Abstract
The anticancer peptide PNC-27, which contains an HDM-2-binding domain corresponding to residues 12-26 of p53 and a transmembrane-penetrating domain, has been found to kill cancer cells (but not normal cells) by inducing membranolysis. We find that our previously determined 3D structure of the p53 residues of PNC-27 is directly superimposable on the structure for the same residues bound to HDM-2, suggesting that the peptide may target HDM-2 in the membranes of cancer cells. We now find significant levels of HDM-2 in the membranes of a variety of cancer cells but not in the membranes of several untransformed cell lines. In colocalization experiments, we find that PNC-27 binds to cell membrane-bound HDM-2. We further transfected a plasmid expressing full-length HDM-2 with a membrane-localization signal into untransformed MCF-10-2A cells not susceptible to PNC-27 and found that these cells expressing full-length HDM-2 on their cell surface became susceptible to PNC-27. We conclude that PNC-27 targets HDM-2 in the membranes of cancer cells, allowing it to induce membranolysis of these cells selectively.
Keywords: HDM-2 binding, membranolysis, three-dimensional structure, transfection
We have found previously that the peptide PNC-27, containing the HDM-2-binding domain of p53, residues 12-26, attached to a transmembrane-penetrating or membrane residency peptide (MRP) on its carboxyl terminal end, and PNC-28 (p53 residues 17-26-MRP) induce tumor cell necrosis by forming pores in cancer cell membranes but have no effects on a number of untransformed cells, including human stem cells from cord blood (1–4), and eradicate tumors in nude mice (5). The p53-HDM-2 complex results in the catabolism of p53 (6).
In contrast, other studies using HDM-2-binding molecules including p53 peptides attached to membrane-penetrating sequences on their amino terminal ends and small molecules that block the p53-HDM-2 interaction found that these agents induce p53-dependent apoptosis of cancer cells (6–11). However, our observations were consistent with the results from a 2D NMR study on the structure of PNC-27 showing that this peptide adopts a strongly amphipathic alpha-helix-loop-alpha-helix structure (12) observed in membrane-active peptides (13, 14). While this study suggested the basis of their lytic action, it did not explain why these peptides lyse the membranes of cancer cells selectively.
Because these peptides contain sequences that are known to bind to HDM-2 in its amino terminal domain, residues 1-109 (15), we now investigate whether they interact with HDM-2 that may be expressed in the membranes of transformed but not untransformed cells.
Results
Superposition of the 2D NMR Structures of the HDM-2-Binding Domain of PNC-27 on the X-Ray Structure of the Isolated Peptide Bound to HDM-2.
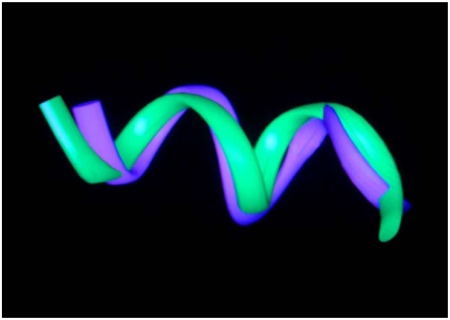
In our prior 2D NMR determination of the solution structure of PNC-27, we obtained 30 structures that fit the NOE constraints (12). When we superimposed these structures on the x-ray structure, all superimposed closely, giving a range of rms deviations of 1.7–2.5 Å. Fig. 1 shows the superposition for the backbone atoms of the structure of lowest rms deviation on the x-ray structure. This agreement occurs despite the fact that the x-ray structure contains a “truncated” peptide beginning at residue 17 and is bound to HDM-2 while PNC-27 contains six additional amino terminal residues (p53 residues 12–16) and Leu 26 attached to the transmembrane-penetrating sequence, not present on the peptide used in the x-ray crystal structure. We conclude that residues 17-26 of PNC-27 fold into a native HDM-2-binding conformation.
Fig. 1.
Ribbon representation for the superimposition of the NMR structure for residues 17–26 of PNC-27 (Blue) on the x-ray crystallographic structure for the same residues of the p53 17–29 peptide (Green) bound to MDM-2. N terminus (Lower Right); C terminus (Upper Left).
Cytotoxicity of PNC-27 to Cancer Cells.
We tested PNC-27 against the cancer cell lines used in this study. As shown in Fig. S1, we found that PNC-27, but not control PNC-29 peptide, is cytotoxic to MIA-PaCa-2 cells, inducing 100% cell death in 90 min and MIA-PaCa-2, TUC-3, and A-2058 cells in a dose-related manner but did not affect untransformed AG13145 primary human fibroblasts. Previously, we found that PNC-27 kills MCF-7 cells by a nonapoptotic (16) mechanism (2).
Blotting for HDM-2 in Cancer Cell Membranes.
