Abstract
The nucleobase modification 5-methylcytosine (m5C) is widespread both in DNA and different cellular RNAs. The functions and enzymatic mechanisms of DNA m5C-methylation were extensively studied during the last decades. However, the location, the mechanism of formation and the cellular function(s) of the same modified nucleobase in RNA still remain to be elucidated. The recent development of a bisulfite sequencing approach for efficient m5C localization in various RNA molecules puts ribo-m5C in a highly privileged position as one of the few RNA modifications whose detection is amenable to PCR-based amplification and sequencing methods. Additional progress in the field also includes the characterization of several specific RNA methyltransferase enzymes in various organisms, and the discovery of a new and unexpected link between DNA and RNA m5C-methylation. Numerous putative RNA:m5C-MTases have now been identified and are awaiting characterization, including the identification of their RNA substrates and their related cellular functions. In order to bring these recent exciting developments into perspective, this review provides an ordered overview of the detection methods for RNA methylation, of the biochemistry, enzymology and molecular biology of the corresponding modification enzymes, and discusses perspectives for the emerging biological functions of these enzymes.
INTRODUCTION
Nucleotide modifications, a ubiquitous feature of life, have first been described in 1948, when the occurrence of m5C was detected in DNA (1). Today, about half a dozen DNA modifications and more than 100 chemically distinct RNA nucleotide modifications, including m5C, are known to be enzymatically synthesized. While research into nucleotide modifications has been hampered by limited performance of analytical methods, the field has recently encountered renewed interest. This is related to the discovery of nucleotide modifications in regulatory RNAs including siRNA, piRNAs, miRNAs and precursors (2–7). Among the various modifications, a privileged position is held by m5C. This residue has been described in several cellular RNAs and a complex enzymatic machinery for its synthesis was found in representative organisms from all kingdoms of life. In addition, recent data strongly suggest its implication in the regulation of various biological processes.
METHODS FOR m5C DETECTION IN RNAs
Detection of nucleotide modifications is based on either of three basic principles: (i) separation according to physicochemical properties, (ii) differential enzymatic turnover and (iii) differential chemical reactivity.
Separation according to physico-chemical properties typically relies on chromatography, i.e. thin layer chromatography (TLC), high-performance liquid chromatography (HPLC), or mass spectrometry, and sometimes on a combination of both (LC-MS). Most widespread is the use of two-dimensional TLC. Because the migration characteristics of nearly 70 chemically distinct modifications have been mapped in three standard solvent systems, identification and quantification of 32P-labeled nucleotides down to the femtomolar level is straightforward (8). Although the use of in vitro substrates containing a single type of labeled nucleotide by nearest neighbor analysis permits the determination of the nucleotide 3′ to the modification, the obligatory nuclease degradation step before TLC analysis destroys most of the modification’s sequence context. For known modification sites, sequence specificity of the analysis can be reintroduced by the use of DNAzymes (9) or oligonucleotide-directed RNase H cleavage (10) prior to labeling and TLC analysis. It has also been shown that chemical conjugation of nucleotides to a fluorescent dyes and subsequent separation and detection by CE-LIF allows quantifications of m5C in DNA (11) in the femtomolar range. A similar approach has also been used for the detection of m5C in RNA (12).
Similar to TLC and CE techniques, a typical LC-MS analysis requires a complete nuclease digestion, which is followed by dephosphorylation prior to separation on a C18 reverse phase column. In addition to the retention times, determination of the nucleosides’ molecular mass, and/or their fragmentation pattern is used to confirm the chemical identity of a given modification. Applications of current LC/MS/MS techniques allow the detection of modified nucleosides even in the sub-picomolar range (13). Modifications can also be detected by LC-MS in oligonucleotides obtained, e.g. by digestion with RNase T1 (14), but sequence information is typically limited to ∼10 nucleotides near the modification site.
Since direct identification through physico-chemical properties prevents the use of PCR amplification it also prevents high throughput applications. While the ability of certain nucleotide modifications to impede enzymatic reactions such as ligations (15) or primer elongations (16,17) can be used for sequence-specific detection, this does not apply to m5C, which behaves normally in terms of Watson–Crick base pairing. However, a promising perspective is offered by differential reactivity of m5C towards chemical reagents, which allows the application of PCR-based amplification steps, or affinity-based enrichment of m5C-containing nucleic acids.
Distinct chemical reactivity of unmodified C and m5C
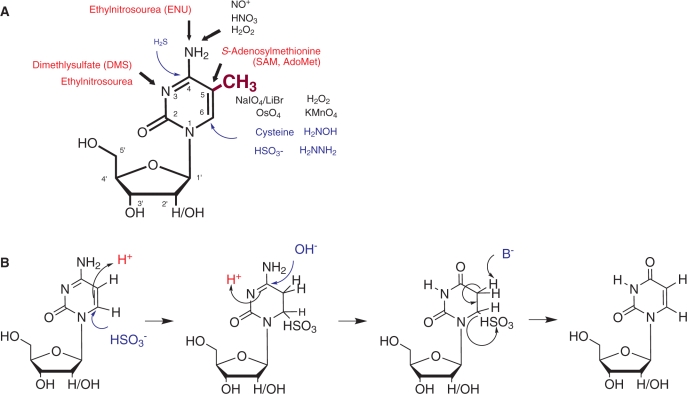
Figure 1A illustrates the relevant reactive sites investigated in 5-methyl and 2′-deoxy-derivatives of cytidine. Of foremost interest are the exocyclic N4-amino function and the C5–C6 double-bond, because reactivities of both sites are significantly affected by the introduction of a methyl group on C5. Methylation generally increases electron density in the aromatic ring by an inductive effect, which can be monitored by chemical shifts in 1H-NMR. Thus, C5-methylation of pyrimidines causes an upfield shift of ∼0.15 p.p.m. in the H6 of both uridine and cytidine (see the SDBSWeb: http://riodb01.ibase.aist.go.jp/sdbs/ (National Institute of Advanced Industrial Science and Technology, 14.7.2009)). This increase in electron density favors reaction types such as oxidation and electrophilic attacks, while disfavoring nucleophilic attacks. There is clearly a strong interest to exploit these features for the detection of methylated pyrimidines by a combination of chemical and biochemical techniques. Some of corresponding approaches will be outlined below.
Figure 1.
(A) Chemical reactivity of cytidine and m5C. The methyl group of m5C is shown in purple. Attack sites on the nucleobase are indicated by arrows, where black indicates oxidizing agents, red indicates alkylating electrophiles and blue arrows indicate nucleophiles. Numbering of the nucleobase atoms 1–6 is indicated inside the ring. Ribose carbon atom numbering is indicated by primed (′) numbers (B) Bisulfite mediated conversion of cytosine to uracil. Nucleophiles, bases and nucleophilic attacks are indicated in blue, acids are indicated in red.
Oxidation of the 5–6 double-bond in pyrimidines to the corresponding glycol has been reported to result from exposure to ionizing radiation or reagents such as KMnO4, OsO4, or Fenton reagents (18–22). The concomitant loss of aromaticity renders the nucleic acid chain susceptible to cleavage by aniline or piperidine in the case of RNA or DNA, respectively (23–25). These reactions are exploited for sequencing, structural probing, or footprinting applications of end-labeled nucleic acids (18,19). Although the permanganate reaction produces insoluble MnO2, and further oxidation of the glycol to the bis-aldehyde may occur under certain conditions, it has been successfully employed in the discrimination of deoxy-m5C versus deoxycytidine (26). The osmium tetroxide reactions do not involve the formation of insoluble products, but the reagent is highly toxic and mutagenic. In the past years, a variety of osmate-based bioconjugate reagents have been developed for the selective detection of m5C in DNA. These reagents exploit the additional electron density brought about by the methyl group for highly selective formation of a stable complex with deoxy-m5C but not with deoxycytidine. However, applications to RNA are still lacking (27).
Compared to cytidine, m5C is more prone to spontaneous deamination of the N4 than cytidine in vivo (28). This reaction is promoted by oxidizing conditions, and is thought to involve the formation of an electron-deficient species, such as NO+, which converts the N4-anilin function into a leaving group which is then hydrolyzed (29,30). Likewise, electrophilic alkylating agents preferentially attack the nitrogen nucleophilic sites. Both the N4- and the N3-aminofunctions are alkylated by highly reactive agents such as ethylnitrosourea (ENU) (31,32). The N3-methylation by dimethylsulfate (DMS) has gained importance in Maxam and Gilbert style sequencing techniques (24,25), as well as in structural probing and footprinting of RNA and DNA (33,34). In these biochemical applications, the N3-methylation renders the aromatic ring electron poor and thus susceptible to nucleophilic addition of hydrazine, which is destroying aromaticity prior to chain scission (24,25). Such nucleophilic additions are otherwise unusual in pyrimidines, and require comparatively more stringent conditions (24). Sodium bisulfite, also a nucleophilic reagent, causes the deamination of cytosine residues in a single-strand DNA through the initial formation a 5,6-dihydrocytosine-6-sulfonate intermediate at acidic pH (Figure 1B). This implies a protonation step at the 5-position in conjunction with a Michael addition of the nucleophile at the 6 position. Subsequent deamination leads to 5,6-dihydrouracil-6-sulfonate from which the 5–6 double-bond is restored by beta-elimination during the subsequent treatment under basic conditions. Because this reaction occurs with selectivity for cytosine over m5C, its application to the detection of m5C sites in DNA has gained outstanding importance in an approach called bisulfite sequencing (27).
Adaptation of DNA bisulfite methylation mapping to RNA
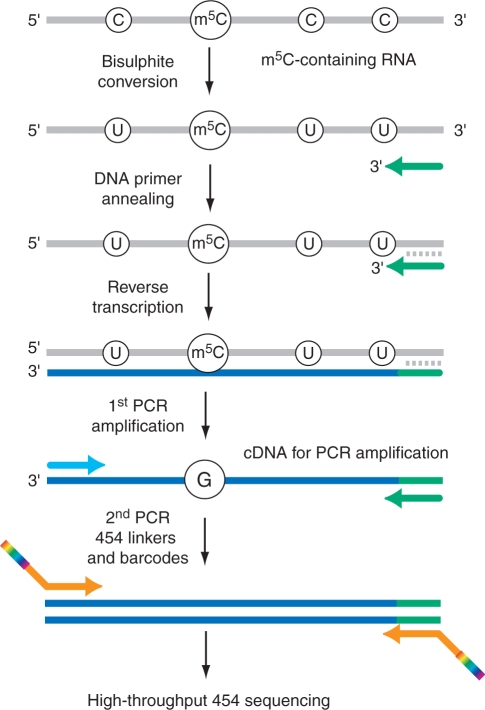
The principle of bisulfite sequencing is outlined in Figure 2. A critical parameter is the fraction of intact analyte nucleic acids surviving the sequential treatment under acidic bisulfite and alkaline conditions (Figure 1B). In these polymers, all cytidines are deaminated to uridines, and will thus behave like thymidines in polymerase-based sequencing or PCR-amplification reactions while m5C residues remain unchanged and behave like cytidines. Typically, the resulting fragments are PCR-amplified before cloning and/or sequencing. Sequence alignments are facilitated by sequencing of untreated samples. In alignments of multiple bisulfite sequence reads, significant methylation is indicated by multiple occurrences of cytidine signals at a given position. The sequencing depth, i.e. the number of individual sequence reads in the alignment, is thus an important quality parameter of the analysis.
