Abstract
The ability to target methylation to specific genomic sites would further the study of DNA methylation’s biological role and potentially offer a tool for silencing gene expression and for treating diseases involving abnormal hypomethylation. The end-to-end fusion of DNA methyltransferases to zinc fingers has been shown to bias methylation to desired regions. However, the strategy is inherently limited because the methyltransferase domain remains active regardless of whether the zinc finger domain is bound at its cognate site and can methylate non-target sites. We demonstrate an alternative strategy in which fragments of a DNA methyltransferase, compromised in their ability to methylate DNA, are fused to two zinc fingers designed to bind 9 bp sites flanking a methylation target site. Using the naturally heterodimeric DNA methyltransferase M.EcoHK31I, which methylates the inner cytosine of 5′-YGGCCR-3′, we demonstrate that this strategy can yield a methyltransferase capable of significant levels of methylation at the target site with undetectable levels of methylation at non-target sites in Escherichia coli. However, some non-target methylation could be detected at higher expression levels of the zinc finger methyltransferase indicating that further improvements will be necessary to attain the desired exclusive target specificity.
INTRODUCTION
Methylation of cytosine residues is a key epigenetic modification of genomes. Methylation patterns appear to play key roles in normal developmental processes and diseases. A variety of evidence indicates that methylation of CpG sites is involved in shaping chromatin structure, silencing of parasitic DNA and regulating gene expression. Genomic methylation patterns are species dependent but mammalian cells normally exhibit global CpG methylation, with small regions of non-methylated DNA (1). These unmethylated sequences correspond to dense regions of CpG sequences (CpG islands) that occur in known promoter regions. Hypomethylated islands correspond to actively transcribed genes, and when these promoter regions become hypermethylated the gene is silenced. Methylation in promoter regions can inhibit the binding of transcription factors (2,3) and may signal chromatin remodeling to further prevent transcription. The ability to specifically target single sites or regions in CpG islands for methylation would be very useful for the study of DNA methylation, for the silencing of genes of interest, and as a potential therapeutic strategy for diseases involving abnormal hypomethylation. Targeted methylation of promoter regions has been demonstrated to reduce transcriptional activity in vitro and in vivo (4,5); however, the issue of specificity (the difference between levels of exogenously derived methylation at target and non-target sites) is paramount. Abnormal methylation patterns have been linked to several diseases, cancer being the most studied. To prevent any unintended or adverse consequences, the amount of non-target methylation must be eliminated or greatly reduced to prevent deleterious effects.
Current methods of targeting DNA methylation include genetically fusing methyltransferases to DNA binding domains to localize the methyltransferase next to a target methylation site (4–11). However, since the methyltransferase domain presumably remains active in the absence of the DNA binding domain associating with its target site, the methyltransferase is free to methylate other sites when not bound to its target site. Thus, the level of specificity that can be achieved by this approach is inherently limited. Accordingly, analyses of the methylation patterns created using this strategy have shown methylation both at target and non-target sites, but with preferential methylation at target sites over non-target sites (5,8,11). This non-target methylation limits the use of these fusion proteins as a research tool and raises the concern that unintended side effects resulting from non-target methylation could be a serious drawback for any therapeutic applications of these fusions in the future (12). The use of mutant methyltransferases with reduced activity has been proposed as an approach for improving specificity (6), but this strategy only serves as a route to achieving the optimum bias such fusions can offer and does not address the fundamental limitation of the approach.
To create a truly site-specific methyltransferase, methyltransferase activity must require the association of the DNA binding domain and its target site. An attractive route to achieving this goal involves splitting naturally monomeric methyltransferases into two fragments and fusing the fragments to different DNA binding domains that bind DNA flanking the target site for methylation (Figure 1A) (13,14). For the success of this strategy, the fragments must not appreciably associate in a functional form in solution (or when only one fragment is bound to DNA). This property could result from the fragments’ dissociation constant being significantly above their cellular concentration. In contrast, when both fragments are bound to proximal sites on the DNA, their local, effective concentration increases above the Kd (and the association also can be facilitated due to entropic reasons if the fragments are oriented correctly) and an active methyltransferase is formed that can only methylate the target site. In addition, the proximity of the two fragments might assist attainment of the correct folded structure, as has been observed with protein complementation assays (15).
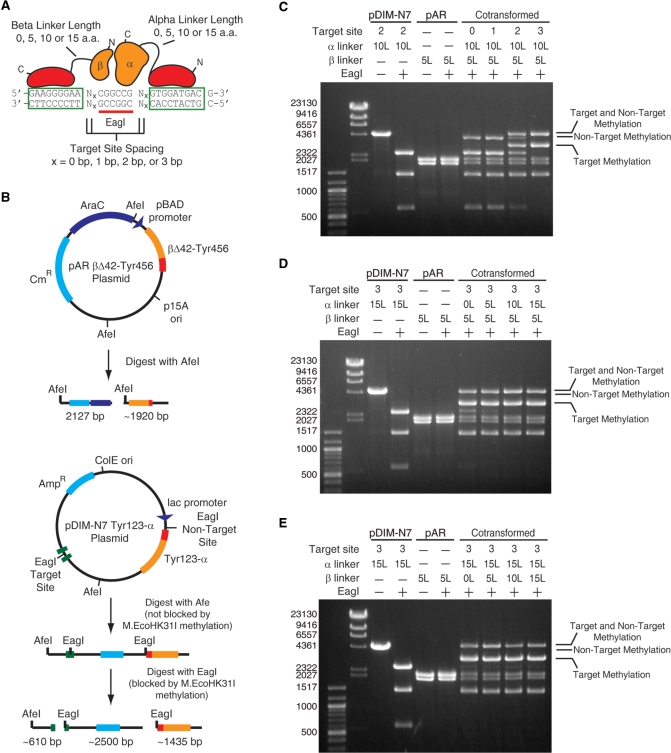
Figure 1.
The effect of fusion linker length and spacing at the target site on the methyltransferase activity and specificity of M.EcoHK31I-derived constructs with a 42-residue deletion on the β fragment. (A) Schematic of the various constructs tested showing the variability in linker length between zinc finger and methyltransferase fragments and the distance between the zinc finger’s binding site and the target site for methylation. (B) The two plasmids encoding the two methyltransferase fragments and the DNA fragments that result from AfeI and EagI digestion if the EagI site is not methylated (methylation blocks EagI but not AfeI digestion). Methylation creates uniquely sized bands depending on if the target or non-target site is protected. Representative results are shown to demonstrate the observed effects of (C) zinc finger–target site distance, (D) linker lengths between the α fragment and Tyr123 and (E) linker lengths between the β fragment and Tyr456. Linker lengths are designated by 0, 5, 10 or 15 L and the spacing at the target site designated by the number of base pairs between the target site and zinc finger site. Individual plasmids digested with AfeI are also shown for size comparisons (pDIM-N7 and pAR) with (+) and without (−) EagI digestion. The locations of bands resulting from only target methylation, only non-target methylation and methylation at both sites are indicated. The plasmids were prepared from cells grown in LB media.
