Abstract
Phytochromes are red- and far-red-sensing photoreceptors that detect the quantity, quality, and duration of light throughout the entire life cycle of plants. Phytochromes accumulate in the cytoplasm in the dark. As one of the earliest responses after light illumination, phytochromes localize to the nucleus where they become associated with discrete nuclear bodies (NBs). Here, we describe the steady-state dynamics of Arabidopsis phytochrome B (phyB) localization in response to different light conditions and define four phyB subnuclear localization patterns: diffuse nuclear localization, small and numerous NBs only, both small and large NBs, and large NBs only. We show that phyB nuclear import is not sufficient for phyB NB formation. Rather, phyB accumulation in NBs is mainly determined by the percentage of the total amount of phyB protein that is in the active phyB conformer, with large NBs always correlating with strong phyB responses. A genetic screen to identify determinants required for subnuclear localization of phyB resulted in several phyB mutants, mutants deficient in phytochrome chromophore biosynthesis, and mutations in at least one previously uninvestigated locus. This study lays the groundwork for future investigations to identify the molecular mechanisms of light-regulated partitioning of plant photoreceptors to discrete subnuclear domains.
Because plants are sessile, they are particularly sensitive to environmental changes. Light is one of the most important environmental cues, controlling plant development throughout the life cycle, from seed germination to floral induction (1). Plants perceive light by using a suite of photoreceptors that absorb light of different wavelengths. Of these, the red (R)/farred (FR)-absorbing phytochromes play a crucial role in every key developmental and growth decision (2-4). During seedling development, phytochromes control the transition from etiolated to photoautotrophic growth by modulating the expression of many light-regulated genes (5-7). During this transition, the seedling develops an optimal body plan for photosynthetic growth, which results in the inhibition of the rate of stem growth and the expansion and greening of cotyledons. Later in development, phytochromes continue to play a role in differential growth responses and ultimately are involved in regulating the floral transition.
Phytochromes are purified as homodimers. Each monomer consists of an ≈125-kDa polypeptide and a covalently linked linear tetrapyrrole chromophore, called phytochromobilin, which is derived from heme. The prototypical phytochrome apoprotein is composed of an N-terminal phytochromobilin binding domain, a C-terminal signaling domain that contains PAS repeats and a histidine-kinase-related domain (HKRD), and a hinge region that links the N-terminal photosensing and C-terminal domains. Some phytochromes have been shown to be light-activated Ser/Thr kinases (8, 9), although the precise function of phytochrome kinase activity in phytochrome signaling is still poorly understood. It has been widely believed that the N-terminal chromophore-binding domain is the photosensing domain and the C-terminal PAS repeat domains and HKRD are involved in signal transmission; however, a recent report suggests that dimers consisting of only the N-terminal half of Arabidopsis phytochrome B (phyB) have phytochrome activity (10). Thus, the C-terminal PAS repeat domain and HKRD may be regulatory domains required for N-terminal function. Phytochromes have two long-lived spectral forms: Pr (λmax, 660 nm) and Pfr (λmax, 730 nm). The Pr form absorbs R light and converts to the Pfr form, which absorbs FR and converts back to Pr. Pfr is correlated with biological responses and is considered to be the active form. The percentage of active Pfr conformers of the total phytochrome enables plants to respond appropriately to different environmental light conditions (3).
In Arabidopsis, phytochromes are encoded by five genes, PHYA-PHYE, with PHYA and PHYB playing the most prominent roles in seedling establishment and flowering (11, 12). Despite their high identity and similar spectral properties, phyA and phyB control plant development by different mechanisms (2). phyA is photolabile and primarily responsible for irreversible responses to FR light, whereas phyB is photostable and responsible for reversible R/FR light responses (2). Both phyA and phyB accumulate in the cytoplasm in the dark and translocate into the nucleus after light illumination (13-15). However, phyA and phyB have different light-quality requirements and kinetics for nuclear import. phyA nuclear import is fast and induced by either FR or R; in contrast, phyB nuclear import is relatively slow, is induced only in R, and can be reversed by FR. Nuclear import of phyB also depends on the fluence rate of R (13-16). In the nucleus, phytochromes localize to discrete subnuclear foci (13-15, 17), which appear similar to nuclear bodies (NBs) defined in other systems (18, 19). The localization of phytochromes to NBs has been reported by using both phy::GFP overexpression lines and in wild-type plants by immunolocalization (15, 17). Several studies support the interpretation that localization to NBs is associated with phytochrome functions. These reports include molecular genetic studies using known phytochrome mutant proteins, which either lead to the loss of phytochrome NBs or result in different NB patterns. Moreover, localization of phyA, phyB, phyC, and phyE to NBs has been shown to be regulated by diurnal oscillations (17).
Despite growing data about regulated phytochrome translocation, detailed and standardized characterization of phytochrome NB patterns or their regulation by light still has not been done. In this report, we characterize the steady-state patterns of localization of Arabidopsis phyB by using a 35S-phyB::GFP (PBG) line. We define four different phyB subnuclear localization patterns that are regulated by the ambient light conditions, including no NBs, small and numerous NBs, both small and large NBs, and large NBs only. We demonstrate that phyB NB formation depends on the percentage of active Pfr conformer of phyB, with large phyB NBs correlated with strong phyB responses. A genetic screen for phyB mislocalization identified a number of phyB mutant alleles, mutations in the enzymes for phytochromobilin biosynthesis, and several mutations that map to loci in which phyB fails to associate with large NBs. Our results suggest that phyB nuclear import is not sufficient for phyB's association with NBs. Moreover, phyB's localization to discrete subnuclear foci can be precisely manipulated by changes in ambient light conditions. These studies thus provide a critical framework for future analyses of the initial events associated with phyB signaling.