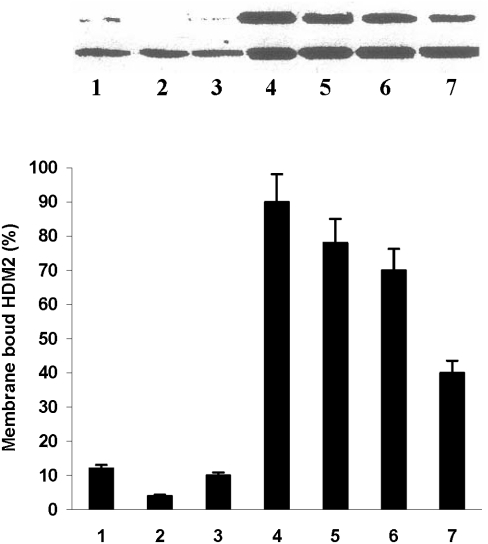
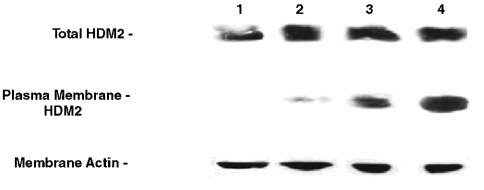
To test whether cancer cells express HDM-2 in their cell membranes, we isolated the membrane fractions (confirmed by electron microscopy) and whole cell lysates from several different cancer and untransformed cell lines shown in Fig. 2 and blotted them for HDM-2. On the lower panel for the blots in Fig. 2, it can be seen that all whole cell lysates blot positively for HDM-2. On the upper panel for the blots in Fig. 2, the membrane fraction of each cancer cell line (lanes 4–7) is seen to contain significant levels of HDM-2. In contrast, the three untransformed cell lines (lanes 1–3) were found to have low levels of HDM-2 in their membrane fractions. The percentage of whole cell lysate of HDM-2 present in the membrane fractions of the cell lines is shown in the bar graph (lowermost in Fig. 2). It can be seen that the fractions present in the membranes of the cancer cell lines are fourfold to ninefold increased over those for the untransformed cells. It should be noted that the HDM-2 bands shown in Fig. 2 were the major band at 92kDa. However, several other less prominent bands of lower Mr representing variants of HDM-2 were also observed in the membrane fractions of cancer cells that were absent in the three untransformed cell lines. We are currently investigating the identity of these variant forms. In other control experiments, we blotted the membrane fractions and whole cell lysates of each cell line for p53 and found that none of the membrane fractions contained this protein while it was present in the cell lysates. Our results suggest that HDM-2 is expressed in the membranes of cancer cells but only minimally in those of untransformed cells.
Fig. 2.
Blots of whole cell lysates (Lower) and membrane fractions (Upper) for H(M)DM-2 in different cell lines as follows: Lane 1, MCF-10-2A; lane 2, BMRPA1; lane 3, AG13145 fibroblasts; lane 4, TUC-3; lane 5, MIA-PaCa-2; lane 6, MCF-7; lane 7, A-2058. The first three cell lines are untransformed; the remainders are different cancer cell lines. Each bar graph shows the percentage of HDM-2 in whole cell lysate that is present in the membrane of each cell line listed above. The numbers on the X axis of the bar graphs correspond to the lane numbers shown in the blots.
Coimmunoprecipitation of HDM-2 with Fluorescent-Labeled PNC-27.
We next sought to determine whether this peptide binds to HDM-2 in cancer cells. We incubated MIA-PaCa-2 cells with the double fluorescent-labeled form of PNC-27 and immunoprecipitated (IP) HDM-2.
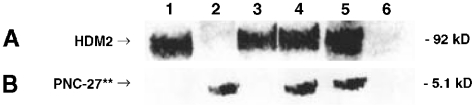
Lane 4 of Fig. 3 shows that IP HDM-2 contains a significant amount of the fluorescent-labeled peptide. In contrast, as shown in lane 3, if the cells are incubated with equimolar concentrations of fluorescent-labeled PNC-27 and unlabeled PNC-27 or closely related PNC-28 (1–3), the 5.1 kDa fluorescent band is no longer present. In contrast, if the cells are incubated with equimolar concentrations of the double-labeled peptide and the negative control peptide PNC-29, the fluorescent band is present as shown in lane 5. These results suggest specific binding of PNC-27 to HDM-2.
Fig. 3.
MIA-PaCa-2 cells were incubated with a double fluorescent-labeled form of PNC-27 (PNC-27**), and the cell lysates were subjected to immunoprecipitation with anti-HDM-2 (sc-13161) and then blotted onto membranes where the presence of fluorescence was directly observed (A); membranes were also blotted with anti-HDM-2 (B; sc-813). Lane 1, untreated cells; lane 2, double fluorescent-labeled PNC-27 control, showing the low molecular weight band for the peptide at 5.1 kDa; lane 3, cells incubated with fluorescent PNC-27 in the presence of “cold” PNC-27; lane 4, cells incubated with fluorescent PNC-27 alone; lane 5, cells incubated with fluorescent PNC-27 in the presence of the control PNC-29 peptide; lane 6, lysate from cells treated with fluorescent PNC-27, immunodepleted with anti-HDM-2, showing absence of fluorescent peptide.