Here, we have taken the approach of starting with a naturally heterodimeric methyltransferase and reducing its association through mutagenesis (by creating deletion mutants) before fusion of the fragments to DNA binding domains. This approach seemed more tractable than using artificially split methyltransferases (13,14) since natural heterodimeric methyltransferases have evolved to be produced as separate polypeptides. We chose the methyltransferase M.EcoHK31I as our model protein. M.EcoHK31I has α and β peptide chains that require association for methyltransferase activity (16). The gene encoding M.EcoHK31I has an unusual architecture. The β chain is encoded entirely within a different reading frame of the α fragment. The large α fragment contains both the DNA binding domain and the catalytic domain (motifs I–VIII and X and TRD), whereas the smaller β fragment contains motif IX that helps stabilize site-specific binding (17). M.EcoHK31I recognizes the sequence 5′-YGGCCR-3′ and methylates the innermost cytosine. Although M.EcoHK31I does not methylate CpG sequences, it is an attractive model protein for testing our strategy. Previous in vitro studies of M.EcoHK31I have shown that deletions of at least 42-amino acid residues from the N-terminus of the β fragment eliminated methylation activity due to a decrease in the association of the two fragments (17). Furthermore, there is growing evidence of the occurrence of non-CpG methylation in mammalian genomes of diseased or cancerous cells (18,19) and embryonic stem cells (20). Any targeted methyltransferase we create from M.EcoHK31I may find use for the study of this type of methylation or for the study of non-CpG methylation found in plants and other organisms.
MATERIALS AND METHODS
Plasmids
Restriction enzymes, T4 DNA ligase and T4 polynucleotide kinase were purchased from New England Biolabs (Ipswich, MA, USA) and used according to manufacturer’s instructions. Agarose gel electrophoresis and PCR were performed essentially as described (21). Escherichia coli K-12 strain ER2267 [F′ proA+B+ lacIq Δ(lacZ)M15 zzf::mini-Tn10 (KanR)/ Δ(argF-lacZ)U169 glnV44 e14−(McrA−) rfbD1? recA1 relA1? endA1 spoT1? thi-1 Δ(mcrC-mrr)114::IS10] was obtained from New England Biolabs (Ipswich, MA, USA) and was used as a host for DNA cloning experiments and methylation protection assays. Cells were grown in LB media supplemented with chloramphenicol (50 µg/ml) and/or ampicillin (100 µg/ml). All DNA primers were obtained from Invitrogen (Carlsbad, CA, USA). Detailed descriptions of plasmid modifications and the creation of methyltransferase fragment constructs can be found in Supplementary Data.
Construction of pDIM-N7 and pAR for expression of M.EcoHK31I fragments
Expression plasmids were derived from pDIM-N7-MHhaI[1–302] and pAR-MHhaI[29–327] (22). Plasmids containing the M.EcoHK31IA and M.EcoHK31IB genes were a gift from Shaw (16). The M.EcoHK31IA and M.EcoHK31IB genes were amplified by PCR and inserted into pDIM-N7 and pAR plasmids, respectively. The truncated M.EcoHK31IB fragments were amplified using the appropriate primers to create truncations ranging from 35- to 50-amino acid residues. The amplified truncated fragment was then inserted into pAR plasmid (see Supplementary Figure S2, Supplementary Data, for detailed plasmid maps).
Construction of pDIM-N7 Tyr123-α and pAR β-Tyr456 plasmids for combinatorial library
The target sites were designed with a methylation site (5′-CGGCCG-3′) flanked by the Tyr123 and Tyr456 zinc finger binding sites (Figure 1A). The zinc finger DNA binding sites are separated from the methylation site by 0, 1, 2 or 3 bp (labeled 0, 1, 2 and 3 bp target sites) with the methylation site centered between the zinc finger binding sites. Oligonucleotides containing the zinc finger binding sites and methylation site were annealed, phosphorylated and ligated into the pDIM-N7 M.EcoHK31IA plasmid.
The zinc finger protein Tyr123 was appended to the N-terminus of the M.EcoHK31IA fragment either directly (linker 0L) or using amino acid linkers of various sizes. Linkers were designed to be flexible and are repeating GGGGS sequences to create linkers of the length 5, 10 or 15 residues long (linkers 5, 10 or 15 L). Zinc finger protein Tyr456 was fused to the C-terminus of the M.EcoHK31IB Δ42 (βΔ42) fragment using identical linkers as the Tyr123-α fusions. Linkers were appended to the appropriate methyltransferase fragments and the Tyr123 and Tyr456 zinc finger proteins by PCR. Overlap extension PCR was then done to fuse the fragments to the zinc fingers, and fused constructs were ligated into pDIM-N7 and pAR plasmids. This created 16 pDIM-N7 Tyr123-M.EcoHK31IA (pDIM-N7 Tyr123-α) plasmids containing every possible combination of the target site (0, 1, 2 or 3 bp) and Tyr123-M.EcoHK31IA linker length (0, 5, 10 or 15 L) and four unique pAR M.EcoHK31IB Δ42-Tyr456 (pAR βΔ42-Tyr456) plasmids with the four different linker sizes. These plasmids were cotransformed into ER2267 cells to form a 64-member combinatorial M.EcoHK31I Tyr123-α/βΔ42-Tyr456 library.
Construction of plasmids for M.EcoHK31IB deletion optimization and control plasmids
The EagI non-target site was modified so that the overlapping NotI site was removed by changing base pairs flanking the EagI site to adenines. This created identical flanking nucleotides as the target EagI site (Figure 2B). We also created several control plasmids to test if the zinc finger binding sites and zinc finger proteins were necessary for targeted methylation. Control target sites lack the zinc finger binding sites but still contain the EagI site with similar flanking base pairs as the 3 bp target site (Figure 2B). The control target site oligonucleotides were annealed, phosphorylated and ligated into the pDIM-N7 Tyr123-15L-α plasmid in place of the original target site. The M.EcoHK31IA gene without Tyr123 was amplified and inserted into the pDIM-N7 plasmids containing the 3 bp target site and the plasmid containing a control target site to create the pDIM-N7 α plasmids.