Figure 2.
Principle of m5C detection by RNA bisulfite sequencing and combination with next-generation sequencing. The DNA primer sequence for reverse transcription is adapted for C to U conversions in the analyzed RNA.
DNA bisulfite sequencing (35,36) has become an indispensable tool for the mapping of m5C in DNA, and its continued development represents a key approach towards characterizing the human epigenome (37,38). An adaptation of the bisulfite treatment protocol to the requirements of RNA showed that individual m5C residues in tRNA could be mapped by primer extension (39). The same study also suggested that complex tRNA modification patterns could be analyzed by bisulfite treatment of RNA samples, followed by reverse transcription into cDNA, PCR amplification, molecular cloning and DNA sequencing of cloned PCR products. However, no experimental details were provided for this approach.
A stringent analysis of cytosine methylation patterns by bisulfite sequencing requires very high cytidine to uridine conversion rates of >95%. This is usually achieved only after prolonged (>1 h) incubation with sodium bisulfite, which promotes degradation of DNA and, particularly, RNA molecules and therefore reduces the efficacy of the ensuing PCR amplification step. Sustaining high deamination rates, and, at the same time, limiting the degradation of RNA molecules thus represented the major challenge for the establishment of an RNA bisulfite sequencing protocol. This problem has recently been solved by reducing the bisulfite reaction temperature from 95 to 60°C, which permitted an extension of the reaction time to 3 h and more, without any major reduction in the efficacy of the RT-PCR reaction (40). Under these conditions, deamination rates of 97% and more could be repeatedly achieved for single-stranded RNA sequences.
High-throughput techniques of m5C mapping and quantification in RNA
Initial tRNA bisulfite sequencing experiments indicated a substantial variation in the cytosine methylation patterns of tRNAs. The robust quantification of cytosine methylation levels at specific positions therefore requires a considerable sequencing depth of 100 (or more) sequence reads. This can be achieved by next-generation sequencing technologies, which permit the massively parallel sequencing of hundreds of thousands single DNA molecules. The sequencing depth provided by next-generation sequencing also permits the analysis of complex methylation patterns. Indeed, 454 sequencing has recently been used for the methylation analysis of various tRNA and rRNAs and has facilitated the quantitative analysis of RNA methylation patterns in these RNA molecules (40).
For 454 RNA bisulfite sequencing, PCR amplicons are generated from deaminated RNA following reverse transcription and then re-amplified with 454 primers (Figure 2). The latter primers contain linker sequences for the 454 sequencing technology and barcode sequences for the bioinformatical identification of individual amplicons. Amplicons are then mixed, sequenced and later subjected to bioinformatical analysis (40).
Importantly, the results obtained with these sequencing approaches can also be used to robustly quantify the methylation level of individual cytosine residues. It is conceivable that RNA methylation can be used as a signal for various cellular processes. In this context, the methyl group would be a dynamic mark that can be added or removed depending on intrinsic or extrinsic cues. Deep sequencing of bisulfite amplicons therefore provides an opportunity to use RNA methylation analysis as a quantifiable assay for various biological questions.
Lastly, deep sequencing of cDNA libraries also permits an additional quality control step that addresses the PCR bias problem associated with most bisulfite-based analytical tools. The biased PCR amplification of certain (c)DNA sequences represents a major problem in the quantitative analysis of bisulfite sequencing experiments, because it can result in a dramatic over- or under-representation of particular PCR products (41). A focused analysis of sequences with unique deamination artifacts or the introduction of random sequence permutations in an additional barcode patch in the reverse transcription primer allows a 1:1 representation of RNA molecules and sequence reads. These are important steps towards the further optimization of the method that will significantly broaden its application potential.
THE PRESENCE OF m5C IN VARIOUS CELLULAR RNAs
m5C is a prevalent modification of many cellular RNAs. Stable and highly abundant RNAs, like tRNAs and rRNAs were extensively analyzed in the past by various techniques. These studies allowed the precise mapping of m5C residues in tRNAs and also in ribosomal RNAs from all three living domains.
tRNAs
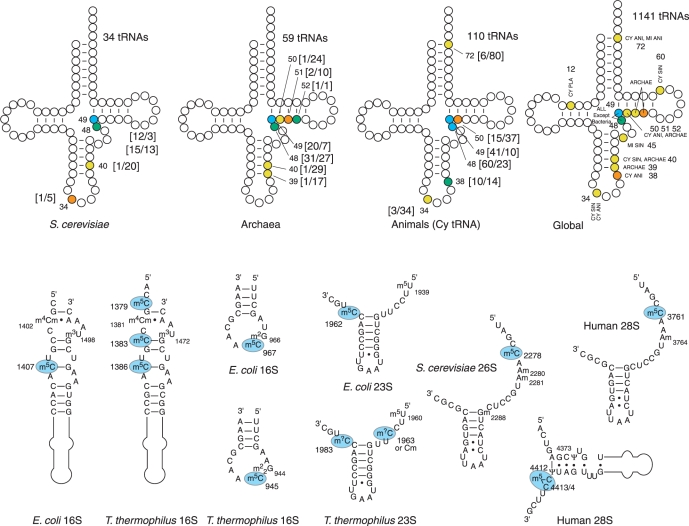
While tRNAs from eubacterial organisms do not contain m5C, this modification was detected in numerous archaeal and eukaryotic tRNAs. For the best studied eukaryotic tRNAs, m5C residues seem to be clustered in the junction between variable region and TΨC-stem, and positions 48 and 49 are most frequently modified. However, other locations in the anticodon loop were also identified. The same tendency exists also for archaeal tRNAs, where the positions 48/49 are most frequently methylated, but other methylation sites have also been reported. In contrast to other organisms, higher eukaryota frequently have an additional m5C residue in the tRNA acceptor stem, at position 72. The overall distribution of m5C-modified positions in yeast, archaeal and higher eukaryotic tRNAs is shown in Figure 3 (42).
Figure 3.
Occurrence of m5C residues in tRNAs and rRNAs. The frequency of m5C occurrence is color-coded in tRNA: yellow, <10%; orange, between 10 and 30%; green, between 30 and 60%; blue >60%. Numbers in square brackets show the number of m5C and number of unmodified or other modified C at a given position [m5C/C] The same color code is used for global distribution in all sequenced tRNAs, numbers are not shown for clarity. Abbreviations: CY, cytoplasm; MI, mitochondria; ANI, animals; SIN, single cell eukaryota; PLA, plants; ARCHAE, Archaea. The lower panel shows rRNA fragments from different species with m5C residues highlighted in blue.
Prokaryotic and eukaryotic rRNAs
The presence of m5C in ribosomal RNAs from different organisms was demonstrated by total digestion of highly purified rRNAs followed by RP-HPLC and MS analysis of the resulting nucleoside mixtures (43–46). Bacteria have a variable number of m5C residues; only three such modifications are present in Escherichia coli rRNAs, while rRNAs from Thermus thermophilus has five or six m5C residues (Figure 3B). Analysis of small and large subunit rRNAs from bacteria and eukaryota similarly demonstrated the presence of several m5C residues in these molecules (see The RNA Modification database http://library.med.utah.edu/RNAmods/). More recently, m5C was also detected in archaeal rRNAs (47). In addition, the locations of this modification in rRNA are rather well conserved from bacteria to humans.
Eukaryota have comparably fewer m5C residues in rRNA. Saccharomyces cerevisiae rRNA methylation has not been exhaustively analyzed, but one m5C residue was mapped in 25S rRNA from S. carlsbergensis (48). Similarly, human 28S rRNA contains only two m5C nucleotides (49) and no m5C was detected in several eukaryotic 18S rRNAs (50). Although very little is known about eukaryotic mitochondrial rRNA methylation, the presence of m5C was reported for 18S mt rRNA from the Syrian hamster Mesocricetus auratus (the SSU rRNA Modification Database, WEB http://134.58.19.20/ssu/ssu.htm).
Analysis of archaeal rRNAs confirmed the presence of m5C residues in SSU and LSU rRNAs, but demonstrated that the number of modifications present in archaeal rRNAs is highly variable. For example, no m5C was detected in Haloferax volcanii 16S rRNA, while its counterpart from Sulfolobus solfataricus has at least one residue in 16S rRNA and one or two m5C in 23S rRNA (47,51). For a comparative methylation analysis of selected prokaryotic and eukaryotic rRNAs see Ref. (52).
m5C in other cellular RNAs
Shortly after the discovery of nucleotide methylation in the cap-structure of eukaryotic mRNAs, numerous investigations of the presence of methylated nucleotides mRNAs and viral RNAs were reported. These were typically based on pre-labeling with 3H-methionine, which is converted to [3H]S-adenosyl-l-methionine (SAM) in living cells followed by incorporation of 3H–CH3 into RNA. Several studies reported the detection of m5C in mRNA from cultured BHK-21 hamster cells (53), in S26 sindbis virus RNA from infected hamster cells (54,55), and in adenovirus RNA from infected HeLa cells (56). However, no m5C was found in mRNA from HeLa cells (57,58), Novikoff hepatoma cells (59), mouse myeloma (60) and SV 40 viral RNA (61).
Certain findings suggest that classical tRNA modification enzymes may also methylate tRNA-like structures in other RNA molecules. Thus, RNA derived from tRNA-related short interspersed nuclear elements (SINEs) was found to be methylated to m5C in vitro (62), although a recent analysis failed to detect m5C methlyation of this RNA in vivo (63). Moreover, the tRNA-like 3′-end of the Turnip Yellow Mosaic Virus (TYMV) RNA was found to be methylated to m5C upon injection into Xenopus oocytes (64). It will be interesting to further investigate the possibility that other cellular RNAs might be methylated in order to fully understand the biological function of this modification (see below).
FAMILIES OF RNA: m5C-MTases
The known enzymes transferring methyl groups from SAM to nucleic acids belong to the SPOUT and MTase superfamilies, the latter containing a Rossmann fold for the accommodation of the cofactor (65). RNA:m5C-MTases represent a large protein family which can be subdivided into several distinct groups, on the basis of sequence similarity and RNA substrate specificity. These subgroups are presented in Table 1.
Table 1.
