Abstract
Down Syndrome cell adhesion molecule(Dscam) genes encode neuronal cell recognition proteins of the immunoglobulin superfamily1,2. In Drosophila, Dscam1 generates 19,008 different ecto-domains by alternative splicing of three exon clusters, each encoding half or a complete variable immunoglobulin domain3. Identical isoforms bind to each other, but rarely to isoforms differing at any one of the variable immunoglobulin domains4,5. Binding between isoforms on opposing membranes promotes repulsion6. Isoform diversity provides the molecular basis for neurite self-avoidance6–11. Self-avoidance refers to the tendency of branches from the same neuron (self-branches) to selectively avoid one another12. To ensure that repulsion is restricted to self-branches, different neurons express different sets of isoforms in a biased stochastic fashion7,13. Genetic studies demonstrated that Dscam1 diversity has a profound role in wiring the fly brain11. Here we show how many isoforms are required to provide an identification system that prevents non-self branches from inappropriately recognizing each other. Using homologous recombination, we generated mutant animals encoding 12, 24, 576 and 1,152 potential isoforms. Mutant animals with deletions encoding 4,752 and 14,256 isoforms14 were also analysed. Branching phenotypes were assessed in three classes of neurons. Branching patterns improved as the potential number of isoforms increased, and this was independent of the identity of the isoforms. Although branching defects in animals with 1,152 potential isoforms remained substantial, animals with 4,752 isoforms were indistinguishable from wild-type controls. Mathematical modelling studies were consistent with the experimental results that thousands of isoforms are necessary to ensure acquisition of unique Dscam1 identities in many neurons. We conclude that thousands of isoforms are essential to provide neurons with a robust discrimination mechanism to distinguish between self and non-self during self-avoidance.
Dscam1 diversity provides the molecular basis of neurite self-avoidance (for review, see ref. 1). Neurite self-avoidance facilitates uniform coverage of target fields by dendritic and axonal branches, while allowing the branches of different neurons to share the same field12. Self-avoidance also has an important role in branch segregation during axon guidance7,8,11. Self-avoidance requires that self-branches repel one another, whereas branches of different neurons do not repel each other. Each neuron expresses a unique combination of Dscam1 isoforms comprising some tens of isoforms in a largely stochastic manner7,13. Thus, isoform-specific homophilic recognition is restricted to self-branches. Binding elicits neurite repulsion6. That Dscam1 diversity underlies the phenomenon of self-avoidance is well documented in the formation of dendritic receptive fields formed by dendritic arborization neurons in the larval body wall6,9,10 and in the segregation of axon branches of mushroom body neurons in the central brain7,11,14,15. Although self-avoidance demands that neurons express different sets of isoforms, the identity of the isoforms they express is not important6,7,11,14 (see later).
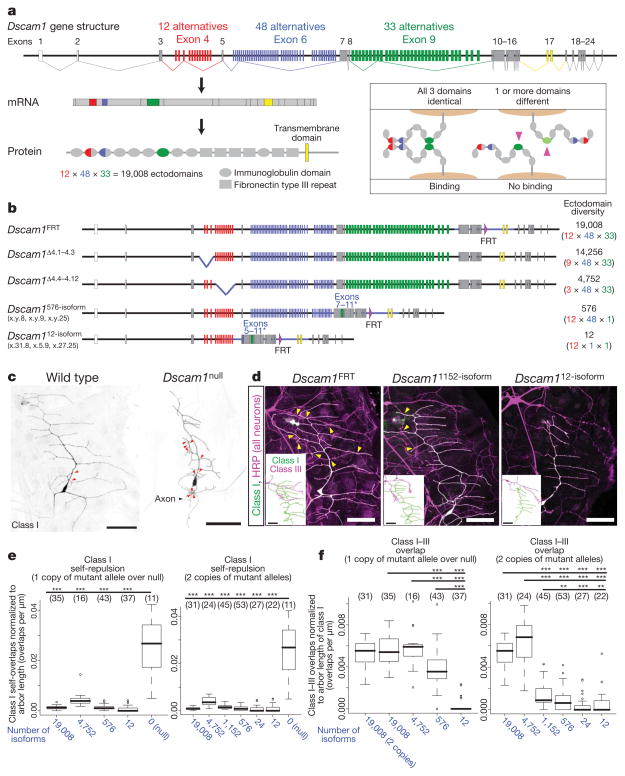
How many isoforms are necessary for each neuron within a group to discriminate between self and non-self? To address this question, we analysed mutants with reduced Dscam1 diversity (Fig. 1a, b). Previously, deletion mutants were generated that removed subsets of exon 4 variants (for example, Dscam1Δ4.1–4.3 and Dscam1Δ4.4–4.12; Fig. 1b)14. The largest deletion (Dscam1Δ4.4–4.12) removes nine out of twelve exon 4 variants and, thus, encodes 4,752 potential isoforms. To reduce Dscam1 diversity further, a series of knock-in mutants encoding between 12 and 1,152 isoforms were generated using homologous recombination (Fig. 1b and Supplementary Fig. 1). These include three different Dscam1576-isoform alleles, each of which contains only one exon 9 variant. Thus, each allele encodes a distinct and non-overlapping set of 576 potential isoforms. Furthermore, three Dscam112-isoform alleles were generated, each of which contains one combination of exon 6 and 9 variants, thus encoding 12 potential isoforms. By combining two different Dscam1576-isoform alleles, animals with 1,152 potential isoforms were generated (referred to as Dscam11152-isoform). In a similar manner, animals with 24 potential isoforms were generated by combining two different Dscam112-isoform alleles (referred to as Dscam124-isoform). Replacement of the genomic regions was confirmed by sequencing ~10 kilobases (kb) spanning the modified segment. Sequence analysis of complementary DNAs isolated from these alleles showed that the fixed exons were expressed with different combinations of the remaining variable exons (Supplementary Fig. 2a). Protein levels and distributions were similar to wild type, as assessed by western blots and immunostaining, respectively (Supplementary Fig. 2b, c). In summary, these alleles encode reduced numbers of isoforms expressed in a similar pattern and level as in wild type.