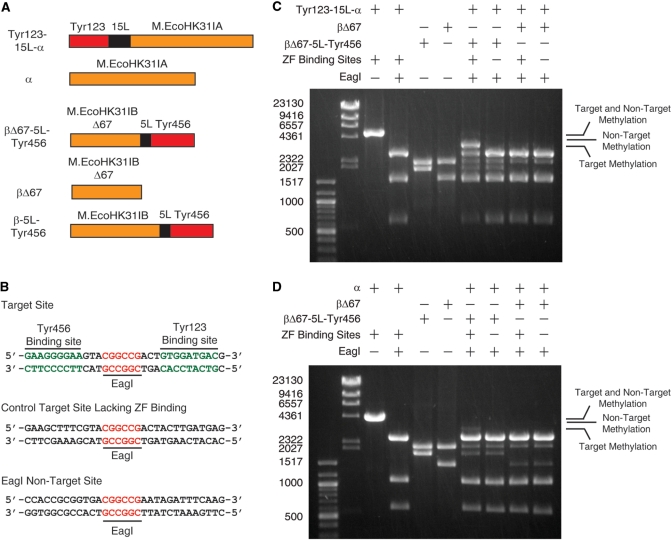
Figure 2.
The effects of removing zinc finger proteins or zinc finger binding sites on the methylation activity and specificity of M.EcoHK31I-derived constructs with a 67-residue deletion on the β fragment. (A) Schematic of the fusion and control constructs used in these experiments. (B) DNA sequences at the target sites (with and without zinc finger binding sites) and non-target site. All pDIM-N7 plasmids contain a non-target site and either the target site containing zinc finger binding sites (+) or containing a control site for which the zinc fingers lack affinity (–). Gel electrophoresis results for plasmids digested with AfeI and EagI (performed as in Figure 1) are shown for plasmids containing the DNA encoding (C) Tyr123-15L-α and (D) α cotransformed with plasmids encoding βΔ67 or βΔ67-5L-Tyr456. Results for AfeI or AfeI/EagI digestions of unmethylated control plasmids are shown for size comparison. The locations of bands resulting from only target methylation, only non-target methylation and methylation at both sites are indicated. The plasmids were isolated from cells grown in LB media.
New M.EcoHK31IB-5L-Tyr456 truncations were created by amplifying the M.EcoHK31IBΔ42-5L-Tyr456 fragment with the appropriate primers. Truncations were created up to 70-amino acid residues and fusions were ligated into the pAR plasmid. M.EcoHK31IB fragments without zinc finger proteins were also amplified and ligated into the pAR plasmid. A full length M.EcoHK31IB-5L-Tyr456 fusion was also created by amplifying the full length M.EcoHK31IB gene and fusing to the Tyrzif456-5L fragment by overlap extension PCR as performed before. The full-length fusion was then inserted in the new pAR plasmid.
Creation of fusion pDIM-N7 plasmids
Fusion plasmids were made by digesting the desired pDIM-N7 plasmid and pAR plasmid with EcoRI and SpeI. The pAR plasmid section containing the desired M.EcoHK31IB fusion gene, pBAD promoter and araC gene is isolated by gel extraction using the QIAquick Gel Extraction kit (Qiagen, Vaencia, CA, USA) and ligated into the pDIM-N7 vector (Figure 3A).
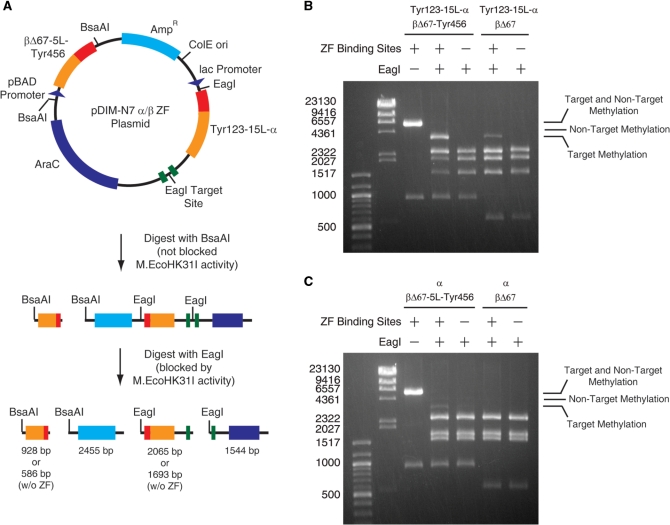
Figure 3.
Methylation activity and specificity resulting from a single plasmid encoding Tyr123-15L-α and βΔ67-5L-Tyr456 (A). Schematic of the plasmid and its digestion with BsaAI (not-blocked by methylation) and EagI (blocked by methylation). Gel electrophoresis results for plasmids digested with BsaAI and EagI are shown for plasmids contain the DNA encoding (B) Tyr123-15L-α and (C) α Combinations with DNA encoding either βΔ67 or βΔ67-5L-Tyr456 and with and without zinc finger binding sites flanking the target site were tested. The locations of bands resulting from only target methylation, only non-target methylation and methylation at both sites are indicated. Plasmids were isolated from cells grown in LB media supplemented with 0.2% glucose.
Restriction endonuclease protection assays
All in vivo assays were performed using E. coli strain ER2267 (New England Biolabs, Ipswich, MA, USA). Plasmids were transformed into ER2267 cells. Cells were grown from a glycerol stock in 25 ml culture tubes in 10 ml of LB media supplemented with antibiotics as necessary to maintain plasmids (100 µg/ml ampicillin and 50 µg/ml chloramphenicol). In some experiments, 0.2–5% w/v glucose was added as indicated to repress expression of both methyltransferase fragments. In other experiments, the media was supplemented with 1 mM IPTG to induce the lac promoter or 1mM arabinose to induce the pBAD promoter. Cultures were incubated overnight (12–14 h) at 37°C and the plasmid DNA was isolated using QIAprep spin miniprep kit (Qiagen, Valencia, CA, USA).