Materials and Methods
Plant Material. The PBG line of Arabidopsis thaliana (ecotype Landsberg er) was described in Yamaguchi et al. (13). Wild-type Arabidopsis (Landsberg er) and the phyB-5 mutant were also used as controls for hypocotyl measurements. Mutant lines from the phyB mislocalization screen were crossed to phyB-9 to distinguish intragenic phyB mutations from extragenic mutations. Mapping of dsf1 was done on the F2 progeny of a cross of dsf1 and phyB-9. Transgenic Arabidopsis lines with phyB-28::YFP were generated in phyB-9.
Growth Conditions and Hypocotyl Measurements. Arabidopsis seeds were surface-sterilized in 50% bleach and 0.01% Triton X-100 for 15 min and washed five times with sterile water before plating on Murashige and Skoog salts (GIBCO), 0.8% (wt/vol) Phytagar (GIBCO), and 1× Gamborg's B5 vitamin (Sigma). The plates were kept in the dark at 4°C for 4 days for stratification. Germination was induced with a 4-h white-light treatment. For both microscopy and hypocotyl measurements, plants were grown at 22°C in a light-emitting diode chamber (Percival Scientific, Perry, IA) under appropriate light conditions for 4 days. Fluence rates of light were measured by using a LiCor LI-1800 spectroradiometer (LiCor, Lincoln, NE). For hypocotyl length measurements, 4-day grown seedlings were scanned, and hypocotyls were measured by using NIH IMAGE (http://rsb.info.nih.gov/nih-image). Biliverdin (BV)-feeding experiments were done according to Parks and Quail (20). Arabidopsis plants were germinated and grown on 0.1 mM BV IXα (Frontier Scientific, Logan, UT) for 4 days under R or FR light before hypocotyls were measured.
Calculation of Percentage of Pfr Conformer. Pfr/P was calculated as k1/(k1 + k2 + kd), where P is the total amount of phyB, k1, k2, and kd are constants for Pfr to Pr photoconversion, Pr to Pfr photoconversion, and dark reversion. The constant of k1 and k2 were calculated according to Kendrick and Kronenberg (ref. 1, chapter 4.7). In brief,
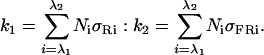 |
In the equations, Ni is the fluence rate at a specific wavelength, and both σRi and σFRi from measurements of phytochromes purified from dark-grown oat and rye seedlings were used (1, 21, 22). The kd of phyB PfrPr to PrPr was calculated from t1/2 of yeast expressed phyB (23, 24). kd = ln(2)/t1/2, t1/2 = 180 s. Therefore, kd = 3.8 × 10-3 s-1. Because PfrPfr conformer of phyB has a much slower dark-reversion rate than that of PfrPr (24), the Pfr percent is underestimated by this method in high-fluence rates of R.
Sequencing of Mutant Lines. Sequencing was done on PCR-amplified fragments at The Salk Institute Sequencing Facility. The PHYB transgene was sequenced in all 25 mutant lines that were less sensitive to R. To avoid sequencing the endogenous PHYB locus, we designed primers across exon/exon junctions. PHYB fragments were amplified by PCR from genomic DNA of the mutant lines by using the following primer pairs: ATGGTTTCCGGAGTCGGGGGTAG and TTCTTTTCCTCGTCCCCTCT; TTCTCGTGCTTTGAGAGGGG and TCTTTAGAACAAATGAACCG; and GAAAGCATTGAAGACGGTTC and CTAATATGGCATCATCAGC. For HY2 sequencing, the coding region was PCR-amplified from genomic DNA by using the following primer sets: TTGAAGAGAGTGTCCGAGGAAG and AGTCATTTACACCTCAGCATTAG, and TCAGGTCTGTATTGTCCAAACTGA and CGTCCTCTTGTGTTGATGACTC. In HY1 sequencing, two primer sets were used to amplify the coding region: GTGCAACACTCAACGCACTGTC and CCTAAGTAGCCAATAGAATCTG and GCACTTCCCATATAGGTACAGTG.
Fluorescence Microscopy. Seedlings grown under different light conditions were mounted on glass slides by using PBS as a mounting medium in dim green light, and then the slides were transferred in a dark box. PhyB::GFP patterns in cells of the upper portion of the hypocotyl were observed by using a DeltaVision deconvolution microscope (Applied Precision, Issaquah, WA). Specific filter sets were used for GFP (490 ± 10 excitation, 510 ± 5 emission) and enhanced yellow fluorescent protein (500 ± 10 excitation, 530 ± 5 emission). Color was artificially added in PHOTOSHOP 6.0 (Adobe Systems, Mountain View, CA). For each condition, at least 20 seedlings were observed; a representative image is presented.
