Abstract
The T-cell receptor (TCR) and immunoglobulin (Ig) genes are unique among vertebrate genes in that they undergo programmed rearrangement, a process that allows them to generate an enormous array of receptors with different antigen specificities. While crucial for immune function, this rearrangement mechanism is highly error prone, often generating frameshift or nonsense mutations that render the rearranged TCR and Ig genes defective. Such frame-disrupting mutations have been reported to increase the level of TCRβ and Igµ pre-mRNA, suggesting the hypothesis that RNA processing is blocked when frame disruption is sensed. Using a chimeric gene that contains TCRβ sequences conferring this upregulatory response, we provide evidence that pre-mRNA upregulation is neither frame- nor translation-dependent; instead, several lines of evidence suggested that it is the result of disrupted cis elements necessary for efficient RNA splicing. In particular, we identify the rearranging VDJβ exon as being uniquely densely packed with exonic-splicing enhancers (ESEs), rendering this exon hypersensitive to mutational disruption. As the chimeric gene that we developed for these studies generates unusually stable nuclear pre-mRNAs that accumulate when challenged with ESE mutations, we suggest it can be used as a sensitive in vivo system to identify and characterize ESEs.
INTRODUCTION
Approximately one-third of inherited genetic disorders are caused by mutations that generate premature termination codons (PTCs). PTCs also arise as a result of biosynthetic errors, including frameshifts and nonsense mutations created by faulty transcription and messenger RNA (mRNA) splicing. Such aberrant mRNAs are recognized and destroyed by nonsense-mediated mRNA decay (NMD), a highly conserved quality-control mechanism (1–3). By rapidly degrading aberrant PTC-bearing transcripts, NMD reduces the translation of C-terminally truncated proteins, some of which possess dominant-negative or deleterious gain-of-function activity. Recently, it has emerged that NMD also regulates the level of mRNAs from ∼5% of wild-type genes, including those generated by alternative splicing (1–4). Thus, NMD is not only an RNA surveillance pathway but also a regulator of normal gene expression.
NMD requires recognition of the stop codon by the translation machinery and a second signal downstream that defines the stop codon as premature. Several different cis elements and trans-acting factors have been identified as NMD second signals in different organisms (1–3). In mammals, a spliceable intron downstream of the stop codon elicits NMD. This intron requirement derives from the fact that the splicing machinery deposits a dynamic assembly of proteins, known as the exon-junction complex (EJC), which interacts with factors deposited on transcripts upon translation termination and elicits NMD (1–3).
Transcripts encoded by the T-cell receptor (TCR) and immunoglobulin (Ig) genes are a unique class of mammalian NMD substrates that acquire PTCs at an extremely high frequency as a result of error-prone programmed gene rearrangements that increase immune receptor diversity (5–7). This frequent acquisition of PTCs may have led to strong selection pressure to efficiently eliminate PTC-bearing TCRβ transcripts. Consistent with this hypothesis, we previously demonstrated that PTCs downregulate TCRβ transcripts more dramatically than transcripts from most nonrearranging genes that have been tested (8,9). We subsequently showed that this robust downregulation is neither specific to T cells, nor does it require a TCRβ promoter; rather it is elicited by TCRβ sequences that promote efficient RNA splicing (10). RNA half-life analysis indicated that the NMD response responsible for this dramatic downregulation occurs in highly purified nuclei that have undetectable levels of cytosolic, endoplasmic reticulum (ER), or processing body (P-body) markers (11). These highly purified nuclei contained high levels of both outer nuclear membrane and chromatin markers, suggesting that PTC-bearing TCRβ transcripts are degraded either at the outer nuclear membrane or in the nucleoplasm (11). A recent study provided evidence that robust downregulation of aberrant TCRβ transcripts is essential for T-cell survival. Analysis of mice conditionally depleted of the NMD gene Upf2 in selected cell populations demonstrated that loss of Upf2 is only lethal for T cells that harbor nonproductively rearranged TCRβ genes harboring PTCs (12). This result implies that NMD is required for the survival of T cells because it dramatically downregulates the level of truncated dominant-negative TCRβ proteins that would otherwise be translated from the nonproductively rearranged TCRβ gene allele.
Nonsense mutations in TCRβ genes have been shown to elicit not only rapid decay of mature TCRβ transcripts but also three other responses. One response is a dramatic shift in the ratio of mature TCRβ transcripts in the nuclear and cytoplasmic fraction of mammalian cells (11). This nonsense codon-induced partitioning shift (NIPS) is specifically triggered by recognition of a disrupted reading frame, as missense mutations do not elicit it and it is reversed by the translation inhibitor cycloheximide (CHX) and a translation-blocking stem–loop. While the underlying mechanism for NIPS has not been clearly defined, several lines of evidence suggest that it is the result of retention of PTC-bearing transcripts in either the outer nuclear membrane or the nucleoplasm of mammalian cells (11).
Another response to nonsense mutations is an increase in the level of alternatively spliced TCRβ transcripts that skip the offending mutation and restore reading frame (13–15). This nonsense-associated altered splicing (NAS) response appears to be elicited by recognition of a disrupted reading frame, as it is also triggered by frameshift mutations and is reversed by compensatory frameshift mutations, suppressor tRNAs and mutation of the start codon or adjacent Kozak consensus sequences (14–16). However, we recently obtained evidence that alternatively spliced TCRβ transcripts can also be upregulated by mutations disrupting exonic splicing enhancers (ESEs) in the VDJ exon (16). Thus, alternatively spliced TCRβ transcripts can be upregulated in response to either reading frame or ESE disruption. Similarly, alternatively spliced fibrillin transcripts have been shown to be upregulated in response to either disruption of reading frame or ESEs (17,18). Other transcripts appear to be upregulated only by reading frame disruption or ESE disruption, not both (19–22).
In this article, we focus on a fourth response to nonsense mutations: nonsense-mediated upregulation of pre-mRNA (NMUP). We previously reported that NMUP has some characteristics in common with NAS. First, NMUP appeared to be specifically triggered by disruption of reading frame, as TCRβ pre-mRNA was upregulated in response to nonsense but not missense mutations (23). Second, NMUP was elicited by a frameshift that generated downstream PTCs, but not when a compensatory frameshift was introduced that prevented the generation of the PTCs (23). Another rearranging gene that we found appears to undergo NMUP is Igµ (23). Comparison of variant plasma cell lines with different mutations in a common Igµ gene revealed that disruption of reading frame correlated with upregulation of Igµ pre-mRNA. This effect was independent of PTC position, as the same Igµ introns were retained, regardless of the location of the nonsense or frameshift mutations tested. However, a subsequent study discovered that the level of PTC-Igµ pre-mRNA varies in plasma cell lines obtained from different sources, casting into doubt whether frame-disrupting mutations really augment Igµ pre-mRNA levels (24). A third rearranging gene that has been reported to undergo NMUP is Igκ. Somatic mutations introduced in this gene during normal B-cell development lead to the generation of PTCs and increased levels of Igκ pre-mRNA by a mechanism that appears to involve inhibited RNA splicing (25,26). A final example of NMUP was reported to occur in the parvovirus minute virus of mice (MVM). PTC-generating mutations introduced at various locations in this viral genome increase the levels of precursor MVM mRNA retaining one of the introns (19,20,27). In contrast, mutations that did not generate PTCs failed to increase MVM precursor mRNA. Similar to many other viruses, MVM generates viral proteins from its partially spliced transcripts and thus NMUP may be a means to increase the production of such proteins.
Here, we investigated the underlying molecular basis for NMUP. We report that the TCRβ exon that undergoes programmed rearrangements—the VDJ exon—is sufficient to trigger a strong NMUP response in a heterologous gene. Using a chimeric gene harboring the VDJ exon, we employed a variety of approaches to determine whether NMUP is elicited by recognition of disrupted reading frame, as previous studies had suggested (19,20,23,25,27). Surprisingly, we obtained several lines of evidence that frame disruption is not responsible for NMUP. Instead, our results strongly suggested that the rearranging VDJ exon is extremely rich in nonredundant ESEs, such that most point and frameshift mutations in the VDJ exon result in inhibited mRNA splicing and consequent pre-mRNA upregulation. As the chimeric gene that we used for our study gives rise to a relatively stable precursor mRNAs that accumulate in the nucleus when splicing elements are impaired, it has the potential to be a useful general tool for elucidating cis elements that regulate both exon inclusion and the magnitude of mRNA splicing in vivo.
MATERIALS AND METHODS
Plasmid constructs
Plasmid constructs A, AN1, AI1+ and AN2, were previously described (plasmids I−, IV+, I5+ and I6+, respectively) (8). Constructs B, BN, C, CN, D and DN were also previously described (plasmids P′, Q, I−, I+, C− and C+, respectively) (10,28). Constructs AN3/M3, AN4/M4, AI2+, AI3, AD1+, AI2D1, AN5/M5, AN6/M6/M6′, AN7/M7/M7′, AN8/M8/M8′ and AD2+ were generated by site-directed mutagenesis of the Bluescript KS+ (Invitrogen) version of construct A (β818), using the primers indicated in Supplementary Table S1. Constructs ΔA, ΔAN2 and ΔAM2 were generated by deletion PCR of the KS+ versions of constructs A (β818) and B+ (β819) using primers indicated in Supplementary Table S1. The mutations were confirmed by sequencing the region surrounding the intended mutation. For mutations in the VDJ exon, the SalI–ClaI fragment containing the mutation were subcloned into the SalI–ClaI site of construct A to generate constructs AN3/M3, AN4/M4, AI2+, AI3, AD1+ and AI2D1. For mutations in exons 5 and 6, the ClaI–BamHI fragments containing the mutations were subcloned into the ClaI–BamHI site of construct A to generate constructs AN5/M5, AN6/M6/M6′ and AN8/M8/M8′. The constructs harboring deletions of IVS-C and IVS-D (ΔA and ΔAN2/M2) were generated by a multistep process involving complementary DNA (cDNA) synthesis [by reverse transcription-polymerase chain reaction (RT-PCR)] of RNA from cells transfected with constructs A, AN2 and AM2, followed by subcloning of the ClaI–BamHI region into pGEM (Promega), and then subcloning the ClaI-BamHI fragment from pGEM into the ClaI–BamHI site of construct A.
