement of E. The arrows in D point to the microspores that show signs of collapsing and those in E and F point to the appressed exines of collapsed microspores. en, endothecium; ep, epidermis; ex, exine; ms, microspore; ob, orbicular wall; ta, tapetum; ub, Ubisch body. (Scale bars = 10 μm.)
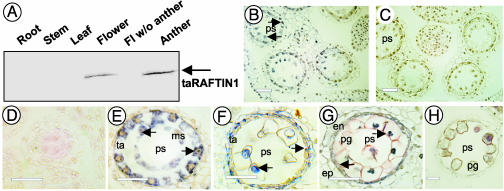
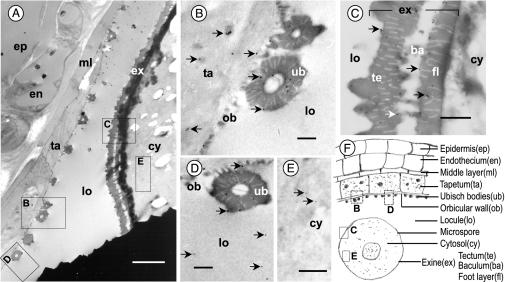
RAFTIN1 Gene Expression and Protein Localization Suggest a Trajectory from the Tapetum to Microspores via the Ubisch Bodies. The anther comprises differentiated tissues such as the epidermis, endothecium, middle layer, tapetum, and other supportive tissues, and developing microspores. In both wheat and rice, in situ hybridization localized RAFTIN1 mRNA distinctly to the tapetum and not to the filament, anther wall, or microspores (Fig. 4). Extended hybridization caused high background reaction without enhancing any specific signal in the microspores, suggesting that if there was any expression in the microspores it was not detectable. Western blots showed the presence of RAFTIN (≈40 kDa) only in young florescence or anther but not in root, stem, leaf, or emasculated florescence (Fig. 5A). In situ immunoanalysis showed RAFTIN in the tapetum and, surprisingly, in the microspore as well (Fig. 5B). RAFTIN was not detectable in other tissues. The presence of RAFTIN was evident from the early free microspore stage (Fig. 5E), becoming abundant in both the tapetum and the microspores as the latter underwent rapid expansion (Fig. 5 E–G). The RAFTIN signal decreased in the three-nucleate pollen grain stage (Fig. 5H). These results suggested temporal and spatial control of RAFTIN production and deposition. Immunogold-labeled antibodies localized RAFTINs to the tapetum, Ubisch bodies, locule, and microspore (Fig. 6; controls are shown in Fig. 10, which is published as supporting information on the PNAS web site). In the microspores, RAFTINs were found to be dispersed in the tectum, baculum, and foot layer (Fig. 6C). We examined overlapping field views of the microspore cytosol in independent sections but did not find a similar occurrence of labeled RAFTINs; a rare section that did have a few gold particles is included in Fig. 6E. This indicates very little expression, if any, of RAFTIN genes in the microspore. For a given surface area in the electronmicrographs, the ratio of gold particles was 44 for the exine, 29 for the Ubisch bodies, and 17 for the tapetum in comparison with the microspore cytosol as the reference at 1 (0.09 particles per μm2 of microspore cytosol area). It has been suggested that Ubisch bodies transport sporopollenin from the tapetum to the developing microspores of grasses and thus are involved in the formation of the sporoderm (15). Accordingly, the locales of RAFTIN suggest that RAFTIN is synthesized in the tapetum, packaged in Ubisch bodies, and transported at appropriate developmental stages to the microspores. Because the Ubisch bodies also contain electron-opaque sporopollenin, it will be interesting to see how RAFTIN is trafficked. Further studies would be required to elucidate the pathways of RAFTIN transport.
Fig. 4.
In situ RNA hybridization of RAFTIN1 transcripts in cross sections of wheat flowers. (A) Hybridized to a taRAFTIN1a antisense probe. (B) Hybridized to a taRAFTIN1a sense probe (control). (Scale bars = 100 μm.)
Fig. 5.