Results
Characterization of phyB Nuclear Bodies in Different Light Conditions. To understand the relationship between the dynamics of Arabidopsis phyB localization and to correlate this with the response of the plant to light, we investigated whether altering the percentage of phyB in its Pfr conformer affected the pattern of phyB-containing NBs. Previous studies examined phyB nuclear import during the transition to photoautotrophic growth. Here, we focused on the steady-state patterns of phyB under constant R, where the ratio of Pfr to total phyB is the important parameter. Two ways exist to manipulate the relative amount of Pfr to total phyB: the R/FR ratio of incident light and the fluence rate of R. It has been shown that Pfr of Arabidopsis phyB is thermally unstable and could convert back to Pr rapidly without FR illumination, a phenomenon called dark reversion. The PfrPr heterodimer of phyB has been reported to convert rapidly to PrPr conformer with a half-life of 1-4 min (24). Thus, under R only and especially low-fluence rates of R, where most phyB is in the PfrPr isoform, the percent of phyB Pfr conformer is the result of the balance between the fluence rate of R light and dark reversion. Because the dark-reversion rate is a constant, different R intensities shift the balance between Pfr and Pr.
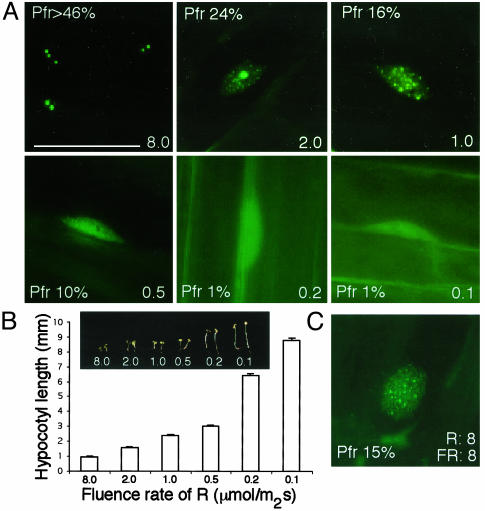
We first examined phyB NB patterns under different fluence rates of R by using a PBG line (13), and measured hypocotyl length as a readout for phyB light response. Conditions where hypocotyl elongation follows an inverse log relationship with light input are well established (1). The PBG line was generated by selecting for a single-copy insertion of a 35S-driven PHYB::GFP transgene into a phyB null allele, phyB-5 (13). As such, phyB responses resulted solely from the 35S-PHYB::GFP transgene. We found four distinctive phyB nuclear and subnuclear patterns under different R intensities, which, to our surprise, were similar to the patterns of phyB::GFP described during etiolated to light-grown transitions (13). At low-fluence rates of R (0.1 and 0.2 μmol·m-2·s-1, 1% of total phyB estimated to be Pfr), phyB::GFP localized to both the nucleus and the cytoplasm, with nuclear phyB::GFP being evenly dispersed throughout the nucleoplasm (Fig. 1A). Under 0.5 μmol·m-2·s-1 of R (10% of total phyB estimated to be Pfr), most phyB::GFP was found to be evenly dispersed in the nucleoplasm, with little cytoplasmic fluorescence. This finding indicated that nuclear import of phyB was the major event under low-fluence rates of R and was R-fluence-rate-dependent. The R-light dependence of phyB nuclear import is consistent with reports using tobacco phyB::GFP (16). In addition, this result clearly demonstrates that nuclear import of phyB is not sufficient for phyB's localization to NBs, even though the seedling is responding to R light in a phyB-dependent way. We defined these phyB patterns with the evenly distributed nuclear phyB::GFP as ”stage I.” At 1 μmol·m-2·s-1 and 15% of phyB estimated to be Pfr, hundreds of small phyB::GFP NBs were observed (Fig. 1 A). These phyB NBs were uniform in size and evenly distributed throughout the nucleoplasm. We called this stage of phyB nuclear localization pattern ”stage II.” In 2 μmol·m-2·s-1of R (≈24% Pfr), a few very large phyB-containing NBs and many small NBs were seen. This pattern of both large and small NBs was defined as ”stage III,” and this stage was never reported by other studies. At fluence rates of R higher than 8 μmol·m-2·s-1 (>43% phyB estimated to be Pfr), only large NBs were observed with little fluorescence in the nucleoplasm. The number of large NBs varied from cell to cell, ranging from 1 to 10 per nucleus. This pattern was defined as ”stage IV.” These results suggest that the amount of Pfr to total phyB must reach a certain threshold to form NBs, and large NBs were formed only when the percent of phyB that is Pfr was high.
Fig. 1.
Characterization of phyB::GFP NB patterns in response to changing fluence rates of R light. (A) Steady-state phyB::GFP localization patterns in hypocotyl cells of 4-day-grown PBG seedlings under different fluence rates of R with different percentages of Pfr in the total. See text for details. (B) Hypocotyl length measurements of 4-day-grown PBG seedlings under different fluence rates of R. (C) NB pattern of phyB::GFP in hypocotyl cells growing in 8 μmol·m-2·s-1 of R and 8 μmol·m-2·s-1 of FR was similar to that of cells growing in 1 μmol·m-2·s-1 of R only. (Bar = 10 μm.) The number labels represent fluence rates with a unit of μmol·m-2·s-1.