Colocalization of PNC-27 with HDM-2 in the Cancer Cell Membrane.
We obtained further evidence for the interaction of PNC-27 with HDM-2 in cancer cell membranes by incubating PNC-27 with two different cancer cell lines, A2058 and MCF-7, and then incubating the cells with FITC- (green-fluorescent-) labeled anti-PNC-27 and TAMRA- (red-fluorescent-) labeled anti-HDM-2. We then viewed the cells using confocal microscopy. Colocalization of PNC-27 with HDM-2 in the cell membrane should result in concurrent green- and red-fluorescent signals from the two antibodies, presenting as yellow fluorescence in the cell membrane.
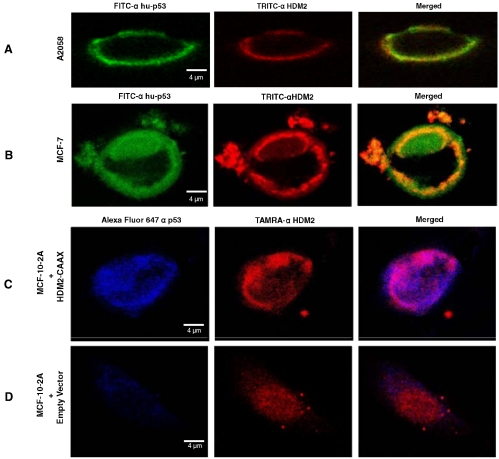
As shown in Fig. 4 A, A-2058 cells treated with PNC-27 show strong green (PNC-27, DO1 antibody) and red (HDM-2) fluorescence uniquely in the membrane of these cells; the latter result confirms the results of Fig. 2. In the third frame of Fig. 4 A, there is pronounced yellow fluorescence in the membrane, suggesting colocalization. Fig. 4 B shows identical results for MCF-7 cells treated with PNC-27. In control experiments, we found that incubation with DO1 antibody to PNC-27/p53 of both cell lines that were not treated with PNC-27 did not show green fluorescence in the cell membrane, indicating that p53 was not present in this fraction. Treatment of two untransformed cell lines, i.e., BMRPA1 and MCF-10-2A, with PNC-27 followed by incubation with the two labeled antibodies resulted in identical patterns of fluorescence in which green fluorescence was diffusely distributed throughout the cells, suggesting that the peptide entered the cells without being held in the membrane, whereas there was no red fluorescence in their membranes, confirming our findings in Fig. 2 showing the absence of HDM-2 in the membrane fractions of untransformed cells by Western blots.
Fig. 4.
(A and B) Confocal microscopic results from the treatment of A2058 (A) and MCF-7 (B) cells with PNC-27, after which the cells were incubated with green fluorescent anti-PNC-27 DO1 antibody and red-fluorescent anti-HDM-2 antibodies. The fluorophores are labeled for each frame of each panel on the top of the figure. (C and D) MCF-10-2A cells transfected with vectors expressing full-length HDM-2-CVVK membrane-localization peptide (C) and empty vector (D). Blue fluorescence is for PNC-27, red fluorescence is for HDM-2.
Effect of PNC-27 on Untransformed Cells that Are Induced to Express HDM-2 on their Cell Membranes.
We then investigated if expression of HDM-2 in the cell membranes of untransformed cells would make them susceptible to PNC-27. To this end, we transfected MCF-10-2A cells with a vector encoding full-length HDM-2 with a membrane-targeting CAAX (Cys-Val-Val-Lys) sequence (17) and green fluorescent protein (GFP) under a constitutive promoter. For controls, we transfected into MCF-10-2A cells vectors encoding full-length HDM-2 without the presence of CAAX or HDM-2 that lacked residues 1-109 that bind to p53/PNC-27 (15) but contained the carboxyl terminal CAAX sequence (del1-109-HDM-2-CVVK) or empty vector. For each transfected cell line, we found the transfection efficiency to be about 45% as computed from the number of GFP-expressing cells divided by the total number of cells in samples.
We then checked the transfected cells for expression of membrane-bound HDM-2-GFP fusion protein (Mr 122 kDa) by Western blotting of the membrane fractions and whole cell lysates as shown in Fig. 5. Empty vector-transfected cells have no detectable HDM-2 in their membranes (lane 1). Full-length HDM-2-expressing cells contain a small amount of HDM-2 expressed in their membranes that are a small fraction of the total HDM-2 expressed (lane 2). In contrast, both full-length HDM-2-CVVK (lane 4) and del1-109-HDM-2-CVVK (lane 3, Mr 110 kDa that migrates closely to the full-length fusion protein) are expressed at significantly higher levels in the cell membrane. These results confirmed expression of HDM-2 proteins with the membrane-localization sequence in the cell membranes.
Fig. 5.