Figure 1. Thousands of isoforms are required for dendrites of dendritic arborization neurons to distinguish between self and non-self.
a, Alternative splicing of Dscam1. The inset shows a schematic of Dscam1 isoform-specific homophilic binding. The crystal structure shows that variable domains pair in an antiparallel configuration23,24. Binding requires matching of variable domains; one mismatch (right, pink arrowheads) disrupts binding. b, Dscam1 alleles with reduced diversity. Alternative exons were replaced with cDNAs encoding exons 7–11 and 5–11 to generate Dscam1576-isoform and Dscam112-isoform alleles, respectively (asterisks). Sequence modified is indicated with blue line. Dscam1FRT is a control for Dscam1576-isoform and Dscam112-isoform knock-in alleles, and contains an FRT site as a result of homologous recombination11. Three different alleles were generated for each mutant. x.y.z refers to combinations of exon 4, 6 and 9, respectively, encoded by alleles. Dscam1Δ4.1–4.3 and Dscam1Δ4.4–4.12 were reported previously14. c, Self-repulsion of dendritic arborization neurons requires Dscam1. Red arrowheads, self-crossing of class I (vpda) dendrites. d, Representative images of dendritic arborization neuron dendrites in different Dscam1 mutants. See Supplementary Fig. 3 for larger images. All neurons were visualized with anti-HRP antibody (magenta), and class I (vpda) neurons were labelled with GFP (green; appears white because they overlap with magenta). Class III (v′pda) neurons were traced from cell bodies16 (insets). Yellow arrowheads, crossing between class I and III dendrites. Scale bars, 50 μm. e, Self-repulsion is intact in all mutants with reduced diversity. Numbers in parenthesis denote class I neurons analysed for each genotype. Data in boxplot format (thick line, median; box, 25–75% quartiles; error bar, data within 1.5× quartile range; circles, outliers). ***P < 0.001; Student’s t-test. f, Increasing the number of available isoforms increases overlaps between class I and class III dendrites. Numbers in parenthesis, class I/III pairs analysed. Data in boxplot format. **P < 0.01, ***P < 0.001; Wilcoxon rank test. Thousands of isoforms are required for normal degree of overlap. For genotypes, see Methods.
We first sought to determine the number of isoforms necessary for patterning dendritic arborization neurons. These neurons elaborate dendrites in a two-dimensional pattern over the larval body wall. Self-dendrites seldom overlap. In contrast, dendrites of different dendritic arborization neurons in the same territory overlap extensively16. Removal of all Dscam1 isoforms results in increased overlap and fasciculation of self-dendrites (Fig. 1c), whereas the overexpression of a single Dscam1 isoform in dendritic arborization neurons with normally overlapping dendrites leads to ectopic avoidance between them6,9,10. In all mutant animals encoding reduced diversity, self-dendrites of class I dendritic arborization neurons avoided each other as in wild-type controls (Fig. 1d, e). In contrast to wild type, however, dendrites of class I and class III neurons, which normally overlap, showed nearly complete avoidance in mutants encoding 12 and 24 isoforms (Fig. 1d, f and Supplementary Fig. 3). This indicates that ectopic repulsion occurred between class I and III dendrites in these mutants.
We next tested the effects of increasing the pool of isoforms to hundreds and thousands on the overlap between class I and III dendrites. We observed increased overlaps as the number of potential isoforms increased (Fig. 1f and Supplementary Fig. 3). Class I and III dendrites overlapped substantially more in Dscam1576-isoform/Dscam1null mutants than in Dscam112-isoform/Dscam1null mutants, although still less than in wild type. Homozygous mutants with two copies of Dscam1576-isoform had fewer dendritic overlaps than the Dscam1576-isoform/Dscam1null mutants. Thus, the number of overlaps between the dendrites of different neurons is sensitive to the overall level of Dscam1 protein with this level of diversity. Dscam11152-isoform mutants showed a modest increase in overlaps compared to that seen in Dscam1576-isoform mutants. In contrast, mutant animals with 4,752 potential isoforms (that is, Dscam1Δ4.4–4.12) had overlaps that were indistinguishable from the wild-type control. A linear regression analysis showed a strong correlation between the number of available isoforms and the degree of overlap between class I and III dendrites (R2 = 0.66, t-test: P < 2 ×10−16). These findings suggest that a substantial fraction of isoforms is shared between class I and III neurons when tens or hundreds of isoforms are available. With thousands of isoforms available, however, different neurons express sufficiently different populations of Dscam1 isoforms to prevent ectopic repulsion. In summary, a pool of thousands of isoforms is necessary for each dendritic arborization neuron to reliably discriminate between self and non-self neurites.