Detection of RAFTIN proteins in wheat anther. (A) Western blot analysis of RAFTIN1 in wheat tissues. The arrow points to a polypeptide of ≈40 kDa, based on reference markers. (B and C) Immunocytochemistry of RAFTIN proteins in wheat young florescence. Specimens were probed with taRAFTIN1a polyclonal antibodies (B) and preimmune sera (control) (C). (D–H) Immunocytochemical detection of RAFTIN protein at different anther development stages. (D) “Tetrad” stage. (E) “Free microspore” stage. (F) “Vacuolated microspore” stage. (G) “Vacuolated pollen grain” stage. (H) “Three-nucleate pollen grain” stage. Root, root tissue; Stem, stem tissue; Leaf, leaf tissue; Flower, developing young flower tissue; Fl w/o anther, developing young flower tissue with anther removed; Anther, developing anther tissue; en, endothecium; ep, epidermis; ms, microspore; pg, pollen grain; ps, pollen sac; ta, tapetum. Arrows point to positive reactions. (Scale bar = 40 μm.)
Fig. 6.
Electron micrographs of wheat anther sections probed with gold-labeled polyclonal antibodies against taRAFTIN1a. (A) Cross section of an anther. (B–E) Magnified views of the boxed zones from A. (F) Diagrammatic representation of an anther cross section pointing out the zones in B–E and a key for the abbreviations. Arrows point to gold particles. (Scale bar = 3 μm in A and 0.5 μm in B–E.)
Organization of RAFTIN1 Genes and Gene Products. The features of the genes are depicted in Fig. 11, which is published as supporting information on the PNAS web site. taRAFTIN1a and taRAFTIN1b ORFs were predicted to encode a 389-aa and a 362-aa product, respectively. These two ORFs were 89% identical to each other at the nucleotide level and 86% at the amino acid level and were derived from three exons. taRAFTIN1a includes a near-perfect tandem repeat of 21 aa from the 96th amino acid, but the corresponding region of taRAFTIN1b lacks much of the amino acid in these repeats (Fig. 12 A, which is published as supporting information on the PNAS web site). Additional cDNA clones that were distinct from the above were not identified in three further screens of the anther cDNA library, even though a shorter genomic fragment (named taRAFTIN1c) from hexaploid wheat that was ≈96% identical to taRAFTIN1a and taRAFTIN1b had been isolated by PCR. taRAFTIN1c was not pursued further. The predicted ORF of osRAFTIN1, also composed of three exons, encodes a 412-aa polypeptide that shares ≈62% identity with its wheat counterparts over the entire length. Southern hybridization of genomic DNA from rice (a diploid) and wheats of hexaploid (AABBDD), tetraploid (AABB), or diploid (AA or DD) genome suggests the presence of only one RAFTIN1 gene per haploid genome equivalent (Fig. 11B).
The wheat and rice RAFTINs contain two predicted transmembrane domains (Fig. 12 A). TBLASTN analysis did not identify any sequences similar to the 168-aa amino domain of taRAFTIN1a, with the exception of ESTs from cereals and a chromosome segment of rice. The remainder (carboxyl domain of 221 aa) was 77% identical to the corresponding portion of osRAFTIN1 (Fig. 12 A) and >90% identical to ESTs from inflorescences of various cereal species. All other significant “hits” (E value of <10-4) were also only from plant accessions, but generally they had only ≈35% identity and included the following BURP domain proteins (29): a seed protein of faba bean (30); the β subunit of polygalacturonase isoenzyme 1 from the developing fruit of tomato (31); an desiccation-induced protein of Arabidopsis (32); an auxin down-regulated protein (33) and an aluminum up-regulated protein (34) from soybean hypocotyl and roots, respectively; microspore embryo development-related protein from Brassica napus (29); an apomixis-specific gene product from the flower buds of guinea grass (35) that showed ≈45% aa sequence identity over the entire carboxy-domain; and a seed coat protein found in developing soybean seed coats (36).
Discussion
We have identified a tapetal protein that packages into Ubisch bodies and microspore exine, and this protein, RAFTIN, from wheat and rice has counterparts only in cereal species. RAFTINs, however, have the features of the enigmatic, plant-specific group of proteins that have been named as BURP domain proteins (29). The most striking commonality of the latter is the presence of ≈30 conserved residues that are dispersed over the carboxyl region of ≈220 aa; the amino region is not conserved and its length is highly variable (68–414 aa). Two C and four CH residues among the 30 conserved amino acids are the hallmarks of the so-called BURP signature. The amino termini in all these proteins also include a predicted signal peptide region. Deduced BURP domains have been noted in genomic/cDNA/EST sequences from both monocots and dicots, and the expression patterns of the corresponding genes include vastly distinct developmental contexts. The essentiality of these proteins to plant form or function has not been previously demonstrated. Our results establish that RAFTIN is essential specifically for the maturation phase of pollen development.