Known and some putative m5C-MTRs and their RNA substrates
| Enzyme family | Enzyme name | Other names | Organism | Life domain | Accession | Identification type | RNA substrate | Modified position(s) | X-ray structure | Reference |
|---|---|---|---|---|---|---|---|---|---|---|
| RsmB family | ||||||||||
| RsmB/Nol1 | RsmB | Fmu/Fmv | Escherichia coli | B | AP_004502 | Biochemical | 16S rRNA | 967 | 1SQF, 1SQG | (68,70) |
| RsmB/Nol1 | P120 | NSUN1 | Homo sapiens | E | P46087 | NO | Unknown | NA | (92) | |
| RsmB/Nol1 | Nop2 | Saccharomyces cerevisiae | E | YNL061W | NO | Unknown | NA | (87) | ||
| RsmF/YebU family | ||||||||||
| YebU | Haloferax volcanii | A | HVO_1594 | Bioinformatics | tRNA | 39/40/48/49 | (75) | |||
| YebU | Pyrococcus abyssi | A | PAB1947 | Biochemical | tRNA | Multiple (mostly 49) | (79) | |||
| YebU | aTrm4 | Pyrococcus horikoshi | A | PH1374 | Bioinformatics | Unknown | NA | 1IXK | (78) | |
| YebU | RsmF | YebU | Escherichia coli | B | P76273 | Biochemical | 16S rRNA | 1407 | 2FRX | (71,72) |
| YebU | hTrm4 | NSUN2/Misu | Homo sapiens | E | NM_017755 | Biochemical | pre-tRNALeu | 34 | (86) | |
| YebU | FLJ22609 | NSUN3 | Homo sapiens | E | Q9H649 | NO | Unknown | NA | (65) | |
| YebU | MGC22920 | NSUN4 | Homo sapiens | E | Q96CB9 | NO | Unknown | NA | (65) | |
| YebU | Trm4 | Ncl1 | Saccharomyces cerevisiae | E | YBL024W | Biochemical | tRNA and pre-tRNA | 48/49 tRNA 34/40 pre-tRNA | (80,81) | |
| Dnmt2 family | ||||||||||
| DNMT2 | Dnmt2 | Drosophila melanogaster | E | Q9U6H7 | Biochemical | tRNA | 38 | (102,107) | ||
| Dnmt2 | trdmt1 | Drosophila rerio | E | Q588C1 | Biochemical | tRNA | 38? | (122) | ||
| DNMT2 | Dnmt2 | Homo sapiens | E | O14717 | Biochemical | tRNA | 38 | 1G55 | (100,102) | |
| DNMT2 | Dnmt2 | Mus musculus | E | O55055 | Bioinformatics | |||||
| DNMT3 | pmt1 | pmt1 | Saccharomyces pombe | E | P40999 | Bioinformatics | ||||
| RlmI family | ||||||||||
| COG1092 | RlmI | YccW | Escherichia coli | B | P75876 | Biochemical | 23S rRNA | 1962 | 3C0K | (73,74) |
| Ynl022 family | ||||||||||
| Ynl022c | WBSCR20A | NSUN5A | Homo sapiens | E | Q96P11 | NO | Unknown | NA | 2B9E | |
| Ynl022c | WBSCR20B | NSUN5B | Homo sapiens | E | Q3KNT7 | NO | Unknown | NA | ||
| Ynl022c | WBSCR20C | NSUN5C | Homo sapiens | E | Q63ZY6 | NO | Unknown | NA | ||
| Ynl022c | WBSCR22 | Homo sapiens | E | O43709 | NO | Unknown | NA | |||
| Ynl022c | NSUN7 | Homo sapiens | E | Q8NE18 | NO | Unknown | NA | |||
| Ynl022c | Ynl022c | Saccharomyces cerevisiae | E | YNL022C | NO | Unknown | NA | |||
| NSUN6 family | ||||||||||
| NSUN6 | NSUN6 | Homo sapiens | E | Q8TEA1 | NO | Unknown | NA |
Abbreviations: A: archaea; B: bacteria; E eukaryota; NA not analyzed.
The first tentative approach towards a whole-genome analysis of m5C-MTases was initiated immediately after the characterization of RsmB as a bacterial m5C-MTase (66). More than 30 homologous proteins were identified by a BLAST search and a classification into eight distinct subgroups was proposed. However, only two E. coli genes (RsmB and YebU) were listed, while the third bacterial protein (YccW = RlmI) showed only little sequence similarity to RsmB.
An exhaustive bioinformatical analysis of structure–function relationships in the m5C-MTase family was later performed by Bujnicki et al. (65). The known m5C-MTases were subdivided into four major sub-families (related to Nop2/Nol1, YebU/Trm4, a large group related to RsmB or Ynl022c and a small group represented by P. horikoshi PH1991 and human NSUN6). This classification is also used in Table 1.
Recently, a careful analysis of m5C-MTase homologs in higher eukaryota (67) suggested the existence of a new, exclusively eukaryotic, subgroup of m5C-MTase-related proteins called RCMT9. These members are distantly related to Trm4 and were detected only in Viridiplantae, Alveolata, Euglenozoa and Mycetozoa taxons.
E. coli m5C-MTases: activity, structure and substrates
All three known m5C-MTases from E. coli were extensively characterized, both biochemically and structurally. Since m5C in bacteria is only found in ribosomal, RNA, all three enzymes are rRNA specific, but recognize their RNA target in a differential manner.
Fmu/fmv (RsmB)
The Fmu/fmv protein from E. coli was the first identified and characterized RNA-specific m5C-MTase (68,69). A partially purified protein fraction possessing 16S rRNA:m5C967–MTase activity was analyzed by N-terminal sequencing and was found to match to the first 23 amino acids of Fmu/fmv (later renamed to RsmB). This is the first and so far still the only example for a biochemical identification of an RNA m5C-MTase. With the exception of YccW(RlmI) and Dnmt2-related proteins, all enzymes discussed below have been identified by sequence similarity with the Fmu protein and their activity was further confirmed by various in vivo and in vitro tests. RsmB from E. coli efficiently catalyzes the methylation of naked 16S rRNA substrates, but assembled 30S ribosomal subunits are not methylated (68). In addition, a small 16-nt stem-loop minisubstrate derived from E. coli 16S rRNA was methylated by RsmB in vitro, albeit less efficiently than the full-size rRNA transcript (69).
The crystal structures of RsmB alone and in complex with SAM have been obtained at high resolution (1.65 and 2.1 Å, respectively) (70). RsmB folds into three almost independent domains, a C-terminal domain that is responsible for the m5C-MTase activity, and two other domains (the N-terminal domain and the central N1 domain) that show significant sequence similarity to known RNA-binding proteins and that may mediate the interaction with the 16S rRNA substrate. The crystal structure of the RsmB–16S rRNA complex has not been published yet, but docking of a 56-nt fragment derived from the 16S rRNA substrate clearly demonstrated structure and charge complementarity between the two molecules. In this predicted conformation, the target cytosine comes into close proximity with the catalytic Cys375 and the activated methyl group of SAM. This overall structure of the complex is consistent with the binding of a completely folded rRNA substrate (70).
YebU (RsmF)
Sequence similarity between RsmB and an E. coli ORF encoded by the yebU gene suggested that YebU also catalyzes m5C formation in E. coli rRNA. YebU was subsequently cloned and expressed, and its activity was studied in vitro and in vivo (71). The results demonstrated that the loss of active YebU protein led to the concomitant loss of m5C1407 in 16S rRNA, while m5C1962 in 23S rRNA remained unaffected. The recombinant YebU protein (later renamed to RsmF) retained its activity, but, in contrast to RsmB, methylated only assembled 30S ribosomal subunits and not naked 16S rRNA or complete 70S E. coli ribosomes.
Crystallized RsmF protein shows two distinct structural domains, the m5C-MTase domain, which resides in the N-terminal part, and the C-terminal domain, which shows significant similarity to the PUA domain frequently found in other RNA modification enzymes, like RNA:pseudouridine synthases and TGT-transglycosylases (72). In silico docking of the 30S ribosomal subunit to the RsmF structure suggests that it interacts not only with 16S rRNA but also with the ribosomal protein S12, providing a plausible explanation as to why naked 16S rRNA is not a substrate for methylation.
YccW (RlmI)
The last putative m5C-MTase in E. coli (YccW, now renamed to RlmI) has recently been characterized. It is worth mentioning that RlmI only distantly relates to two other E. coli m5C-MTases (RsmB and RsmF), and its sequence is closer to known m5U-MTases. Consequently, RlmI does not contain recognizable PC- and TC- sequence motifs present in other m5C-MTases and the catalytic Cys residue is located in the SCS motif close to the C-terminus. On this basis, RlmI was predicted to have pyrimidine-C5 MTase activity. However, mass-spectrometric analysis of 23S rRNA extracted from a YccW-deficient E. coli strain clearly linked RlmI activity to the unique m5C1962 found in E. coli 23S rRNA (73). Recombinant RlmI methylates naked 23S rRNA, but not assembled 50S ribosomal subunits, in spite of the rather high accessibility of the target m5C1962 at the subunit interface.
The crystal structure of RlmI has also become available recently (74). The protein folds into three independent domains, an N-terminal domain resembling the PUA domain of other RNA modification enzymes, a central EEHEE domain that is common to other modification enzymes including m5U-MTases and a C-terminal catalytic m5C-MTase domain. A model of the ternary complex containing RlmI, SAM and an rRNA fragment was analyzed by in silico modeling and subsequent minimization of the structure. The results predicted an interaction between RlmI and single-stranded 23S rRNA, rather than the folded and highly constrained two-dimensional structure. This observation provides an explanation for the inability of RlmI to methylate 50S ribosomal subunits, even if the target cytosine residue is highly accessible.
Archaeal m5C-MTases
The number of putative m5C MTases in Archaea is quite variable. Only one related protein seems to be present in mesophilic archaebacteria H. volcanii (75), while many hyperthermophilic organisms display several homologous proteins. The presence of m5C in bulk tRNA fractions extracted from P. furiosus and other hyperthermophilic Archaea was first demonstrated by HPCL/MS-ESI analysis (76). Later on, activity tests performed with cell-free P. furiosus extracts in vitro allowed the detection of at least m5C49- and, probably, m5C40-forming activities acting on heterologous tRNAIle from H. volcanii and yeast tRNAAsp and tRNAPhe substrates (77).
The analysis of numerous archaeal genomic sequences demonstrated the presence of multiple putative m5C-MTases in many genomes. One of these proteins, the ORF PH1374-encoded protein from P. horikoshi showed significant sequence similarity to the putative human p120 m5C-MTase (see below). The protein was then recombinantly expressed and its structure was determined at 1.9 Å resolution (78). The RNA substrate of this archaeal m5C-MTase is not known, but its overall fold is very similar to the central domain of bacterial RsmB and RsmF, suggesting similar activity (72).
In the genome sequence of hyperthermophilic P. abyssi, the ORF PAB1947 encodes a potential tRNA:m5C-MTase and forms a bicistronic operon with the gene encoding archaease, a protein known to stabilize several other archaeal proteins. ORF PAB1947 has been expressed in E. coli and its activity was characterized in vitro. The major methylation sites corresponded to the expected positions 48 and 49 in a P. furiosus tRNAAsp, but surprisingly, other cytosines were also methylated, especially at high incubation temperatures. Lower temperatures or the presence of archaease increased the specificity of tRNA substrate recognition (79). These data strongly suggest that archaeal m5C-MTases may have a rather broad specificity, compared to their yeast or human counterparts.
Classes of eukaryotic m5C-MTases
At least four distinct groups of RNA:m5C-MTases, differing in their sequence and RNA substrate specificity, were found in different eukaryotic organisms. Only three of them were detected in S. cerevisiae, but all four groups seem to be present in most eukaryotic genomes.