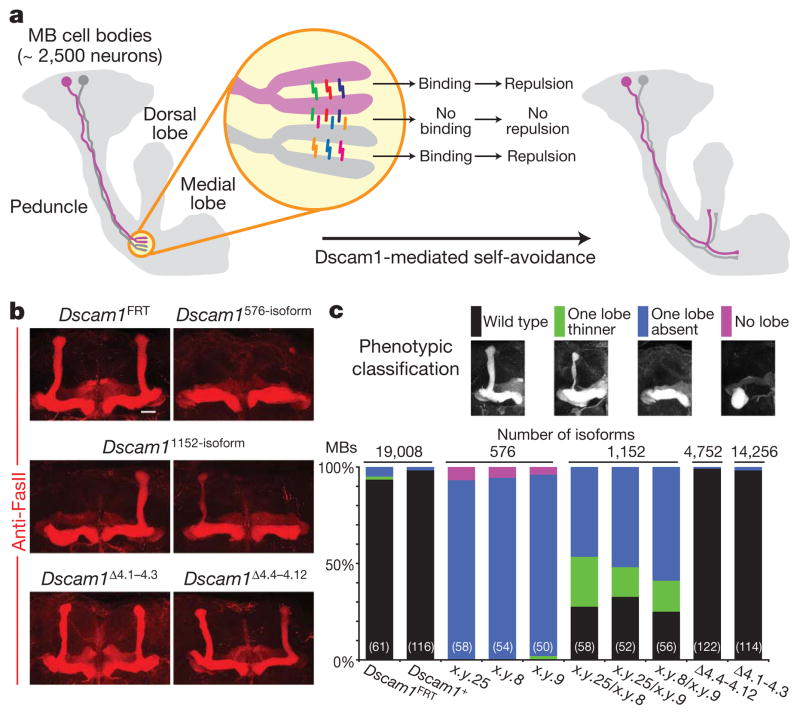
We next assessed the extent of diversity required for mushroom body development. The mushroom body comprises some 2,500 neurons and most axons form two branches projecting into two orthogonal paths, which result in the formation of two distinct lobes17. Dscam1-mediated self-avoidance is required for self-branches to segregate with high fidelity (Fig. 2a)7,11. Mushroom bodies in mutants with only a single ectodomain encoded at the Dscam1 locus (Dscam1single) invariably form only one lobe, indicating that mushroom body axons do not segregate appropriately11. In contrast, self-branches of a single mushroom body neuron expressing a single isoform surrounded by wild-type neighbours in a genetically mosaic animal segregate with high fidelity7,11,14. Together, these data suggest that the failure to form two lobes in Dscam1single mutants reflects inappropriate recognition between the branches of different neurons11. All Dscam1576-isoform mutant animals form only a single lobe, similar to that seen in Dscam1single mutants (Fig. 2b, c). Increasing the potential pool of isoforms to 1,152 by combining two different alleles of Dscam1576-isoform, partially improved mushroom body lobe segregation (Fig. 2b, c). In contrast, the morphology of mushroom bodies in mutants with 4,752 potential isoforms (that is, Dscam1Δ4.4–4.12) was indistinguishable from wild type (Fig. 2b, c). These results show that between 1,152 and 4,752 isoforms are required for mushroom body axon branches to distinguish between self and non-self, thereby supporting orderly axon branch segregation and the formation of the bi-lobed mushroom body structure.
Figure 2. Thousands of isoforms are required for mushroom body lobe development.
a, Schematic of mushroom body (MB) development. The call-out circle represents a model of selective recognition between self-branches mediated by Dscam1 isoform-specific homophilic binding. Dscam1 isoforms are shown as coloured bars. b, Mushroom body lobe morphology in mutant animals, visualized with monoclonal antibody 1D4 (anti-FasII, red). Scale bar, 20 μm. c, Quantification of the mushroom body lobe phenotypes. The numbers in the parenthesis represent the number of mushroom bodies (that is, the number of brain hemispheres) examined for each genotype. See Methods for genotypes.
In summary, thousands of isoforms are essential for generating sufficiently different Dscam1 cell-surface identities to support a robust discrimination mechanism for dendrites of dendritic arborization neurons and axon branches of mushroom body neurons to distinguish between self and non-self. This self-avoidance function of Dscam1 does not depend on the identity of the isoforms, as different mutants with the same degree of diversity showed similar spectra of phenotypes, and deletion mutants collectively removing all isoforms (Dscam1Δ4.1–4.3 and Dscam1Δ4.4–4.12) showed normal mushroom body lobe segregation and patterns of dendrites of dendritic arborization neurons (Fig. 2b, c, Supplementary Fig. 3 and ref. 6).