For initial in vivo deletion studies with pDIM-N7 M.EcoHK31IA plasmids (Supplementary Figure S2A, Supplementary Data), 500 ng of DNA was incubated with 3 U of NotI. For cotransformed samples containing the pDIM-N7 Tyr123-α or pDIM-N7 α plasmids, 2 µg of purified DNA were first digested with 20 U AfeI to linearize pDIM-N7 plasmids and purified using the DNA Clean and Concentrator kit (Zymo Research Corp., Orange, CA, USA). Then, 500 ng of linearized DNA was digested with 12.5 U of EagI and analyzed by gel electrophoresis using a 0.8% agarose gel.
For cases in which both methyltransferase fragments were encoded on the same plasmid, 500 ng of DNA was incubated with 3.75 U BsaAI and 12.5 U of EagI to determine methylation at the target and non-target EagI sites. Alternatively, plasmids were digested with 15 U of XbaI and 3.75 U of EaeI overnight at 37°C to probe protection at all potential methylation sites. After digestion, 500 ng of DNA was analyzed by agarose gel electrophoresis using a 0.8% agarose gel.
Bisulfite sequencing
Cultures of ER2267 containing the pDIM-N7-Tyr123-15L-α/βΔ67-5L-Tyr456 vector were grown in 10 ml LB, 100 µg/ml ampicillin and 0.2% w/v glucose. Plasmid DNA was recovered and digested to linearize DNA. Bisulfite conversion was performed using the Epitect Bisulfite Kit (Qiagen, Valencia, CA, USA) according to manufacturer’s instructions. After the bisulfite conversion, the target region (using primers Bisulfite Top Target-for and Bisulfite Top Target-rev or Bisulfite Bottom Target-for and Bisulfite Bottom Target-rev) and the non-target region containing an EagI site (using primers Bisulfite Non-Target-for and Bisulfite Non-Target-rev) were amplified by PCR using Qiagen’s HotStarTaq DNA polymerase. Primer sequences can be found in Supplementary Table S4. The fragments were then inserted into the pAR plasmid and transformed into ER2267 cells. Random individual colonies were selected for sequencing. Sequencing was performed by Genewiz (South Plainfield, NJ, USA).
RESULTS
M.EcoHK31I is functional in vivo when the two fragments are encoded as separate genes.
The β fragment of M.EcoHK31I is entirely encoded in an alternate reading frame within the DNA encoding the α fragment (16). We placed DNA encoding the α- and β-fragment of M.EcoHK31I on separate compatible plasmids. The DNA encoding M.EcoHK31I was placed downstream from the lac promoter on plasmid derived from pDIM-N7-MHhaI[1–302] (22). This plasmid contained an M.EcoHK31I methylation site that overlaps a NotI site (Supplementary Figure S2A, Supplementary Data); thus, methylation can be detected by observation of protection from NotI digestion. The start codon for the β protein was removed by silently mutating in the frame of the α-subunit. However, this gene still conferred some methylation activity to cells, since partial protection from NotI digestion of the plasmid was observed. Since the β fragment does not require the first 42 N-terminal amino acids for activity in vitro (17), we suspected that methionine codons at positions 17 or 26 in the β fragment reading frame were serving as internal translation initiation sites. A gene in which these two sites were removed by silently mutating in the α reading frame no longer conferred any methylation activity as judged by a lack of protection at the NotI site (Supplementary Figure S2B, Supplementary Data). We refer to this mutated gene as M.EcoHK31IA. DNA encoding just the β fragment (referred to as M.EcoHK31IB) was placed downstream from the pBAD promoter on a compatible plasmid. Co-transformation of these plasmids into cells resulted in plasmid DNA that showed complete protection from NotI digestion (Supplementary Figure S2B, Supplementary Data) demonstrating that our split gene version of M.EcoHK31I methylates DNA in vivo.
Removal of 50 amino acids from the N-terminus of the β fragment severely reduces but does not eliminate methylation activity in vivo
We have previously shown that fragments of M.HhaI that can assemble into a functional methyltransferase without assistance retain the ability to methylate non-target sites when fused to zinc finger domains (13). In order to engineer our desired site-specific methyltransferase, we required mutants of M.EcoHK31I that can no longer assemble into a functional methyltransferase at cellular concentrations. Studies on purified M.EcoHK31I have shown that progressive deletion of amino acids from the N-terminus of the β fragment results in a progressive decrease in the association constant of the two fragments (17). This correlates with a loss of methylation activity as demonstrated by in vitro methylation assays. Almost no activity was observed when 41 amino acids were deleted from the β fragment and the protein was assayed at 1 µM concentration (the Kd for fragment interaction was 2.4 µM) (17). Deletions of 42, 46 (Kd = 5.3 µM) and 50 (Kd = 7.1 µM) amino acids showed no activity at this same concentration (17).
We created a series of variants of M.EcoHK31IB in which up to 50 codons were deleted from the 5′ end and tested their ability to confer methylation activity to cells expressing an intact α fragment. Cells were grown without the addition of IPTG or arabinose (i.e. under non-inducing conditions); thus, activity is dependent on the low-level, leaky expression of the two fragments. Reduced protection from NotI digestion was observed starting with the βΔ37 truncation fragment and protection progressively decreased with increasing truncation of the β fragment (Supplementary Figure S2B, Supplementary Data). In contrast to previous in vitro experiments (17), we observed some protection with βΔ42, βΔ45 and βΔ50. Potentially, cellular concentrations of the fragments are higher than concentrations used in the in vitro experiments.
Design of the methylation target site and fusion of the α and βΔ42 fragments to zinc fingers
Even though we did not obtain a truncation mutant that completely lacked methylation activity in vivo, we thought that the severely reduced activity obtained with βΔ42 was sufficient for testing whether fusing zinc fingers to these fragments could be used to bias the methylation to a target site located between the zinc fingers’ binding sites. The plasmid encoding the α fragment was modified to contain a target site for M.EcoHK31I methylation flanked by zinc finger binding sites (Figure 1A and B). Four different target sites were created differing only in the number of bp between the M.EcoHK31I site and the zinc finger binding sites (Figure 1A). In all cases, this target site was also an EagI site. EagI activity is blocked by methylation—affording an easy assay for methylation at this site. A second ‘non-target’ M.EcoHK31I methylation site with an EagI site was located elsewhere on the plasmid such that digestion with EagI could be used to determine which of the two sites was methylated (Figure 1B).