Yeast and human Trm4 (Ncl1)
The S. cerevisiae Ncl1 protein was initially characterized as a nuclear protein of unknown function (80). Further biochemical studies demonstrated its role in the modification of yeast tRNAs and some tRNA precursors at positions 34, 40, 48 and 49. Positions 34 and 40 are modified only in the intron-containing pre-tRNA and are specific for tRNALeu(CAA) and tRNAPhe(GAA), respectively (81). On the basis of this finding, Ncl1 (encoded by ORF YBL024) was renamed as Trm4 for tRNA-specific MTase 4.
Structure–function relationships in yeast Trm4 were further studied by various approaches like site-directed mutagenesis and limited trypsin digestion. By these methods, the catalytic role of Cys310 in motif IV was confirmed and the importance of the N-terminal domain for cytosine methylation was identified. The C-terminal domain is not required for tRNA binding and for catalysis and seems to fold independently from the N-terminal part of the protein. However, the presence of the C-terminal domain of Trm4 considerably stimulates the MTase activity of the N-terminal catalytic domain (82,83).
Sequence similarity with yeast Trm4 also allowed the cloning and characterization of a human homologue called hTrm4 or Misu/NSUN2. Misu/NSUN2 was found to be involved in Myc-mediated proliferation of cancer cells. It is expressed at low levels in normal epidermis but is up-regulated upon Myc activation (84). Misu/NSUN2 is phosphorylated at Ser139 by the cell-cycle protein kinase Aurora-B, and this phosphorylation affects its m5C-MTase activity and its association with nucleophosmin and nucleolin, two nucleolar interaction partners (85). Despite its high similarity with the yeast protein, the tRNA-MTase activity of hTrm4 seems to be restricted to position 34 of the intron containing human tRNALeu(CAA) (86). Human tRNA methyltransferases mediating m5C48/49 methylation still remain to be identified.
Yeast and human Nop2/p120
The second member of the family, the yeast S. cerevisiae nucleolar protein Nop2 (encoded by ORF YNL061) was shown to be involved in biogenesis of 60S ribosomal subunit and in the maturation of 26S rRNA (87,88). These functions of Nop2, and not the putative RNA-MTase activity, are probably important for the viability of yeast cells. Furthermore, an RNA:m5C-MTase activity or an RNA substrate for Nop2 remain to be identified. Site-directed mutagenesis of yeast Nop2 demonstrated that it has a Cys residue (Cys424) essential for viability. All other tested amino acid substitutions did not affect Nop2 functions in vivo (89). Further analysis showed that this Cys residue is important for enzyme regeneration at the second step of the reaction and its substitution by alanine or serine led to the accumulation of a covalent intermediate attached to the second catalytic cysteine (90) (see below). Genetic analysis of Nop2 thermosensitive (ts) alleles allowed to distinguish between the function as a 26S rRNA maturation factor and a potential RNA:m5C-MTase. These data further confirmed the role of Nop2 in 26S rRNA maturation, even though the absence or reduced levels of Nop2 did not affect 18S rRNA maturation or 40S subunit biogenesis (91).
The strong sequence similarity of human proliferation associated nucleolar protein p120 with yeast Nop2 and the already characterized bacterial and eukaryotic m5C-MTases, together with its implication in rRNA biogenesis, strongly suggested that p120 possesses a similar activity (92,93). Experimental analysis of p120 delineated its N-terminal Arg-rich region as a high-affinity binding domain for rRNA. In addition, human p120 associates with 60S–80S pre-ribosomal particles in HeLa nuclear extracts (93). However, the significance of this physical association and the exact functions of human p120 protein in rRNA maturation and processing remain elusive.
The family of NSUN/NOP2/NOL1 related proteins in humans is represented by at least nine different members (NSUN1 to NSUN7, genes NSUN5A, B and C probably resulted from a recent gene duplication). Most genes of this group are highly conserved in mammals. At the protein level, the number of related protein variants is very important since multiple mRNA splicing isoforms were characterized and some variants may lack important catalytic or RNA-binding domains. One of the NSUN proteins (NSUN5A fragment 127–429) was crystallized and its structure was determined at high resolution (PDB accession 2B9E). In most cases, these NSUN proteins retained the putative m5C-MTase domain bearing two catalytic cysteines. However, with the exception of hTrm4/NSUN2, neither the activity nor the substrate specificity of these potential m5C-MTases have been investigated so far. NSUN5A, B and C are deleted in Williams–Beuren syndrome, which is a complex developmental disorder with multisystemic manifestations including supravalvular aortic stenosis and a specific cognitive phenotype (94). Of note, NSUN5 also shows considerable sequence similarity to the third putative m5C-MTase in S. cerevisiae (encoded by ORF YNL022C), which has not been characterized in detail yet. Lastly, the human NSUN7 gene provides another illustration for the implication of putative m5C-MTases in various disorders, because a point mutation in this gene has been shown to cause reduced sperm motility and infertility in male mice (95).
Higher eukaryotic Dnmt2 and homologs
A relatively new addition to the eukaryotic RNA:m5C-MTase family are the Dnmt2-related proteins. The first Dnmt2-like protein (pmt1) was described in the fission yeast S. pombe (96) and recognized by its substantial sequence homologies to DNA:m5C-MTases. It was also noted that the pmt1 protein sequence contained a serine insertion in the catalytic proline–cysteine (PC) motif, which was interpreted to represent an inactivating mutation, consistent with the absence of DNA methylation in fission yeast (96). Several years later, human and mouse Dnmt2 were discovered in screens for sequences with homologies to known DNA:m5C-MTases (97,98). The corresponding protein sequences contained all the catalytic signature motifs of DNA:m5C-MTases, but no corresponding enzymatic activity could be described initially. Dnmt2 has been widely conserved during evolution and the use of more sensitive methods has allowed the identification of a residual DNA-MTase activity for Dnmt2. However, the biological relevance of this activity has remained contentious (99).
When the three-dimensional structure of human Dnmt2 was first analyzed, the results demonstrated a high similarity to the protein structure of the bacterial M.HhaI DNA:m5C-MTase (100), thus again suggesting that Dnmt2 might have DNA-MTase activity. However, an independent examination of the structural data also identified a tyrosine residue in the target recognition domain of Dnmt2 that was not present in other DNA:m5C-MTases (101). It was proposed that this tyrosine residue might impede the binding of DNMT2 to DNA substrates and that the enzyme might thus favor alternative substrates. This notion was confirmed when it was shown that Dnmt2 from various species methylates C38 of tRNAAsp (102). The tRNA-MTase activity has now been confirmed in several other laboratories and appears to be substantially higher than the residual DNA-MTase activity described initially. In light of the findings discussed above, it appears reasonable to assume that Dnmt2 is predominantly a tRNA-MTase and that the weak and highly distributive DNA-MTase activity is a consequence of a secondary enzyme activity with potentially little biological relevance. A more recent publication suggesting a role of Dnmt2-mediated DNA methylation in the epigenetic regulation of transposable elements in Drosophila (103) seems to contradict this notion. However, the results from this study indicate a robust and highly processive catalytic activity of Dnmt2 in the methylation of genomic retroelements. These findings are surprising because they suggest that a substantial DNA-MTase activity of Dnmt2 has been overlooked previously. It will therefore be important to confirm these results in independent assays and in other model systems.
Subcellular localization of eukaryotic m5C MTases
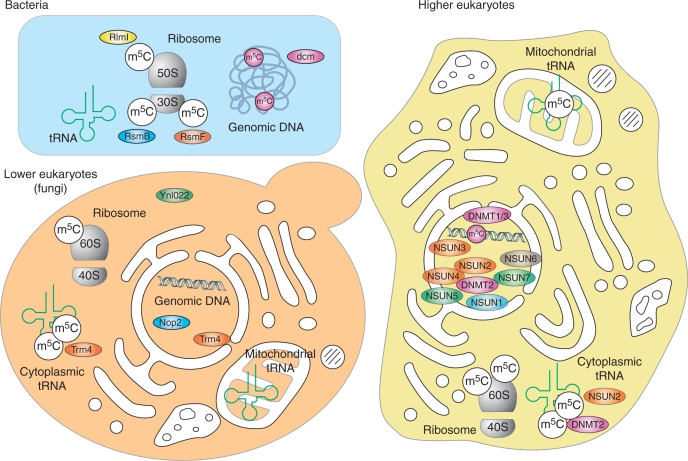
The subcellular localization of RNA modification enzymes can provide important indications about their biological functions. Figure 4 gives an overview of the current knowledge on the subcellular localization of methylated RNAs and modification enzymes. Most m5C-MTases are predicted to be nuclear or nucleolar proteins which corresponds well to their functions in tRNA and rRNA processing. Subcellular enzyme localization has been studied experimentally, by using various immunostaining and direct fluorescence approaches, but can also be inferred from the modification pattern of RNA in different cellular compartments. This latter method is particularly useful for mitochondrial localization, since these small and very dynamic organelles are difficult to observe by microscopy and because mitochondrial tRNAs and rRNAs are produced in the mitochondrial matrix and may be modified only by mitochondria-associated RNA modification enzymes. For example, yeast mitochondrial tRNAs do not contain m5C even though they contain many potential target cytosine residues for Trm4 activity, thus indicating the absence of Trm4 from this compartment.
Figure 4.
Subcellular localization of m5C residues in RNAs and m5C-MTases in bacteria (upper left), lower eukaryota (lower left) and higher eukaryota (right). Known m5C residues in RNAs are shown in white circles, together with the corresponding enzyme (if known). DNA-MTases and Dnmt2 are colored in purple, the RsmB/Nol1 family in blue, the YebU (RsmF) family in orange, the Ynl022 family in green and NSUN6 in grey.
Little is known about precise localization of various m5C-MTases in eukaryota. Yeast Ncl1 (Trm4) was localized at nuclear periphery (80), while Nop2 is a well-known nucleolar protein (88). Similarly, human p120 is also nuclear and nucleolar (104). The subcellular localization of Dnmt2 has been discussed controversially with various studies providing indications for either a nuclear or a cytoplasmic localization. A recent analysis of the subcellular localization of Dnmt2 in Drosophila suggested that the enzyme localizes both to the nucleus and to the cytoplasm (105). This may be reflective of the complex activities of Dnmt2.
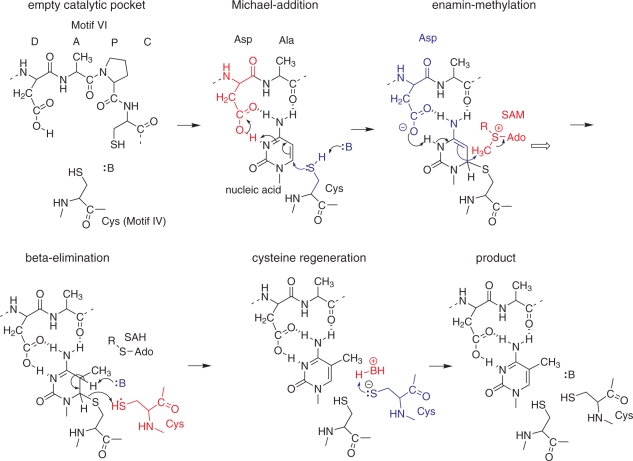
THE CATALYTIC MECHANISM OF m5C-MTases
The catalytic mechanism of m5C-MTases bears strong parallels to that of m5U-MTases, including in particular Trm2 which generates m5U (also known as rT) at position 54 of most tRNAs. As opposed to most other RNA-MTases, which methylate nitrogen atoms, the target position of m5-pyrimidine-MTases is a non-nucleophilic carbon atom. Activation of this position proceeds via Michael addition of a catalytic cysteine to the 6-position, which, after protonation, results in an enamin in the case of m5C-MTases (shown in Figure 5A), or an enol in the case of m5U-MTases (not shown). The 5-carbon in these intermediates, frequently referred to as ‘enolate’, is a nucleophile and as such easily methylated by the electrophilic SAM. Methyl-group transfer leads to another key intermediate, a covalent adduct of enzyme and nucleic acid, from which the enzyme is regenerated by beta-elimination of cysteine and the hydrogen on C5, which is thought to be base-mediated. The similarities among pyrimidine-MTases extend to inhibition of the elimination step by 5-fluoro-pyrimidine targets. Nucleic acids containing 5-flurouracil or 5-fluorocytidine frequently form covalent adducts with cognate MTases (83).
