Abstract
Site-specific modification of RNA is of great significance to investigate RNA structure, function and dynamics. Recently, we reported a new method for sequence- and cytosine-selective chemical modification of RNA based on the functional group transfer reaction of the 1-phenyl-2-methylydene-1,3-diketone unit of the 6-thioguanosine base incorporated in the oligodeoxynucleotide probe. In this study, we describe that the functionality transfer rate is greatly enhanced and the selectivity is shifted to the guanine base when the reaction is performed under alkaline conditions. Detailed investigation indicated that the 2-amino group of the enolate form of rG is the reactant of the functionality transfer reaction. As a potential application of this efficient functionality transfer reaction, a pyrene group as a relatively large fluorescent group was successfully transferred to the target guanine base of RNA with a high guanine and site selectivity. This functionality transfer reaction with high efficiency and high site-selectivity would provide a new opportunity as a unique tool for the study of RNA.
INTRODUCTION
RNA is implicated in critical functions within cells and possibly also in as-yet unknown functions. A number of genetic tools are used to study the new RNA functions. Nevertheless, to understand the molecular basis of their functions there is increasing demand for tools for characterizing their chemical and physical properties. The site-specific modifications with a variety of molecules, such as a fluorescent labeling group and a tagging molecule, are of great significance to investigate RNA structure, function and dynamics (1,2). Chemically modified short RNA is synthesized by a conventional solid-phase synthesizer. However, this method is not directly applicable to modify long RNA. Despite a number of methods (3–20), introducing tags or probes onto RNA at unique sites is still very difficult and a new approach is highly desired.
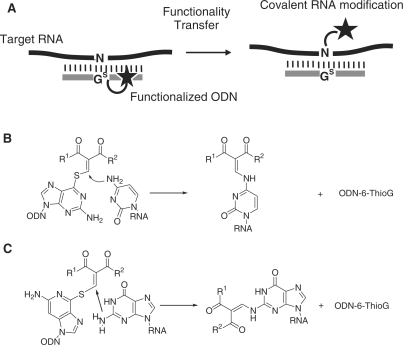
Recently, we reported a unique approach to modify RNA molecules by a functionality-transfer reaction utilizing the oligodeoxynucleotide (ODN) containing 2′-deoxy-6-thioguanosine (Figure 1A). The first demonstration was the site-specific NO-transfer reaction from S-nitroso-6-deoxythioguanosine to the amino group of the target cytosine in the ODN (21). The nitrosylated amino group of cytosine caused deamination, and its potential application as an artificial tool for RNA editing is now under investigation. This strategy was further expanded to the functionality-transfer reaction of a relatively large organic entity with the amino group of cytosine in RNA. After a number of structure–reactivity correlations, the 1-phenyl-2-methylydene-1,3-diketone unit of the 6-thioguanosine base was determined to be a structure unit suitable for efficient transfer (Figure 1B) (22). This is a unique method for DNA/RNA-directed transfer of reporter groups except for a few examples (23,24). During investigation for further improvement of this method for the tagging and labeling of a large RNA, we surprisingly found that the functionality transfer rate was greatly enhanced and the selectivity was shifted to the guanine base when the reaction was performed under alkaline conditions (Figure 1C). As a potential application of this efficient functionality transfer reaction, a pyrene group as a relatively large fluorescent group was successfully transferred to the target guanine base of RNA with a high guanine and site selectivity. We now describe in detail the functionality transfer reaction with the guanine base of RNA.
Figure 1.
(A) The general strategy of the site-specific RNA modification. (B) Functionality transfer reaction to modify rC using S-modified thioguanosine-containing ODN probe. (C) rG-Selective transfer reaction described in this study.
MATERIALS AND METHODS
General
1H NMR (400, 500 MHz) spectra were recorded by Varian UNITY-400 and INOVA-500 spectrometers, respectively. IR spectra were obtained using a PerkinElmer FTIR-SpectrumOne. High-resolution mass spectra (ESI) were recorded on an Applied Biosystems Mariner System 5299 spectrometer. MALDI-TOF mass spectra were recorded on a Bruker Daltonics Microflex instrument.
Synthesis of oligodeoxynucleotides and oligonucleotides
All oligonucleotides were synthesized by an automated DNA synthesizer (Applied Biosystems 394 DNA/RNA synthesizer) according to the conventional amidite chemistry. The cleaved oligonucleotides were purified by HPLC equipped with an ODS column using a linear gradient between the 0.1 M TEAA buffer and CH3CN. The MALDI-TOF/MS data are summarized in Supplementary Table S2.