M.EcoHK31IA and M.EcoHK31IBΔ42 were fused to zinc fingers Tyr123 and Tyr456 (23), respectively, using amino acid linkers of 0, 5, 10 or 15 residues consisting of repeats of the sequence GGGGS. Tyr123 binds to the sequence 5′-GTGGATGAC-3′ and Tyr456 binds 5′-GAAGGGGAA-3′. Tyr123 was fused to the N-terminus of the M.EcoHK31IA fragment and Tyr456 was attached to the C-terminus of M.EcoHK31IB Δ42. In total, 16 different plasmids containing variants of the α fragment were created (4 different linkers between the α fragment and Tyr123 × 4 different spacing between the target site and the zinc finger binding sites). These were cotransformed with plasmids encoding the βΔ42 fragment (four different linkers) into ER2267 cells resulting in 64 combinations of target site spacing and linker lengths. Glycerol cell stocks of these 64 different cells were made from overnight inocula grown in LB containing 0.2% glucose (to repress the lac and pBAD promoters).
Site-biased methylation is dependent on the relative position of the zinc finger binding sites and the target methylation site
LB media lacking glucose was inoculated with the cotransformants and incubated overnight at 37°C. The absence of glucose allowed leaky expression of the Tyr123-α and βΔ42-Tyr456 proteins. Plasmid DNA was recovered, digested with AfeI restriction enzyme to linearize the plasmid (AfeI is not blocked by M.EcoHK31I activity), digested with EagI and analyzed by agarose gel electrophoresis. Methylation at the target and non-target sites can be distinguished by the size of the bands that result (Figure 1B). Methylation at the non-target site band results in a unique ∼3935 bp band, and methylation at the target site results in a unique ∼3110 bp band. Methylation at both sites results in a ∼4550 bp band. A lack of methylation at either site results in three bands all ≤2500 bp.
Representative results illustrating the major trends in methylation activity and specificity observed in the 64 different construct combinations are shown in Figure 1C–E. The distance between the zinc finger binding sites and the target methylation site had the greatest affect on methylation specificity (Figure 1C). Short distances (0 and 1 bp) inhibited methylation at the target site and resulted in a methylation only at the non-target site. Longer distances (2 and 3 bp) results in a mix of both target and non-target site methylation, with a definite bias toward target site methylation occurring with the 3 bp spacing. We suspect that at short distances, zinc finger binding to its cognate sites on the DNA may block the methyltransferase from methylating at the target site.
To a lesser extent, the length of the peptide linker joining Tyr123 and the α subunit impacted methylation specificity. This was most noticeable with the 3 bp distance between the zinc finger binding site and the target site. Incomplete methylation at the target site is evident with shorter linker lengths (0 and 5 amino acids) whereas nearly complete methylation at the target site is observed with the longer linkers (10 and 15 amino acids) (Figure 1D). Methylation levels at the non-target site appear unaffected by the linker length. These trends are most evident by observing the gradual disappearance of the band at ∼2500 bp (indicating increasing methylation at the target site) and the invariable levels of the 1435 bp band (indicating unchanging levels of non-target methylation) as the linker length increases. The length of the linkers between Tyr456 and βΔ42 did not have a significant effect on the methylation patterns in any context (Figure 1E).
In all constructs, higher levels of methylation were observed compared to constructs lacking zinc fingers. This is potentially due to the stabilization of one or both of the M.EcoHK31I fragments by the zinc finger domain, which would result in increased fragment concentrations in the cells. The highest levels of methylation and specificity for the target site was observed with a 3 bp spacing between the zinc finger binding site and the target site and longer linker lengths (10 or 15 residues) in the Tyr123-α fragment. However, the occurrence of significant levels of non-target methylation prompted us to pursue further optimization.
Further optimization by longer truncations of the N-terminus of M.EcoHK31IB
To reduce non-specific methylation we created larger deletions of M.EcoHK31IB. Since the addition of zinc fingers to the M.EcoHK31I fragments increased overall methylation activity in the previous set of experiments, the effects of further truncations were performed with the methyltransferase fragments fused to zinc fingers. Since the βΔ42-Tyr456 linker lengths had little if any effect on target site methylation, a 5-residue linker (GGGGS) was chosen for the M.EcoHK31IB-Tyr456 fusions. A pDIM-N7 plasmid containing the 3 bp spacing between the zinc finger binding site and the target site and the Tyr123-15L-M.EcoHK31IA α subunit was used. A control plasmid in which the zinc finger binding sites were removed (control target site, Figure 2B) was used to test for the level of methylation independent of zinc finger binding.
Fusions of Tyr456 to M.EcoHK31IB containing deletions of 50, 55, 60, 65 and 70 amino acids from the N-terminus of the M.EcoHK31IB were constructed and cotransformed with pDIM-N7 Tyr123-15L-α. Assays for in vivo methylation were performed as before. Initial experiments indicated that in the absence of the zinc finger binding sites there was a slight and unexpected bias toward methylation at the target site (data not shown). Flanking base pairs to the cognate DNA recognition sites can affect restriction enzyme and methylation activities, and we suspected that this was occurring with M.EcoHK31I. In order to minimize these effects, the flanking base pairs at the non-target site were changed to match the target site. Changing the flanking sites eliminated the intrinsic methylation bias for the target site and all subsequent experiments used plasmids containing this modified non-target site (EagI non-target site, Figure 2B).
Using the pDIM-N7 plasmids with this new non-target site, we evaluated the methylation activities of the more extensive truncations of the β fragment. These experiments were performed in the context of both a target and non-target site so that both methylation activity and methylation specificity could be evaluated. A progressive decrease in methylation activity up to the deletion of 70 amino acids (Supplementary Figure S3A, Supplementary Data) was observed. Target site methylation decreased starting at the 65 amino acid deletion, and a significant decrease in methylation occurred between βΔ65-5L-Tyr456 and βΔ70-5L-Tyr456. Truncation lengths intermediate between 65 and 70 amino acids were also tested (Supplementary Figure S3B, Supplementary Data) and the βΔ67-5L-Tyr456 showed similar target methylation levels to βΔ65-5L-Tyr456, but non-target methylation appeared to be eliminated. Overall, non-target methylation decreased more rapidly than target methylation and seemed to disappear around βΔ66-5L-Tyr456. Deletions larger than Δ67 displayed a decrease in target methylation with a significant drop in methylation between Δ69 and Δ70. We decided to perform further studies on the Tyr123-15L-α and βΔ67-5L-Tyr456 combination since this pair seemed to almost completely lack non-target methylation activity while exhibiting significant target site methylation activity.