Figure 5.
Catalytic mechanism of m5C-MTases. Nucleophiles, bases and nucleophilic attacks are indicated in blue; acids and electrophiles are indicated in red.
The elucidation of the catalytic mechanism(s) of m5C-MTases continues to be critically important for understanding the substrate specificity and biological function of these enzymes. As outlined below, structural biology with particular emphasis on the residues in active sites has been combined with insights from bioinformatics and biochemistry. The resulting picture discriminates typical DNA- versus RNA-MTases, and identified Dnmt2 as a hybrid RNA-MTase, which uses a catalytic cysteine typical of DNA-MTase (see below).
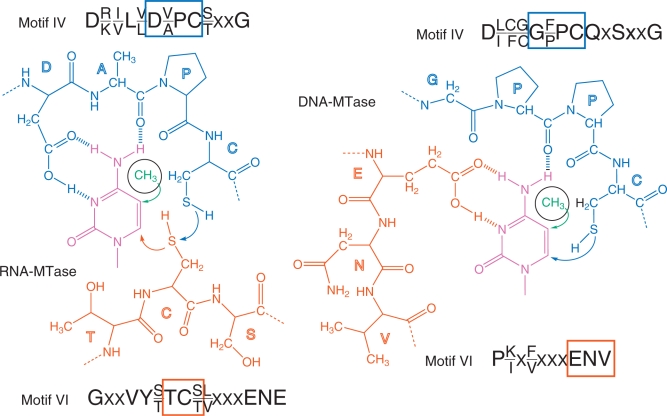
Mechanism with two cysteines
Comparative studies of RNA- and DNA-specific m5C-MTases resulted in the conclusion that DNA and RNA m5C-methylation are being mediated by similar yet distinct enzymatic mechanisms, which involve different amino acid residues (106). DNA and RNA-MTases exhibit sequence permutations, resulting in a variable linear order of the conserved motifs, which are numbered I through X. Typical catalytic pockets of RNA and DNA MTases are shown in Figure 6. Typical RNA:m5C-MTases contain two Cys-containing characteristic motifs (motif IV and VI, referred to as PC- and TC-sequences, respectively). However, only the PC-sequence (motif IV) is found to be conserved in DNA:m5C-MTases, while motif VI contains a conserved ENV-sequence. Site-directed mutagenesis of the E. coli Fmu (RsmB) protein showed that only the replacement of Cys375 (in motif VI, TC-sequence), but not Cys325 (in motif IV, PC-sequence) abolishes the RNA m5C-MTase activity in vitro and the capacity to form a covalent adduct with 5-fluoro-cytidine containing RNA. These in vitro data came in apparent contradiction with the results of site-directed mutagenesis of Nop2 in vivo (89). Indeed, in this case, only a substitution of Cys424 (motif IV, PC-sequence) affected the viability of yeast cells. All other substitutions in Nop2 (including replacement of Cys478 in motif VI) did not affect Nop2 functions in vivo. This apparent contradiction was resolved by further studies (90) demonstrating that Nop2 and Trm4 (and probably other RNA:m5C-MTases, like RsmB) use both conserved cysteine residues for catalysis. The initial activation of the cytosine ring is performed by the cysteine residue of motif VI (TC-sequence, Cys478 in the case of Nop2), while the release of the RNA substrate from the covalent reaction intermediate requires the presence of the second Cys in motif IV (PC-sequence, Cys424 in Nop2). In the absence of the ‘regeneration’ Cys residue, the RNA substrate is not released from the intermediate covalent Nop2–RNA complex as indicated by the presence of covalent adducts of higher molecular mass accumulating in yeast cells and in E. coli. In the crystal structure of E. coli RsmB, both Cys325 and Cys375 are located in close proximity to each other and Cys325 aligns well with Cys81, which is located in motif IV of the HpaI DNA:m5C-MTase. The spatial locations of these conserved residues are thus fully compatible with the proposed two-cysteine catalytic mechanism of RNA:m5C-MTases (70). More recently, the catalytic mechanism involving both conserved Cys residues was also confirmed for yeast Trm4, which forms a stable covalent intermediate with 5-fluoro-cytidine containing RNA (83).
Figure 6.
Catalytic pocket of RNA- (left) and DNA-MTases (right). The target cytidine residue is indicated in purple, the sequences of conserved motifs IV and VI are given for both classes of enzymes. Motif IV is in blue and motif VI is in orange. Boxes highlight residues belonging to the respective sequence motifs.
A recent and intriguing addition to this field is the catalytic mechanism of the Dnmt2 m5C-MTase. Dnmt2 is an enzyme that has long been regarded as pure DNA-MTase, based on sequence similarity with other DNA-MTases, including a linear arrangement of motifs I through X, which is characteristic for Dnmt enzymes. The discovery of RNA-MTase activity in Dnmt2 was thus a surprise (102). In subsequent studies of human Dnmt2, mutational analysis identified essential residues for analysis which include E119 (ENV-motif VI), as well as R160 and R162 (motif VIII). The essential catalytic cysteine, which conducts the initial nucleophilic attack for tRNA methylation at position C38 was identified as C79, which is situated in the PC-motif IV. It was thus concluded, that C38 methylation of tRNA by Dnmt2 utilizes a catalytic mechanism typical for DNA-MTases (107). These findings uncover a previously unexpected link between RNA and DNA methylation which may be important for understanding the biological function(s) of Dnmt2 (see below).
Covalent complex formation with the RNA substrate
Covalent complexes formed between RNA substrate and mutant forms of RNA:m5C-MTases have been observed in several studies, but their detailed biochemical characterization was not reported until recently. Site-directed mutagenesis of yeast Trm4 confirmed the importance of motif VI Cys310 (TC-sequence) for catalysis and motif IV Cys260 (PC-sequence) for substrate release. Trm4 C260A or C260S mutants efficiently form covalent complexes both in vitro and in vivo, when expressed in E. coli. SDS–PAGE analysis showed that SAM is required for covalent complex formation and that the RNA substrate is present in a covalent complex that contains the CH3-group, an indication that the release of the final reaction product is indeed impeded by the mutation in the PC-sequence (108).
THE BIOLOGICAL SIGNIFICANCE OF m5C IN RNA
General stabilization of tRNA molecules and Mg2+ binding
A known general role of modified nucleotides in tRNA is their structural and metabolic stabilization (109). Structural stabilization of RNAs by m5C was demonstrated by various physico-chemical techniques, like circular dichroism and NMR. One of the m5C residues present in the S. cerevisiae tRNAPhe at position 40 was reported to be important for appropriate Mg2+ binding and appears to induce conformational transitions of the whole anticodon loop (110–112).
Modified residues in tRNAs are also known to influence the stability of these molecules in vivo, and there are at least two pathways that are known to degrade hypomodified tRNA (113–115). An exosome-independent pathway was found to degrade m5C-deficient tRNA, but only in double mutants lacking both TRM4 and the methyltransferase activity responsible for the formation of 7-methylguanosine. The accelerated degradation of hypomodified tRNA thus appears not to be specific to the lack of single m5C residues. Rather, it is hypothesized that the combined lack of several modifications lowers the conformational stability of the tRNA below a certain threshold, which leads to its degradation by a mechanism that is yet to be elucidated in detail (114,115).
Importance of m5C for mRNA translation
Very little is known about specific role of m5C residues in tRNA and rRNA during mRNA decoding on the ribosome. The presence of m5C at position 34 in eukaryotic tRNALeu seems to be important for its function in translation, specifically for suppressor function in vivo (116).
Deletion of the non-essential yeast TRM4 gene does not affect cell growth or steady-state levels of large and small ribosome subunits, monoribosomes and polyribosomes. However, disruption of TRM4 leads to increased sensitivity to the antibiotic paromomycin (80). Paromomycin is known to affect the precision of the ribosome decoding, thus the presence of m5C in yeast tRNA may be important for maintenance of low error translation.
On the other hand, m5C in ribosomal RNA may also participate in tRNA recognition and peptidyl transfer. Despite their distant locations in the rRNA sequence, all three m5C residues present in bacterial 16S and 23S rRNAs are localized in close proximity to each other in the three-dimensional structure of the ribosome. In addition, the paromomycin binding site has been mapped precisely to this region (117). It is not clear if yeast rRNA contains a m5C residue at a similar position, but the sensitivity of the ΔYBL024 mutant to paromomycin may indicate the relations between ribosomal decoding and m5C functions in rRNA (80).
Other biological functions of m5C residues
Because the phenotypes of RNA:m5C-MTase mutants are comparably weak, it has generally been difficult to pinpoint the biological function of RNA cytosine methylation. However, there is some direct evidence that the methylation of RNA molecules can have a modulatory effect on the innate human immune system. Unmodified RNA molecules have been shown to strongly stimulate the immune system through activation of toll-like receptors (118). This effect was greatly reduced when either of several methylated nucleotides, including m5C, was incorporated into the stimulatory RNAs, suggesting that the methylation prevents recognition of endogenous RNAs by the innate immune system.
Another, currently more speculative role for RNA methylation is the regulation of epigenetic inheritance patterns. While it is clear that most phenotypic traits are inherited through DNA, there is also evidence for RNA-dependent inheritance of certain phenotypes (119). The methylation of cytosine residues in DNA has long been known to play an important role in the modulation of genetic information (120). It will be fascinating to explore the possibility whether RNA methylation plays a similar role.
CONCLUSION AND PERSPECTIVES
Recent developments in the field of RNA methylation open a large perspective for further analysis of this important biological process. Enzymatic activity, catalytic mechanisms and RNA-recognition specificity of the corresponding RNA:m5C-MTases clearly require better characterization. The biological role of m5C residues in RNA has been largely ignored and further studies using genetically engineered models are needed to investigate this point. High-throughput sequencing techniques will be extremely helpful in the methylation profiling of low abundance species [e.g. mRNA (53)], or even newly discovered RNA species, including various regulatory RNAs. While some of the latter are known to be methylated at other positions such as the 2′-OH, (6,121), verification of the presence or absence of m5C in these species should further deepen our understanding of the biological meaning of m5C in RNA.