To assess this hypothesis with a different experiment, we examined phyB::GFP NB patterns after growing seedlings under different R/FR ratios of light. We found that PBG plants growing under different R/FR schemes resulted in similar phyB::GFP subnuclear NB patterns as growing plants under different fluence rates of R as long as the percent of phyB that is Pfr was kept similar. As examples, plants growing either under 1 μmol·m-2·s-1 of R alone or growing under 8 μmol·m-2·s-1 of R and 8 μmol·m-2·s-1 of FR each have about 15% of the total phyB in the Pfr conformer, and in each case, phyB::GFP showed the stage II pattern (Fig. 1C). In all R treatment experiments, hypocotyl length decreased with increasing fluence rate of R, which results in more Pfr phyB (Fig. 1B). This finding suggests a correlation between phyB NB pattern and phyB responses, with large phyB NBs associated with the strongest response to light.
PhyB::GFP Mislocalization Screen. To get a handle on the physiological significance of phyB's localization to nuclear bodies and to determine whether other proteins are required for this localization, we designed a mutant screen. PBG seeds (20,000) were mutagenized with ethylmethanesulfonate. We screened visually for mutants with elongated hypocotyls compared with the PBG control line growing in 1 μmol·m-2·s-1 of R for 4 days. From, 40,000 M2 seedlings, 60 R-light-hyposensitive lines were selected. A secondary screen was done subsequently on M3 plants by examining phyB::GFP nuclear localization patterns on 4-day-grown seedlings under high (8 μmol·m-2·s-1) R conditions, in which phyB::GFP localized to a few very large NBs (stage IV pattern). Of the 60 lines from the primary screen, 35 lacked any GFP signal, and we did not carry these lines forward. For the remaining 25 lines with GFP signal, two experiments were done for further characterization. First, seedlings were grown under continuous FR to examine whether they were also hyposensitive to FR. Five lines were found to be less sensitive to both FR light and R. The remaining 20 lines were hyposensitive to R light specifically. Second, phyB::GFP NB patterns were examined in the 25 M3 lines. Various phyB::GFP patterns were observed in these lines, including seven lines with diffuse phyB::GFP nucleoplasmic fluorescence (stage I), five lines with similarly few but much smaller NBs than the PBG control, six lines with intermediate NB patterns of stage III or II, and seven lines with the same NB pattern as the parental PBG line. These latter lines, with normal phyB NBs, may represent downstream components of the phyB-signaling pathway and are beyond the scope of this investigation.
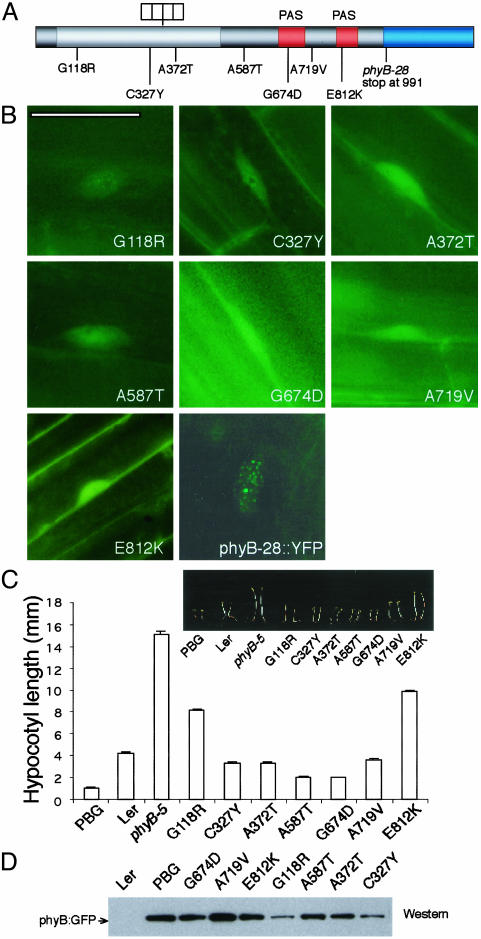
Intragenic phyB Mutations. To distinguish intragenic phyB mutations from second-site mutations, we sequenced the phyB::GFP transgene in all 25 mutant lines. Seven missense phyB alleles were recovered: G118R, C327Y, A372T, A587T, G674D, A719V, and E812K (Fig. 2 A and C). Two alleles (G118R and E812K) have been reported before (25, 26); the other five (C327Y, A372T, A587T, G674D, and A719V) are new alleles that have not been described in previous phyB mutant screens. In all these lines, no phyB::GFP NBs could be detected (Fig. 2B), although phyB::GFP was present as diffuse fluorescence in the nucleus. These phyB mutations were localized throughout the phyB protein, in the N-terminal chromophore-binding domain (G118R, C327Y, and A372T), the hinge region (A587T), and the C-terminal PAS domains (G674D, A719V, and E812K) (Fig. 2 A). PhyB::GFP levels were detected by Western blot by using GFP antibody. Although the three N-terminal and hinge domain mutants showed a reduction in phyB::GFP protein levels, the rest of the lines contained a similar amount of phyB::GFP protein compared with the PBG line (Fig. 2D).
Fig. 2.
Mutants with defective phyB apoproteins do not form NBs under R. (A) phyB mutations were found throughout the PHYB gene. (B) Evenly distributed phyB::GFP pattern in phyB mutants under 8 μmol·m-2·s-1 of R. PhyB-28::YFP showed small NBs under 8 μmol·m-2·s-1 of R. (C) Hypocotyl length of phyB mutants under the same fluence rates of R. (D) PhyB::GFP levels were determined by Western blot analysis with a GFP antibody. (Bar = 10 μm.)