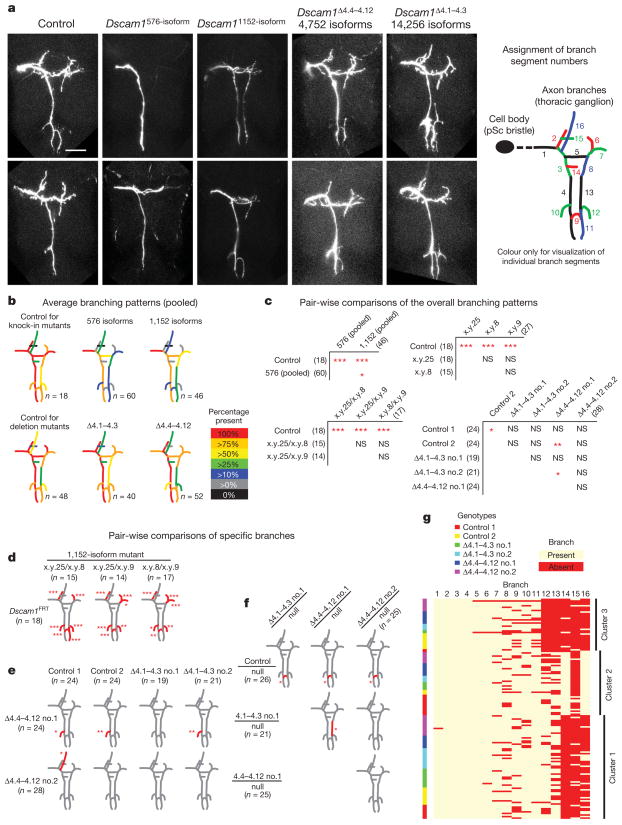
A fundamentally different role for Dscam1 diversity in regulating branching was proposed in a previous study18, where the complex branching patterns of posterior scutellar mechanosensory neurons were analysed in two different, although partially overlapping, exon 4 deletion mutants with the same degree of diversity (11,088 potential isoforms). Differences in the branching patterns of posterior scutellar neurons between deletions and between deletions and control were described. These included an increase in the number of animals with branches that are present in a fraction of control animals, and the appearance of some ectopic branches unique to each mutant. These findings led to the proposal that, in contrast to self-avoidance, it is the identity of specific isoforms that has a role in regulating the formation of specific branches of posterior scutellar neurons. Here we revisit the role of Dscam1 diversity in branching of posterior scutellar neurons. In contrast to previous work18, our data demonstrate that there is no requirement for specific isoforms, and that it is the number of isoforms that is crucial in regulating branching.
All Dscam1576-isoform mutant animals had marked defects in the axonal branching patterns of posterior scutellar neurons (Fig. 3a–c). The branching patterns were improved in Dscam11152-isoform mutants. The spectra of phenotypes were indistinguishable between different Dscam1576-isoform mutants and between different Dscam11152-isoform mutants encoding different sets of isoforms (Fig. 3c, d and Supplementary Figs 5 and 6). Thus, the improvement seen in Dscam11152-isoform mutants derives solely from the increase in the number of potential isoforms, and not from acquisition of ‘correct’ isoforms.
Figure 3. Branching of posterior scutellar neurons requires thousands of isoforms independent of their identity.
a, Representative images of posterior scutellar (pSc) neuron axon branch patterns in homozygous Dscam1 mutants with different degrees of diversity (for Dscam1 deletion mutants over Dscam1null alleles, see Supplementary Fig. 4). Scale bar, 50 μm. Right panel, branch segment numbers were assigned arbitrarily for scoring (scoring method, Supplementary Fig. 4). b, Average branch patterns for each genotype (for Dscam1 deletion mutants over Dscam1null alleles, and raw data, see Supplementary Fig. 5). c, Pair-wise comparisons of overall branch patterns. P values were determined using permutation test with Bonferroni correction accounting for 30 different pair-wise comparisons (see Methods). Numbers in parenthesis indicate the number of animals analysed. See Supplementary Fig. 6 for raw statistical data. d–f, Branches that show difference in frequency between each pair are red. P values were calculated using Fisher’s exact test with Bonferroni correction accounting for 16 different branch segments. g, The result of partitioning cluster analysis (homozygous deletion mutants). Each individual sample was clustered based on its branching pattern. Each of the three clusters contains controls, Dscam1Δ4.1–4.3 mutants, and Dscam1Δ4.4–4.12 mutants in similar ratios. See Supplementary Fig. 6 for results for knock-in mutants. NS, not significant; *P < 0.05, **P < 0.01, ***P < 0.001.
That isoform identity is not essential for elaborating the branching pattern of posterior scutellar neurons was demonstrated by examining branching patterns in Dscam1Δ4.1–4.3 and Dscam1Δ4.4–4.12 mutants, which collectively remove all Dscam1 isoforms. A few branch segments exhibited nominally significant differences between different genotypes. However, these differences did not persist in the same mutants in different genetic backgrounds (Fig. 3c, e and Supplementary Figs 5 and 6), or in mutants with one copy of a deletion allele and one copy of a protein-null allele (Fig. 3f and Supplementary Figs 4–6). Furthermore, cluster analysis showed that the branching patterns of Dscam1Δ4.1–4.3 and Dscam1Δ4.4–4.12 mutants were indistinguishable from controls (Fig. 3g and Supplementary Fig. 6). Thus, it is the number of isoforms, not their identity, that is crucial in regulating the branching pattern of posterior scutellar neurons.