Methylation specificity for the target site depends on zinc finger binding
We next examined whether specificity for the target site depended on the zinc finger proteins and the zinc finger binding sites. All combinations of constructs (with and without zinc finger binding sites flanking the target site, M.EcoHK31IA with and without fusion to the Tyr123 zinc finger, M.EcoHK31IB Δ67 with and without fusion to the Tyr456 zinc finger) were created (Figure 2A and B). These constructs were cotransformed and tested for target and non-target methylation. A combination of high levels of target methylation (>50%) and a virtual lack of methylation at the non-target site was seen only with zinc fingers fused to the fragments and the presence of both zinc finger binding sites flanking the target site. Removal of the zinc finger binding sites eliminated methylation at the target site as did removal of Tyr456 or both Tyr456 and Tyr123 (Figure 2C and D). Removal of only Tyr123 resulted in low, but detectable, levels of target site methylation (Figure 2D). This suggests that although the Tyr456 zinc finger may be more important than Tyr123 in this context, the effect of the two zinc fingers is synergistic for targeting methylation.
Methylation specificity is affected by gene copy number and induction levels
For the next set of experiments, we combined the Tyr123-15L-α and βΔ67-5L-Tyr456 genes onto a single plasmid. A one-vector system is preferable to maintaining two plasmids in cells. The pDIM-N7 and pAR plasmids were designed to allow the facile construction of a single expression vector containing the Tyr123-15L-α and βΔ67-5L-Tyr456 and other fragment variants. The pDIM-N7 and pAR plasmids were digested and the pAR plasmid section containing the methyltransferase gene, pBAD promoter and AraC gene were ligated into the pDIM-N7 plasmid to form the pDIM-N7 α/β ZF plasmids. The plasmids contain the same target and non-target EagI sites, but a different enzyme was needed to linearize the larger fragment to create unique-size target and non-target bands for analysis. The plasmid was first digested with BsaAI, which excised the β fragment gene (Figure 3A). Subsequent digestion with EagI was used to distinguish between target and non-target methylation (Figure 3A).
The pAR plasmid has a lower copy number than the pDIM-N7 plasmid. Thus, an increase in the protein levels of the M.EcoHK31IB fragments was expected to result from placing the β fragment gene together with the α fragment gene on the pDIM-N7 plasmid. Growth of cells bearing the pDIM-N7 α/β ZF plasmids under the same conditions as the previous experiments with the cotransformed plasmids (i.e. leaky expression) resulted in continued high level of methylation at the target site, but detectable protection at the non-target site. However, by repressing expression with 0.2% glucose, non-target methylation could be eliminated, although low levels of methylation were detected in some experiments (Supplementary Figure S4A, Supplementary Data). Increasing the concentration from 0.2% to 5% (w/v) did not result in the loss of any target methylation but prevented non-target methylation (Supplementary Figure S4B, Supplementary Data). This indicates that high levels of glucose can be used to ensure minimal non-target methylation. Induction with either 1mM IPTG, 1mM arabinose or both increased non-target methylation but did not have a significant effect on target methylation (Supplementary Figure S4A, Supplementary Data). In the absence of the zinc finger binding sites flanking the target site and in the absence of any glucose repression, equally low levels of methylation occurred at the target and non-target sites. Although induction reduced the level of specificity for the desired target site, under all conditions methylation at the target site flanked by zinc finger binding sites was preferred over methylation at the non-target site. Maintenance of bias for target methylation over a wide range of expression conditions would not be expected for fusions of intact methyltransferase domains to zinc fingers. High level of expression of such fusions would result in the opportunity for the methyltransferase domain in the fusion to extensively methylate non-target sites.
We also created a single plasmid encoding full-length fragments β-5L-Tyr456 (i.e. no truncation of the β fragment) and Tyr123-15L-α and tested them for methylation activity in cultures grown in media with different glucose levels. Plasmids recovered from cells grown in 0.2% w/v and 5% w/v glucose were fully protected from EagI digestion (Supplementary Figure S5, Supplementary Data) indicating no specificity, as expected. This indicates that the reduction in methylation at the non-target site is due to the deletions made to the M.EcoHK31IB fragment.
We next sought to probe the effects of the zinc fingers and zinc finger binding sites in the new one-plasmid systems, similar to experiments performed with cotransformed plasmids in Figure 2. We performed these experiments at the glucose levels at which faint non-target methylation was sometimes previously observed: LB supplemented with 0.2% glucose. Methylation occurred only when zinc finger binding sites were present (Figure 3B) and non-target methylation was not observed under any conditions. Full protection at the target site was not observed. Analysis of the band intensities indicated that 50–60% of the target sites were methylated. Small amounts (∼10%) of target site methylation were observed when either the Tyr123 or Tyr456 zinc fingers (but not both) were removed (Figure 3B and C). As was observed using the two-plasmid system, both zinc fingers were not required for some bias toward methylation at the target site. However, the two zinc fingers synergistically acted to direct methylation to the target site, since protection with both zinc fingers exceeded the level expected by adding the level of protection seen with each zinc finger individually. Such a synergistic effect is the basis for the increase specificity that the split methyltransferase strategy offers.
Confirmation of methylation specificity by bisulfite sequencing
To confirm that the protection observed in restriction endonuclease assays resulted from methylation at the EagI sites, we performed bisulfite sequencing on the pDIM-N7 Tyr123-15L-α/βΔ67-5L-Tyr456 plasmid isolated from cells grown in LB media supplemented with 0.2% glucose. We sequenced both the target site (n = 10 on each strand) and the non-target site (n = 10). None of the non-target site clones sequenced was methylated. We sequenced the target site and probed both the top and bottom strands (of the sequence depicted in Figure 2B). We found that 4 out of 10 bottom strand clones and 1 out of 10 top strand clones showed methylation at the target site. These results confirm that of the restriction enzyme protection assays and suggest that there may be a bias toward methylating one strand over the other.