Western blots for expression of HDM-2 in the membrane fractions of each of the four sets of transfected MCF-10-2A cells as follows: lane 1, empty vector-transfected cells; lane 2, cells transfected with plasmid expressing full-length HDM-2; lane 3, cells transfected with plasmid expressing del 1-109 HDM-2-CVVK; lane 4, cells transfected with plasmid expressing full-length HDM-2-CVVK. (Top) Blots for HDM-2 from whole cell lysates. (Middle) Blots for HDM-2 from the membrane fractions. (Bottom) Blots for actin in the membrane fractions.
We then incubated membrane-associated HDM-2-CVVK-expressing cells and HDM-2-expressing cells with PNC-27 to determine if PNC-27 colocalized with HDM-2 in the cell membrane. Fig. 4 C shows that PNC-27 (blue fluorescence) binds to the membrane of HDM-2-CVVK-expressing cells that express high levels of membrane-bound HDM-2 (red fluorescence), confirming the Western blot results in Fig. 5. The last frame of Fig. 4 C shows that there is extensive colocalization of PNC-27 with HDM-2-CVVK in the cell membrane (lavender fluorescence). Fig. 4 D shows that the PNC-27 signal in the membranes of control cells transfected with empty vector is only minimally present and that the HDM-2 signal is diffuse in the cell and not present in the cell membrane. No colocalization signal is present.
Susceptibility of Transfected Cells to PNC-27.
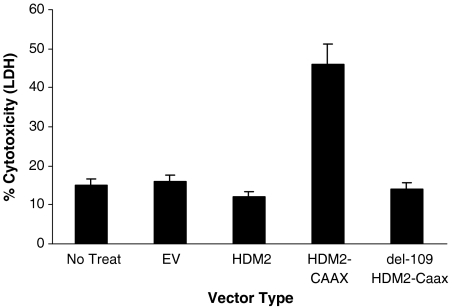
We further plated each set of transfected cells and treated each with PNC-27 for 24 hr. As shown in Fig. 6, the cells expressing membrane-bound HDM-2-CVVK are induced to release LDH over twice the background value for untreated or empty-vector-transfected cells, and none of the other control cells were found to release LDH above this background value. We also observed a major decrease in cell viability only in the full-length HDM-2-CVVK-expressing cells (Fig. S2A). This decrease was threefold compared with that for untreated cells and almost threefold compared with that for empty-vector-transfected controls. It was also at least 2.5-fold reduced compared with the cell viability of cells expressing HDM-2 on their membranes but lacking the binding site for HDM-2 despite the presence of HDM-2 in the cell membrane fraction (Fig. S2A). None of these sets of cells was induced to undergo apoptosis as revealed by the absence of enhanced caspase activity above background (Fig. S2B), consistent with our prior findings that PNC-27 induces tumor cell necrosis and not apoptosis (1–5). Control peptide PNC-29 (200 μg/ml), had no effect on either LDH release or the viability of any of the four sets of transfected cells, suggesting that PNC-27 binds specifically to HDM-2 on the cell membrane, allowing for its cytotoxicity.
Fig. 6.
Results of LDH assays for each of the sets of MCF-10-2A cells transfected with Precision Shuttle vectors as labeled on the abscissa for A–C. EV is empty vector; HDM-2 is overexpressed HDM-2; HDM-2 + CAAX is full-length HDM-2 with the membrane-targeting CVVK sequence on the carboxyl terminus; del1-109 HDM-2 + CAAX is HDM-2 without its p53 (PNC-27)-binding domain, residues 1-109, attached on its C terminus to the CVVK membrane-localization sequence.
Discussion
Our present studies suggest that PNC-27 induces cancer cell cytotoxicity by a previously undescribed mechanism in which the peptide induces cancer cell membranolysis that appears to depend on the binding of the peptide to HDM-2 in the cancer cell membrane. HDM-2 is present in the membranes of four different cancer cell lines but is only minimally present in the membranes of three different untransformed cell lines, providing a rationale for the specificity of action of this peptide. These observations are consistent with the results of prior studies on HDM-2 expression in mammalian cells. HDM-2is overexpressed in fast-growing tumors, and the levels of expression of HDM-2 correlated with metastatic potential in primary tumor cell cultures from patients with breast cancer (18). Importantly, HDM-2 was found to colocalize with E-cadherin in the cancer cells’ plasma membranes and to induce its ubiquitination and subsequent degradation, leading to increased cancer cell motility (18). This latter study suggests that HDM-2 is expressed in the membranes of cancer cells and points to a functional role for its presence in the cell membrane.
Binding of PNC-27 to HDM-2 in Cell Membranes Is Critical to its Membranolytic Action.
That PNC-27 interacts with membrane-bound HDM-2 is supported by our confocal microscopy studies on PNC-27-treated cancer cells that show that PNC-27 colocalizes with HDM-2 in the membranes of these cells to which PNC-27 is lethal, a phenomenon that does not occur with untransformed cells that do not express HDM-2 in their membranes and that are not affected by PNC-27.