FUNDING
M.H. and F.L. acknowledge funding by the DFG Dnmt2 (FOR 1082). Funding for open access charge: DFG HE 3397/6-1.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
M.H. thanks Andres Jäschke for constant support.
REFERENCES
- 1.Hotchkiss RD. The quantitative separation of purines, pyrimidines, and nucleosides by paper chromatography. J. Biol. Chem. 1948;175:315–332. [PubMed] [Google Scholar]
- 2.Luciano DJ, Mirsky H, Vendetti NJ, Maas S. RNA editing of a miRNA precursor. RNA. 2004;10:1174–1177. doi: 10.1261/rna.7350304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blow MJ, Grocock RJ, van Dongen S, Enright AJ, Dicks E, Futreal PA, Wooster R, Stratton MR. RNA editing of human microRNAs. Genome Biol. 2006;7:R27. doi: 10.1186/gb-2006-7-4-r27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang Z, Ebright YW, Yu B, Chen X. HEN1 recognizes 21-24 nt small RNA duplexes and deposits a methyl group onto the 2′ OH of the 3′ terminal nucleotide. Nucleic Acids Res. 2006;34:667–675. doi: 10.1093/nar/gkj474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu B, Yang Z, Li J, Minakhina S, Yang M, Padgett RW, Steward R, Chen X. Methylation as a crucial step in plant microRNA biogenesis. Science. 2005;307:932–935. doi: 10.1126/science.1107130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Horwich MD, Li C, Matranga C, Vagin V, Farley G, Wang P, Zamore PD. The Drosophila RNA methyltransferase, DmHen1, modifies germline piRNAs and single-stranded siRNAs in RISC. Curr. Biol. 2007;17:1265–1272. doi: 10.1016/j.cub.2007.06.030. [DOI] [PubMed] [Google Scholar]
- 7.Saito K, Sakaguchi Y, Suzuki T, Siomi H, Siomi MC. Pimet, the Drosophila homolog of HEN1, mediates 2′-O-methylation of Piwi- interacting RNAs at their 3′ ends. Genes Dev. 2007;21:1603–1608. doi: 10.1101/gad.1563607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grosjean H, Droogmans L, Roovers M, Keith G. Detection of enzymatic activity of transfer RNA modification enzymes using radiolabeled tRNA substrates. Methods Enzymol. 2007;425:55–101. doi: 10.1016/S0076-6879(07)25003-7. [DOI] [PubMed] [Google Scholar]
- 9.Hengesbach M, Meusburger M, Lyko F, Helm M. Use of DNAzymes for site-specific analysis of ribonucleotide modifications. RNA. 2008;14:180–187. doi: 10.1261/rna.742708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao X, Yu YT. Detection and quantitation of RNA base modifications. RNA. 2004;10:996–1002. doi: 10.1261/rna.7110804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stach D, Schmitz OJ, Stilgenbauer S, Benner A, Dohner H, Wiessler M, Lyko F. Capillary electrophoretic analysis of genomic DNA methylation levels. Nucleic Acids Res. 2003;31:E2. doi: 10.1093/nar/gng002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cornelius MG, Schmeiser HH. RNA analysis by MEKC with LIF detection. Electrophoresis. 2007;28:3901–3907. doi: 10.1002/elps.200700127. [DOI] [PubMed] [Google Scholar]
- 13.Li HY, Wang SM, Liu HM, Li J, Han D, Bu SS, Zhang MZ. Analysis of modified nucleosides in the urine of patients with malignant cancer by liquid chromatography/electrospray ionization mass spectrometry. Rapid Commun. Mass Spectrom. 2008;22:3161–3171. doi: 10.1002/rcm.3721. [DOI] [PubMed] [Google Scholar]
- 14.Wagner TM, Nair V, Guymon R, Pomerantz SC, Crain PF, Davis DR, McCloskey JA. A novel method for sequence placement of modified nucleotides in mixtures of transfer RNA. Nucleic Acids Symp. Ser. 2004;48:263–264. doi: 10.1093/nass/48.1.263. [DOI] [PubMed] [Google Scholar]
- 15.Dai Q, Fong R, Saikia M, Stephenson D, Yu YT, Pan T, Piccirilli JA. Identification of recognition residues for ligation-based detection and quantitation of pseudouridine and N6-methyladenosine. Nucleic Acids Res. 2007;35:6322–6329. doi: 10.1093/nar/gkm657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu YT, Shu MD, Steitz JA. A new method for detecting sites of 2′-O-methylation in RNA molecules. RNA. 1997;3:324–331. [PMC free article] [PubMed] [Google Scholar]
- 17.Motorin Y, Muller S, Behm-Ansmant I, Branlant C. Identification of modified residues in RNAs by reverse transcription-based methods. Methods Enzymol. 2007;425:21–53. doi: 10.1016/S0076-6879(07)25002-5. [DOI] [PubMed] [Google Scholar]
- 18.Lambrinakos A, Humphrey KE, Babon JJ, Ellis TP, Cotton RG. Reactivity of potassium permanganate and tetraethylammonium chloride with mismatched bases and a simple mutation detection protocol. Nucleic Acids Res. 1999;27:1866–1874. doi: 10.1093/nar/27.8.1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bui CT, Rees K, Cotton RG. Permanganate oxidation reactions of DNA: perspective in biological studies. Nucleosides Nucleotides Nucleic Acids. 2003;22:1835–1855. doi: 10.1081/NCN-120023276. [DOI] [PubMed] [Google Scholar]
- 20.Cotton RG, Rodrigues NR, Campbell RD. Reactivity of cytosine and thymine in single-base-pair mismatches with hydroxylamine and osmium tetroxide and its application to the study of mutations. Proc. Natl Acad. Sci. USA. 1988;85:4397–4401. doi: 10.1073/pnas.85.12.4397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rubin CM, Schmid CW. Pyrimidine-specific chemical reactions useful for DNA sequencing. Nucleic Acids Res. 1980;8:4613–4619. doi: 10.1093/nar/8.20.4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zuo S, Boorstein RJ, Teebor GW. Oxidative damage to 5-methylcytosine in DNA. Nucleic Acids Res. 1995;23:3239–3243. doi: 10.1093/nar/23.16.3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tabone T, Sallmann G, Chiotis M, Law M, Cotton R. Chemical cleavage of mismatch (CCM) to locate base mismatches in heteroduplex DNA. Nat. Protoc. 2006;1:2297–2304. doi: 10.1038/nprot.2006.352. [DOI] [PubMed] [Google Scholar]
- 24.Maxam AM, Gilbert W. A new method for sequencing DNA. Proc. Natl. Acad. Sci. USA. 1977;74:560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peattie DA. Direct chemical method for sequencing RNA. Proc. Natl Acad. Sci. USA. 1979;76:1760–1764. doi: 10.1073/pnas.76.4.1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fritzsche E, Hayatsu H, Igloi GL, Iida S, Kossel H. The use of permanganate as a sequencing reagent for identification of 5-methylcytosine residues in DNA. Nucleic Acids Res. 1987;15:5517–5528. doi: 10.1093/nar/15.14.5517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Okamoto A. Chemical approach toward efficient DNA methylation analysis. Org. Biomol. Chem. 2009;7:21–26. doi: 10.1039/b813595a. [DOI] [PubMed] [Google Scholar]
- 28.Rideout WM, 3rd, Coetzee GA, Olumi AF, Jones PA. 5-Methylcytosine as an endogenous mutagen in the human LDL receptor and p53 genes. Science. 1990;249:1288–1290. doi: 10.1126/science.1697983. [DOI] [PubMed] [Google Scholar]
- 29.Suzuki T, Nakamura T, Yamada M, Ide H, Kanaori K, Tajima K, Morii T, Makino K. Isolation and characterization of diazoate intermediate upon nitrous acid and nitric oxide treatment of 2′-deoxycytidine. Biochemistry. 1999;38:7151–7158. doi: 10.1021/bi982803t. [DOI] [PubMed] [Google Scholar]
- 30.Ali MM, Alam MR, Kawasaki T, Nakayama S, Nagatsugi F, Sasaki S. Sequence- and base-specific delivery of nitric oxide to cytidine and 5-methylcytidine leading to efficient deamination. J. Am. Chem. Soc. 2004;126:8864–8865. doi: 10.1021/ja0498888. [DOI] [PubMed] [Google Scholar]
- 31.Singer B. All oxygens in nucleic acids react with carcinogenic ethylating agents. Nature. 1976;264:333–339. doi: 10.1038/264333a0. [DOI] [PubMed] [Google Scholar]
- 32.Singer B. The chemical effects of nucleic acid alkylation and their relation to mutagenesis and carcinogenesis. Prog. Nucleic Acid Res. Mol. Biol. 1975;15:219–284. [PubMed] [Google Scholar]
- 33.Peattie DA, Gilbert W. Chemical probes for higher-order structure in RNA. Proc. Natl Acad. Sci. USA. 1980;77:4679–4682. doi: 10.1073/pnas.77.8.4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peattie DA, Herr W. Chemical probing of the tRNA–ribosome complex. Proc. Natl Acad. Sci. USA. 1981;78:2273–2277. doi: 10.1073/pnas.78.4.2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Frommer M, McDonald LE, Millar DS, Collis CM, Watt F, Grigg GW, Molloy PL, Paul CL. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc. Natl Acad. Sci. USA. 1992;89:1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clark SJ, Harrison J, Paul CL, Frommer M. High sensitivity mapping of methylated cytosines. Nucleic Acids Res. 1994;22:2990–2997. doi: 10.1093/nar/22.15.2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eckhardt F, Lewin J, Cortese R, Rakyan VK, Attwood J, Burger M, Burton J, Cox TV, Davies R, Down TA, et al. DNA methylation profiling of human chromosomes 6, 20 and 22. Nat. Genet. 2006;38:1378–1385. doi: 10.1038/ng1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meissner A, Gnirke A, Bell GW, Ramsahoye B, Lander ES, Jaenisch R. Reduced representation bisulfite sequencing for comparative high-resolution DNA methylation analysis. Nucleic Acids Res. 2005;33:5868–5877. doi: 10.1093/nar/gki901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gu W, Hurto RL, Hopper AK, Grayhack EJ, Phizicky EM. Depletion of Saccharomyces cerevisiae tRNAHis guanylyltransferase Thg1p leads to uncharged tRNAHis with additional m5C. Mol. Cell Biol. 2005;25:8191–8201. doi: 10.1128/MCB.25.18.8191-8201.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schaefer M, Pollex T, Hanna K, Lyko F. RNA cytosine methylation analysis by bisulfite sequencing. Nucleic Acids Res. 2009;37:e12. doi: 10.1093/nar/gkn954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Warnecke PM, Stirzaker C, Melki JR, Millar DS, Paul CL, Clark SJ. Detection and measurement of PCR bias in quantitative methylation analysis of bisulphite-treated DNA. Nucleic Acids Res. 1997;25:4422–4426. doi: 10.1093/nar/25.21.4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sprinzl M, Vassilenko KS. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 2005;33:D139–D140. doi: 10.1093/nar/gki012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith JE, Cooperman BS, Mitchell P. Methylation sites in Escherichia coli ribosomal RNA: localization and identification of four new sites of methylation in 23S rRNA. Biochemistry. 1992;31:10825–10834. doi: 10.1021/bi00159a025. [DOI] [PubMed] [Google Scholar]