None of the isolated phyB mutations mapped to the HKRD at the C terminus of phyB. To investigate whether phyB's HKRD is required for phyB's localization to NBs, we determined localization patterns of a previously characterized phyB allele, phyB-28 (25), which lacks most of the HKRD of phyB. We generated a 35S-driven phyB-28::YFP transgene and transformed it into the phyB-9 background. The NB dynamics of phyB28::YFP was studied in transgenic lines under different fluence rates of R light. At low-fluence rates of R, phyB28::YFP was in stage I similar to that of PBG lines, which suggested that nuclear import of phyB28 was not likely impaired. However, phyB28::YFP did not localize to large NBs under high-fluence rate R light. Only small NBs were observed even in high-fluence rate R (>8 μmol·m-2·s-1) (Fig. 2 A). These results suggest that the HKRD is required for phyB to associate with large NBs.
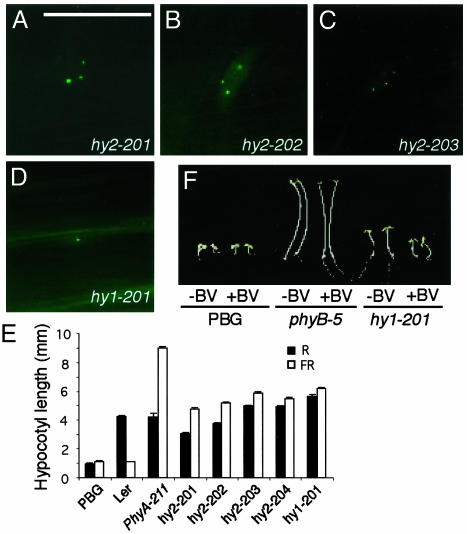
Chromophore-Deficient Mutants. Five mutants with smaller phyB NBs were less sensitive to both R and FR light (Fig. 3 A-E). Previous studies have shown that mutants with this phenotype are defective in phytochromobilin synthesis. Two enzymes are involved in phytochromobilin biosynthesis: a heme oxygenase, which converts heme to BV IXα, and a phytochromobilin synthase, which converts BV into phytochromobilin. Arabidopsis has two genes encoding heme oxygenases, HY1/HO1 and HO2 (27, 28). Phytochromobilin synthase is encoded by the HY2 gene (29). One way to distinguish heme oxygenase mutants from hy2 is by feeding with BV. Mutants of heme oxygenases can be rescued by BV, whereas hy2 alleles cannot. When the five mutant lines were grown on plates containing BV, only one line was partially rescued (Fig. 3F). This finding suggested that this line was defective in either the HO1 or HO2 gene. We sequenced the coding region of both genes in this line. A G-to-A change was found in the HO1 (HY1) gene, which led to a change of amino acid 265 from Glu to Lys. Glu-265 has been shown to be highly conserved in heme oxygenases from Arabidopsis, cyanobacteria, algae, and animals (30). We named this line hy1-201. For the remaining four lines, we sequenced the HY2 locus and found a mutation in each mutant line. We named these four lines hy2-201 to hy2-204. hy2-201 had a premature stop codon at R319, resulting from a single nucleotide deletion. hy2-202 had a change in amino acid 319 from Arg to Cys. Both hy2-203 and hy2-204 carried the same missense mutation P128S, which was similar to previously reported hy2-1 and hy2-104 (P128L) alleles. Therefore, all five lines in this group were chromophore-deficient. The size of phyB NBs were much smaller in these lines than that of the PBG line, which was most likely due to less phyB holoprotein. However, the pattern of phyB NBs in these lines was still the same as that of the PBG line. Under high-fluence rates of R, all the chromophore-deficient lines contained 1-10 relatively large NBs and no smaller NBs. This result suggests that the dynamics of phyB NB formation was not affected by the amount of phyB holoprotein.
Fig. 3.
Phytochrome chromophore mutants contain smaller NBs in high-fluence rates of R. (A-D) PhyB::GFP patterns in R/FR hyposensitive lines under 8 μmol·m-2·s-1 of R. These mutants have a similar number of NBs; however, the size of the NBs was much smaller than that of the PBG line. (E) Hypocotyl-length measurements under 8 μmol·m-2·s-1 ofRor16 μmol·m-2·s-1 of FR. (F) Mutant hy1-201 is rescued by BV feeding. (Bar = 10 μm.)