Cell culture, transfection and RNA interference
HeLa and NIH-3T3 cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and penicllin/streptomycin (6). HeLa cells cultured to 40–60% confluency in six-well plates were transiently transfected with reporter plasmids (30 ng) and β-globin (15 ng) as a control using lipofectamine reagent (Invitrogen) according to manufacturer protocols. Cells were harvested 42–48 h after transient transfection. For stable selection of reporter or UPF1 short hairpin RNA (shRNA) plasmids, HeLa cells transfected as above were treated with G418 (700 μg/ml) for 2 weeks. For NIH-3T3 cell transfections, cells cultured to 70% confluence in six-well plates were transfected with reporter plasmids (1.5–2 μg) using lipofectamine 2000 reagent (Invitrogen). Eight hours post-transfection, plates were washed with serum-free media and serum starved for 24 h, after which the cells were provided with serum before harvesting.
RNA interference (RNAi) against UPF1 was performed with the small interfering RNA (siRNA) oligo 5′-GAUGCAGUUCCGCUCCAUU-3′ (Ambion). Firefly luciferase-specific siRNA (Ambion) as used by Chan et al. (4) was used as a negative control (29). HeLa cells, cultured as above, were grown to 15% confluency in six-well plates, and transfected with siRNA oligos (100 nM final concentration) using Lipofectamine 2000 reagent (Invitrogen). After ∼20 h, cells were transfected with reporter plasmids (300 ng) and β-globin (15 ng) or TCRβ (50 ng) internal control plasmids using Lipofectamine reagent as per manufacturer protocol. Cells were harvested 42–48 h after reporter transfection.
RNA isolation and analysis
Total cellular RNA was isolated as described before (8), or by using Trizol reagent (Invitrogen) (4). RNA isolation from subcellular fractions was performed as in Bhalla et al. (11). RNase protection analysis was performed as described previously (10), using 5–10 μg RNA. Probe 1 for RNase protection analysis was described previously (4,9,10). Probes 2, 3 and 4 were made by PCR amplification of sequences in construct A, using primers given in Supplementary Table S2. All probes generated were transcribed from PCR products subcloned into the pGEM-T easy vector (Promega). Quantification of RNA levels was determined using a Storm Imager (GE) and Total Lab (Nonlinear) TL100 image quantitation software.
Northern blot analysis was performed as described previously (8,30) using 10 μg RNA. Northern probes E, C and D, were made by PCR amplification of sequences in construct A corresponding to the locations of each probe (given in the figures), using primers given in Supplementary Table S3. The PCR products were subcloned into the pGEM-T easy vector (Promega Inc.). Northern probes A and B were generated by digesting construct A with either AgeI and BglII (probe A) or with ScaI (probe B). Quantification of Northern RNA levels was determined using an Instant Imager (Packard Instruments).
RESULTS
TCRβ sequences sufficient to confer NMUP
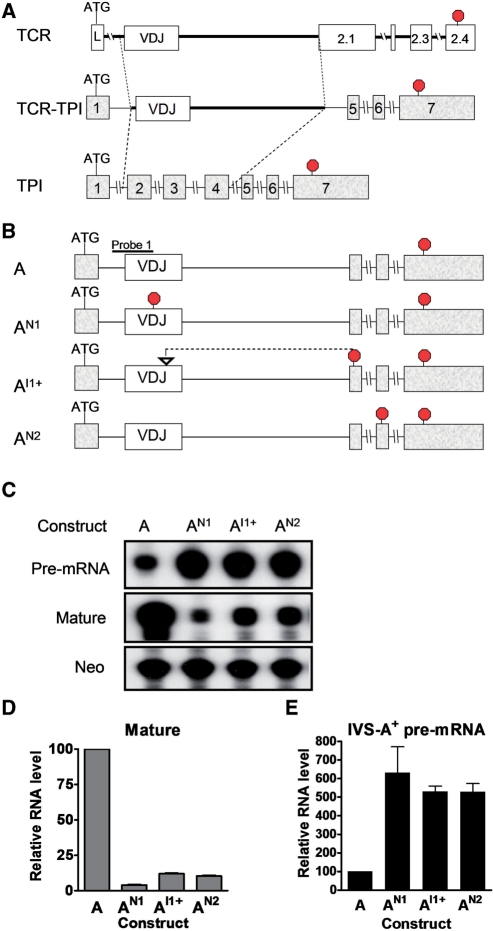
We previously identified a segment of TCRβ that recapitulates all four of its known responses to PTCs: (i) robust mature mRNA downregulation (8); (ii) boundary-independent mature mRNA downregulation (i.e., in response to PTCs closer than 55 nt from the 3′ exon–exon junction) (28); (iii) polar mRNA downregulation (i.e. 5′ PTCs elicit stronger downregulation than do 3′ PTCs) (28); and (iv) the NIPS response (11). The TCRβ region conferring these four responses consisted of the VDJ exon (354 nt), the 3′-end of the upstream intron (IVS-LV; 23 nt) and the 5′-end of the downstream intron (IVS-JC; 674 nt). To determine whether this TCRβ region was also sufficient to trigger NMUP, we inserted it into the triose phosphate isomerase (TPI) gene, which was previously shown to give rise to precursor transcripts that do not undergo NMUP (31). We substituted this TCRβ region for the TPI region shown in Figure 1A to generate a chimeric pre-mRNA that should be spliced into an mRNA similar in size to that of normal TPI mRNA.
Figure 1.
Nonsense and frameshift mutations upregulate pre-mRNA. (A) Schematic diagram showing how the TCR/TPI chimeric gene construct was generated from the parental TCRβ and TPI gene constructs. (B) Schematic diagram of the TCR/TPI chimeric constructs used for the transfection experiments in (C). The position of the nonsense and frameshift mutations in AN1, AI1+ and AN2 are indicated (at codons 91 and 100 and 194, respectively). Stop signs indicate the position of in-frame termination codons. (C) RNase protection analysis of total cellular RNA (10 µg) from HeLa cells transiently transfected with the constructs described in (B). Probe 1 (B) was used to detect both pre- and mature mRNA. Neomycin (Neo) mRNA, an independent transcription unit expressed from the TCR/TPI plasmids, serves as an internal control for transfection efficiency. (D, E) Quantification of mature mRNA (D) and pre-mRNA (E) levels from cells transfected as in (C). The values were determined from three or more experiments and normalized to Neo mRNA level. Pre-mRNA and mature mRNA levels from construct A were arbitrarily set to 100. Error bars indicate standard error.
PTC-generating mutations were introduced at three different positions in this chimeric construct: (i) a nonsense mutation in the VDJ exon (codon 91; construct AN1); (ii) a frameshift resulting from a 10-nt insertion in the VDJ exon (generates a PTC in exon 5; construct AI1+); and (iii) a nonsense mutation in exon 6 (codon 194; construct AN2) (Figure 1B). These mutant constructs, as well as a PTC-lacking control (construct A), were transiently transfected into HeLa cells and total cellular RNA was prepared. Given that all three mutations introduce PTCs in the mature mRNA, we predicted that all three mutations should trigger NMD. Indeed, RNase protection analysis showed that all three mutations strongly reduced the level of mature mRNA (Figure 1C and D). Conversely, we predicted that if the chimeric pre-mRNA is regulated by NMUP, it would be increased in level by the three mutations. In agreement with this prediction, we found that all three mutations increased the level of the pre-mRNA (Figure 1C and E).
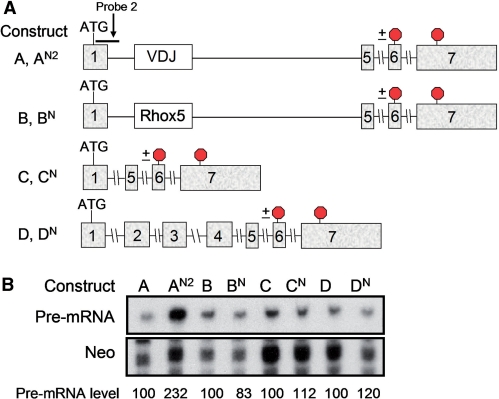
To examine the specificity of this upregulatory response, we generated and tested several control constructs: (i) a chimeric gene harboring a heterologous exon of similar length as the VDJ exon [exon 4 (357 nt) from the Rhox5 homeobox gene (28)] (constructs B and BN in Figure 2A); (ii) a TPI ‘minigene’ that lacked the same TPI sequences deleted in the TCRβ/TPI chimeric construct (constructs C and CN); and (iii) a TPI full-length gene (constructs D and DN) (Figure 2A). A PTC was introduced in the penultimate exon (codon 194), as this exon is common to all these gene constructs. RNase protection analysis showed that only the TCR/TPI chimeric construct (AN2) gave rise to a pre-mRNA upregulated by the nonsense mutation (Figure 2B). None of the other constructs (BN, CN and DN) generated pre-mRNAs upregulated by the nonsense mutation (Figure 2B). These data supported an earlier study showing that TPI pre-mRNA is not subject to NMUP (31) and it provided evidence that this upregulatory response is uniquely conferred by TCRβ sequences.
Figure 2.
Pre-mRNA upregulation requires the VDJ exon. (A) Schematic diagrams of the constructs transfected in (B) (see text for further details of these constructs). Note that AN2, BN, CN and DN all have the same nonsense mutation at the same position in exon 6 (indicated with a stop sign). (B) RNase protection analysis of total cellular RNA (10 µg) harvested from HeLa cells transiently transfected with the constructs shown. Probe 2 (A) was used to detect the pre-mRNA; quantification was done as in Figure 1 (average of two experiments). The values below the gel were quantified as in Figure 1D and E from two experiments. Pre-mRNA levels for each wild-type construct were arbitrarily set to 100.