PNC-27 does not kill MCF-10-2A cells (2). However, transfected cells that express full-length HDM-2-CVVK in their membranes become susceptible to the action of this peptide. In contrast, cells expressing either full-length HDM-2 that does not localize to the cell membrane or HDM-2 that does localize to the cell membrane but does not contain the p53 (PNC-27)-binding domain are much less susceptible to the action of this peptide. These results suggest that the membranolytic action of PNC-27 requires its interaction with HDM-2 in the cancer cell membrane. Similar results to ours have been reported in another study in which plasmid-driven overexpression of MDM-2 in rodent cardiomyocytes rendered these cells susceptible to PNC-28 (19).
Unique Membranolytic Action of PNC-27 May be Due to Its Membrane-Active Conformation.
Using double fluorophore-labeled PNC-27 (as in Fig. 3), we found that the peptide remains intact during tumor cell membranolysis (20). In solution, PNC-27 adopts a membrane-active conformation in which the HDM-2-binding subdomain adopts the x-ray HDM-2-binding conformation. If the membrane-active conformation is retained when the peptide binds to HDM-2 in the cell membrane, then the peptide would remain membrane-active and form pores in the cancer cell membrane, as has been found to occur for other membrane-active peptides (14, 21). When double fluorophore-labeled PNC-27 is incubated with untransformed cells, a small amount of peptide remains in the cell membrane while only the amino terminal domain is found in the nucleus, i.e., the carboxyl terminus is absent, signifying peptide cleavage in the cell (20). This pattern was also found in the colocalization experiments for untransformed cells incubated with PNC-27 (Fig. 4 D). Interestingly, agents, like p53 peptides, that block p53-HDM-2 binding do not induce apoptosis in untransformed cells (4, 6–11). Thus it appears that HDM-2 in the cell membrane blocks the transit of PNC-27 into the cell, allowing it to remain in the membrane where it can undergo pore formation.
This hypothesis can explain why other agents, including small molecules like the nutlins (11), that bind to HDM-2 competitively with p53 in cancer cells, and therefore should also bind to HDM-2 in the cell membrane, induce apoptosis, and not necrosis, of these cells. These agents would not be expected to adopt membrane-active conformations. This includes HDM-2-binding peptides that contain membrane-penetrating sequences on their amino terminal ends (8–10), which we previously found are much less alpha-helical (1) and therefore are not likely to adopt membrane-active conformations.
Materials and Methods
Materials.
Peptides.
PNC-27, H-Pro-Pro-Leu-Ser-Gln-Glu-Thr-Phe-Ser-Asp-Leu-Trp-Lys-Leu-Leu-Lys-Lys-Trp-Lys-Met-Arg-Arg-Asn-Gln-Phe-Trp-Val-Lys-Val-Gln-Arg-Gly-OH (1 g), was synthesized using solid phase methods (Shaanxi Zhongbang Pharma-Tech Corp., NanguanZhengjie, Xi’an, China) and was > 95% pure by HPLC and mass spectrographic analysis. The bold sequence corresponds to amino acid residues 12-26 of the HDM-2-binding domain of human p53 while the italicized sequence corresponds to the MRP segment that allows entry of the whole peptide into cells. PNC-28, which contains p53 residues 17-26 linked to the MRP, was likewise synthesized by solid phase methods. The negative control peptide, PNC-29 (1–5), containing the X13 peptide from cytochrome P450 (bold) attached to the MRP (italics), H-Met-Pro-Phe-Ser-Thr-Gly-Lys-Arg-Ile-Met-Leu-Gly-Glu- Lys-Lys-Trp-Lys-Met-Arg-Arg-Asn-Gln-Phe-Trp-Val-Lys-Val-Gln-Arg-Gly-OH, was likewise synthesized by solid phase methods and was likewise > 95% pure.
FITC- and TAMRA-double fluorophore-labeled PNC-27 peptide.
This peptide was synthesized at the Biopeptide Corp., La Jolla, CA, with two fluorescent labels: amino terminal 5,6-carboxy-fluorescein (green fluorescence) and carboxyl terminal 5-tetramethyl Rhodamine (TAMRA) (red fluorescence) and was found to be > 95% pure (20).
Plasmids.
Untransformed cells that do not express HDM-2 in their cell membranes are not susceptible to PNC-27 (1–5). To determine if these cells become susceptible to this peptide if they contain HDM-2 in their cell membranes, we have transfected into these cells a plasmid that expresses full-length HDM-2 attached on its carboxyl terminal end to a CAAX localization signal peptide sequence, i.e., Cys-Val-Val-Lys (CVVK), called HDM-2-CVVK. The plasmid was a Precision ShuttleVector with a green fluorescent protein (GFP) tag and a specified open reading frame sequence, in our case, HDM-2-CVVK (Origene, Rockville, MD). Full-length HDM-2 with the carboxyl terminal CAAX sequence, called pHDM2-CVVK, was inserted into this plasmid between Sgf1 and Mlu 1 endonuclease restriction sites. Both the HDM-2 construct and GFP were under a constitutive CMV expression promoter that also induced expression of amp and neo resistance genes.