Tyr123-15L-α/βΔ67-5L-Tyr456 preferentially methylates the target site over other non-target sites
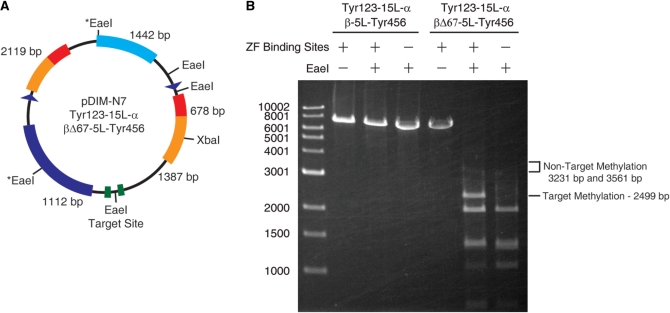
We next examined whether methylation was occurring at other M.EcoHK31I sites on the pDIM-N7 α/β ZF plasmid. There are five potential methylation sites on this plasmid and EaeI can digest all sites (5′-YGGCCR-3′), but only if they are not methylated. The EaeI restriction enzyme was not generally useful in previous experiments since the enzyme digests some sequences slower than others (24)—requiring DNA to be incubated overnight to ensure complete digestion. EaeI was useful here for probing methylation at previously unassayed non-target sites, since protection at either of two such sites (marked with an * in Figure 4A) would result in bands clearly distinguishable from the target methylation band. Analysis of the EaeI digestion of plasmids encoding Tyr123-15L-α and βΔ67-5L-Tyr456 showed a clear band where the target site band would occur and only a very faint band at about the size expected for methylation at these two non-target sites (Figure 4B). Thus, our construct exhibits specificity for the target site over other M.EcoHK31I sites on the plasmid in addition to the previously studied non-target site. In contrast, full-length fragments attached to zinc fingers (Tyr123-5L-α and β-5L-Tyr456) fully protect plasmids from digestion with EaeI (Figure 4B).
Figure 4.
Tyr123-15L-α/βΔ67-5L-Tyr456 has a high preference for the target site over other M.EcoHK31I methylation sites. (A) The pDIM-N7 plasmid contains five methylation sites for M.EcoHK31I that overlap EaeI restriction enzyme sites (EaeI is blocked by methylation) and one XbaI site used to linearize the plasmid and facilitate analysis. The sizes of the five largest DNA fragments that result from XbaI/EaeI digestion of unmethylated DNA are shown. (B) Agarose gel electrophoresis results of XbaI/EaeI digestions of plasmids expressing Tyr123-15L-α/βΔ67-5L-Tyr456 or Tyr123-15L-α/β-5L-Tyr456 (i.e. control protein with no truncation of the β fragment). Methylation at the target site yields a 2499 bp band if the flanking sites are not methylated. Methylation at the two EaeI sites marked with an asterisk would result in bands at either 3231 bp or 3561 bp if their flanking sites are not methylated. These two EaeI sites are non-target sites that were not tested in previous experiments. Plasmids were isolated from cells grown in LB media supplemented with 0.2% glucose.
DISCUSSION AND CONCLUSIONS
Several studies have examined the effect of zinc finger–methyltransferase fusions on the methylation patterns in cells that have endogenous mechanisms for methylating their DNA (4,5,7). The strength of these studies lies in the examination of the fusions’ effect in the cell type for which they are intended. However, these studies are limited in their ability to address the fusions’ inherent methylation specificity in a cellular environment, since it is hard to distinguish between methylation in a region caused solely by an exogenous methyltransferase acting non-specifically and methylation resulting from very specific exogenous methylation spreading in a region due to endogenous methyltransferase activity. The study of such fusions’ effects in yeast (11,25) and E. coli (6), which lack endogenous methylation at the target site for exogenous methylation, have not shown that such fusions can achieve both high levels of target site methylation and high specificity for the target site. This is not surprising since methylation by these fusions does not inherently require association of the zinc finger to its cognate site. Optimal site-biased methylation in such fusions should occur when the fusion concentration is kept low enough such that few fusion molecules exist that are not bound to the target site through the zinc finger. Reduction of methyltransferase activity (6) can be used for a similar purpose when fusion protein concentrations is higher (i.e. although there are more free fusion molecules, their intrinsic ability to methylate is decreased). However, these optimizations do not address the inherent limitation of the approach.
The split methyltransferase strategy described here addresses this problem by incorporating a mechanism for limiting the activity of free methyltransferase proteins and requiring binding to the desired target site for the assembly of an active methyltransferase. We have engineered a construct capable of methylating a high fraction of the target site (>50%) with undetectable levels of non-target methylation. This activity depended on the proper location of the zinc fingers’ cognate binding site relative to the target site for methylation. The synergistic effect of the two zinc fingers on target site methylation is exactly the advantageous property this strategy promises. Our results are a significant step forward toward the goal of methylating unique sites within a genome.
However, limitations of our fusions are apparent. Although it is encouraging that a bias for target methylation could be maintained over a wide range of induction conditions, expression of the fragments needed to be kept low to completely avoid non-target site methylation. In addition, specific methylation of a unique site in the human genome will require that both zinc finger fusions be bound to their cognate sites for targeted methylation to occur. This is reminiscent of the mechanism of the action of zinc finger nucleases (ZFNs), which requires that two ZFNs be bound to their cognate sites for dimerization of the nuclease domain and efficient cleavage of the target site (23). Our constructs with only one zinc finger still biased methylation to the target site. One possible explanation is that non-targeted methylation results from the assembly of free Tyr123-15L-α and βΔ67-5L-Tyr456 in solution. Alternatively, an assembled methyltransferase domain bound through its zinc fingers at the target site on one plasmid may be capable of methylating a non-target site on a second nearby plasmid. For ultimate success of the strategy, not only do we need to minimize activity in the absence of binding of the zinc finger domain to its cognate site on the DNA but we also need to constrain the reassembled methyltransferase such that it can only methylate its target site and cannot methylate DNA that might come in proximity to the methyltransferase. This might be difficult to overcome with a split enzyme such as M.EcoHK31I for which structural information is lacking. Further modification of the M.EcoHK31I-zinc finger fragments using random mutagenesis or other directed evolution methods might offer the best approach for creating fragments with higher specificity. However, M.EcoHK31I is limited by the methyltransferase’s inherent specificity. Ultimately it might be best to apply this approach for well-studied, structurally characterized CpG methyltransferases that have been artificially split such as HhaI methyl transferase (13,22) but whether the artificiality of such split methyltransferases will present additional challenges remains to be seen.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (GM072794 to M.O., GM077291 to S.C.); Maryland Stem Cell Research Fund (to S.C.). Funding for open access charge: National Institutes of Health and Johns Hopkins University.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank P.C. Shaw for providing plasmids pET3a-M38 and pET3a-C23 containing the M.EcoHK31I α and β fragments. We thank Brian Chaikind for reading and commenting on the manuscript.