PTCs are distinguished from normal termination codons by the presence of a ‘second signal’ downstream (‘Introduction’ section). In mammalian cells, this second signal is typically a downstream intron. Following intron splicing from the pre-mRNA, the resulting exon–exon junction recruits a set of NMD-promoting molecules that are collectively called the ‘EJC’ (1–3). This EJC interacts with factors recruited at the site of premature termination, leading to rapid mRNA decay (1–3). Most normal mRNAs are exempt from this regulation, as normal termination codons are typically in the final exon and so all EJCs are stripped from the mRNA by translating ribosomes prior to stop codon recognition. To test whether NMUP also depends on exon–exon junctions, we deleted the introns surrounding the final exon in the chimeric construct (Supplementary Figure S1A). As a control, we first examined whether NMD was abolished by the removal of these introns. We found that indeed this prevented the ability of the nonsense mutation in TPI exon 6 to trigger NMD (Supplementary Figure S1B), consistent with past studies examining intron deletions in the parental TCR and TPI genes (32,33). To determine whether NMUP also depends on exon–exon junctions, we examined the effect of intron removal on the level of IVS-A and -B-containing pre-mRNA. We found that intron removal abolished the ability of the nonsense mutation to upregulate pre-mRNA harboring either IVS-A or -B (Supplementary Figure S1C and D). As another control, we examined the effect of a missense mutation and found that it did not elicit either NMD or NMUP (Supplementary Figure S1B–D), nor did it significantly affect mRNA splicing rate, as judged by pre-mRNA/mRNA ratio (Supplementary Figure S1E). We conclude that, similar to NMD, the NMUP response depends on the presence of introns.
The NMUP substrate is a stable, partially spliced nuclear RNA
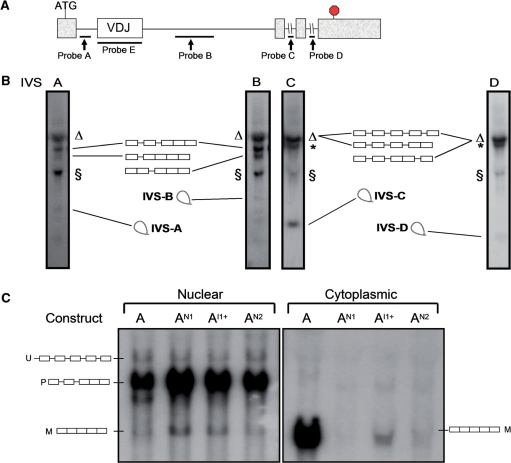
To characterize the pre-mRNA upregulated by nonsense and frameshift mutations, we first used northern blot analysis in conjunction with probes that specifically recognized introns IVS-A, -B, -C or -D in the chimeric construct (Figure 3A). Analysis of cells transfected with the wild-type chimeric construct revealed transcripts migrating at sizes expected for the unspliced pre-mRNA and several partially spliced mRNAs (Figure 3B). In addition, we detected small RNAs with sizes consistent with that of the spliced introns themselves (Figure 3B). The introduction of nonsense and frameshift mutations (Figures 1B and 3A) leads to an increase in the level of partially spliced transcripts that contained IVS-A and/or -B, but not IVS-C or -D (Supplementary Figure S2). This demonstrated that ‘the NMUP substrate’ is a partially spliced pre-mRNA that harbors IVS-A, IVS-B or both. The most plausible explanation for the accumulation of these partially spliced pre-mRNA species in response to nonsense and frameshift mutations is that these mutations inhibited IVS-A and -B splicing.
Figure 3.
The upregulated pre-mRNAs are partially spliced. (A) Schematic diagram of construct A, indicating the position of the intron probes used for the northern blot analysis in (B) and (C). (B) Northern blot analysis of total cellular RNA (10 µg) isolated from HeLa cells stably transfected with construct A. The schematics indicate the introns present in the pre-mRNAs in each band, based on band migration and their hybridization with the different probes. The bands corresponding to spliced IVS-A, -B, -C and -D migrated at a position consistent with their expected sizes (∼0.5, ∼0.8, ∼0.25 and ∼0.1 nt, respectively). The Δ denotes 28S rRNA that cross-hybridized with all probes and was present in nontransfected HeLa cells (data not shown); it is a broad band that co-migrated with the high-molecular-weight TCR/TPI pre-mRNAs. The § denotes ∼1.8–2.0-kb transcripts that hybridized with all the intron probes; their size is consistent with them being 3′ cleavage intermediates that have either IVS-A and -B at their 5′ terminus (as a result of 5′ splice-site cleavage but not 3′ splice-site cleavage). IVS-C and -D are present in a fraction of these § transcripts presumably because these small introns are sometimes retained. The asterisk indicates a transcript whose size and hybridization characteristics suggest it is a partially spliced TCR/TPI pre-mRNA lacking the β-actin intron, which is upstream of IVS-A (data not shown). (C) Northern blot analysis of nuclear and cytoplasmic fraction RNA from HeLa cells stably transfected with the constructs shown and hybridized with probe E. U, unspliced pre-mRNA; P, partially spliced pre-mRNA; M, mature mRNA. The blots shown in (B) and (C) are representative of two or more independent blots.
To determine whether the upregulated TCR/TPI pre-mRNA is confined to the nucleus (where it is synthesized) or is exported to the cytoplasm, we analyzed nuclear and cytoplasmic RNA. Northern blot analysis showed that the partially spliced pre-mRNA species upregulated by nonsense and frameshift mutations was exclusively in the nucleus (the band labeled ‘P’ in Figure 3C). Likewise, a low-abundance transcript that migrated at a size (∼4.1 kb) consistent with its being the unspliced precursor (labeled ‘U’) was also nuclear (band ‘U’ in Figure 3C). Only the partially spliced pre-mRNAs, not the unspliced pre-mRNA, were increased in level in response to the nonsense and frameshift mutations in the nucleus (Figure 3C), consistent with our results obtained with total cellular RNA (Figure 3B). In contrast, the RNA species predicted by size to be the TCR/TPI mature mRNA (∼1.6 kb) was primarily in the cytoplasm (the band labeled ‘M’ in Figure 3C), indicating that upon completion of splicing, the mature mRNA is efficiently exported to the cytoplasm. The mature mRNA was shorter in the cytoplasm than in the nucleus (Figure 3C), probably because of shortening of the poly(A) tail in the cytoplasm.
Together, our results indicated that TCR/TPI precursor transcripts rapidly splice IVS-C and -D and then they accumulate as partially spliced nuclear pre-mRNAs that slowly splice the remaining two introns: IVS-A and -B. This predicts that partially spliced pre-mRNAs harboring IVS-A and/or -B have a long half-life. Indeed, we found that pre-mRNA harboring either IVS-A or -B had a half-life of ∼90 min., as judged using either the transcriptional inhibitor actinomycin D (Supplementary Figure S3A) or a c-fos promoter-driven inducible system (Supplementary Figure S3B). In contrast, pre-mRNA harboring IVS-D had a shorter half-life of ∼30 min (Supplementary Figure S3C). Actin and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) pre-mRNAs had half-lives of <10 min (Supplementary Figure S3E). We conclude that IVS-A and -B are inefficiently spliced introns that are retained in nuclear pre-mRNA for remarkably long periods of time in vivo.
Frame-disrupting mutations upregulate the partially spliced pre-mRNA
The finding that three independent PTC-generating mutations upregulated the TCR/TPI pre-mRNA (Figure 1C) suggested the possibility that the signal responsible for this upregulation is disruption of reading frame. Indeed, our previous analysis of a limited number of nonsense, missense and frameshift mutations provided evidence that TCRβ pre-mRNA is upregulated specifically in response to frame disruption (23). However, an alternative explanation for mutations increasing the level of the TCR/TPI pre-mRNA is that these mutations inhibit pre-mRNA splicing by virtue of their ability to disrupt ESEs. While we are not aware of studies showing that nonsense mutations increase pre-mRNA levels as a result of splicing inhibition, several studies have demonstrated that nonsense mutations disrupting ESEs crucial for normal splicing can trigger the use of alternative splice sites (‘Discussion’ section).
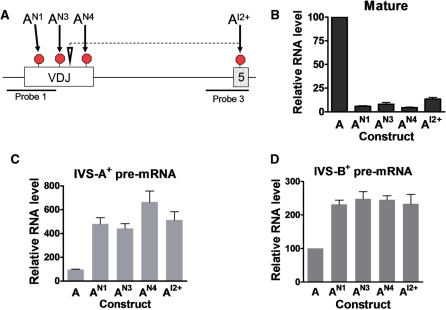
As a first step to distinguish whether the TCR/TPI pre-mRNA is upregulated as a result of reading-frame or ESE disruption, we introduced nonsense and frameshift mutations at several positions in the VDJ exon (Figure 4A). If NMUP is induced by reading-frame disruption, this predicts that all these mutations would elicit pre-mRNA upregulation. Alternatively, if NMUP is induced by ESE disruption, this predicts that only a subset of the mutations would elicit pre-mRNA upregulation, as past studies have shown that ESEs are only at specific sites in exons (‘Discussion’ section). We found that each of the nonsense and frameshift mutations decreased the level of mature mRNA, demonstrating that NMD was triggered (Figure 4B; compare constructs AN1, AN3, AN4 and AI2+ with construct A). Each of the mutations also upregulated the pre-mRNA, indicating that NMUP was also elicited (Figure 4C and D). Together with the data described earlier showing that nonsense and frameshift mutations at other positions also increased pre-mRNA level (Figure 1), this leads to the conclusion that all PTC-generating mutations (four of four nonsense mutations and two of two frameshift mutations) triggered upregulation of the pre-mRNA. These data supported the notion that NMUP is elicited by disruption of reading frame.
Figure 4.