In addition to this plasmid, we prepared two other plasmids encoding proteins that served as controls: full-length HDM-2 without the CAAX sequence, called pHDM2 and HDM-2-CVVK, that lacks amino acid residues 1-109 that constitute the binding site for p53 and for PNC-27, called pdel1-109-HDM2-CVVK. All plasmids were prepared at Origene. The following primers were employed to construct the DNA sequences encoding each of the HDM-2 proteins. For these sequences all nuclease sites are given in bold; the start (ATG) and stop (TTA) codons are italicized; and the carboxyl terminal codons for the membrane-localization signal sequence, CVVK (CAAX box), are underlined: Full-length HDM-2:
CTACAGCGATCGCCATGGTGAGGAGCAGGCAAATGTGC (+strand),
ACGAGACGCGTGGGGAAATAAGTTAGCACAATCATTTG (-strand); Full-Length HDM-2 with C-terminal CVVK membrane-attaching CAAX sequence
CTACAGCGATCGCCATGGTGAGGAGCAGGCAAATGTGC (+strand)
GCGTACGCGTTTACATAATTACACACTTGGGGAAATAAGTTAGCACAATCATTTG G (-strand); HDM-2-CVVK with residues 1-109 deleted
CTACAGCGATCGCCATCTACAGGAACTTGGTAGTAGTC (+STRAND)
GCGTACGCGTTTACATAATTACACACTTGGGGAAATAAGTTAGCACAATCATTTGG (-STRAND)
After digestion with the cloning restriction enzyme Sgf-I and Mlu I, the PCR products were cloned into the Origene Precision Shuttle plasmid pCMV6-AN-GFP with an N-terminal fused GFP-tag. Final constructs were sequenced with VP1.5 5′ GGACTTTCCAAAATGTCG 3′ and XL39 5′ ATTAGGACAAGGCTGGTGGG 3′ primers.
Cell lines.
The following cell lines were obtained from the American Type Culture Collection (Manassas, VA): MIA-PaCa-(human pancreatic cancer), MCF-7 (human breast cancer), A2058 (human melanoma), A-2058 (human melanoma), MCF-10-2A (normal human breast epithelial cells). Ag13145 cells (primary human fibroblasts, 46 chromosome, XY) were obtained from the Coriell Institute for Medical Research (Camden, NJ) and cultured in DMEM supplemented with 10% fetal bovine serum (FBS; Atlanta Biologicals, Atlanta, GA). In addition, we employed two cell lines that we have developed, i.e., BMRPA1, a normal rat pancreatic acinar cell line, and its k-ras-transformed counterpart pancreatic cancer cell line, called TUC-3 (1), both of which were grown in cRPMI medium as described previously (1).
Methods
Superposition of 2D NMR structures for PNC-27 on that for the p53 17-29 peptide bound to HDM-2.
We superimposed the coordinates for the backbone atoms of the 30 structures that fit the NOE constraints for residues 17-26 of PNC-27 (12) on those for the x-ray structure (15) as described previously (3).
Assays.
Lactate dehydrogenase (LDH) assay using the LDH Cytotoxicity Assay (Promega, Madison, WI); casapse assay for apoptosis [positive control: 2 × 104 cells treated with staurosporine (Sigma, St Louis, MO) (45 μg/ml) (16)]; and MTT cell viability assay were all performed as described previously (2–4). Protein concentrations were performed using the Bradford assay (Pierce, Rockford, IL).
Western blots.
Lysates of 2 × 106cells were either used directly or employed for preparation of purified plasma membranes (22). To assure that the final preparations contained plasma membranes, samples were immunoblotted for membrane β-catenin and by transmission electron microscopy (4). Blotting of both fractions for H(M)DM-2 was performed in a manner similar to that described previously (1–4). Anti-HDM-2 polyclonal antibody (Santa Cruz Biotechnology, Santa Cruz, CA) was used at a dilution of 1∶4,000. Secondary antibody was HRP-conjugated donkey anti-mouse IgG (HRP-anti-M IgG) (Jackson ImmunoResearch, West Grove, PA) used at a dilution of 1∶1,000 in 0.1% milk in TBS-T. Immun-Star HRP Peroxide Buffer + Immun-Star HRP Luminol/Enhancer (ratio 1∶1) (BioRad) was added to the nitrocellulose membranes.
Immunoprecipitation experiments.
To determine if fluorophore-labeled PNC-27 binds to HDM-2 in cancer cells treated with this peptide, we incubated double-fluorophore-labeled PNC-27 (50 μg/ml) (20) with 1 × 106 MIA-PaCa-2 cells for 4 hr. The cells were then lysed (1–4), and 500 μg protein in lysate samples were subjected to IP with 0.5 ug anti-MDM2 mouse monoclonal antibody (D-7, Santa Cruz Biotechnology), 2 μg biotinylated horse antimouse IgG (Pierce) and 30 μl 50% suspension of UltraLink-immobilized NeutrAvidin Biotin-Binding Protein beads. The samples were then electrophoresed in 12.5% PAAG gel (BioRad). The position of the fluorescent peptide was analyzed in the gel with Kodak Image Station 2000R. The proteins were transferred to a PVDF membrane, immunobloted with polyclonal anti-MDM2 antibody (Santa Cruz, N-20), and developed with ECL (Pierce).