- 44.Kowalak JA, Pomerantz SC, Crain PF, McCloskey JA. A novel method for the determination of post-transcriptional modification in RNA by mass spectrometry. Nucleic Acids Res. 1993;21:4577–4585. doi: 10.1093/nar/21.19.4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kowalak JA, Bruenger E, McCloskey JA. Posttranscriptional modification of the central loop of domain V in Escherichia coli 23 S ribosomal RNA J. Biol. Chem. 1995;270:17758–17764. doi: 10.1074/jbc.270.30.17758. [DOI] [PubMed] [Google Scholar]
- 46.Suzuki T, Ikeuchi Y, Noma A, Sakaguchi Y. Mass spectrometric identification and characterization of RNA-modifying enzymes. Methods Enzymol. 2007;425:211–229. doi: 10.1016/S0076-6879(07)25009-8. [DOI] [PubMed] [Google Scholar]
- 47.Noon KR, Bruenger E, McCloskey JA. Posttranscriptional modifications in 16S and 23S rRNAs of the archaeal hyperthermophile Sulfolobus solfataricus J. Bacteriol. 1998;180:2883–2888. doi: 10.1128/jb.180.11.2883-2888.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Veldman GM, Klootwijk J, de Regt VC, Planta RJ, Branlant C, Krol A, Ebel JP. The primary and secondary structure of yeast 26S rRNA. Nucleic Acids Res. 1981;9:6935–6952. doi: 10.1093/nar/9.24.6935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maden BE. Locations of methyl groups in 28 S rRNA of Xenopus laevis and man. Clustering in the conserved core of molecule. J. Mol. Biol. 1988;201:289–314. doi: 10.1016/0022-2836(88)90139-8. [DOI] [PubMed] [Google Scholar]
- 50.Maden BE. Identification of the locations of the methyl groups in 18 S ribosomal RNA from Xenopus laevis and man. J. Mol. Biol. 1986;189:681–699. doi: 10.1016/0022-2836(86)90498-5. [DOI] [PubMed] [Google Scholar]
- 51.Kowalak JA, Bruenger E, Crain PF, McCloskey JA. Identities and phylogenetic comparisons of posttranscriptional modifications in 16S ribosomal RNA from Haloferax volcanii. J. Biol. Chem. 2000;275:24484–24489. doi: 10.1074/jbc.M002153200. [DOI] [PubMed] [Google Scholar]
- 52.Piekna-Przybylska D, Decatur WA, Fournier MJ. The 3D rRNA modification maps database: with interactive tools for ribosome analysis. Nucleic Acids Res. 2008;36:D178–D183. doi: 10.1093/nar/gkm855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dubin DT, Taylor RH. The methylation state of poly A-containing messenger RNA from cultured hamster cells. Nucleic Acids Res. 1975;2:1653–1668. doi: 10.1093/nar/2.10.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dubin DT, Stollar V. Methylation of Sindbis virus “26S” messenger RNA. Biochem. Biophys. Res. Commun. 1975;66:1373–1379. doi: 10.1016/0006-291x(75)90511-2. [DOI] [PubMed] [Google Scholar]
- 55.Dubin DT, Stollar V, Hsuchen CC, Timko K, Guild GM. Sindbis virus messenger RNA: the 5′-termini and methylated residues of 26 and 42 S RNA. Virology. 1977;77:457–470. doi: 10.1016/0042-6822(77)90471-8. [DOI] [PubMed] [Google Scholar]
- 56.Sommer S, Salditt-Georgieff M, Bachenheimer S, Darnell JE, Furuichi Y, Morgan M, Shatkin AJ. The methylation of adenovirus-specific nuclear and cytoplasmic RNA. Nucleic Acids Res. 1976;3:749–765. doi: 10.1093/nar/3.3.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Salditt-Georgieff M, Jelinek W, Darnell JE, Furuichi Y, Morgan M, Shatkin A. Methyl labeling of HeLa cell hnRNA: a comparison with mRNA. Cell. 1976;7:227–237. doi: 10.1016/0092-8674(76)90022-2. [DOI] [PubMed] [Google Scholar]
- 58.Furuichi Y, Morgan M, Shatkin AJ, Jelinek W, Salditt-Georgieff M, Darnell JE. Methylated, blocked 5 termini in HeLa cell mRNA. Proc. Natl Acad. Sci. USA. 1975;72:1904–1908. doi: 10.1073/pnas.72.5.1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Desrosiers R, Friderici K, Rottman F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc. Natl Acad. Sci. USA. 1974;71:3971–3975. doi: 10.1073/pnas.71.10.3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Adams JM, Cory S. Modified nucleosides and bizarre 5′-termini in mouse myeloma mRNA. Nature. 1975;255:28–33. doi: 10.1038/255028a0. [DOI] [PubMed] [Google Scholar]
- 61.Lavi S, Shatkin AJ. Methylated simian virus 40-specific RNA from nuclei and cytoplasm of infected BSC-1 cells. Proc. Natl Acad. Sci. USA. 1975;72:2012–2016. doi: 10.1073/pnas.72.6.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sakamoto K, Okada N. 5-Methylcytidylic modification of in vitro transcript from the rat identifier sequence; evidence that the transcript forms a tRNA-like structure. Nucleic Acids Res. 1985;13:7195–7206. doi: 10.1093/nar/13.20.7195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rozhdestvensky TS, Crain PF, Brosius J. Isolation and posttranscriptional modification analysis of native BC1 RNA from mouse brain. RNA Biol. 2007;4:11–15. doi: 10.4161/rna.4.1.4306. [DOI] [PubMed] [Google Scholar]
- 64.Brule H, Grosjean H, Giege R, Florentz C. A pseudoknotted tRNA variant is a substrate for tRNA (cytosine-5)-methyltransferase from Xenopus laevis. Biochimie. 1998;80:977–985. doi: 10.1016/s0300-9084(99)80003-0. [DOI] [PubMed] [Google Scholar]
- 65.Bujnicki JM, Feder M, Ayres CL, Redman KL. Sequence-structure-function studies of tRNA:m5C methyltransferase Trm4p and its relationship to DNA:m5C and RNA:m5U methyltransferases. Nucleic Acids Res. 2004;32:2453–2463. doi: 10.1093/nar/gkh564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Reid R, Greene PJ, Santi DV. Exposition of a family of RNA m5C methyltransferases from searching genomic and proteomic sequences. Nucleic Acids Res. 1999;27:3138–3145. doi: 10.1093/nar/27.15.3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pavlopoulou A, Kossida S. Phylogenetic analysis of the eukaryotic RNA (cytosine-5)-methyltransferases. Genomics. 2009;93:350–357. doi: 10.1016/j.ygeno.2008.12.004. [DOI] [PubMed] [Google Scholar]
- 68.Tscherne JS, Nurse K, Popienick P, Michel H, Sochacki M, Ofengand J. Purification, cloning, and characterization of the 16S RNA m5C967 methyltransferase from Escherichia coli. Biochemistry. 1999;38:1884–1892. doi: 10.1021/bi981880l. [DOI] [PubMed] [Google Scholar]
- 69.Gu XR, Gustafsson C, Ku J, Yu M, Santi DV. Identification of the 16S rRNA m5C967 methyltransferase from Escherichia coli. Biochemistry. 1999;38:4053–4057. doi: 10.1021/bi982364y. [DOI] [PubMed] [Google Scholar]
- 70.Foster PG, Nunes CR, Greene P, Moustakas D, Stroud RM. The first structure of an RNA m5C methyltransferase, Fmu, provides insight into catalytic mechanism and specific binding of RNA substrate. Strucure. 2003;11:1609–1620. doi: 10.1016/j.str.2003.10.014. [DOI] [PubMed] [Google Scholar]
- 71.Andersen NM, Douthwaite S. YebU is a m5C methyltransferase specific for 16 S rRNA nucleotide 1407. J. Mol. Biol. 2006;359:777–786. doi: 10.1016/j.jmb.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 72.Hallberg BM, Ericsson UB, Johnson KA, Andersen NM, Douthwaite S, Nordlund P, Beuscher AEt, Erlandsen H. The structure of the RNA m5C methyltransferase YebU from Escherichia coli reveals a C-terminal RNA-recruiting PUA domain. J. Mol. Biol. 2006;360:774–787. doi: 10.1016/j.jmb.2006.05.047. [DOI] [PubMed] [Google Scholar]
- 73.Purta E, O'C;onnor M, Bujnicki JM, Douthwaite S. YccW is the m5C methyltransferase specific for 23S rRNA nucleotide 1962. J. Mol. Biol. 2008;383:641–651. doi: 10.1016/j.jmb.2008.08.061. [DOI] [PubMed] [Google Scholar]
- 74.Sunita S, Tkaczuk KL, Purta E, Kasprzak JM, Douthwaite S, Bujnicki JM, Sivaraman J. Crystal structure of the Escherichia coli 23S rRNA:m5C methyltransferase RlmI (YccW) reveals evolutionary links between RNA modification enzymes. J. Mol. Biol. 2008;383:652–666. doi: 10.1016/j.jmb.2008.08.062. [DOI] [PubMed] [Google Scholar]
- 75.Grosjean H, Gaspin C, Marck C, Decatur WA, de Crecy-Lagard V. RNomics and Modomics in the halophilic archaea Haloferax volcanii: identification of RNA modification genes. BMC Genomics. 2008;9:470. doi: 10.1186/1471-2164-9-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kowalak JA, Dalluge JJ, McCloskey JA, Stetter KO. The role of posttranscriptional modification in stabilization of transfer RNA from hyperthermophiles. Biochemistry. 1994;33:7869–7876. doi: 10.1021/bi00191a014. [DOI] [PubMed] [Google Scholar]
- 77.Constantinesco F, Motorin Y, Grosjean H. Transfer RNA modification enzymes from Pyrococcus furiosus: detection of the enzymatic activities in vitro. Nucleic Acids Res. 1999;27:1308–1315. doi: 10.1093/nar/27.5.1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ishikawa I, Sakai N, Tamura T, Yao M, Watanabe N, Tanaka I. Crystal structure of human p120 homologue protein PH1374 from Pyrococcus horikoshii. Proteins. 2004;54:814–816. doi: 10.1002/prot.10645. [DOI] [PubMed] [Google Scholar]
- 79.Auxilien S, El Khadali F, Rasmussen A, Douthwaite S, Grosjean H. Archease from Pyrococcus abyssi improves substrate specificity and solubility of a tRNA m5C methyltransferase. J. Biol. Chem. 2007;282:18711–18721. doi: 10.1074/jbc.M607459200. [DOI] [PubMed] [Google Scholar]
- 80.Wu P, Brockenbrough JS, Paddy MR, Aris JP. NCL1, a novel gene for a non-essential nuclear protein in Saccharomyces cerevisiae. Gene. 1998;220:109–117. doi: 10.1016/s0378-1119(98)00330-8. [DOI] [PubMed] [Google Scholar]
- 81.Motorin Y, Grosjean H. Multisite-specific tRNA:m5C-methyltransferase (Trm4) in yeast Saccharomyces cerevisiae: identification of the gene and substrate specificity of the enzyme. RNA. 1999;5:1105–1118. doi: 10.1017/s1355838299982201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Walbott H, Auxilien S, Grosjean H, Golinelli-Pimpaneau B. The carboxyl-terminal extension of yeast tRNA m5C methyltransferase enhances the catalytic efficiency of the amino-terminal domain. J. Biol. Chem. 2007;282:23663–23671. doi: 10.1074/jbc.M703818200. [DOI] [PubMed] [Google Scholar]