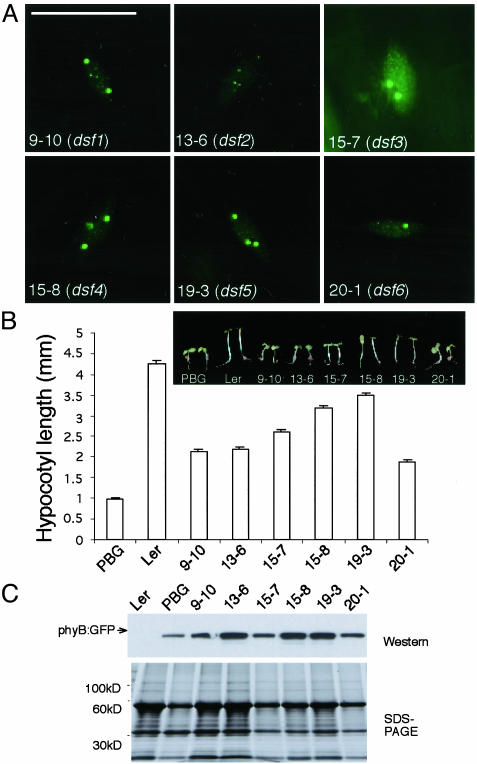
Dsf (Deficient in Speckle Formation) Mutants. The last group of six R-specific hyposensitive mutants showed stage II or III patterns under very high fluence rates of R light (Fig. 4 A and B). None of them had mutations in the PHYB::GFP transgene, and they complemented a phyB null allele, phyB-9, indicating that they were not allelic to phyB. In line 13-6, no large phyB-containing NBs were formed. Rather, this line appeared to be arrested in stage II with only small NBs. The rest of the mutants had both large and small phyB:GFP NBs in high-fluence rates of R (stage III). phyB::GFP protein levels were not affected in these mutants, suggesting that the phenotype was caused by deficiency in phyB signaling rather than reduced phyB protein (Fig. 4C). We named this class of mutant dsf (Deficient in Speckle Formation). These DSF genes thus define determinants that are required for the proper response of phyB to high-fluence rates of R light. As an initial attempt to clone these genes, we rough-mapped dsf1 (9, 10) to chromosome III between SSLP marker GAPAB and CAPS marker AFC1. To our knowledge, no known phyB-signaling component maps to this interval of chromosome III, indicating that DSF1 is a previously uninvestigated phyB-signaling component.
Fig. 4.
dsf mutants with big and small NBs define extragenic factors required for phyB's localization to stage IV NBs. (A) phyB::GFP NB pattern in dsf mutants grown in 8 μmol·m-2·s-1 of R. (B) dsf mutants were specifically less sensitive to R. (C) phyB::GFP levels were similar to that of the PBG line by Western blot analysis with GFP antibodies. (Bar = 10 μm.)
Discussion
In recent years, it has been largely accepted that nuclear import of phytochromes is a prerequisite for the majority of their functions (3, 31). One current model for phytochrome's signaling mechanism involves phytochrome's association with transcription factors on DNA to regulate the expression of genes in response to light (32-34). Although it has been shown that phyB or phyB::GFP is imported to the nucleus in a red-light-dependent manner, and that it localizes to NBs that are dynamic in nature in terms of the diurnal cycle, the physiological significance of phyB's localization to discrete nuclear sites is still unclear. In this report, we show that we can predictably and precisely manipulate the size and number of phyB-containing NBs by changes in light quality or fluence rate, which is correlated with the percentage of phyB that is active (Pfr). Large phyB NBs always appear at higher fluence rates of R when phyB responses are the strongest, whereas at low-fluence rates, phyB:GFP was imported into the nucleus but remained dispersed throughout the nucleoplasm. These results were corroborated by a genetic screen, which also showed a correlation between phyB responses and NB formation. Specifically, mutations that resulted in changes in the phyB apoprotein, deletion of the HKRD, and extragenic dsf mutants did not form stage IV NBs and were less sensitive to high-fluence rates of R. In addition, among the phyB mutations recovered from our screen, E812K, which is the same as phyB-101, was isolated and extensively characterized (26, 35). The E812K mutation causes instability of the Pfr form. This is consistent with our observation that Pfr appears to be required for NB formation.
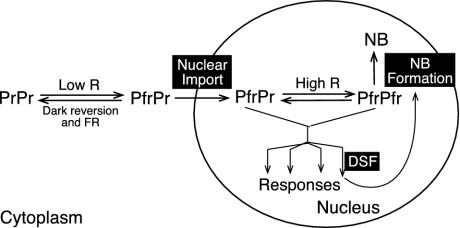
Our results suggest that phyB nuclear import and its localization to discrete subnuclear foci are two independent processes (Fig. 5). In this model, we take into account two observations: (i) phytochromes are dimers, and (ii) no reports exist that photoconversion of one monomer to Pfr affects the other monomer. Thus, in low-fluence-rate R light, phyB might exist as a PfrPr heterodimer, which is capable of moving into the nucleus, but unable to localize to NBs. In contrast, phyB localizes to NBs under high-fluence rates of R, conditions in which phyB is likely to be PfrPfr homodimers. This finding suggests that a PfrPr heterodimer is sufficient for nuclear import, but may be insufficient for localization to NBs. Based on this notion, we had expected to isolate mutations affecting either phyB nuclear import or NB formation. However, it is somewhat surprising that all phyB alleles and dsfs from our screen affected phyB NB formation rather than nuclear import. One explanation is that our genetic screen is unlikely to be saturated. However, this might also indicate that the regulation of phyB NB formation is complex, requiring more cis and trans elements than those for nuclear import.
Fig. 5.
Model of Arabidopsis phyB-regulated translocation. Red-light-induced phyB subnuclear localization comprises two steps, nuclear import and localization to NBs. Whereas PfrPr is sufficient for nuclear import, a higher percent of Pfr (likely PfrPfr) is required for phyB localization to NBs. NB formation may serve as a desensitization mechanism to control the amount of active nucleoplasmic phyB. This process could also be feedback-enhanced by phyB responses. DSFs may be components in the phyB-signaling pathways leading to the feedback loop. In dsf mutants, defective phyB NB formation could be due to reduced phyB responses.