In summary, we conclude that the branching of posterior scutellar neurons requires between 1,152 and 4,752 isoforms, similar to that observed for dendritic arborization and mushroom body neurons, and that no specific isoforms are required for this process. Although the details of branch formation of posterior scutellar neurons in the developing thoracic ganglion are not known, the target areas of branches contain neurites of many neurons. Thus, it is likely that the branching defects observed in mutants with reduced diversity reflect the failure of branches to adequately discriminate between self and non-self neurites. That is, insufficient diversity leads to inappropriate repulsive interactions between neurites of different neurons. Notably, thousands of isoforms are required for normal patterning in all three systems analysed in this paper.
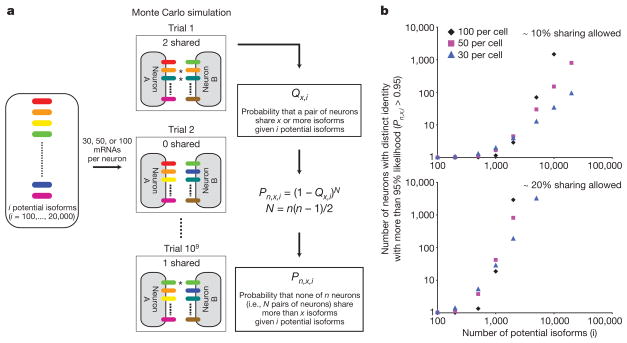
To address why so many isoforms are required to prevent interactions between self and non-self, we mathematically modelled the relationship between the number of potential isoforms and the number of distinct Dscam1 identities that can be generated. Although isoform expression in dendritic arborization and posterior scutellar neurons has not been analysed, previous molecular studies on mushroom body neurons, as well as other neurons, showed that each neuron expresses some tens of isoforms in a largely stochastic fashion7,13. Accordingly, we used a Monte Carlo simulation and binomial distribution to estimate the number of neurons that reliably acquire ‘unique’ Dscam1 identities (that is, arrays of isoforms expressed in a pair of neurons that do not trigger ectopic repulsion between them), given different numbers of potential isoforms (Fig. 4a). We found that the probabilities that a pair of neurons express the same isoform(s) were rather high (Supplementary Fig. 7a). This finding is analogous to the ‘birthday problem’ in which the likelihood of two individuals sharing the same birthday from a randomly chosen population is counter-intuitively high (for example, there is a 50% likelihood that at least two individuals share the same birthday in a group of 23 people). For example, there is 4.4% chance that a pair of neurons shares at least one isoform out of 30 messenger RNAs even with a pool of 20,000 isoforms. Thus, if one isoform shared between two neurons was sufficient to trigger repulsion, then fewer than three neurons would reliably acquire unique identities, even from a pool of 20,000 isoforms (Supplementary Fig. 7b). This indicates that some degree of overlap in the isoforms expressed between two neurons is probably permissive. When sharing some fraction of isoforms (for example, ~20%) between two neurons is permitted, a nonlinear increase in the number of neurons acquiring unique identities was observed as the pool of isoforms increases from hundreds to thousands, regardless of the number of isoforms expressed per neuron (Fig. 4b). Notably, thousands of isoforms are necessary to reliably generate tens of neurons or more with unique Dscam1 identities. Thus, this modelling study provides a mathematical explanation for why thousands of isoforms are necessary to promote repulsion between self-branches with minimal risk of ectopic recognition between non-self branches.
Figure 4. Mathematical model showing that thousands of isoforms are required to give rise to many unique Dscam1 identities.
a, A schematic representation of mathematical modelling used to determine number of neurons that acquire unique Dscam1 identities. Tables of Qx,i for each condition are shown in Supplementary Fig. 7a. b, The number of neurons that obtain unique identities at more than 95% likelihood with varying degrees of diversity. Both axes are in log scale. See Supplementary Fig. 7b for the condition in which no sharing of isoforms between neurons is allowed.
In conclusion, we demonstrate a profound requirement for thousands of Dscam1 isoforms in patterning neural circuits. The importance of this level of diversity is underscored by the observation that all arthropod species for which genome sequence is available encode thousands of different Dscam1 ectodomains (for example, ~3,000 isoforms in water flea19, ~6,000 in honey bee20, and up to 20,000 in different Drosophila species20). We show that around 5,000 isoforms are sufficient to pattern all three systems analysed in this paper. Further diversification of Drosophila Dscam1 beyond 5,000 isoforms may reflect differential splicing mechanisms in different neuronal subclasses (for example, some neurons may show more extensive bias in splicing thus limiting the access to the full diversity), a function mediating recognition between neurons in other developmental contexts (for example, to promote adhesive interactions between synaptic partners) or functions elsewhere such as in the immune system21. Notably, our study argues that the demand for robust discrimination between self and non-self neurites underlying self-avoidance alone is sufficient to account for thousands of isoforms encoded at the Dscam1 locus in Drosophila.