- 83.Walbott H, Husson C, Auxilien S, Golinelli-Pimpaneau B. Cysteine of sequence motif VI is essential for nucleophilic catalysis by yeast tRNA m5C methyltransferase. RNA. 2007;13:967–973. doi: 10.1261/rna.515707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Frye M, Watt FM. The RNA methyltransferase Misu (NSun2) mediates Myc-induced proliferation and is upregulated in tumors. Curr. Biol. 2006;16:971–981. doi: 10.1016/j.cub.2006.04.027. [DOI] [PubMed] [Google Scholar]
- 85.Sakita-Suto S, Kanda A, Suzuki F, Sato S, Takata T, Tatsuka M. Aurora-B regulates RNA methyltransferase NSUN2. Mol. Biol. Cell. 2007;18:1107–1117. doi: 10.1091/mbc.E06-11-1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Brzezicha B, Schmidt M, Makalowska I, Jarmolowski A, Pienkowska J, Szweykowska-Kulinska Z. Identification of human tRNA:m5C methyltransferase catalysing intron-dependent m5C formation in the first position of the anticodon of the pre-tRNALeu(CAA) Nucleic Acids Res. 2006;34:6034–6043. doi: 10.1093/nar/gkl765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.de Beus E, Brockenbrough JS, Hong B, Aris JP. Yeast NOP2 encodes an essential nucleolar protein with homology to a human proliferation marker. J. Cell. Biol. 1994;127:1799–1813. doi: 10.1083/jcb.127.6.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hong B, Brockenbrough JS, Wu P, Aris JP. Nop2p is required for pre-rRNA processing and 60S ribosome subunit synthesis in yeast. Mol. Cell. Biol. 1997;17:378–388. doi: 10.1128/mcb.17.1.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.King M, Ton D, Redman KL. A conserved motif in the yeast nucleolar protein Nop2p contains an essential cysteine residue. Biochem. J. 1999;337:29–35. [PMC free article] [PubMed] [Google Scholar]
- 90.King MY, Redman KL. RNA methyltransferases utilize two cysteine residues in the formation of 5-methylcytosine. Biochemistry. 2002;41:11218–11225. doi: 10.1021/bi026055q. [DOI] [PubMed] [Google Scholar]
- 91.Hong B, Wu K, Brockenbrough JS, Wu P, Aris JP. Temperature sensitive nop2 alleles defective in synthesis of 25S rRNA and large ribosomal subunits in Saccharomyces cerevisiae. Nucleic Acids Res. 2001;29:2927–2937. doi: 10.1093/nar/29.14.2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Koonin EV. Prediction of an rRNA methyltransferase domain in human tumor-specific nucleolar protein P120. Nucleic Acids Res. 1994;22:2476–2478. doi: 10.1093/nar/22.13.2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gustafson WC, Taylor CW, Valdez BC, Henning D, Phippard A, Ren Y, Busch H, Durban E. Nucleolar protein p120 contains an arginine-rich domain that binds to ribosomal RNA. Biochem. J. 1998;331:387–393. doi: 10.1042/bj3310387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Doll A, Grzeschik KH. Characterization of two novel genes, WBSCR20 and WBSCR22, deleted in Williams-Beuren syndrome. Cytogenet. Cell Genet. 2001;95:20–27. doi: 10.1159/000057012. [DOI] [PubMed] [Google Scholar]
- 95.Harris T, Marquez B, Suarez S, Schimenti J. Sperm motility defects and infertility in male mice with a mutation in Nsun7, a member of the Sun domain-containing family of putative RNA methyltransferases. Biol. Reprod. 2007;77:376–382. doi: 10.1095/biolreprod.106.058669. [DOI] [PubMed] [Google Scholar]
- 96.Wilkinson CR, Bartlett R, Nurse P, Bird AP. The fission yeast gene pmt1+ encodes a DNA methyltransferase homologue. Nucleic Acids Res. 1995;23:203–210. doi: 10.1093/nar/23.2.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yoder JA, Bestor TH. A candidate mammalian DNA methyltransferase related to pmt1p of fission yeast. Hum. Mol. Genet. 1998;7:279–284. doi: 10.1093/hmg/7.2.279. [DOI] [PubMed] [Google Scholar]
- 98.Okano M, Xie S, Li E. Dnmt2 is not required for de novo and maintenance methylation of viral DNA in embryonic stem cells. Nucleic Acids Res. 1998;26:2536–2540. doi: 10.1093/nar/26.11.2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Jeltsch A, Nellen W, Lyko F. Two substrates are better than one: dual specificities for Dnmt2 methyltransferases. Trends Biochem. Sci. 2006;31:306–308. doi: 10.1016/j.tibs.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 100.Dong A, Yoder JA, Zhang X, Zhou L, Bestor TH, Cheng X. Structure of human DNMT2, an enigmatic DNA methyltransferase homolog that displays denaturant-resistant binding to DNA. Nucleic Acids Res. 2001;29:439–448. doi: 10.1093/nar/29.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu. Rev. Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- 102.Goll MG, Kirpekar F, Maggert KA, Yoder JA, Hsieh CL, Zhang X, Golic KG, Jacobsen SE, Bestor TH. Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science. 2006;311:395–398. doi: 10.1126/science.1120976. [DOI] [PubMed] [Google Scholar]
- 103.Phalke S, Nickel O, Walluscheck D, Hortig F, Onorati MC, Reuter G. Retrotransposon silencing and telomere integrity in somatic cells of Drosophila depends on the cytosine-5 methyltransferase DNMT2. Nat. Genet. 2009;41:696–702. doi: 10.1038/ng.360. [DOI] [PubMed] [Google Scholar]
- 104.Ochs RL, Reilly MT, Freeman JW, Busch H. Intranucleolar localization of human proliferating cell nucleolar antigen p120. Cancer Res. 1988;48:6523–6529. [PubMed] [Google Scholar]
- 105.Schaefer M, Steringer JP, Lyko F. The Drosophila cytosine-5 methyltransferase Dnmt2 is associated with the nuclear matrix and can access DNA during mitosis. PLoS ONE. 2008;3:e1414. doi: 10.1371/journal.pone.0001414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Liu Y, Santi DV. m5C RNA and m5C DNA methyl transferases use different cysteine residues as catalysts. Proc. Natl Acad. Sci. USA. 2000;97:8263–8265. doi: 10.1073/pnas.97.15.8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Jurkowski TP, Meusburger M, Phalke S, Helm M, Nellen W, Reuter G, Jeltsch A. Human DNMT2 methylates tRNAAsp molecules using a DNA methyltransferase-like catalytic mechanism. RNA. 2008;14:1663–1670. doi: 10.1261/rna.970408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Redman KL. Assembly of protein-RNA complexes using natural RNA and mutant forms of an RNA cytosine methyltransferase. Biomacromolecules. 2006;7:3321–3326. doi: 10.1021/bm051012l. [DOI] [PubMed] [Google Scholar]
- 109.Helm M. Post-transcriptional nucleotide modification and alternative folding of RNA. Nucleic Acids Res. 2006;34:721–733. doi: 10.1093/nar/gkj471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Chen Y, Sierzputowska-Gracz H, Guenther R, Everett K, Agris PF. 5-Methylcytidine is required for cooperative binding of Mg2+ and a conformational transition at the anticodon stem-loop of yeast phenylalanine tRNA. Biochemistry. 1993;32:10249–10253. doi: 10.1021/bi00089a047. [DOI] [PubMed] [Google Scholar]
- 111.Basti MM, Stuart JW, Lam AT, Guenther R, Agris PF. Design, biological activity and NMR-solution structure of a DNA analogue of yeast tRNAPhe anticodon domain. Nat. Struct. Biol. 1996;3:38–44. doi: 10.1038/nsb0196-38. [DOI] [PubMed] [Google Scholar]
- 112.Stuart JW, Koshlap KM, Guenther R, Agris PF. Naturally-occurring modification restricts the anticodon domain conformational space of tRNAPhe. J. Mol. Biol. 2003;334:901–918. doi: 10.1016/j.jmb.2003.09.058. [DOI] [PubMed] [Google Scholar]
- 113.Kadaba S, Krueger A, Trice T, Krecic AM, Hinnebusch AG, Anderson J. Nuclear surveillance and degradation of hypomodified initiator tRNAMet in S. cerevisiae. Genes Dev. 2004;18:1227–1240. doi: 10.1101/gad.1183804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Alexandrov A, Chernyakov I, Gu W, Hiley SL, Hughes TR, Grayhack EJ, Phizicky EM. Rapid tRNA decay can result from lack of nonessential modifications. Mol. Cell. 2006;21:87–96. doi: 10.1016/j.molcel.2005.10.036. [DOI] [PubMed] [Google Scholar]
- 115.Chernyakov I, Whipple JM, Kotelawala L, Grayhack EJ, Phizicky EM. Degradation of several hypomodified mature tRNA species in Saccharomyces cerevisiae is mediated by Met22 and the 5′-3′ exonucleases Rat1 and Xrn1. Genes Dev. 2008;22:1369–1380. doi: 10.1101/gad.1654308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Strobel MC, Abelson J. Effect of intron mutations on processing and function of Saccharomyces cerevisiae SUP53 tRNA in vitro and in vivo. Mol. Cell. Biol. 1986;6:2663–2673. doi: 10.1128/mcb.6.7.2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Vicens Q, Westhof E. Crystal structure of paromomycin docked into the eubacterial ribosomal decoding A site. Strucure. 2001;9:647–658. doi: 10.1016/s0969-2126(01)00629-3. [DOI] [PubMed] [Google Scholar]
- 118.Kariko K, Buckstein M, Ni H, Weissman D. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity. 2005;23:165–175. doi: 10.1016/j.immuni.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 119.Rassoulzadegan M, Grandjean V, Gounon P, Vincent S, Gillot I, Cuzin F. RNA-mediated non-mendelian inheritance of an epigenetic change in the mouse. Nature. 2006;441:469–474. doi: 10.1038/nature04674. [DOI] [PubMed] [Google Scholar]
- 120.Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat. Rev. Genet. 2008;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 121.Huang Y, Ji L, Huang Q, Vassylyev DG, Chen X, Ma JB. Structural insights into mechanisms of the small RNA methyltransferase HEN1. Nature. 2009;461:823–827. doi: 10.1038/nature08433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Rai K, Chidester S, Zavala CV, Manos EJ, James SR, Karpf AR, Jones DA, Cairns BR. Dnmt2 functions in the cytoplasm to promote liver, brain, and retina development in zebrafish. Genes Dev. 2007;21:261–266. doi: 10.1101/gad.1472907. [DOI] [PMC free article] [PubMed] [Google Scholar]