3′, 5′-O-Bis(tert-butyldimethylsilyl)-2′-deoxy-2-N-modified guanosine (15)
To a solution of 14 (83 mg, 0.13 mmol) and 12 (41 mg, 0.20 mmol) in anhydrous DMF (1.5 ml) was added K2CO3 (27 mg, 0.20 mmol) at room temperature, and the mixture was stirred. After 1.5 h, the reaction mixture was quenched with saturated aqueous NH4Cl (15 ml), and extracted with diethyl ether (15 ml × 3). The organic phases were washed with brine (20 ml), and dried over Na2SO4. The solvent was evaporated under reduced pressure, and the residue was purified by silica gel chromatography (CHCl3/MeOH = 49:1) to give 15 as a yellow solid (13 mg, 15%). IR (cm−1): 3200, 1710, 1647, 1554, 1258, 836. 1H NMR (400 MHz, CDCl3) δ: 12.53 (1H, d, J = 11.2 Hz), 8.34 (1H, d, J = 11.2 Hz), 8.00 (1H, s), 7.77 (2H, d, J = 7.3 Hz), 7.56 (1H, t, J = 7.3 Hz), 7.47 (2H, t, J = 7.3 Hz), 6.09 (1H, t, J = 6.4 Hz), 4.48–4.45 (1H, m), 3.92–3.89 (1H, m), 3.79–3.71 (2H, m), 2.45 (3H, s), 2.62 (1H, ddd, J = 13.1, 6.4, 6.1 Hz), 2.23 (1H, ddd, J = 12.8, 6.1, 4.0 Hz), 0.89 (9H, s), 0.88 (9H, s), 0.07 (3H, s), 0.06 (3H, s), 0.05 (6H, s). High-resolution MS (ESI): Calcd for C33H49N5O6Si2 (M + H)+: 668.3294. Found: 668.3310.
2′-Deoxy-2-N-modified guanosine (16)
To a solution of 15 (8.3 mg, 0.012 mmol) in anhydrous THF (10 ml) was added TBAF (946 mg, 3.0 mmol) at room temperature, and the mixture was stirred. After 1.5 h, the reaction mixture was evaporated under reduced pressure. The residue was purified by silica gel chromatography (CHCl3/MeOH = 19:1) to give 16 (1.8 mg, 33%) as a white solid. After further purification by reverse-phase HPLC (Column: Nacalai Tesque: COSMOSIL 5C18-AR- II, 10 × 250 mm; solvent: A: 50 mM HCOONH4, B: CH3CN, B: 5 to 40%/30 min, linear gradient; flow rate, 3.0 ml/min; monitored by UV detector at 254 nm), the sample was used for NMR measurement. IR (cm−1): 1695, 1645, 1555, 1272, 836. 1H NMR (500 MHz, CD3OD) δ: 8.62 (1H, s), 7.96 (1H, s), 7.69 (2H, d, J = 7.3 Hz), 7.59 (1H, t, J = 7.3 Hz), 7.51 (2H, t, J = 7.3 Hz), 6.20 (1H, m), 4.37 (1H, m), 3.96 (1H, m), 3.62 (2H, m), 2.72 (1H, m), 2.45 (3H, s), 2.27 (1H, m). High-resolution MS (ESI): calcd for C21H21N5O6 (M + H)+: 440.1565. Found: 440.1553.
The synthesis of the functionalized ODN1F [X = GS(Me,Ph)] as the general procedure for the synthesis of ODN1F-4F [X = GS(R1,R2)]
A solution of 12 (5 mM in CH3CN, 0.38 µl, 1.9 nmol) was added to a solution of thioguanosine containing ODN1 (750 µM, 0.25 µl, 188 pmol) in carbonate buffer (30 mM, 3.12 µl, pH 10), and the mixture was kept at room temperature. After 10 min, the synthesized ODN1F [X = GS(Me,Ph)] was purified by HPLC for the MALDIMS measurement. On the other hand, the reaction mixture containing ODN1F [X = GS(Me,Ph)] was used for the functionality transfer reaction without HPLC purification.