Colocalization experiments and confocal microscopy.
These experiments were performed on two different cancer cell lines, A-2058 human melanoma and MCF-7 human breast cancer cells, and on two untransformed cell lines, BMRPA1 rat pancreatic acinar cells (1) and MCF-10-2A untransformed human breast epithelial cells. Cells grown on glass cover slips to 50–60% density were treated for up to 15 min at 37 °C in a humidified 5% CO2—95% air incubator chamber with PNC-27 or PNC-29 (control) at 50 μg/ml incubation medium and were then washed and fixed in 3% paraformaldehyde in PBS (pH 7.2) supplemented with 0.01% glutaraldehyde for 1.5 h followed by extensive washing and transfer into PBS for storage until mounting on glass slides for microscopy. Free aldehyde groups were quenched by incubating cells with glycine (0.2 M) and sodium borohydride (75 mM) followed by washing in PBS. Cells were then stained (direct staining) for 2 h, 4 °C, with fluorescein-labeled mouse monoclonal antibody against p53 [FITC- mAbα-p53 (DO-1)] (5 μg/ml) and rhodamine-labeled (TRITC-) mAbα-against H/R/MDM-2 (5 μg/ml) (both labeled antibodies, Pierce) in 1% FBS-PBS. After removal of nonreactive Ab and extensive washing, the cover slips were mounted on glass slides over Prolong Gold Antifade (Molecular Probes, Invitrogen, Carlsbad, CA) and examined with a laser-equipped Olympus Confocal microscope 1 × 76 (Olympus America Inc, Center Valley, PA). Results were digitally recorded. The colocalization of the two antibodies was confirmed by overlapping green (anti-PNC-27) and red (antiH/R/MDM-2) fluorescent labels that produced yellow fluorescence (combined green and red fluorescence).
Dose response experiments.
LDH assays were performed on different cancer cell lines (n = 3–5) that were incubated for 30 min with PNC-27 over a concentration range of 10 μg.ml/1 mg/ml as described previously (2–4).
Transfection of HDM-2 constructs into untransformed cells.
The plasmids that were constructed as described in Materials above were transfected into untransformed MCF-10-2A cells using the procedures described previously (4). The transfection efficiency was evaluated by analyzing the GFP fluorescence at 480 nm.
Treatment of transfected cells with PNC-27 or PNC-29.
The transfected cells in media were then incubated at 37 °C with 5% CO2 for 24 h, at which time they were treated with PNC-27 or PNC-29 peptide (sonicated briefly prior to addition) such that the final concentration was 300 μg/ml. Samples were assayed for LDH, MTT, and caspase. In addition, samples were processed for confocal microscopy as described above except for the following modification. Because the cells contained GFP, to localize PNC-27, it was necessary to use another fluorescent probe other than green-fluorescent FITC-labeled DO1 antibody. Cells were incubated with unlabeled DO1 and anti-HDM-2 as described above. The cells were then washed and incubated with Alexa Fluor 647 goat antimouse IgG (1∶200) (against DO1 mouse) (Invitrogen–Molecular Probe, Eugene, OR) and TAMRA-labeled goat antirabbit IgG (1∶200) (against anti-HDM-2 rabbit polyclonal IgG) (Sigma, St. Louis, MO). The cells were processed for confocal microscopy, and the membrane fractions and whole cell lysates were blotted for either HDM-2 or actin [rabbit anti-actin-42 polyclonal antibody (1∶5000)] (Sigma).
Supplementary Material
Acknowledgments.
This work was supported in part by a grant from Innomab Inc. (to M.R.P. and J.M) and by National Institutes of Health Grant CA42500, a Veterans Administration Merit Grant (to M.R.P.), a Lustgarten Foundation for Pancreatic Cancer Research Grant (to M.R.P. and J.M.), and a Veterans Administration Grant and an American College of Surgeons Faculty Research Fellowship Award (to W.B.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0909364107/DCSupplemental.