Pollen development may be generalized to occur through the following stages: meiosis/tetrad, young free microspore, vacuolated and expanded microspore, vacuolated pollen grain and three-nucleate pollen grain. The tapetum provides nutrients, proteinaceous and lipidic precursors for developing microspores and it secretes the callase that is required for resolving the postmeiotic tetrads (12, 18, 20, 37–39). Continuing with the supportive role, the tapetum ultimately degenerates in a programmed manner at the advanced stages of microspore development, releasing its contents into the locule and leaving behind the remnants and orbicular wall-attached Ubisch bodies (28). It is known in Arabidopsis, for example, that pollen wall lipids affect fertility and afford specificity in pollen–pistil interactions (40–43). In Brassicaceae members, proteins and lipids from tapetum become a part of the pollen exine and the outer coatings (20, 44). There are no Ubisch bodies in Brassicaceae (15), and some of the tapetal lipids and proteins are packaged into tapetosomes and elaioplasts (21, 45). These organelles have not been described in wheat or rice, and the transport of tapetal contents by mode(s) other than secretion or tapetal lysis has not been shown for these species. Ubisch bodies originate as pro-orbicular cores within the tapetum and they are extruded to locular side where the cores become surrounded by sporopollenin polymer, which is similar to the exine- and peritapetal wall-associated sporopollenin in electron-opacity (12, 17, 46, 47). Ubisch bodies have not been isolated from any species. Although the Ubisch body-associated RAFTIN is surmised to be in transit from the tapetal cells to the microspore exine, independent embedding of RAFTINs within the sporopollenin matrices of Ubisch bodies and the exine cannot be ruled out.
Sporopollenin comprises phenylpropanoids and fatty acid derivatives, but its exact composition is unknown (48). It is synthesized in young microspores and also in the sporophytic tapetum. The microspore-synthesized sporopollenin is used for exine formation in the early stages when the microspores are shielded from the locular fluid by a callose wall, and exine maturation occurs in conjunction with deposition of tapetally derived sporopollenin (17, 48). The physico-biochemical aspects of the transformation of sporopollenin as a recalictrant polymer remain to be elucidated. We have found RAFTINs in orbicular/ peritapetal wall, Ubisch bodies and the exine, the three locations where polymerized sporopollenin occurs. Based on the spatio-temporal aspects of the formation of Ubisch bodies, the orbicular wall and the exine with reference to pollen development (17, 47), and the collapse of late-stage microspores in RAFTIN-silenced lines, it seems possible that RAFTINs could play a guiding role in proper “fixation” of sporopollenin polymers from its precursors of tapetal origin. The lack of any gross changes in the electron-opacity of the exine of the collapsed microspores suggests that the changes in the exine are subtle and yet the effects on microspore maturation beyond the stages of cell vacuolation and expansion are dramatic. A gametocidal agent that affects sporopollenin deposition has a similar effect on wheat microspores (12). Clearly, the formation of nearly normal vacuolated microspores in the RAFTIN-silenced lines shows that the gametophytic contribution to exine ontogeny is unaffected and that RAFTIN has a role pertaining to maturation.
Supplementary Material
Acknowledgments
We gratefully acknowledge the assistance of Sara R. Caldwell and Elfriede Moya with microscopy; Rozina Hirji and Eugen Kurylo with EST generation; Kevin Koh with EST analysis; Ping Zheng with rice transformation; Darrin Klassen, Barry Panchuk, and Inge Roewer with DNA sequencing; and Don Schwab with oligonucleotide synthesis. We thank Dayakar Pareddy and Tonya Strange for advice on rice transformation, Dr. Takuji Sasaki for providing the initial rice seed stock, and the anonymous reviewers for their very helpful comments. This work was funded by the Strategic Initiatives Fund and the National Research Council of Canada. This is National Research Council of Canada Publication 45260.
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AJ575662–AJ575668).