Functionality transfer reaction from the functionalized ODN1F [X = GS(Me,Ph)] to ORN5 as the general procedure for the functionality transfer reaction
A solution of ODN1F [X = GS(Me,Ph), 50 µM, 0.75 µl, 37.5 pmol] and a solution of the target ORN5 (50 µM, 0.5 µl, 25 pmol) were mixed in a carbonate buffer (50 mM, pH 9.4) containing NaCl (100 mM), and the mixture was kept at 25°C. The reaction mixture was analyzed by HPLC (Column: SHISEIDO C18, 4.6 × 250 mm; Solvent: A: 0.1 M TEAA Buffer, B: CH3CN, B: 10–30%/20 min, linear gradient; flow rate, 1.0 ml/min; monitored by the fluorescence at 518 nm with emission at 494 nm or by UV detector at 254 nm).
Enzymatic hydrolysis of the modified ODN6
The modified ODN6 (100 µM) was hydrolyzed with BAP (30 U/ml), Nuclease P1 (30 U/ml) and SVPD (0.6 U/ml) in a buffer (50 mM Tris–HCl, 1 mM MgCl2, pH 9.0) at 37°C for 1 h, and analyzed by HPLC (solvent A: 50 mM HCOONH4, buffer B: CH3CN, B: 5–40%/30 min, linear gradient; flow rate, 1.0 ml/min; monitored by UV detector at 254 nm).
UV melting temperature measurements
UV melting temperatures were measured using 1.3 µM of each ODN and ORN strands in 50 mM carbonate buffer (pH 9.8) or MES buffer (pH 7.0) containing 100 mM NaCl with monitoring at 260 nm by UV-Vis spectrophotometer, DU 800 (Beckman Coulter).
Circular dichroism measurements
Circular dichroism (CD) spectra were measured by using 2.5 µM each ODN and ORN strands in 50 mM carbonate buffer (pH 9.8) or MES buffer (pH 7.0) containing 100 mM NaCl at room temperature with a JASCO J-720 W spectrophotometer in a cylindrical quartz cell with a path length of 0.1 cm.
RESULTS AND DISCUSSION
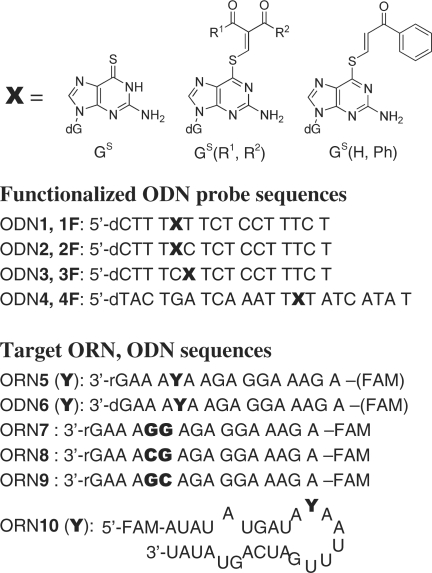
The sequences of the functionalized ODN and the target ORN used in this study are summarized in Figure 2. S-Functionalized ODN probes [ODN1F-4F: X = GS(R1,R2)] were prepared by mixing the corresponding 2-chloromethylydene derivative with the respective starting ODN (ODN1-4, X = GS) containing 2′-deoxy-6-thioguanosine at pH 10, as already reported (22,25). The ODNs containing the monoketone unit GS(H,Ph) were prepared as the non-reactive control (Figure 2).
Figure 2.
Structure of S-functionalized 6-thioguanosine (GS) and the sequences of the ODN probes, the target ORN and ODN.