METHODS SUMMARY
Dscam1576-isoform and Dscam112-isoform mutant alleles were generated essentially as described11 (Supplementary Fig. 1). At least two independent recombinants were generated for each allele encoding distinct variants, and these were used to generate homozygous mutants to avoid effects from potential second site mutations (for example, Dscam1x.y.25a/Dscam1x.y.25b). Molecular characterization of these mutants was performed essentially as described11. Immunostaining was performed essentially as described6,11. The branching patterns of posterior scutellar mechanosensory neurons were analysed by DiI tracing as described18,22. The branching patterns of different exon 4 deletion mutants were scored blindly to avoid potential bias in scoring (for details of the scoring method, see Supplementary Fig. 4 and Methods). Pair-wise comparisons of branch occurrence between genotypes were performed by Fisher’s exact test. Comparisons between genotypes that account for all branches were conducted using a permutation-based test. Because a large number of comparisons are carried out in this study, we used the Bonferroni criteria to correct for multiple testing.
METHODS
Generation of Dscam1576-isoform and Dscam112-isoform alleles
Dscam1576-isoform and Dscam112-isoform alleles were generated through a two-step gene targeting strategy previously described11. Approximately 4-kb homology arms were PCR-amplified from the reference strain for the Drosophila genome sequencing project25. These genomic PCR products were combined with cDNA encoding a single exon 9 variant (for Dscam1576-isoform) or a single exon 6 and a single exon 9 variant (for Dscam112-isoform) by overlap-extension PCR and cloned into a custom vector. These constructs were introduced into flies by a standard transgenic technique. At least two independent inserts of these constructs were used for gene targeting crosses. At least two independent Dscam15′HR alleles were used to generate each knock-in allele and independent knock-in alleles were used to make homozygous animals (for example, Dscam1x.y.25a/Dscam1x.y.25b) to avoid effects of potential second site mutations.
Molecular characterization of Dscam1576-isoform and Dscam112-isoform alleles
The molecular characterization of the Dscam1576-isoform and Dscam112-isoform alleles was performed essentially as described11. cDNA sequencing was performed from RNA extracted from animals with one copy of mutant Dscam1 allele over a deficiency allele, Dscam1Df6055.
Genetics
Flies with the following genotypes were used in this study. Dscam121 and Dscam123 are protein-null alleles of Dscam1. For dendritic arborization neuron analysis: (1) Dscam1FRT/Dscam123; 221-Gal4,UAS-mCD8GFP/+, (2) Dscam121/Dscam123; 221-Gal4,UAS-mCD8GFP/+, (3) Dscam112-isoform/Dscam123; 221-Gal4,UAS-mCD8GFP/+, (4) Dscam1576-isoform/Dscam123; 221-Gal4,UAS-mCD8GFP/+, (5) Dscam1Δ4.4–4.12/Dscam123; 221-Gal4,UAS-mCD8GFP/+, (6) Dscam1FRT; 221-Gal4,UAS-mCD8GFP/+, (7) Dscam112-isoform; 221-Gal4,UAS-mCD8GFP/+, (8) Dscam1576-isoform; 221-Gal4,UAS-mCD8GFP/+, (9) Dscam1x.27.25/Dscam1x.31.8; 221-Gal4,UAS-mCD8GFP/+, (10) Dscam1x.27.25/Dscam1x.5.9; 221-Gal4,UAS-mCD8GFP/+, (11) Dscam1x.y.25/Dscam1x.y.8; 221-Gal4,UAS-mCD8GFP/+, (12) Dscam1x.y.25/Dscam1x.y.9; 221-Gal4,UAS-mCD8GFP/+, (13) Dscam1x.y.8/Dscam1x.y.9; 221-Gal4,UAS-mCD8GFP/+, and (14) Dscam1Δ4.4–4.12; 221-Gal4,UAS-mCD8GFP/+.
For mushroom body neuron analysis: (1) Dscam1FRT, (2) FRTG13,Dscam1+/Dscam1FRT, (3) FRTG13,Dscam1+/Dscam1+, (4) Dscam1576-isoform, (5) Dscam1x.y.25/Dscam1x.y.8, (6) Dscam1x.y.25/Dscam1x.y.9, (7) Dscam1x.y.8/Dscam1x.y.9, (8) FRTG13,Dscam1Δ4.4–4.12/Dscam1Δ4.4–4.12, (9) Dscam1Δ4.4–4.12, (10) FRTG13,Dscam1Δ4.1–4.3*/FRTG13,Dscam1Δ4.1–4.3, and (11) FRTG13,Dscam1Δ4.1–4.3.
For posterior scutellar neuron analysis: (1) Dscam1FRT/Dscam1+, (2) control 1: FRTG13,Dscam1+/Dscam1+, (3) control 2: FRTG13,Dscam1+/Dscam1FRT, (4) Dscam1576-isoform, (5) Dscam1x.y.25/Dscam1x.y.8, (6) Dscam1x.y.25/Dscam1x.y.9, (7) Dscam1x.y.8/Dscam1x.y.9, (8) Δ4.4–4.12 no.1: FRTG13,Dscam1Δ4.4–4.12/Dscam1Δ4.4–4.12, (9) Δ4.4–4.12 no.2: Dscam1Δ4.4–4.12, (10) Δ4.1–4.3 no.1:FRTG13,Dscam1Δ4.1–4.3*/FRTG13,Dscam1Δ4.1–4.3, (11) Δ4.1–4.3 no.2: FRTG13,Dscam1Δ4.1–4.3, (12) FRTG13,Dscam1+/Dscam121, (13) Δ4.4–4.12 no.1 (heterozygous): FRTG13,Dscam1Δ4.4–4.12/Dscam121, (14) Δ4.4–4.12 no.2 (heterozygous): Dscam1Δ4.4–4.12/Dscam121, and (15) FRTG13,Dscam1Δ4.1–4.3*/Dscam121.