Nonsense and frameshift mutations in the VDJ exon trigger pre-mRNA upregulation. (A) Schematic diagram indicating the location of the nonsense and frameshift mutations introduced into construct A. Constructs AN1, AN3 and AN4 have nonsense mutations at codons 91, 112 and 146, respectively. Construct AI2+ has a 1-nt insertion at codon 114 (indicated by an inverted triangle) that generates the downstream PTC shown. (B–D) Quantification of RNase protection analysis performed on total cellular RNA (10 µg) harvested from HeLa cells transiently transfected with the constructs shown. Probes 1 and 3 (A) were used to detect mature/IVS-A+ pre-mRNA and IVS-B+ pre-mRNA, respectively. Values were quantified from three or more independent experiments by the approach described in Figure 1. Error bars indicate standard error.
Frame-independent NMUP
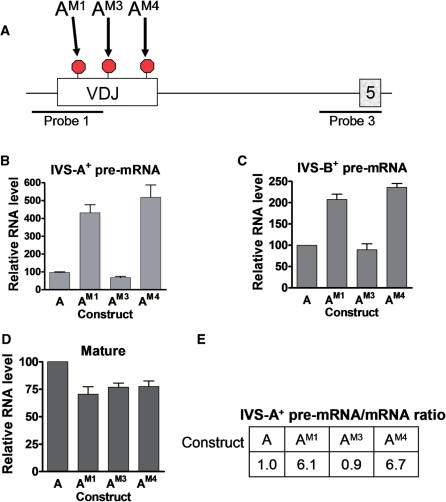
While the data described above supported the frame-disruption model, they did not rule out the ESE-disruption model. In other words, it remained possible that that the VDJ exon is particularly rich in ESEs and, thus, all six of the nonsense and frameshift mutations that we introduced in this exon disrupted ESEs. To test this possibility, we introduced three missense mutations in the VDJ exon (Figure 5A). The premise behind testing missense mutations is that they have the potential to disrupt ESEs but, by definition, they cannot disrupt reading frame. Analysis of these three missense mutations demonstrated that two of them upregulated the pre-mRNA (Figure 5A–C; constructs AM1 and AM4). This result clearly indicated that upregulation of the pre-mRNA does not result only from reading-frame disruption.
Figure 5.
Missense mutations in the VDJ exon trigger pre-mRNA upregulation. (A) Schematic diagram indicating the location of missense mutations (AM1, AM3 and AM4) introduced at the same nucleotide positions as the nonsense mutations in AN1, AN3 and AN4, respectively (Figure 4A). (B–D) Quantification of RNase protection analysis performed as described in Figure 4. (E) Splicing rate as measured by pre-mRNA-to-mRNA ratio. The ratios are calculated from the values in (B) and (D); the ratio for construct A is arbitrarily set to 1.
The two missense mutations that increased the level of pre-mRNA also modestly decreased the level of mature mRNA (Figure 5D), suggesting that they inhibited RNA splicing [note that the decrease was much less than that triggered by nonsense mutations at the same sites (Figure 4B), as expected since the latter, but not the former, trigger NMD]. To quantify the extent of inhibited spicing, we calculated the pre-mRNA/mature mRNA ratio (10). The ratios from each construct were compared against that of the wild-type construct (construct A), which was assigned a value of 1. Values >1 indicate a splicing defect. We found that the missense-containing constructs AM1 and AM4 had ratios of 6.1 and 6.7, respectively, indicating that splicing was strongly inhibited by these missense mutations (Figure 5E). In contrast, the missense construct AM3 had a pre-mRNA/mature-mRNA ratio of 0.9, suggesting that the mutation at this location did not significantly affect splicing efficiency and hence did not disrupt an ESE. These data clearly indicated that at least some mutations in the VDJ exon upregulate the chimeric pre-mRNA by a reading frame-independent mechanism.
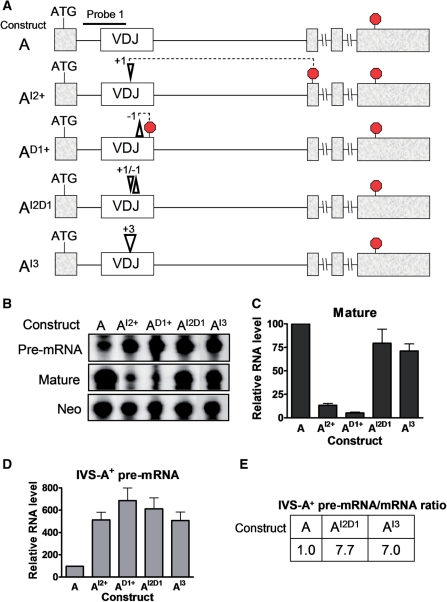
To further test whether frame disruption is responsible for eliciting NMUP, we introduced both frame-correcting and -neutral mutations. For the former, we introduced the following frameshifts into the TCR/TCPI construct: (i) a 1-nt insert (+1; construct AI2+), (ii) a 1-nt deletion (−1; construct AD1+),or (iii) both the +1 and −1 frameshifts (+1/−1; construct AI2D1) (Figure 6A). As a first test of these mutations, we examined their ability to elicit NMD. The +1 and −1 frameshifts both create PTCs and thus would be predicted to trigger NMD, while the +1/−1 double frameshift has restored reading frame and hence lacks a PTC and should not trigger NMD. Indeed, we found that both the +1 and −1 frameshifts downregulated the mature mRNA, while the +1/−1 double frameshift did not (compare constructs AI2+ and AD1+ with AI2D1 in Figure 6B and C).
Figure 6.
Both frame-disrupting and frame-neutral insertions and deletions elicit pre-mRNA upregulation. (A) Schematic diagrams of construct A variants harboring either a 1-nt insertion at codon 114 (+1, construct AI2+), a 1-nt deletion at codon 146 (−1, construct AD1+), both the 1-nt insertion and deletions (+1/−1, construct AI2D1) or a 3-nt insertion (+3, construct AI3). The location of the downstream PTC generated by the +1 and −1 mutations are shown. (B) RNase protection analysis performed using probe 1 (A) and a Neo probe (Figure 1) on total cellular RNA (10 µg) harvested from HeLa cells transiently transfected with constructs shown. (C, D) Quantification of mature mRNA (C) and pre-mRNA (D) levels from cells analyzed as in (B). Values are the average of three experiments, determined as described in Figure 1. Error bars indicate standard error. (E) Splicing rate as measured by pre-mRNA-to-mRNA ratio, determined as in Figure 5E, using the values in (C) and (D).
We next examined the ability of these frameshifts to trigger NMUP. If reading-frame disruption is responsible for triggering NMUP, both the +1 and −1 frameshifts should upregulate the pre-mRNA, while the +1/−1 double frameshift should not. We found that all three frameshifts upregulated the pre-mRNA, including the double frameshift restoring reading frame (constructs AI2+, AD1+, and AI2D1 in Figure 6B and D). The +1/−1 construct had a very high pre-mRNA/mature-mRNA ratio (construct AI2D1 in Figure 6E), indicative of strongly inhibited splicing. This did not support the reading frame-disruption model and instead supported the ESE-disruption model. It also provided more evidence that the VDJ exon is extremely rich in ESEs, as the location of the +1 and −1 frameshifts that we introduced were different than that of the mutations described above that upregulated the pre-mRNA (Figures 1 and 4).
As another test of reading-frame dependence, we introduced a 3-nt insert into the VDJ exon. This +3 (‘frame-neutral’) insert maintains the reading frame and thus, as expected, it did not trigger NMD (mature mRNA from construct AI3 in Figure 6B and C). In marked contrast, it strongly upregulated the pre-mRNA (Figure 6B and D), leading to a greatly elevated pre-mRNA/mature-mRNA ratio (Figure 6E). Thus, as with the data from the frame-restoring mutant, the data from the frame-neutral mutant supported the ESE-disruption model.
NMUP is independent of protein synthesis and UPF1
While the results described above clearly demonstrated that pre-mRNA can be upregulated by mutations that do not disrupt reading frame, they did not rule out that pre-mRNA can also be upregulated by a mechanism that detects frame disruption. Indeed, our finding that eight of eight frame-disrupting mutations elicited pre-mRNA upregulation (Figures 1, 4 and 6) provided correlative support for the notion that frame disruption is a signal that elicits NMUP. Given that the only sensor known to detect reading frame is the translation apparatus, this predicts that a frame-dependent NMUP mechanism should require translation. In support of this, other frame-dependent mechanisms, including NMD and frame-dependent NAS, have been shown to require translation (6,9,14,15,32,33).
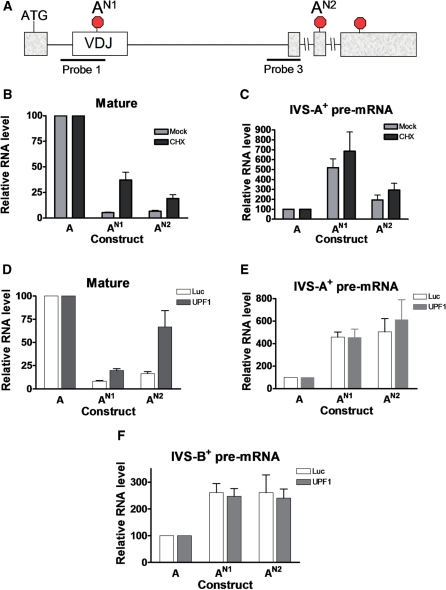
As one means to assess whether NMUP depends on translation, we determined whether the protein synthesis inhibitor CHX reversed the upregulation of pre-mRNA in response to the nonsense mutations. As a positive control, we examined the effect of CHX on NMD, as we have previously shown that CHX is a potent NMD inhibitor (6,32). As expected, CHX increased the level of mature mRNA harboring either of two independent nonsense mutations in the TCR/TPI construct (AN1 and AN2 in Figure 7A and B), indicating that CHX reversed NMD. In contrast, CHX had no significant effect on the level of pre-mRNA harboring these same two nonsense mutations (Figure 7C). This result provided evidence that the NMUP response elicited by these nonsense mutations does not require protein synthesis.