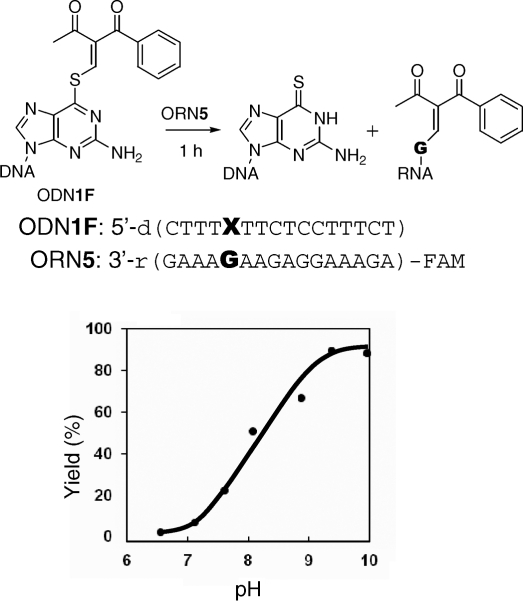
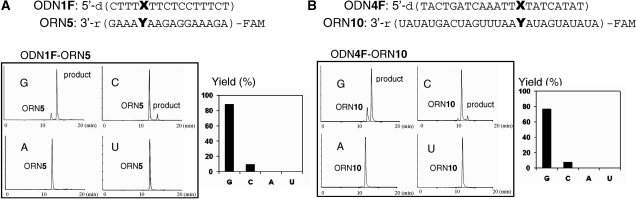
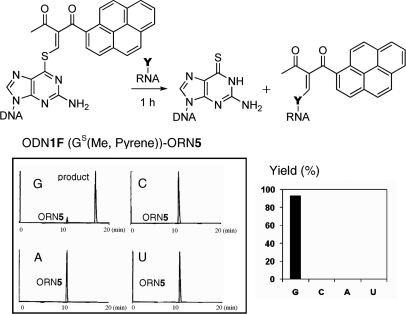
The transfer reaction between ODN1F [X = GS(Me,Ph)] and the target ORN5 (Y = rC) produced the rC-modified product selectively at pH 7.0, as previously reported (22). In contrast, the same transfer reaction between ODN1F [X = GS(Me,Ph)] and ORN5 (Y = rG) was found to take place at higher rates under alkaline conditions compared to that under neutral conditions (Figure 3). The base selectivity of the transfer reaction under alkaline conditions was investigated using ODN1F [X = GS(Me,Ph)] and ORN5 (Y = rG, rC, rA, U). The transfer reactions were performed at pH 9.6 for 1 h and were followed by monitoring the FAM fluorescence. It should be noted that the reaction efficiency with the guanine base was very high compared with the cytosine base, and no reaction was observed for the adenine and uracil bases (Figure 4A). The reaction mechanism and the origin of the selectivity shift from rC to rG will be discussed later in this article. The generality of this functionality-transfer reaction was investigated using ODN4F [X = GS(Me,Ph)] and ORN10 (Y = rG, rC, rA, U) with a loop structure. Similar to the reaction between the ODN1F [X = GS(Me,Ph)]-ORN5, selective transfer to the guanine base was confirmed (Figure 4B). A larger fluorescent functional group (R1, R2 = Me, pyrene) was also successfully transferred to the target guanine with a higher selectivity (Figure 5), suggesting potential application of this method for fluorescent labeling of RNA.
Figure 3.
The pH dependency of the functionality transfer reaction. The transfer reactions were performed using 1.5 µM of S-functionalized ODN1F [GS(Me,Ph)] and 1.0 µM of the target ORN5 (G) in 50 mM phosphate or carbonate buffer (pH > 9) containing 100 mM NaCl at 25°C for 1 h, followed by HPLC.
Figure 4.
Selectivity for the guanine base in the RNA substrates. The transfer reaction was performed in 50 mM carbonate buffer containing 100 mM NaCl at pH 9.6 for 1 h, followed by HPLC monitored by fluorescence at 518 nm with emission at 494 nm. (A) 1.5 µM of S-functionalized ODN1F [GS(Me,Ph)] and 1.0 µM of the target ORN5 (Y = rG, rC, rA, U), (B) 1.5 µM of S-functionalized ODN4F [GS(Me,Ph)], (B) 1.0 µM of the target ORN10 (Y = rG, rC, rA, U).
Figure 5.
Selectivity to the guanine base in transfer reaction of the pyrene derivative. The transfer reactions were performed in 50 mM carbonate buffer containing 100 mM NaCl at pH 9.6 for 1 h, followed by HPLC monitored by fluorescence at 518 nm with emission at 494 nm. Using 1.5 µM of S-functionalized ODN1F [GS(Me, Pyrene)] and 1.0 µM of the target ORN5 (Y).
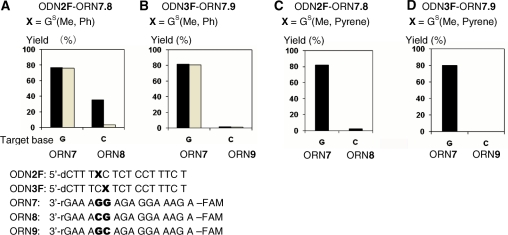
The site selectivity was checked using ODN2F and 3F in the reaction with ORN7 consisting of two continuous rGs and ORN8 and 9 consisting of the mutated versions at the target site, CG or GC, respectively (Figure 6). ODN2F and 3F were designed to react with rG at each complementary position of the 3′ and 5′ sides of ORN7-9, respectively. ODN2F [GS(Me,Ph)] gave the transferred products with the target ORN7; in contrast, a lower yield was obtained with ORN8 (Figure 6A, black bars). The adduct with rC was labile due to the treatment with aqueous NH3, whereas that with dG was stable under the same conditions (Figure 6A, grey bars). This result indicated that the functionality group did not transfer to rG but to rC of ORN8 at the complementary site of X of ODN2F. On the other hand, ODN3F [GS(Me,Ph)] formed the transferred product with ORN7 not with ORN9, in which rC is located at the complementary site with X of ODN3F (Figure 6B, black and grey bars). Similar results were obtained with the functionalized ODN containing the pyrene unit [GS(Me, Pyrene)] (Figure 6C and D). These results clearly indicated that the transfer reaction took place site-selectively with rG of the ORN at the complementary position toward X of the functionalized ODN probe. Thus, a high efficiency and site selectivity for the guanine base have been achieved by the functionality-transfer reaction under alkaline conditions.