REFERENCES
- 1.Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat. Rev. Genet. 2008;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 2.Zhu WG, Srinivasan K, Dai Z, Duan W, Druhan LJ, Ding H, Yee L, Villalona-Calero MA, Plass C, Otterson GA. Methylation of adjacent CpG sites affects Sp1/Sp3 binding and activity in the p21(Cip1) promoter. Mol. Cell. Biol. 2003;23:4056–4065. doi: 10.1128/MCB.23.12.4056-4065.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deng G, Chen A, Pong E, Kim YS. Methylation in hMLH1 promoter interferes with its binding to transcription factor CBF and inhibits gene expression. Oncogene. 2001;20:7120–7127. doi: 10.1038/sj.onc.1204891. [DOI] [PubMed] [Google Scholar]
- 4.Smith AE, Hurd PJ, Bannister AJ, Kouzarides T, Ford KG. Heritable gene repression through the action of a directed DNA methyltransferase at a chromosomal locus. J. Biol. Chem. 2008;283:9878–9885. doi: 10.1074/jbc.M710393200. [DOI] [PubMed] [Google Scholar]
- 5.Li F, Papworth M, Minczuk M, Rohde C, Zhang Y, Ragozin S, Jeltsch A. Chimeric DNA methyltransferases target DNA methylation to specific DNA sequences and repress expression of target genes. Nucleic Acids Res. 2007;35:100–112. doi: 10.1093/nar/gkl1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith AE, Ford KG. Specific targeting of cytosine methylation to DNA sequences in vivo. Nucleic Acids Res. 2007;35:740–754. doi: 10.1093/nar/gkl1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Minczuk M, Papworth MA, Kolasinska P, Murphy MP, Klug A. Sequence-specific modification of mitochondrial DNA using a chimeric zinc finger methylase. Proc. Natl Acad. Sci. USA. 2006;103:19689–19694. doi: 10.1073/pnas.0609502103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McNamara AR, Hurd PJ, Smith AE, Ford KG. Characterisation of site-biased DNA methyltransferases: specificity, affinity and subsite relationships. Nucleic Acids Res. 2002;30:3818–3830. doi: 10.1093/nar/gkf501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu GL, Bestor TH. Cytosine methylation targetted to pre-determined sequences. Nat. Genet. 1997;17:376–378. doi: 10.1038/ng1297-376. [DOI] [PubMed] [Google Scholar]
- 10.van Steensel B, Henikoff S. Identification of in vivo DNA targets of chromatin proteins using tethered dam methyltransferase. Nat. Biotechnol. 2000;18:424–428. doi: 10.1038/74487. [DOI] [PubMed] [Google Scholar]
- 11.Carvin CD, Parr RD, Kladde MP. Site-selective in vivo targeting of cytosine-5 DNA methylation by zinc-finger proteins. Nucleic Acids Res. 2003;31:6493–6501. doi: 10.1093/nar/gkg853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kiss A, Weinhold E. Functional reassembly of split enzymes on-site: a novel approach for highly sequence-specific targeted DNA methylation. Chembiochem. 2008;9:351–353. doi: 10.1002/cbic.200700662. [DOI] [PubMed] [Google Scholar]
- 13.Meister GE, Chandrasegaran S, Ostermeier M. An engineered split M.HhaI-zinc finger fusion lacks the intended methyltransferase specificity. Biochem. Biophys. Res. Commun. 2008;377:226–230. doi: 10.1016/j.bbrc.2008.09.099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nomura W, Barbas CF., 3rd In vivo site-specific DNA methylation with a designed sequence-enabled DNA methylase. J. Am. Chem. Soc. 2007;129:8676–8677. doi: 10.1021/ja0705588. [DOI] [PubMed] [Google Scholar]
- 15.Michnick SW, Remy I, Campbell-Valois FX, Vallee-Belisle A, Pelletier JN. Detection of protein-protein interactions by protein fragment complementation strategies. Methods Enzymol. 2000;328:208–230. doi: 10.1016/s0076-6879(00)28399-7. [DOI] [PubMed] [Google Scholar]
- 16.Lee KF, Kam KM, Shaw PC. A bacterial methyltransferase M.EcoHK311 requires two proteins for in vitro methylation. Nucleic Acids Res. 1995;23:103–108. doi: 10.1093/nar/23.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fung WT, Sze KH, Lee KF, Shaw PC. Functional studies of the small subunit of EcoHK31I DNA methyltransferase. Biol. Chem. 2006;387:507–513. doi: 10.1515/BC.2006.066. [DOI] [PubMed] [Google Scholar]
- 18.Barres R, Osler ME, Yan J, Rune A, Fritz T, Caidahl K, Krook A, Zierath JR. Non-CpG methylation of the PGC-1alpha promoter through DNMT3B controls mitochondrial density. Cell Metab. 2009;10:189–198. doi: 10.1016/j.cmet.2009.07.011. [DOI] [PubMed] [Google Scholar]
- 19.Kouidou S, Agidou T, Kyrkou A, Andreou A, Katopodi T, Georgiou E, Krikelis D, Dimitriadou A, Spanos P, Tsilikas C, et al. Non-CpG cytosine methylation of p53 exon 5 in non-small cell lung carcinoma. Lung Cancer. 2005;50:299–307. doi: 10.1016/j.lungcan.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 20.Ramsahoye BH, Biniszkiewicz D, Lyko F, Clark V, Bird AP, Jaenisch R. Non-CpG methylation is prevalent in embryonic stem cells and may be mediated by DNA methyltransferase 3a. Proc. Natl Acad. Sci. USA. 2000;97:5237–5242. doi: 10.1073/pnas.97.10.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sambrook J, Russell D. Molecular Cloning: A Laboratory Manual. 3rd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 22.Choe W, Chandrasegaran S, Ostermeier M. Protein fragment complementation in M.HhaI DNA methyltransferase. Biochem. Biophys. Res. Commun. 2005;334:1233–1240. doi: 10.1016/j.bbrc.2005.07.017. [DOI] [PubMed] [Google Scholar]
- 23.Mani M, Kandavelou K, Dy FJ, Durai S, Chandrasegaran S. Design, engineering, and characterization of zinc finger nucleases. Biochem. Biophys. Res. Commun. 2005;335:447–457. doi: 10.1016/j.bbrc.2005.07.089. [DOI] [PubMed] [Google Scholar]
- 24.Jacobs D, Brown NL. Isolation and characterization of the M.EaeI modification methylase. Biochem. J. 1986;238:613–615. doi: 10.1042/bj2380613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carvin CD, Dhasarathy A, Friesenhahn LB, Jessen WJ, Kladde MP. Targeted cytosine methylation for in vivo detection of protein-DNA interactions. Proc. Natl Acad. Sci. USA. 2003;100:7743–7748. doi: 10.1073/pnas.1332672100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.