Figure 7.
Evidence that the pre-mRNA upregulatory response is independent of protein synthesis and UPF1. (A) Schematic diagram denoting the locations of the nonsense mutations in constructs AN1 and AN2 (also in Figure 1). (B, C) Quantification of RNase protection analysis performed on total cellular RNA (10 µg) harvested from HeLa cells incubated for 6 h with cycloheximide (CHX). Prior to CHX treatment, the cells were transiently transfected with the constructs shown, as well as a β-globin expression vector as an internal control, and cultured for 2 days. Probe 1 (A) was used to detect both the pre-mRNA and mature mRNA. The values shown are the average of two independent experiments that were normalized with β-globin mRNA, the internal control. Error bars indicate standard error. (D–F) Quantification of RNase protection analysis performed on total cellular RNA (10 µg) harvested from HeLa cells transiently transfected with a UFP1 siRNA to deplete UPF1 levels or a Luciferase (Luc) siRNA as a negative control. Probes 1 and 3 (A) were used to detect mature/IVS-A+ pre-mRNA and IVS-B+ pre-mRNA, respectively. Values were quantified from three independent experiments by the approach described in Figure 1.
As a second approach to assess whether NMUP depends on translation, we determined whether suppressor tRNAs reversed the upregulation of the pre-mRNA. Suppressor tRNAs recognize specific stop codons, but rather than terminating translation, they cause the incorporation of an amino acid, and thus they suppress events triggered by translation termination, including TCRβ NMD and NAS (15,34). To examine the effect of suppressor tRNAs on NMUP, we co-transfected suppressor tRNA expression constructs with TCR/TPI constructs harboring either a UAA or UGA premature stop codon or no premature stop codon (constructs AN1, AN2 and A, respectively, in Supplementary Figure S4A). As a positive control, we examined the effect of the suppressor tRNAs on NMD. We found that the UAA-specific suppressor tRNA inhibited NMD elicited by the UAA stop codon (construct AN1 in Supplementary Figure S4B). In contrast, the UAG- and UGA-specific suppressor tRNAs did not significantly affect NMD elicited by the UAA stop codon (Supplementary Figure S4B). Likewise, the NMD response of a construct with a UGA stop codon was specifically affected by the UGA-specific suppressor tRNA, but not the other suppressor tRNAs (construct AN2; Supplementary Figure S4B). In contrast to the reversal of NMD, the NMUP response was not reversed by the suppressor tRNAs (Supplementary Figure S4C and D). In fact, two of the suppressor tRNAs appeared to further increase the level of pre-mRNA harboring IVS-B (Supplementary Figure S4D). These data provided more evidence that NMUP is not a response elicited by PTC recognition during translation.
Finally, we examined whether NMUP depends on the RNA helicase UPF1. While not directly involved in frame recognition, UPF1 is recruited by release factors to the site of translation termination and is found at higher levels in NMD target mRNAs than other mRNAs and hence UPF1 is likely to be involved in premature translation termination (1–3). Indeed, UPF1 has been shown to be essential for both NMD and frame-dependent NAS (13,15,16). To determine the role of UPF1 in NMUP, we depleted UPF1 using RNAi. Using the UPF1-specific siRNA described in Mendell et al. (13), we were able to achieve over 80% UPF1 protein depletion (data not shown). This depletion of UPF1 was sufficient to reverse NMD, as shown by the upregulation of mature mRNA harboring nonsense mutations (Figure 7D). In contrast, we found that UPF1 depletion had no significant effect on the level of pre-mRNA harboring a nonsense mutation at either of two different positions, regardless of whether total cellar RNA (Figure 7E and F) or nuclear RNA (data not shown) was examined. We conclude that the upregulation of pre-mRNA in response to the two nonsense mutations that we tested does not depend on either UPF1 or translation. This strongly suggests that NMUP is not elicited as a result of frame disruption, at least not for the two nonsense mutations that we examined. Instead, our results are most simply explained by these nonsense mutations disrupting ESEs, which leads to splicing inhibition and hence the accumulation of partially spliced pre-mRNA.
NMUP is selectively elicited in response to mutations in the VDJ exon
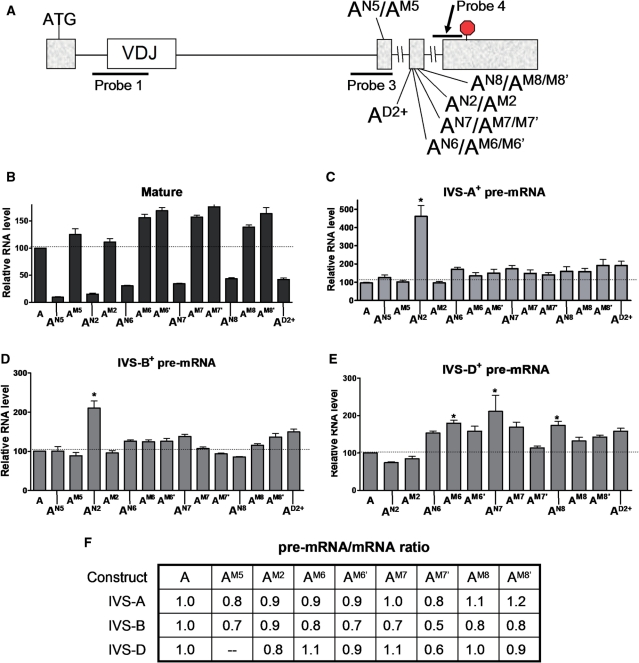
The finding that almost all the mutations (11 of 12) that we introduced into the VDJ exon, regardless of position, elicited upregulation of pre-mRNA harboring IVS-A and -B (Figures 1, 4, 5 and 6), suggested that the VDJ exon is remarkably ESE rich. To determine whether this is a property unique to the VDJ exon, we examined the effect of mutations in other exons. We first examined exon 5, as it is directly adjacent to an intron affected by VDJ mutations: IVS-B. We introduced a nonsense mutation in exon 5 (construct AN5 in Figure 8A) and found that while it strongly elicited NMD (Figure 8B), but it did not significantly affect the level of pre-mRNA (Figure 8C and D). A missense mutation at the same position in exon 5 (construct AM5 in Figure 8A) also did not have a significant effect on pre-mRNA level (Figure 8C and D), nor did it significantly increase pre-mRNA/mRNA ratio (Figure 8F), indicating that it did not impair RNA splicing.
Figure 8.
Nonsense mutations in exons 5 and 6 trigger strong NMD but modest or no pre-mRNA upregulation. (A) Schematic diagram indicating the location of the nonsense and missense mutations introduced into construct A. Constructs AN5/M5, AN6/M6/M6′, AN7/M7/M7′, AN8/M8/M8′ and AD2+ have nonsense (N) or missense (M or M′) mutations at codons 164, 191, 192, 195 and 190, respectively. M and M′ are distinct missense mutations. (B–E) Quantification of RNase protection analysis performed on total cellular RNA (10 µg) harvested from HeLa cells transiently transfected with the constructs shown. Probes 1, 3 and 4 (A) were used to detect mature/IVS-A+ pre-mRNA, IVS-B+ pre-mRNA and IVS-D+ pre-mRNA, respectively. Values were quantified from three or more independent experiments by the approach described in Figure 1. Error bars indicate standard error. (F) Splicing rate as measured by pre-mRNA-to-mRNA ratio, determined as in Figure 5E, using the values in (B–E).
Next, we examined the effect of mutations in exon 6. The introduction of nonsense mutations at various locations of exon 6 (AN2, AN6, AN7 and AN8 in Figure 8A) elicited NMD (Figure 8B), but only one of these nonsense mutations upregulated IVS-A+ or -B+ pre-mRNA (Figure 8C and D). The only nonsense mutation that triggered IVS-A+ or -B+ pre-mRNA upregulation was AN2, which is one of the three originals mutations in construct A that we tested (Figures 1 and 2). We also tested seven missense mutations in exon 6 (Figure 8A) and found that none of these significantly upregulated the IVS-A+ or -B+ pre-mRNAs (Figure 8C and D). Thus, only one of the 11 point mutations we introduced in exon 6 elicited a significant increase in the level of IVS-A+ or -B+ pre-mRNA. This clearly distinguished exon 6 from the VDJ exon and it suggested that exon 6 has few nonredundant ESEs. To determine the effect of exon 6 mutations on an intron directly adjacent to exon 6, we examined the levels of pre-mRNA harboring IVS-D. RNase protection analysis showed that three of the mutations modestly upregulated IVS-D+ pre-mRNA, whereas the other mutations had no significant effect (Figure 5E). As most of the exon-6 mutations had no effect, and those that did had only a modest effect, this confirmed that exon 6 is not ESE rich.
DISCUSSION
This article examines the relationship between frame-disrupting mutations and RNA splicing. Several groups have proposed that frame-disrupting mutations perturb or otherwise alter RNA splicing by triggering RNA surveillance mechanisms that scrutinize reading frame (13–16,18–20,22,25,27,35). Consistent with such a mechanism, we previously reported that TCRβ pre-mRNA level is selectively increased by mutations that disrupt reading frame (23). Here, we investigated the underlying mechanism for this NMUP response using a chimeric pre-mRNA that undergoes robust upregulation in response to nonsense and frameshift mutations in a TCRβ exon that naturally acquires such mutations during T-cell development. Surprisingly, we obtained several lines of evidence that the upregulatory response is not the result of recognition of disrupted reading frame. First, not only frame-disrupting mutations (nonsense and frameshift) but also missense mutations elicited pre-mRNA upregulation (Figure 5B and C). By contrast, these missense mutations did not elicit NMD, a reading frame-dependent event (Figure 5D). Second, restoration of reading frame of a frameshift mutant by introduction of a second frameshift did not restore normal levels of pre-mRNA (Figure 6B and D). Third, a suppressor tRNA specific for an introduced nonsense mutation did not dampen the pre-mRNA upregulatory response (Supplementary Figure S4). Fourth, inhibition of protein synthesis also did not dampen the pre-mRNA upregulatory response (Figure 7C). Finally, depletion of UPF1, a factor recruited upon translation termination and an essential NMD factor (1–3), also did not reverse the pre-mRNA upregulatory response (Figure 7E). As a control, we examined the effect of these perturbations on NMD. We found that NMD was inhibited or abolished by depletion of UPF1, protein synthesis inhibition, addition of a suppressor tRNA or restoration of reading frame (Figures 6C, 7B and 7D; Supplementary Figure S4). Together, these results distinguish NMUP from NMD and strongly suggest that nonsense and frameshift mutations can elicit pre-mRNA upregulation by a mechanism not involving recognition of a disrupted reading frame.