References
- 1.Koltunow, A. M., Truettner, J., Cox, K. H., Wallroth, M. & Goldberg, R. B. (1990) Plant Cell 2, 1201-1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bedinger, P. A. (1992) Plant Cell 4, 879-887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chaudhury, A. M. (1993) Plant Cell 5, 1277-1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goldberg, R. B., Beals, T. P. & Sanders, P. M. (1993) Plant Cell 5, 1217-1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hamilton, D. A. & Mascarenhas, J. P. (1997) in Pollen Biotechnology for Crop Production and Improvement, eds. Shivanna, K. R. & Sawhney, V. K. (Cambridge Univ. Press, Cambridge, U.K.), pp. 41-58.
- 6.McCormick, S. (1993) Plant Cell 5, 1266-1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shivanna, K. R., Cresti, M. & Ciampolini, F. (1997) in Pollen Biotechnology for Crop Production and Improvement, eds. Shivanna, K. R. & Sawhney, V. K. (Cambridge Univ. Press, Cambridge, U.K.), pp. 15-39.
- 8.Yang, W. C. & Sundaresan, V. (2000) Curr. Opin. Plant Biol. 3, 53-57. [DOI] [PubMed] [Google Scholar]
- 9.D'Arcy, W. G. (1996) in The Anther: Form, Function, and Phylogeny, eds. D'Arcy, W. D. & Keating, R. C. (Cambridge Univ. Press, Cambridge, U.K.), pp. 1-24.
- 10.De Block, M., Debrouwer, D. & Moens, T. (1997) Theor. Appl. Genet. 95, 125-131. [Google Scholar]
- 11.Mariani, C., Gossele, V., de Beuckeleer, M., de Block, M., Goldberg, R. B., de Greef, W. & Leemans, J. (1992) Nature 357, 384-387. [Google Scholar]
- 12.Mizelle, M. B., Sethi, R., Ashton, M. E. & Jensen, W. A. (1989) Sex. Plant Reprod. 2, 231-253. [Google Scholar]
- 13.Bhojwani, S. S. & Bhatnagar, S. P. (1992) The Embryology of Angiosperms (Vikas, New Delhi), pp. 11-32.
- 14.Furness, C. A. & Rudall, P. J. (2001) Int. J. Plant Sci. 162, 375-392. [Google Scholar]
- 15.Huysmans, S., El-Ghazaly, G. & Smets, E. (1998) Bot. Rev. 64, 240-272. [Google Scholar]
- 16.Heslop-Harrison, J. (1968) Science 161, 230-237. [DOI] [PubMed] [Google Scholar]
- 17.Dickinson, H. G. & Bell, P. R. (1972) Planta 107, 205-215. [DOI] [PubMed] [Google Scholar]
- 18.Sanders, P. M., Bui, A. Q., Weterings, K., McIntire, K. N., Hsu, Y.-C., Lee, P. Y., Truong, M. T., Beals, T. P. & Goldberg, R. B. (1999) Sex. Plant Reprod. 11, 297-322. [Google Scholar]
- 19.Piffanelli, P., Ross, J. H. E. & Murphy, D. J. (1997) Plant J. 11, 549-652. [DOI] [PubMed] [Google Scholar]
- 20.Piffanelli, P., Ross, J. H. E. & Murphy, D. J. (1998) Sex. Plant Reprod. 11, 65-80. [Google Scholar]
- 21.Wu, S. S. H., Platt, K. A., Ratnayake, C., Wang, T.-W., Ting, J. T. L. & Huang, A. H. (1997) Proc. Natl. Acad. Sci. USA 94, 12711-12716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang, A., Xia, Q., Xie, W., Dumonceaux, T., Zou, J., Datla, R. & Selvaraj, G. (2002) Plant J. 30, 613-623. [DOI] [PubMed] [Google Scholar]
- 23.Chen, L., Zhang, S., Beachy, R. N. & Fauquet, C. M. (1998) Plant Cell Rep. 18, 25-31. [DOI] [PubMed] [Google Scholar]
- 24.Wang, A. & Sanfacon, H. (2000) J. Gen. Virol. 81, 2771-2781. [DOI] [PubMed] [Google Scholar]
- 25.Wan, L., Xia, Q., Qiu, X. & Selvaraj, G. (2002) Plant J. 30, 1-10. [DOI] [PubMed] [Google Scholar]