Two chromosomes containing the Δ4.1–4.3 mutation were kept in two different genetic backgrounds (one with a balancer and one without balancers). Asterisk indicates the chromosome that was maintained with a balancer.
Immunostaining
Immunostaining for the embryo and the mushroom body was performed essentially as described11. Dendritic arborization neuron analysis was performed essentially as described6. The antibodies were used at the following dilutions for immunostaining: monoclonal antibody 1D4 (anti-FasII) (1:10), monoclonal antibody 11G4 (anti-Dscam1) (1:2), and rabbit anti-GFP (Molecular Probes, 1:1,000). The following secondary antibodies were used (at 1:200 dilution): Alexa488-goat-anti-rabbit IgG, Alexa568-goat-anti-mouse IgG, and Cy5-conjugated anti-HRP (Molecular Probes).
Mechanosensory neuron analysis
Branching patterns of posterior scutellar mechanosensory neurons were visualized essentially as described previously18,22. In brief, head and abdominal segments were separated, and thoracic segments were fixed in 3.7% formaldehyde solution in 0.2 M sodium carbonate buffer (pH 9.5) overnight at 4 °C. DiI crystals were dissolved in 100% ethanol at 10 mg ml−1 concentration, and a paste of DiI was made by mixing this stock solution with 0.2 M sodium carbonate buffer (pH 9.5). Paste was injected into the socket of a posterior scutellar mechanosensory bristle in the scutellar segment of the thorax and DiI was allowed to transfer at room temperature for 3 days in a humidified chamber in 0.2 M sodium carbonate buffer with 0.05% sodium azide. Preparations were dissected further to isolate thoracic ganglia and imaged immediately using LSM510 confocal microscopy. All individual experiments were conducted with appropriate control genotypes for ‘wild type’ (that is, alleles with the Dscam1 locus intact in different backgrounds). DiI injection and imaging were done blindly of the genotypes in all experiments involving deletion mutants. Each deletion was confirmed by PCR and sequencing. Only images with high signal to noise ratio containing one labelled posterior scutellar neuron were used for branching pattern analysis. All images of deletion mutants and wild-type controls were coded and scored blindly, independently by D.H. and Y.C., to avoid potential bias in scoring.
Statistical analysis of branching patterns of posterior scutellar neurons
The details of statistical analysis on branching patterns of posterior scutellar neurons are available on request. In brief, branching patterns were compared in a pairwise fashion between individual genotypes using Fisher’s exact test for each branch segment. To test differences in the overall branching patterns between two genotypes, we used the minimum P value of the Fisher’s exact test across each branch. The significance of these statistics is evaluated using permutation and results are reported as ‘Permutation P values’. Both when looking at branch-specific and global tests, one must account for the large number of comparisons (that is, 16 branches and many genotype pairs). We suggest using a Bonferroni correction. In general, given the large number of tests carried out for this paper, a simplistic approximation would suggest that only tests with P < 0.001 should be considered significant. To perform cluster analysis, we use Partition Around Medoids26, a partitioning algorithm. Similarity between neurons was evaluated using Gower’s distance, which is well suited for the 0–1 values indicating the presence or absence of branches. Silhouette is a statistical measure of separation between clusters, which takes on values between 0 and 1. Silhouette values larger than 0.5 indicate reasonably reliable cluster structures.
Stochastic modelling
The details of stochastic modelling are available on request. In brief, a stochastic simulation algorithm was used to generate a table of probabilities (Qx,i) that a pair of neurons share x or more isoforms when each neuron stochastically chooses 30, 50 or 100 Dscam1 mRNAs from different numbers of potential isoforms (i). Then the probabilities that none of N pairs of neurons share x or more isoforms is represented by: Pn,x,i = (1 − Qx,i)N. The number of neurons (n) contained in N pairs of neurons was calculated by solving: N = n(n − 1)/2. The number of neurons that obtain unique identities at more than 95% likelihood was calculated by solving: Pn,x,i ≥ 0.95.
Supplementary Material
Acknowledgments
We thank T. Lee and B. Dickson for fly stocks, B. Dickson and E. Demir for advice on homologous recombination procedures, S. Miura for staining of embryos, and N. Nocera and other members of the Zipursky laboratory for help in screening homologous recombinants. We thank D. Eisenberg for helpful discussion on mathematical modelling. We thank R. Axel, A. Nern, W. Wojtowicz and other members of the Zipursky and Grueber laboratories for discussion and comments on the manuscript. This work was supported by a grant from the NIH (S.L.Z.), and NRSA from NIH/NINDS (B.J.M.).W.B.G. is a Searle Scholar, McKnight Scholar and Klingenstein Fellow. S.L.Z. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions D.H. generated knock-in mutants. B.J.M. and W.B.G. analysed dendritic arborization neuron patterning. D.H. and Y.C. performed analyses of mushroom body and posterior scutellar neurons. L.S. and D.H. conducted mathematical modelling. C.S. performed statistical analysis on the branching of posterior scutellar neuron and linear regression analysis on dendritic arborization neuron data. D.H. and S.L.Z. designed the study and wrote the paper.