If it is not disrupted reading frame, then what is responsible for pre-mRNA upregulation in response to nonsense and frameshift mutations in the VDJ exon? We suggest the simplest explanation is that such mutations disrupt ESEs, leading to inhibited RNA splicing and accumulation of pre-mRNA. In support of this, ESEs have been shown to be crucial for the normal splicing of a wide variety of pre-mRNAs (36,37). However, to our knowledge, all ESE mutations that have been described elicit alternative splicing; e.g. they trigger exon skipping or a switch to alternative splice sites. We know of no study that found ESE mutations cause an accumulation of pre-mRNA, as in our study. What is responsible for this unique response? We suggest that ESE disruption causes pre-mRNA accumulation if three criteria are met: (i) the pre-mRNA has sequences that allow it to avoid rapid mRNA decay by nuclear RNA surveillance mechanisms that normally would degrade inefficiently processed mRNAs; (ii) the pre-mRNA avoids export from the nucleus; and (iii) the pre-mRNA’s architecture does not favor alternative splicing events. With regard to the first and second criteria, we found that the chimeric pre-mRNA is extremely long-lived compared to most pre-mRNAs (Supplementary Figure S3) and remains in the nucleus (Figure 3C). With regard to the third criterion, the chimeric transcripts may be less susceptible to alternative splicing than wild-type TCRβ transcripts because the alternative splice acceptor that we previously identified upstream of the VDJ exon (14) has been mutated in the TCR/TPI chimeric construct. While the alternative splice donor that we previously defined in the 5′-end of the VDJ exon (14) is retained in the TCR/TPI construct, we found that mutations in the VDJ exon do not increase its usage (data not shown).
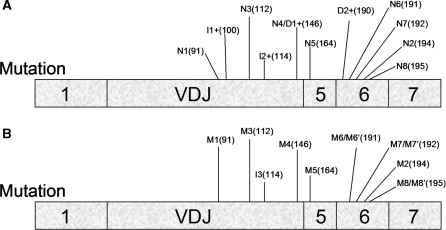
The ability of mutations distributed through much of the VDJ exon to trigger pre-mRNA upregulation strongly suggests that the VDJ exon is remarkably rich in ESEs. Indeed, we found that 11 of 12 mutations in this exon elicited pre-mRNA upregulation (Figure 9). As described above, many of these mutations did not disrupt reading frame (e.g. missense mutations, frame-restoration mutations and a frame-neutral 3-nt insertion), demonstrating that frame disruption is not responsible for pre-mRNA upregulation, at least in these cases. Other evidence that inhibited splicing is responsible for pre-mRNA upregulation was our finding that the pre-mRNA/mature mRNA ratio was dramatically increased by the missense and frame-restoration mutations that we introduced (Figures 5E and 6E). We cannot rule out that some of mutations do not disrupt classical ESEs, but instead perturb secondary structure crucial for efficient RNA splicing. However, we think this is probably not the mechanism of action of most of the mutations that we introduced, as most of them are point mutations that are unlikely to significantly disrupt secondary structure.
Figure 9.
Location of all mutations introduced into the TCR/TPI chimeric gene. (A) Nonsense and frameshift mutations. Figures 1, 4, 6 and 8 indicate their codon location. Note that the schematic diagram is of the mature mRNA and thus introns are not shown. (B) Missense and frame-neutral mutations. Figures 5, 6 and 8 indicate their codon location.
How frequent are ESEs in exons in other genes? A large number of ESEs—most of which are ∼6 nt in length—have been computationally defined and empirically tested by several different laboratories (38–40). While sequences conforming to these computationally defined ESE consensus sequences have been found in some vertebrate exons, their frequency in most exons is not known. This stems from the fact that an empirical analysis has only been conducted on a limited number of exons and most studies have not examined the frequency of ESEs in a systematic way to accurately determine ESE frequency. Instead, many ESE-rich exons have been discovered by virtue of their association with genetic diseases; e.g. HPRT exon 8 and TAU exon 10 undergo skipping as a result of naturally occurring mutations at various sites in these exons (41,42). An unbiased approach to determine ESE frequency was conducted by Chasin and Chen (43), who used mutagenesis followed by drug selection to identify cell mutants that skip DHFR exon 2. They were only able to recover one mutant with a mutation in this exon; all other mutations were in the splice sites and/or branch sites of the adjacent introns, suggesting that DHFR exon 2 is largely devoid of ESEs. In contrast, an unbiased analysis of CFTR exon 12 indicated that it is ESE rich. About one-quarter of the synonymous codon mutations introduced into this exon elicit exon 12 skipping in transfected cells (44).
Most pre-mRNAs are rapidly degraded if they are not efficiently spliced (45–47). Several factors, identified in Saccharomyces cerevisiae, have been found to mediate this decay, including the endonuclease Rnt1p, the nuclear exosome component Rrp6p, the nuclear Lsm complex and factors involved in transcription termination (46,48–50). Whether mammals have homologous proteins that mediate nuclear mRNA decay is poorly understood. In the future, it will be crucial to define the mammalian factors that mediate pre-mRNA decay and to elucidate how the chimeric pre-mRNA that we have defined in this paper largely avoids their action.
The retention of the partially spliced chimeric pre-mRNA in the nucleus is consistent with past studies showing that mRNAs harboring processing defects are retained in the nucleus. The factors that mediate nuclear RNA retention, similar to those that mediate nuclear RNA decay, have been defined primarily in S. cerevisiae. One retention factor is the perinuclear localized, myosin-like protein, Mlp1p (51). Other retention factors have been identified that act within the nucleoplasm; in some cases, they appear to elicit retention by promoting the interaction of pre-mRNA with spliceosomal components (48,52,53). Identification of the equivalent mammalian retention complexes would permit analysis of whether they mediate nuclear retention of the partially spliced chimeric pre-mRNA described in this article. We previously reported that TCRβ and Igµ pre-mRNAs upregulated by nonsense and frameshift mutations accumulate at or near the site of transcription (23).
We found that the introns directly adjacent to the VDJ exon splice very slowly. The half-life of pre-mRNA harboring IVS-A or -B was ∼90 min, indicating that these introns typically require an hour or more to splice in vivo (Supplementary Figure S3A–B). This is much longer than typical introns; previous studies have shown that mammalian introns splice in ∼1–10 min (54–56) consistent with what we observed for β-actin and GAPDH introns (Supplementary Figure S3C). It is possible that IVS-A and -B splice slowly simply because they are artificial introns containing both TCRβ and TPI sequences (Figure 1A). We believe this is unlikely, as normal TCRβ introns also splice inefficiently. We previously showed that partially spliced TCRβ transcripts accumulate to extremely high levels—easily detectable by northern blot analysis—in T-cell lines and normal thymocytes (30,57). Among the partially spliced TCRβ mRNAs that accumulate in T cells are 3′ splicing intermediates, which are indicative of a slow second step of splicing, as they result from 5′ splice-site cleavage (the first step of splicing) in the absence of 3′ splice-site cleavage (the second step of splicing) (30). IVS-A and -B in the TCR/TPI pre-mRNA may also undergo a slow second step of splicing, as we identified IVS-A- and -B-containing pre-mRNAs that migrated at a size consistent with being 3′ splicing intermediates (Figure 3B and Supplementary Figure S2).
Why is the VDJ exon ESE rich? One possibility is that a dense array of nonredundant ESEs is required for efficient inclusion of the large VDJ exon in mature mRNA. The length of the Vβ8.1Jβ2.3Cβ2 exon used in our study is 354 nt, which is at the upper limit for vertebrate exons (58). Further selection pressure for the VDJ exon being ESE rich may come from the fact that the downstream TCRβ exon (either the Cβ2.1 or Cβ2.2 exon, depending on the programmed rearrangement event) is also over 300 nt in length. The presence of two adjacent very large exons is likely to be an unfavorable configuration for splicing, given that splicing events are typically coupled (58). Another nonmutually exclusive explanation for why the VDJ exon is ESE rich is that the high density of ESEs promotes TCR pre-mRNA retention in the nucleus, a crucial property given that TCRβ pre-mRNA is highly susceptible to premature export to the cytoplasm by virtue of its slow splicing rate. Support for this possibility is a recent report demonstrating that ESEs mediate retention of pre-mRNAs in the nucleus, probably through the ability of ESEs to bind to nuclear-restricted SR proteins (59). Finally, we suggest that ESE richness could be a consequence of the unique way that VDJ exons are generated. Unlike most exons, which have a fixed sequence, VDJ exons are generated by programmed rearrangements that juxtapose V, D and J elements during lymphocyte development (5,7). While this diversity is crucial for generating a large reservoir of receptors that interact with foreign antigens, we suggest it necessitates selection for multiple strong ESEs to permit efficient VDJ exon inclusion in the face of adverse splicing cis elements (e.g. exonic splicing silencers) that will sometimes be created at the junctions of V, D and J elements.