Figure 6.
Site-selectivity of the functionality transfer reaction. The transfer reactions were performed using 1.5 µM of S-functionalized ODN2F, 3F and 1.0 µM of the target ORN7, 8 or 9 in 50 mM carbonate buffer containing 100 mM NaCl at pH 9.6 for 1 h, followed by HPLC monitored by fluorescence at 518 nm with emission at 494 nm. (A) ODN2F-ORN7.8, X = GS(Me,Ph). Grey bars indicate the reaction yield after treatment of aqueous NH3. (B) ODN3F-ORN7.9, X = GS(Me,Ph). Grey bars indicate the reaction yield after treatment of aqueous NH3. (C) ODN2F-ORN7.8, X = GS(Me, Pyrene). (D) ODN3F-ORN7.9, GS(Me, Pyrene).
In order to confirm that the transfer reaction takes place within the duplex under alkaline conditions, the CD spectra and the UV melting profiles of the duplex were measured using the non-reactive ODN1F-4F [X = GS(H,Ph)] at pH 9.8 (Supplementary Figure S1 and Table S1). The duplex formed with ODN1F [X = GS(H,Ph)] and ORN5 (X = G) showed a Tm value of 42°C under alkaline conditions, which is lower than the Tm value of 47°C at pH 7. The Tm values of the duplexes of all the other combinations at pH 9.8 were in the range of 33–45°C. It was confirmed by these results that the transfer reactions were performed at lower temperature than the Tm values. The CD spectra of the duplex formed between ODN1F [X = GS(H,Ph)] and ORN5 (Y = G) under alkaline conditions were similar to those taken at pH 7, indicating that the duplex was in a conformation between the A and B types (26). These results, together with the fact that the reaction with the corresponding monomer substrates did not produce the transferred product, have supported that the functionality-transfer proceeds within the duplex due to the close proximity between the S-functionalized thioguanosine and the guanine base at the target site of the complementary strand within the duplex.
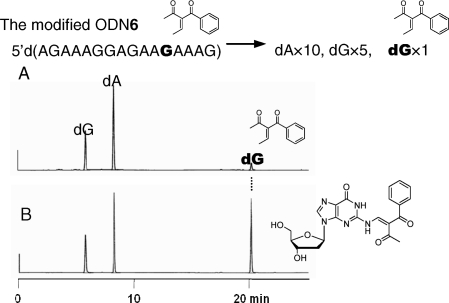
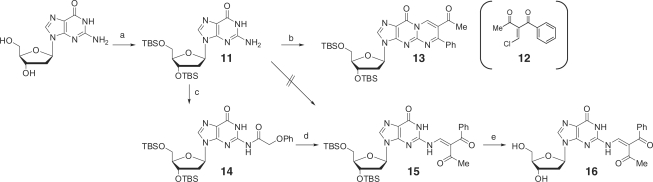
As the transfer reaction proceeded with the ODN targets in a fashion similar to the ORN targets with respect to the pH dependency (Supplementary Figure S2) and guanine selectivity (Supplementary Figure S3), the ODN targets were used for structure determination of the modified guanine and investigation on structure–activity relationship. The modified ODN6 was subjected to enzymatic hydrolysis with bacterial alkaline phosphatase (BAP), Nuclease P1 and snake venom phosphodiesterase (SVPD), and the hydrolysates were analyzed by HPLC. In addition to the major peaks corresponding to dG and dA, a new peak was observed (Figure 7A), which was isolated and subjected to ESI-HRMS measurements [calcd for C21H21N5O6 (M + H)+: 440.1565, found: 440.1597] and 1H NMR measurements (Supplementary Figure S4). The authentic 2-N-modified dG shown in Figure 7B was synthesized using the 2′-deoxyguanosine monomer (Scheme 1). The reaction of 2′-deoxyguanosine protected with tert-butyldimethylsilyl (TBDMS) groups with 12 proceeded to afford the cyclized product 13. In contrast, the reaction of 12 with 2-N-phenoxyacetyl-2′-deoxyguanosine protected with TBDMS groups (14) produced the 2-N-modified dG derivative (15), which was subjected to deprotection of the TBDMS groups to give the 2-N-modified dG (16). 1H NMR and HPLC profiles of 16 were identical to those of the isolated product from the functionality transfer reaction.