References
- 1.Kanovsky M, et al. Peptides from the amino terminal mdm-2 binding domain of p53, designed from conformational analysis, are selectively cytotoxic to transformed cells. Proc Natl Acad Sci USA. 2001;98:12438–12443. doi: 10.1073/pnas.211280698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Do TN, et al. Preferential induction of necrosis in human breast cancer cells by a p53 peptide derived from the mdm-2 binding site. Oncogene. 2003;22:1431–1444. doi: 10.1038/sj.onc.1206258. [DOI] [PubMed] [Google Scholar]
- 3.Pincus MR, Michl J, Bowne W, Zenilman M. Anti-cancer peptides from the ras-p21 and p53 proteins. In: Mohan R, editor. Research Advances in Cancer. Kerala, India: Global Research Network Publishers; 2007. pp. 65–90. [Google Scholar]
- 4.Bowne WB, et al. The penetratin sequence in the anticancer PNC-28 peptide causes tumor necrosis rather than apoptosis of human pancreatic cancer cells. Ann Surg Oncol. 2008;15:3588–3600. doi: 10.1245/s10434-008-0147-0. [DOI] [PubMed] [Google Scholar]
- 5.Michl J, et al. PNC-28, a p53 peptide that is cytotoxic to cancer cells, blocks pancreatic cancer cell growth in vivo. Int J Cancer. 2006;119:1577–1585. doi: 10.1002/ijc.22029. [DOI] [PubMed] [Google Scholar]
- 6.Picksley SM, Spicer JF, Barnes DM, Lane DP. The p53-MDM2 interaction in a cancer-prone family, and the identification of a novel therapeutic target. Acta Oncol. 1996;35:429–434. doi: 10.3109/02841869609109917. [DOI] [PubMed] [Google Scholar]
- 7.Bottger V, et al. Identification of novel mdm2 binding peptides by phage display. Oncogene. 1996;13:2141–2147. [PubMed] [Google Scholar]
- 8.Wasylyk C, et al. p53 mediated death of cells overexpressing MDM2 by an inhibitor of MDM2 interaction with p53. Oncogene. 1999;18:1921–1934. doi: 10.1038/sj.onc.1202528. [DOI] [PubMed] [Google Scholar]
- 9.Chene P, Fuchs J, Carena I, Furet P, Garcia-Echeverria C. Study of the cytotoxic effect of a peptidic inhibitor of the p53-hdm2 interaction in tumor cells. Febs Lett. 2002;529:293–297. doi: 10.1016/s0014-5793(02)03362-8. [DOI] [PubMed] [Google Scholar]
- 10.Harbour JW, Worley L, Ma D, Cohen M. Transducible peptide therapy for uveal melanoma and retinoblastoma. AMA Arch Ophthalmol. 2002;120:1341–1346. doi: 10.1001/archopht.120.10.1341. [DOI] [PubMed] [Google Scholar]
- 11.Vassilev LT, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 12.Rosal R, et al. NMR solution structure of a peptide from the mdm-2 binding domain of the p53 protein that is selectively cytotoxic to cancer cells. Biochemistry. 2004;43:1754–1861. doi: 10.1021/bi035718g. [DOI] [PubMed] [Google Scholar]
- 13.Pincus MR. In: The Physiological Structure and Function of Proteins in Principles of Cell Physiology. 3rd Ed. Sperelakis N, editor. New York: Academic; 2001. pp. 19–42. [Google Scholar]
- 14.Dathe M, Wieprecht T. Structural features of helical anti-microbial peptides: Their potential to modulate activity on model membranes and biological cells. Biochem Biophys Acta. 1999;1462:71–87. doi: 10.1016/s0005-2736(99)00201-1. [DOI] [PubMed] [Google Scholar]
- 15.Kussie PH, et al. Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science. 1996;274:921–922. doi: 10.1126/science.274.5289.948. [DOI] [PubMed] [Google Scholar]
- 16.Bertrand R, Solary E, O’Connor P, Kohn KW, Pommier Y. Induction of a common pathway of apoptosis by staurosporine. Exp Cell Res. 1994;211:314–321. doi: 10.1006/excr.1994.1093. [DOI] [PubMed] [Google Scholar]
- 17.Manne V, et al. Ras farnesylation as a target for novel antitumor agents: Potent and selective farnesyl diphosphate analogue inhibitors of farnesyltransferase. Drug Dev Res. 1995;34:121–137. [Google Scholar]
- 18.Yang J-Y, et al. MDM2 promotes cell motility and invasiveness by regulating E-cahedrin degradation. Mol Cell Biol. 2006;26:7269–7282. doi: 10.1128/MCB.00172-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Toth A, Nickson P, Qin LL, Erhardt P. Differential regulation of cardiomyocyte survival and hypertrophy by MDM2, an E3 ubiquitin ligase. J Biol Chem. 2006;281:3679–3689. doi: 10.1074/jbc.M509630200. [DOI] [PubMed] [Google Scholar]
- 20.Sookraj K, et al. The anticancer peptide, PNC-27, induces tumor cell lysis as the intact peptide. Cancer Chemoth Pharm. 2009 doi: 10.1007/s00280-009-1166-7. in press. [DOI] [PubMed] [Google Scholar]
- 21.Palmer M, Valeva A, Kehoe M, Bhakdi S. Kinetics of streptolysin O self-assembly. Eur J Biochem. 1995;231:388–395. doi: 10.1111/j.1432-1033.1995.tb20711.x. [DOI] [PubMed] [Google Scholar]
- 22.Ramjeesingh M, Kidd JF, Huan LJ, Wang Y, Bear CE. Dimeric cystic fibrosis transmembrane conductance regulator exists in the plasma membrane. Biochem J. 2003;374:797–797. doi: 10.1042/BJ20030683. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.