While our paper provides several lines of evidence that ESE disruption triggers an increase in the steady-state level of pre-mRNA, this does not rule out that nonsense and frameshift mutations may, in some instances, elicit pre-mRNA upregulation as a result of reading frame disruption. Muhlemann et al. (23) provided evidence for frame-dependent pre-mRNA accumulation using two different immune-system genes that undergo rearrangement: endogenously expressed Igμ transcripts in mouse plasma cells and TCRβ transcripts derived from TCRβ transgenes stably transfected in HeLa cells. Frame-disrupting mutations elicited TCRβ and Igµ pre-mRNA upregulation even when the mutations were quite distant from the introns present in the pre-mRNA that accumulates. This is unlikely to be caused by ESE disruption, as the effectiveness of splicing enhancers tends to decrease as their distance from a given intron increases (60). In further support of this, we found that a nonsense mutation in exon 6 of the chimeric TCRβ/TPI transcript (AN2) elicited strong upregulation of pre-mRNA retaining introns far upstream of exon 6 (IVS-A and -B) but not an immediately adjacent intron (IVS-D) (Figures 1, 2 and 8). A missense mutation at the same codon (AM2) did not elicit this response (Figure 8). While this suggests the existence of a frame-dependent mechanism, we found that nonsense mutations at other positions in exon 6 did not significantly upregulate pre-mRNA retaining the upstream introns (Figure 8). It remains for future investigations to determine whether such divergent effects reflect the existence of frame-dependent and -independent mechanisms that compete and/or mask each other.
How could disruption of reading frame elicit upregulation of pre-mRNA? Models to explain this can be divided into two basic types (61,62). One model posits that it is triggered by recognition of a PTC by the cytoplasmic translation apparatus, which elicits a signal that travels to the nucleus and inhibits RNA splicing in a transcript-specific manner. The other model posits that frame-dependent NMUP is triggered by a nuclear RNA surveillance pathway that detects frame disruption. The cytoplasmic recognition model has the advantage that stop codon recognition occurs by a well-established mechanism in the cytoplasm (translation), whereas the nuclear recognition model necessarily involves a controversial frame-reading nuclear scanning mechanism that has not been proven (61,62). A disadvantage of the cytoplasmic recognition model is that it requires an unprecedented signal transduction mechanism initiated in the cytoplasm that leads to transcript-specific splicing inhibition in the nucleus, whereas the nuclear recognition model posits that stop codon recognition and splicing inhibition both occur in the same compartment—the nucleus—thereby simplifying the mechanism by which it would occur.
We suggest that the chimeric gene that we described in this report has several unique characteristics that make it a useful model system to identify and characterize splicing elements. In part, this stems from its ability to give rise to a partially spliced pre-mRNA that remains stable in the nucleus for long time periods, despite being spliced very slowly. As the pre-mRNA is extremely stable, its accumulation (and its level relative to mature mRNA) allows accurate quantification of splicing rate. By introducing test sequences of interest into this chimeric gene, cis elements and trans-acting factors can be identified that promote exon inclusion and control RNA splicing efficiency in vivo.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The National Institutes of Health (GM586595 to M.F.W.). Funding for open access charge: GM586595.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Yao-fu ‘Joseph’ Chang for his helpful comments in writing this manuscript.
REFERENCES
- 1.Stalder L, Muhlemann O. The meaning of nonsense. Trends Cell Biol. 2008;18:315–321. doi: 10.1016/j.tcb.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 2.Chang YF, Imam JS, Wilkinson MF. The nonsense-mediated decay RNA surveillance pathway. Annu. Rev. Biochem. 2007;76:51–74. doi: 10.1146/annurev.biochem.76.050106.093909. [DOI] [PubMed] [Google Scholar]
- 3.Isken O, Maquat LE. The multiple lives of NMD factors: balancing roles in gene and genome regulation. Nat. Rev. Genet. 2008;9:699–712. doi: 10.1038/nrg2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chan WK, Huang L, Gudikote JP, Chang YF, Imam JS, MacLean JA, II, Wilkinson MF. An alternative branch of the nonsense-mediated decay pathway. EMBO J. 2007;26:1820–1830. doi: 10.1038/sj.emboj.7601628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bruce SR, Wilkinson MF. Nonsense-Mediated Decay: A Surveillance Pathway that Detects Faulty TCR and BCR Transcripts. Trivandrum, India: Research Signpost; 2003. [Google Scholar]
- 6.Carter MS, Doskow J, Morris P, Li S, Nhim RP, Sandstedt S, Wilkinson MF. A regulatory mechanism that detects premature nonsense codons in T-cell receptor transcripts in vivo is reversed by protein synthesis inhibitors in vitro. J. Biol. Chem. 1995;270:28995–29003. doi: 10.1074/jbc.270.48.28995. [DOI] [PubMed] [Google Scholar]
- 7.Li S, Wilkinson MF. Nonsense surveillance in lymphocytes? Immunity. 1998;8:135–141. doi: 10.1016/s1074-7613(00)80466-5. [DOI] [PubMed] [Google Scholar]
- 8.Gudikote JP, Wilkinson MF. T-cell receptor sequences that elicit strong down-regulation of premature termination codon-bearing transcripts. EMBO J. 2002;21:125–134. doi: 10.1093/emboj/21.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang J, Vock VM, Li S, Olivas OR, Wilkinson MF. A quality control pathway that down-regulates aberrant T-cell receptor (TCR) transcripts by a mechanism requiring UPF2 and translation. J. Biol. Chem. 2002;277:18489–18493. doi: 10.1074/jbc.M111781200. [DOI] [PubMed] [Google Scholar]
- 10.Gudikote JP, Imam JS, Garcia RF, Wilkinson MF. RNA splicing promotes translation and RNA surveillance. Nat. Struct. Mol. Biol. 2005;12:801–809. doi: 10.1038/nsmb980. [DOI] [PubMed] [Google Scholar]
- 11.Bhalla AD, Gudikote JP, Wang J, Chan WK, Chang YF, Olivas OR, Wilkinson MF. Nonsense codons trigger an RNA partitioning shift. J. Biol. Chem. 2009;284:4062–4072. doi: 10.1074/jbc.M805193200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weischenfeldt J, Damgaard I, Bryder D, Theilgaard-Monch K, Thoren LA, Nielsen FC, Jacobsen SE, Nerlov C, Porse BT. NMD is essential for hematopoietic stem and progenitor cells and for eliminating by-products of programmed DNA rearrangements. Genes Dev. 2008;22:1381–1396. doi: 10.1101/gad.468808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mendell JT, ap Rhys CM, Dietz HC. Separable roles for rent1/hUpf1 in altered splicing and decay of nonsense transcripts. Science. 2002;298:419–422. doi: 10.1126/science.1074428. [DOI] [PubMed] [Google Scholar]
- 14.Wang J, Chang YF, Hamilton JI, Wilkinson MF. Nonsense-associated altered splicing: a frame-dependent response distinct from nonsense-mediated decay. Mol. Cell. 2002;10:951–957. doi: 10.1016/s1097-2765(02)00635-4. [DOI] [PubMed] [Google Scholar]
- 15.Wang J, Hamilton JI, Carter MS, Li S, Wilkinson MF. Alternatively spliced TCR mRNA induced by disruption of reading frame. Science. 2002;297:108–110. doi: 10.1126/science.1069757. [DOI] [PubMed] [Google Scholar]
- 16.Chang YF, Chan WK, Imam JS, Wilkinson MF. Alternatively spliced T-cell receptor transcripts are up-regulated in response to disruption of either splicing elements or reading frame. J. Biol. Chem. 2007;282:29738–29747. doi: 10.1074/jbc.M704372200. [DOI] [PubMed] [Google Scholar]
- 17.Caputi M, Kendzior RJ, Jr, Beemon KL. A nonsense mutation in the fibrillin-1 gene of a Marfan syndrome patient induces NMD and disrupts an exonic splicing enhancer. Genes Dev. 2002;16:1754–1759. doi: 10.1101/gad.997502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dietz HC, Kendzior RJ., Jr Maintenance of an open reading frame as an additional level of scrutiny during splice site selection. Nat. Genet. 1994;8:183–188. doi: 10.1038/ng1094-183. [DOI] [PubMed] [Google Scholar]
- 19.Gersappe A, Burger L, Pintel DJ. A premature termination codon in either exon of minute virus of mice P4 promoter-generated pre-mRNA can inhibit nuclear splicing of the intervening intron in an open reading frame-dependent manner. J. Biol. Chem. 1999;274:22452–22458. doi: 10.1074/jbc.274.32.22452. [DOI] [PubMed] [Google Scholar]
- 20.Gersappe A, Pintel DJ. A premature termination codon interferes with the nuclear function of an exon splicing enhancer in an open reading frame-dependent manner. Mol. Cell. Biol. 1999;19:1640–1650. doi: 10.1128/mcb.19.3.1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu HX, Cartegni L, Zhang MQ, Krainer AR. A mechanism for exon skipping caused by nonsense or missense mutations in BRCA1 and other genes. Nat. Genet. 2001;27:55–58. doi: 10.1038/83762. [DOI] [PubMed] [Google Scholar]
- 22.Maquat LE. NASty effects on fibrillin pre-mRNA splicing: another case of ESE does it, but proposals for translation-dependent splice site choice live on. Genes Dev. 2002;16:1743–1753. doi: 10.1101/gad.1014502. [DOI] [PubMed] [Google Scholar]