Figure 7.
(A) HPLC analysis of the enzymatic hydrolysate of the modified ODN6, monitored by UV at 254 nm. (B) Co-injection of the hydrolysate and the synthesized N2-modified dG.
Scheme 1.
Synthesis of 2′-deoxy-2-N-modified guanosine. (a) TBDMSCl, imidazole, DMF, rt, 94%; (b) 12, K2CO3, rt, 55%; (c) phenoxyacethyl chloride, 1-HOBt, pyridine, CH3CN, rt, 37%; (d) 12, K2CO3, rt, 15%; (e) TBAF, THF, rt, 33%.
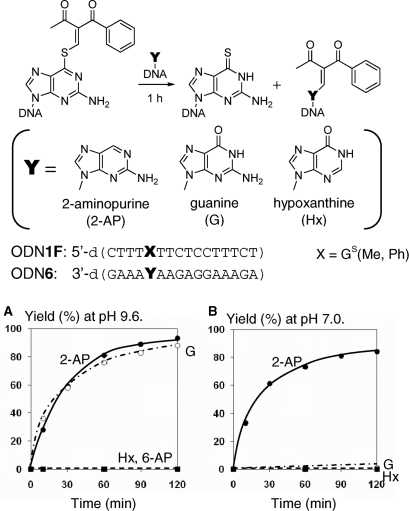
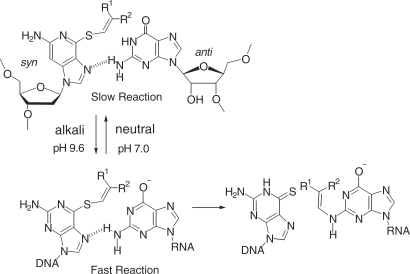
The following additional experiments were performed using the functionalized ODN1F and the target ODN6 to confirm that the 2-amino modified structure is the initial product and not a secondary product, such as those formed from the 1-N-modified one. 2-Aminopurine showed a similar transfer rate with guanine, but hypoxanthine and 6-aminopurine (adenine) did not give a transferred product (Figure 8A). 2-Aminopurine did not show any pH-dependency (Figure 8B). Accordingly, it is apparent that the 2-amino group of the purine base is responsible for the transfer reaction. Because guanine should be predominantly in its enolate form at pH 9.6 according to its pKa value (9.2) (27), the 2-amino group of the enolate form of the guanine base is responsible for the nucleophilic attack to cause the transfer reaction (Figure 9). The selectivity shift from rC at pH 7 to rG at pH 9.6 is reasonably explained by increase in nucleophilicity of the enolate form of the guanine base under alkaline conditions. The facts that the 6-amino group of adenine and the 4-amino group of cytosine did not induce the transfer reaction under the alkaline conditions used in this study suggest that the 2-amino group of the purine base must be in close proximity to the transfer group. As the vinyl group of the modified thioguanosine and the 2-amino group of the guanine base are directed away from each other in the anti functionalized thioguanosine and syn guanosine conformation, it may be speculated that the reaction occurs in a complex formed between the syn conformation of the S-functionalized thioguanosine and the anti conformation of guanosine (Figure 9). In such a hypothesized syn-anti complex resembling the natural guanine-guanine base pair, the hydrogen bonding between the 2-amino group of guanine and the 7-N of the thioguanine in the syn conformation might assist their reaction.
Figure 8.
Comparison of the reactivity of the purine analogs. (A) Reaction at pH 9.6. (B) Reaction at pH 7.0. The transfer reactions were performed using 30 µM of S-functionalized ODN1F [GS(Me,Ph)] and 25 µM of the target ODN6 in 50 mM carbonate buffer containing 100 mM NaCl followed by HPLC. Abbreviations of nucleobase in the graph represent 2-AP for 2-aminopurine, 6-AP for 6-aminopurine (adenine), G for guanine and Hx for hypoxanthine.