- 26.Venglat, S. P., Dumonceaux, T., Rozwadowski, K., Parnell, L., Babic, V., Keller, W., Martienssen, R., Selvaraj, G. & Datla, R. (2002) Proc. Natl. Acad. Sci. USA 99, 4730-4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith, N. A., Singh, S. P., Wang, M.-B., Stoutjesdijk, P. A., Green, A. G. & Waterhouse, P. M. (2000) Nature 407, 319-320. [DOI] [PubMed] [Google Scholar]
- 28.Raghavan, V. (1988) Am. J. Bot. 75, 183-196. [Google Scholar]
- 29.Hattori, J., Boutilier, K. A., van Lookeren Campagne, M. M. & Miki, B. L. (1998) Mol. Gen. Genet. 259, 424-428. [DOI] [PubMed] [Google Scholar]
- 30.Bäumlein, H., Boerjan, W., Nagy, I., Bassüner, R., Van Montagu, M., Inzé, D. & Wobus, U. (1991) Mol. Gen. Genet. 225, 459-467. [DOI] [PubMed] [Google Scholar]
- 31.Zheng, L., Heupel, R. C. & DellaPenna, D. (1992) Plant Cell 4, 1147-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamaguchi-Shinozaki, K. & Shinozaki, K. (1993) Mol. Gen. Genet. 238, 17-25. [DOI] [PubMed] [Google Scholar]
- 33.Datta, N., LaFayette, P. R., Kroner, P. A., Nagao, R. T. & Key, J. L. (1993) Plant Mol. Biol. 21, 859-869. [DOI] [PubMed] [Google Scholar]
- 34.Ragland, M. & Soliman, K. M. (1997) Plant Physiol. 114, 395.9159957 [Google Scholar]
- 35.Chen, L., Miyazaki, C., Kojima, A., Saito, A. & Adachi, T. (1999) J. Plant Physiol. 154, 55-62. [Google Scholar]
- 36.Batchelor, A. K., Boutilier, K., Miller, S. S., Hattori, J., Bowman, L. A., Hu, M., Lantin, S., Johnson, D. A. & Miki, B. L. A. (2002) Planta 215, 523-532. [DOI] [PubMed] [Google Scholar]
- 37.Clément, C., Laporte, P. & Audran, J. C. (1998) Sex. Plant Reprod. 11, 94-106. [Google Scholar]
- 38.Mascarenhas, J. P. (1990) Annu. Rev. Plant Physiol. Plant Mol. Biol. 41, 317-338. [Google Scholar]
- 39.Pacini E. (1990) in Microspores: Evolution and Ontogeny, eds. Blackmore, S. & Knox, R. B. (Academic, London), pp. 213-237.
- 40.Mayfield, J. A., Fiebig, A., Johnstone, S. E. & Preuss, D. (2001) Science 292, 2482-2485. [DOI] [PubMed] [Google Scholar]
- 41.Pruitt, R. E., Vielle-Calzada, J.-P., Ploense, S. E., Grossniklaus, U. & Lolle, S. J. (2000) Proc. Natl. Acad. Sci. USA 97, 1311-1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stintzi, A. & Browse, J. (2000) Proc. Natl. Acad. Sci. USA 97, 10625-10630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wolters-Arts, M., Lush, W. M. & Mariani, C. (1998) Nature 392, 818-821. [DOI] [PubMed] [Google Scholar]
- 44.Staigher, D., Kappeler, S., Müller, M., Apel, K. (1994) Planta 192, 221-231. [PubMed] [Google Scholar]
- 45.Hernández-Pinzón, I., Ross, J. H. E., Barnes, K. A., Damant, A. P. & Murphy, D. J. (1999) Planta 208, 588-598. [DOI] [PubMed] [Google Scholar]
- 46.El-Ghazaly, G. & Jensen, W. A. (1986) Grana 25, 1-29. [Google Scholar]
- 47.Heslop-Harrison, J. & Dickinson, H. G. (1969) Planta 84, 199-214. [DOI] [PubMed] [Google Scholar]
- 48.Scott, R. J. (1994) in Molecular and Cellular Aspects of Plant Reproduction, eds. Scott, R. J. & Stead, A. D. (Cambridge Univ. Press, Cambridge, U.K.), pp. 49-81.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.