- 23.Muhlemann O, Mock-Casagrande CS, Wang J, Li S, Custodio N, Carmo-Fonseca M, Wilkinson MF, Moore MJ. Precursor RNAs harboring nonsense codons accumulate near the site of transcription. Mol. Cell. 2001;8:33–43. doi: 10.1016/s1097-2765(01)00288-x. [DOI] [PubMed] [Google Scholar]
- 24.Lytle JR, Steitz JA. Premature termination codons do not affect the rate of splicing of neighboring introns. RNA. 2004;10:657–668. doi: 10.1261/rna.5241404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aoufouchi S, Yelamos J, Milstein C. Nonsense mutations inhibit RNA splicing in a cell-free system: recognition of mutant codon is independent of protein synthesis. Cell. 1996;85:415–422. doi: 10.1016/s0092-8674(00)81119-8. [DOI] [PubMed] [Google Scholar]
- 26.Lozano F, Maertzdorf B, Pannell R, Milstein C. Low cytoplasmic mRNA levels of immunoglobulin kappa light chain genes containing nonsense codons correlate with inefficient splicing. EMBO J. 1994;13:4617–4622. doi: 10.1002/j.1460-2075.1994.tb06783.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Naeger LK, Schoborg RV, Zhao Q, Tullis GE, Pintel DJ. Nonsense mutations inhibit splicing of MVM RNA in cis when they interrupt the reading frame of either exon of the final spliced product. Genes Dev. 1992;6:1107–1119. doi: 10.1101/gad.6.6.1107. [DOI] [PubMed] [Google Scholar]
- 28.Wang J, Gudikote JP, Olivas OR, Wilkinson MF. Boundary-independent polar nonsense-mediated decay. EMBO Rep. 2002;3:274–279. doi: 10.1093/embo-reports/kvf036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miyagishi M, Taira K. U6 promoter-driven siRNAs with four uridine 3′ overhangs efficiently suppress targeted gene expression in mammalian cells. Nat. Biotechnol. 2002;20:497–500. doi: 10.1038/nbt0502-497. [DOI] [PubMed] [Google Scholar]
- 30.Qian L, Theodor L, Carter M, Vu MN, Sasaki AW, Wilkinson MF. T cell receptor-beta mRNA splicing: regulation of unusual splicing intermediates. Mol. Cell. Biol. 1993;13:1686–1696. doi: 10.1128/mcb.13.3.1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cheng J, Maquat LE. Nonsense codons can reduce the abundance of nuclear mRNA without affecting the abundance of pre-mRNA or the half-life of cytoplasmic mRNA. Mol. Cell. Biol. 1993;13:1892–1902. doi: 10.1128/mcb.13.3.1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carter MS, Li S, Wilkinson MF. A splicing-dependent regulatory mechanism that detects translation signals. EMBO J. 1996;15:5965–5975. [PMC free article] [PubMed] [Google Scholar]
- 33.Maquat LE. When cells stop making sense: effects of nonsense codons on RNA metabolism in vertebrate cells. RNA. 1995;1:453–465. [PMC free article] [PubMed] [Google Scholar]
- 34.Li S, Leonard D, Wilkinson MF. T cell receptor (TCR) mini-gene mRNA expression regulated by nonsense codons: a nuclear-associated translation-like mechanism. J. Exp. Med. 1997;185:985–992. doi: 10.1084/jem.185.6.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wachtel C, Li B, Sperling J, Sperling R. Stop codon-mediated suppression of splicing is a novel nuclear scanning mechanism not affected by elements of protein synthesis and NMD. RNA. 2004;10:1740–1750. doi: 10.1261/rna.7480804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zheng ZM. Regulation of alternative RNA splicing by exon definition and exon sequences in viral and mammalian gene expression. J. Biomed. Sci. 2004;11:278–294. doi: 10.1159/000077096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blencowe BJ. Exonic splicing enhancers: mechanism of action, diversity and role in human genetic diseases. Trends Biochem. Sci. 2000;25:106–110. doi: 10.1016/s0968-0004(00)01549-8. [DOI] [PubMed] [Google Scholar]
- 38.Zhang XH, Arias MA, Ke S, Chasin LA. Splicing of designer exons reveals unexpected complexity in pre-mRNA splicing. RNA. 2009;15:367–376. doi: 10.1261/rna.1498509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cartegni L, Wang J, Zhu Z, Zhang MQ, Krainer AR. ESEfinder: A web resource to identify exonic splicing enhancers. Nucleic Acids Res. 2003;31:3568–3571. doi: 10.1093/nar/gkg616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fairbrother WG, Yeh RF, Sharp PA, Burge CB. Predictive identification of exonic splicing enhancers in human genes. Science. 2002;297:1007–1013. doi: 10.1126/science.1073774. [DOI] [PubMed] [Google Scholar]
- 41.Steingrimsdottir H, Rowley G, Dorado G, Cole J, Lehmann AR. Mutations which alter splicing in the human hypoxanthine-guanine phosphoribosyltransferase gene. Nucleic Acids Res. 1992;20:1201–1208. doi: 10.1093/nar/20.6.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ballatore C, Lee VM, Trojanowski JQ. Tau-mediated neurodegeneration in Alzheimer's; disease and related disorders. Nat. Rev. Neurosci. 2007;8:663–672. doi: 10.1038/nrn2194. [DOI] [PubMed] [Google Scholar]
- 43.Chen IT, Chasin LA. Direct selection for mutations affecting specific splice sites in a hamster dihydrofolate reductase minigene. Mol. Cell. Biol. 1993;13:289–300. doi: 10.1128/mcb.13.1.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pagani F, Raponi M, Baralle FE. Synonymous mutations in CFTR exon 12 affect splicing and are not neutral in evolution. Proc. Natl Acad. Sci. USA. 2005;102:6368–6372. doi: 10.1073/pnas.0502288102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hilleren PJ, Parker R. Cytoplasmic degradation of splice-defective pre-mRNAs and intermediates. Mol. Cell. 2003;12:1453–1465. doi: 10.1016/s1097-2765(03)00488-x. [DOI] [PubMed] [Google Scholar]
- 46.Bousquet-Antonelli C, Presutti C, Tollervey D. Identification of a regulated pathway for nuclear pre-mRNA turnover. Cell. 2000;102:765–775. doi: 10.1016/s0092-8674(00)00065-9. [DOI] [PubMed] [Google Scholar]
- 47.Reed R, Hurt E. A conserved mRNA export machinery coupled to pre-mRNA splicing. Cell. 2002;108:523–531. doi: 10.1016/s0092-8674(02)00627-x. [DOI] [PubMed] [Google Scholar]
- 48.Das B, Das S, Sherman F. Mutant LYS2 mRNAs retained and degraded in the nucleus of Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA. 2006;103:10871–10876. doi: 10.1073/pnas.0604562103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Torchet C, Bousquet-Antonelli C, Milligan L, Thompson E, Kufel J, Tollervey D. Processing of 3′-extended read-through transcripts by the exosome can generate functional mRNAs. Mol. Cell. 2002;9:1285–1296. doi: 10.1016/s1097-2765(02)00544-0. [DOI] [PubMed] [Google Scholar]
- 50.Danin-Kreiselman M, Lee CY, Chanfreau G. RNAse III-mediated degradation of unspliced pre-mRNAs and lariat introns. Mol. Cell. 2003;11:1279–1289. doi: 10.1016/s1097-2765(03)00137-0. [DOI] [PubMed] [Google Scholar]
- 51.Galy V, Gadal O, Fromont-Racine M, Romano A, Jacquier A, Nehrbass U. Nuclear retention of unspliced mRNAs in yeast is mediated by perinuclear Mlp1. Cell. 2004;116:63–73. doi: 10.1016/s0092-8674(03)01026-2. [DOI] [PubMed] [Google Scholar]
- 52.Scherrer FW, Jr, Spingola M. A subset of Mer1p-dependent introns requires Bud13p for splicing activation and nuclear retention. RNA. 2006;12:1361–1372. doi: 10.1261/rna.2276806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Trowitzsch S, Weber G, Luhrmann R, Wahl MC. An unusual RNA recognition motif acts as a scaffold for multiple proteins in the pre-mRNA retention and splicing complex. J. Biol. Chem. 2008;283:32317–32327. doi: 10.1074/jbc.M804977200. [DOI] [PubMed] [Google Scholar]
- 54.Audibert A, Weil D, Dautry F. In vivo kinetics of mRNA splicing and transport in mammalian cells. Mol. Cell. Biol. 2002;22:6706–6718. doi: 10.1128/MCB.22.19.6706-6718.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Singh J, Padgett RA. Rates of in situ transcription and splicing in large human genes. Nat. Struct. Mol. Biol. 2009;11:1123–1124. doi: 10.1038/nsmb.1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kessler O, Jiang Y, Chasin LA. Order of intron removal during splicing of endogenous adenine phosphoribosyltransferase and dihydrofolate reductase pre-mRNA. Mol. Cell. Biol. 1993;13:6211–6222. doi: 10.1128/mcb.13.10.6211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Qian L, Vu MN, Carter MS, Doskow J, Wilkinson MF. T cell receptor-beta mRNA splicing during thymic maturation in vivo and in an inducible T cell clone in vitro. J. Immunol. 1993;151:6801–6814. [PubMed] [Google Scholar]
- 58.Berget SM. Exon recognition in vertebrate splicing. J. Biol. Chem. 1995;270:2411–2414. doi: 10.1074/jbc.270.6.2411. [DOI] [PubMed] [Google Scholar]
- 59.Taniguchi I, Masuyama K, Ohno M. Role of purine-rich exonic splicing enhancers in nuclear retention of pre-mRNAs. Proc. Natl Acad. Sci. USA. 2007;104:13684–13689. doi: 10.1073/pnas.0704922104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tian M, Maniatis T. A splicing enhancer exhibits both constitutive and regulated activities. Genes Dev. 1994;8:1703–1712. doi: 10.1101/gad.8.14.1703. [DOI] [PubMed] [Google Scholar]
- 61.Dahlberg JE, Lund E, Goodwin EB. Nuclear translation: what is the evidence? RNA. 2003;9:1–8. doi: 10.1261/rna.2121703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wilkinson MF, Shyu AB. RNA surveillance by nuclear scanning? Nat. Cell. Biol. 2002;4:E144–E147. doi: 10.1038/ncb0602-e144. [DOI] [PubMed] [Google Scholar]
- 63.Maclean JA, II, Chen MA, Wayne CM, Bruce SR, Rao M, Meistrich ML, Macleod C, Wilkinson MF. Rhox: a new homeobox gene cluster. Cell. 2005;120:369–382. doi: 10.1016/j.cell.2004.12.022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.