Figure 9.
A hypothesized syn-anti base pair involving the enolate form of the guanine base.
CONCLUSIONS
In this study, we have revealed that the transfer reaction of the 1-aryl-2-methylydene-1,3-diketone unit of the 6-thioguanosine base incorporated in the ODN probe is greatly accelerated under alkaline conditions and that the rC selectivity at pH 7.0 is shifted to rG at pH 9.6. Detailed investigation indicated that the 2-amino group of the enolate form of rG is the reactant in the functionality transfer reaction. It was postulated from the high selectivity that the syn conformation of the S-functionalized thioguanosine base may be included in the reaction of the anti conformation of rG in the target RNA. Combined with the previous protocol (22), the modification site of RNA by the S-functionalized ODN probe can be controlled by changing the pH of the reaction buffer, i.e. cytosine modification at pH 7 and guanine modification at pH 9.6. As a variety of molecules, such as fluorescent labeling groups and tagging molecules, may be potentially site-specifically transferred to large RNAs, this functionality transfer reaction would provide a new opportunity as a unique tool for the study of RNA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
CREST from the Japan Science and Technology Agency. Funding for open access charge: Grant-in-Aid for Scientific Research (S) from the Japan Society for the Promotion of Science (JSPS).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank the Research Fellowship of the Japan Society for the Promotion of Science (JSPS) for Young Scientists (to K.O.).
REFERENCES
- 1.Chow CS, Mahto SK, Lamichhane TN. Combined approaches to site-specific modification of RNA. ACS Chem. Biol. 2008;3:30–37. doi: 10.1021/cb7002225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Das SR, Fong R, Piccirilli JA. Nucleotide analogues to investigate RNA structure and function. Curr. Opin. Chem. Biol. 2005;9:585–593. doi: 10.1016/j.cbpa.2005.10.009. [DOI] [PubMed] [Google Scholar]
- 3.England TE, Uhlenbeck OC. 3′-Terminal labeling of RNA with T4 RNA ligase. Nature. 1978;275:560–561. doi: 10.1038/275560a0. [DOI] [PubMed] [Google Scholar]
- 4.Hausch F, Jäschke A. Libraries of multifunctional RNA conjugates for the selection of new RNA catalysts. Bioconjugate. Chem. 1997;8:885–890. doi: 10.1021/bc9701151. [DOI] [PubMed] [Google Scholar]
- 5.Pitulle C, Kleineidam RG, Sproat B, Krupp G. Initiator oligonucleotides for the combination of chemical and enzymatic RNA synthesis. Gene. 1992;112:101–105. doi: 10.1016/0378-1119(92)90309-d. [DOI] [PubMed] [Google Scholar]
- 6.Huang F. Efficient incorporation of CoA, NAD and FAD into RNA by in vitro transcription. Nucleic Acids Res. 2003;31:e8. doi: 10.1093/nar/gng008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fiammengo R, Musilek K, Jäschke A. Efficient preparation of organic substrate-RNA conjugates via in vitro transcription. J. Am. Chem. Soc. 2005;127:9271–9276. doi: 10.1021/ja051179m. [DOI] [PubMed] [Google Scholar]
- 8.Zhang L, Sun L, Cui Z, Gottlieb RL, Zhang B. 5′-sulfhydryl-modified RNA: initiator synthesis, in vitro transcription, and enzymatic incorporation. Bioconjugate. Chem. 2001;12:939–948. doi: 10.1021/bc015504g. [DOI] [PubMed] [Google Scholar]
- 9.Fusz S, Eisenführ A, Srivatsan SG, Heckel A, Famulok M. A ribozyme for the aldol reaction. Chem. Biol. 2005;12:941–950. doi: 10.1016/j.chembiol.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 10.Schlatterer JC, Jäschke A. Universal initiator nucleotides for the enzymatic synthesis of 5′-amino- and 5′-thiol-modified RNA. Biochem. Biophys. Res. Commun. 2006;344:887–892. doi: 10.1016/j.bbrc.2006.03.218. [DOI] [PubMed] [Google Scholar]