Both our data and data of others indicate that nucleoplasmic dispersed phyB is active. Under low-fluence rates of R, conditions in which phyB is evenly dispersed throughout the nucleoplasm, plants still respond to R light (Fig. 1). Published studies (16, 20) and our own results from the phyB mislocalization screen further corroborate this conclusion. Mutations in the phyB apoprotein do not form NBs in high-fluence rates of R. However, all these mutants retain some phyB function (Fig. 2). Moreover, recently published results demonstrate that nucleoplasm-dispersed N-terminal phyB is even more active than full-length phyB (10). Thus, it is clear that some phyB functions are performed in the nucleoplasm.
The function of phyB NBs is still unclear. The NB may be a site of sequestration of plant photoreceptors or a signaling site for some phyB functions. In the first model (Fig. 5), phyB is inactive when sequestered in NBs. NB formation might serve as a desensitization mechanism to draw phyB from where it functions under high-light conditions. This is consistent with the data of Matsushita et al. (10) who suggested that the C terminus of phyB antagonizes the N-terminal signaling functions. These authors also showed that phyB's C terminus is both necessary and sufficient for NB localization and that nuclear-localized truncations of phyB that eliminate the C terminus are more active than full-length phyB. As such, phyB's C terminus may negatively regulate N-terminal function by directing phyB NB formation. This model is also supported by our observation that the localization of phyB shifts from nucleoplasm to NBs with an increase in the intensity of R, i.e., an enhanced phyB response. Such a high correlation between phyB response and NB formation implies a feedback enhancement of phyB NB localization from phyB responses. In this model, DSFs might be phyB-signaling components leading to the feedback loop, and the deficiency of phyB NB formation in dsfs would be caused by reduced phyB responses. Although this model is attractive, it should be noted that no direct evidence exists that phyB is inactive in NBs.
Several observations still cannot be explained by this model. For example, this model does not explain why most phyB PAS domain mutations from our studies and reports (17, 36) do not form phyB NBs but are loss-of-function alleles and why no loss-of-function mutation has been isolated with normal or enhanced phyB NB formation. Therefore, we propose an alternative model, in which phyB is active both in the nucleoplasm and in NBs with different signaling mechanisms to respond to low and high intensities of R, respectively. The correlation between strong phyB responses and large NB formation supports this model. This model is also consistent with the results from our genetic screen, from which mutations in phyB or other factors affecting NB formation would lead to deficiencies in phyB responses under high intensities of R.
Numerous functions of NBs have been suggested in the higher eukaryotic nucleus, including transcription regulation and premRNA splicing and processing (18, 37, 38). One possible function for phyB NBs may be to regulate downstream-signaling components, such as transcription factors. Recently, LAF1, a putative transcription factor involved in phyA signaling, was demonstrated to colocalize with COP1 in NBs, and COP1 has been shown to recruit a separate transcription factor, HY5, to NBs (39, 40). COP1 is an E3 ubiquitin ligase (40). It has been proposed that COP1 NBs are the site for LAF1 degradation (40), which may be required for the transcriptional response initiated by plant photoreceptors. However, it is unknown whether phyB colocalizes with COP1 in the same NBs. Moreover, it has also been shown that phyB enhances the plant response to blue light (by cryptochrome), perhaps by direct interactions of phyB and CRY, and phyB and CRY2 have been shown to colocalize to NBs in vivo (41). Regardless of the model, phyB NBs appear to be unique and dynamic with respect to the ambient-light environment and are crucial to the regulation of phyB signaling. Our ability to reliably manipulate the size and number of phyB NBs provides a unique opportunity to unravel the mechanisms of light signaling in plants and to gain insight into the molecular basis of function and regulation of NBs in eukaryotes.
Acknowledgments
We thank Jennifer Nemhauser, Kunhua Chen, and Pablo Cerdan for critical discussions and comments on this manuscript. This work was supported by National Institutes of Health Grant GM52413 (to J.C.). J.C. is an Investigator of the Howard Hughes Medical Institute.
Abbreviations: NB, nuclear body; phy, phytochrome; R, red; FR, far-reds; Pr, red-absorbing form of phytochrome; Pfr, far-red-absorbing form of phytochrome; HKRD, histidinekinase-related domain; PBG, 35S-phyB-GFP; BV, biliverdin.
References
- 1.Kendrick, R. E. & Kronenberg, G. H. M. (1994) Photomorphogenesis in Plants (Kluwer Academic, Dordrecht, The Netherlands).