- 11.Pfander S, Fiammengo R, Kirin SI, Nolte N-M, Jäschke A. Reversible site-specific tagging of enzymatically synthesized RNAs using aldehyde-hydrazine chemistry and protease-cleavable linkers. Nucleic Acids Res. 2007;35:e25. doi: 10.1093/nar/gkl1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moore MJ, Query CC. Joining of RNAs by splinted ligation. Methods Enzymol. 2000;317:109–123. doi: 10.1016/s0076-6879(00)17009-0. [DOI] [PubMed] [Google Scholar]
- 13.Kurschat WC, Muller J, Wombacher R, Helm M. Optimizing splinted ligation of highly structured small RNAs. RNA. 2005;11:1909–1914. doi: 10.1261/rna.2170705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kawai R, Kimoto M, Ikeda S, Mitsui T, Endo M, Yokoyama S, Hirao I. Site-specific fluorescent labeling of RNA molecules by specific transcription using unnatural base pairs. J. Am. Chem. Soc. 2005;127:17286–17295. doi: 10.1021/ja0542946. [DOI] [PubMed] [Google Scholar]
- 15.Moriyama K, Kimoto M, Mitsui T, Yokoyama S, Hirao I. Site-specific biotinylation of RNA molecules by transcription using unnatural base pairs. Nucleic Acids Res. 2005;33:e129. doi: 10.1093/nar/gni128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hirao I, Kimoto M, Mitsui T, Fujiwara T, Kawai R, Sato A, Harada Y, Yokoyama S. An unnatural hydrophobic base pair system: site-specific incorporation of nucleotide analogs into DNA and RNA. Nat. Methods. 2006;3:729–735. doi: 10.1038/nmeth915. [DOI] [PubMed] [Google Scholar]
- 17.Kimoto M, Mitsui T, Harada Y, Sato A, Yokoyama S, Hirao I. Fluorescent probing for RNA molecules by an unnatural base-pair system. Nucleic Acids Res. 2007;35:5360–5369. doi: 10.1093/nar/gkm508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Proudnikov D, Mirzabelkov A. Chemical methods of DNA and RNA fluorescent labeling. Nucleic Acids Res. 1996;24:4535–4542. doi: 10.1093/nar/24.22.4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baum DA, Silverman SK. Deoxyribozyme-catalyzed labeling of RNA. Angew. Chem. Int. Ed. 2007;46:3502–3504. doi: 10.1002/anie.200700357. [DOI] [PubMed] [Google Scholar]
- 20.Yoshimura Y, Noguchi Y, Fujimoto K. Highly sequence specific RNA terminal labeling by DNA photoligation. Org. Biomol. Chem. 2007;5:139–142. doi: 10.1039/b615715g. [DOI] [PubMed] [Google Scholar]
- 21.Ali MM, Alam MR, Kawasaki T, Nakayama S, Nagatsugi F, Sasaki S. Sequence- and base-specific delivery of nitric oxide to cytidine and 5-methylcytidine leading to efficient deamination. J. Am. Chem. Soc. 2004;126:8864–8865. doi: 10.1021/ja0498888. [DOI] [PubMed] [Google Scholar]
- 22.Onizuka K, Taniguchi Y, Sasaki S. Site-specific covalent modification of RNA guided by functionality-transfer oligodeoxynucleotides. Bioconjugate Chem. 2009;20:799–803. doi: 10.1021/bc900009p. [DOI] [PubMed] [Google Scholar]
- 23.Grossmann TN, Seitz O. DNA-catalyzed transfer of a reporter group. J. Am. Chem. Soc. 2006;128:15596–15597. doi: 10.1021/ja0670097. [DOI] [PubMed] [Google Scholar]
- 24.Grossmann TN, Röglin L, Seitz O. Target-catalyzed transfer reactions for the amplified detection of RNA. Angew. Chem. Int. Ed. 2008;47:7119–7122. doi: 10.1002/anie.200801355. [DOI] [PubMed] [Google Scholar]
- 25.Onizuka K, Taniguchi Y, Sasaki S. A new odorless procedure for the synthesis of 2′deoxy-6-thioguanosine and its Incorporation Into Oligonucleotides. Nucleosides, Nucleotides and Nucleic Acids. 2009;28:752–760. doi: 10.1080/15257770903155576. [DOI] [PubMed] [Google Scholar]
- 26.Hung SH, Yu Q, Gray DM, Ratliff RL. Evidence from CD spectra that d(purine)-r(pyrimidine) and r(purine)-d(pyrimidine) hybrids are in different structural classes. Nucleic Acids Res. 1994;22:4326–433. doi: 10.1093/nar/22.20.4326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burkard ME, Turner DH. NMR Structures of r(GCAGGCGUGC)2 and determinants of stability for single guanosine-guanosine base pairs. Biochemistry. 2000;39:11748–11762. doi: 10.1021/bi000720i. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.