- 2.Quail, P. H. (2002) Nat. Rev. Mol. Cell Biol. 3, 85-93. [DOI] [PubMed] [Google Scholar]
- 3.Nagy, F. & Schafer, E. (2002) Annu. Rev. Plant Biol. 53, 329-355. [DOI] [PubMed] [Google Scholar]
- 4.Fankhauser, C. & Staiger, D. (2002) Planta 216, 1-16. [DOI] [PubMed] [Google Scholar]
- 5.Tepperman, J. M., Zhu, T., Chang, H. S., Wang, X. & Quail, P. H. (2001) Proc. Natl. Acad. Sci. USA 98, 9437-9442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang, H., Ma, L., Habashi, J., Li, J., Zhao, H. & Deng, X. W. (2002) Plant J. 32, 723-733. [DOI] [PubMed] [Google Scholar]
- 7.Ma, L., Li, J., Qu, L., Hager, J., Chen, Z., Zhao, H. & Deng, X. W. (2001) Plant Cell 13, 2589-2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yeh, K. C. & Lagarias, J. C. (1998) Proc. Natl. Acad. Sci. USA 95, 13976-13981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fankhauser, C., Yeh, K. C., Lagarias, J. C., Zhang, H., Elich, T. D. & Chory, J. (1999) Science 284, 1539-1541. [DOI] [PubMed] [Google Scholar]
- 10.Matsushita, T., Mochizuki, N. & Nagatani, A. (2003) Nature 424, 571-574. [DOI] [PubMed] [Google Scholar]
- 11.Sharrock, R. A. & Quail, P. H. (1989) Genes Dev. 3, 1745-1757. [DOI] [PubMed] [Google Scholar]
- 12.Clack, T., Mathews, S. & Sharrock, R. A. (1994) Plant Mol. Biol. 25, 413-427. [DOI] [PubMed] [Google Scholar]
- 13.Yamaguchi, R., Nakamura, M., Mochizuki, N., Kay, S. A. & Nagatani, A. (1999) J. Cell Biol. 145, 437-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kircher, S., Kozma-Bognar, L., Kim, L., Adam, E., Harter, K., Schafer, E. & Nagy, F. (1999) Plant Cell 11, 1445-1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hisada, A., Hanzawa, H., Weller, J. L., Nagatani, A., Reid, J. B. & Furuya, M. (2000) Plant Cell 12, 1063-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gil, P., Kircher, S., Adam, E., Bury, E., Kozma-Bognar, L., Schafer, E. & Nagy, F. (2000) Plant J. 22, 135-145. [DOI] [PubMed] [Google Scholar]
- 17.Kircher, S., Gil, P., Kozma-Bognar, L., Fejes, E., Speth, V., Husselstein-Muller, T., Bauer, D., Adam, E., Schafer, E. & Nagy, F. (2002) Plant Cell 14, 1541-1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spector, D. L. (2001) J. Cell Sci. 114, 2891-2893. [DOI] [PubMed] [Google Scholar]
- 19.Dundr, M. & Misteli, T. (2001) Biochem. J. 356, 297-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parks, B. M. & Quail, P. H. (1991) Plant Cell 3, 1177-1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kelly, J. M. & Lagarias, J. C. (1985) Biochemistry 24, 6003-6010. [Google Scholar]
- 22.Lagarias, J. C., Kelly, J. M., Cyr, K. L. & Smith, W. O. (1987) Photochem. Photobiol. 46, 5-13. [Google Scholar]
- 23.Eichenberg, K., Baurle, I., Paulo, N., Sharrock, R. A., Rudiger, W. & Schafer, E. (2000) FEBS Lett. 470, 107-112. [DOI] [PubMed] [Google Scholar]
- 24.Hennig, L. & Schafer, E. (2001) J. Biol. Chem. 276, 7913-7918. [DOI] [PubMed] [Google Scholar]
- 25.Krall, L. & Reed, J. W. (2000) Proc. Natl. Acad. Sci. USA 97, 8169-8174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Elich, T. D. & Chory, J. (1997) Plant Cell 9, 2271-2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muramoto, T., Kohchi, T., Yokota, A., Hwang, I. & Goodman, H. M. (1999) Plant Cell 11, 335-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davis, S. J., Bhoo, S. H., Durski, A. M., Walker, J. M. & Vierstra, R. D. (2001) Plant Physiol. 126, 656-669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kohchi, T., Mukougawa, K., Frankenberg, N., Masuda, M., Yokota, A. & Lagarias, J. C. (2001) Plant Cell 13, 425-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Davis, S. J., Kurepa, J. & Vierstra, R. D. (1999) Proc. Natl. Acad. Sci. USA 96, 6541-6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huq, E., Al-Sady, B. & Quail, P. H. (2003) Plant J. 35, 660-664. [DOI] [PubMed] [Google Scholar]
- 32.Ni, M., Tepperman, J. M. & Quail, P. H. (1998) Cell 95, 657-667. [DOI] [PubMed] [Google Scholar]
- 33.Ni, M., Tepperman, J. M. & Quail, P. H. (1999) Nature 400, 781-784. [DOI] [PubMed] [Google Scholar]
- 34.Huq, E. & Quail, P. H. (2002) EMBO J. 21, 2441-2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wagner, D., Koloszvari, M. & Quail, P. H. (1996) Plant Cell 8, 859-871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yanovsky, M. J., Luppi, J. P., Kirchbauer, D., Ogorodnikova, O. B., Sineshchekov, V. A., Adam, E., Kircher, S., Staneloni, R. J., Schafer, E., Nagy, F. & Casal, J. J. (2002) Plant Cell 14, 1591-1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spector, D. L. (1993) Annu. Rev. Cell Biol. 9, 265-315. [DOI] [PubMed] [Google Scholar]
- 38.Eskiw, C. H. & Bazett-Jones, D. P. (2002) Biochem. Cell Biol. 80, 301-310. [DOI] [PubMed] [Google Scholar]
- 39.Ballesteros, M. L., Bolle, C., Lois, L. M., Moore, J. M., Vielle-Calzada, J. P., Grossniklaus, U. & Chua, N. H. (2001) Genes Dev. 15, 2613-2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seo, H. S., Yang, J. Y., Ishikawa, M., Bolle, C., Ballesteros, M. L. & Chua, N. H. (2003) Nature 424, 995-999. [DOI] [PubMed] [Google Scholar]
- 41.Mas, P., Devlin, P. F., Panda, S. & Kay, S. A. (2000) Nature 408, 207-211. [DOI] [PubMed] [Google Scholar]