Author Information Reprints and permissions information is available at www.nature.com/reprints.
References
- 1.Hattori D, Millard SS, Wojtowicz WM, Zipursky SL. Dscam-mediated cell recognition regulates neural circuit formation. Annu Rev Cell Dev Biol. 2008;24:597–620. doi: 10.1146/annurev.cellbio.24.110707.175250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schmucker D, Chen B. Dscam and DSCAM: complex genes in simple animals, complex animals yet simple genes. Genes Dev. 2009;23:147–156. doi: 10.1101/gad.1752909. [DOI] [PubMed] [Google Scholar]
- 3.Schmucker D, et al. Drosophila Dscam is an axon guidance receptor exhibiting extraordinary molecular diversity. Cell. 2000;101:671–684. doi: 10.1016/s0092-8674(00)80878-8. [DOI] [PubMed] [Google Scholar]
- 4.Wojtowicz WM, Flanagan JJ, Millard SS, Zipursky SL, Clemens JC. Alternative splicing of Drosophila Dscam generates axon guidance receptors that exhibit isoform-specific homophilic binding. Cell. 2004;118:619–633. doi: 10.1016/j.cell.2004.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wojtowicz WM, et al. A vast repertoire of Dscam binding specificities arises from modular interactions of variable Ig domains. Cell. 2007;130:1134–1145. doi: 10.1016/j.cell.2007.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matthews BJ, et al. Dendrite self-avoidance is controlled by Dscam. Cell. 2007;129:593–604. doi: 10.1016/j.cell.2007.04.013. [DOI] [PubMed] [Google Scholar]
- 7.Zhan XL, et al. Analysis of Dscam diversity in regulating axon guidance in Drosophila mushroom bodies. Neuron. 2004;43:673–686. doi: 10.1016/j.neuron.2004.07.020. [DOI] [PubMed] [Google Scholar]
- 8.Zhu H, et al. Dendritic patterning by Dscam and synaptic partner matching in the Drosophila antennal lobe. Nature Neurosci. 2006;9:349–355. doi: 10.1038/nn1652. [DOI] [PubMed] [Google Scholar]
- 9.Hughes ME, et al. Homophilic Dscam interactions control complex dendrite morphogenesis. Neuron. 2007;54:417–427. doi: 10.1016/j.neuron.2007.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Soba P, et al. Drosophila sensory neurons require Dscam for dendritic self-avoidance and proper dendritic field organization. Neuron. 2007;54:403–416. doi: 10.1016/j.neuron.2007.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hattori D, et al. Dscam diversity is essential for neuronal wiring and self-recognition. Nature. 2007;449:223–227. doi: 10.1038/nature06099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kramer AP, Stent GS. Developmental arborization of sensory neurons in the leech Haementeria ghilianii. II Experimentally induced variations in the branching pattern. J Neurosci. 1985;5:768–775. doi: 10.1523/JNEUROSCI.05-03-00768.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Neves G, Zucker J, Daly M, Chess A. Stochastic yet biased expression of multiple Dscam splice variants by individual cells. Nature Genet. 2004;36:240–246. doi: 10.1038/ng1299. [DOI] [PubMed] [Google Scholar]
- 14.Wang J, et al. Transmembrane/juxtamembrane domain-dependent Dscam distribution and function during mushroom body neuronal morphogenesis. Neuron. 2004;43:663–672. doi: 10.1016/j.neuron.2004.06.033. [DOI] [PubMed] [Google Scholar]
- 15.Wang J, Zugates CT, Liang IH, Lee CH, Lee T. Drosophila Dscam is required for divergent segregation of sister branches and suppresses ectopic bifurcation of axons. Neuron. 2002;33:559–571. doi: 10.1016/s0896-6273(02)00570-6. [DOI] [PubMed] [Google Scholar]
- 16.Grueber WB, Jan LY, Jan YN. Tiling of the Drosophila epidermis by multidendritic sensory neurons. Development. 2002;129:2867–2878. doi: 10.1242/dev.129.12.2867. [DOI] [PubMed] [Google Scholar]
- 17.Lee T, Lee A, Luo L. Development of the Drosophila mushroom bodies: sequential generation of three distinct types of neurons from a neuroblast. Development. 1999;126:4065–4076. doi: 10.1242/dev.126.18.4065. [DOI] [PubMed] [Google Scholar]
- 18.Chen BE, et al. The molecular diversity of Dscam is functionally required for neuronal wiring specificity in Drosophila. Cell. 2006;125:607–620. doi: 10.1016/j.cell.2006.03.034. [DOI] [PubMed] [Google Scholar]
- 19.Brites D, et al. The Dscam homologue of the crustacean Daphnia is diversified by alternative splicing like in insects. Mol Biol Evol. 2008;25:1429–1439. doi: 10.1093/molbev/msn087. [DOI] [PubMed] [Google Scholar]
- 20.Graveley BR, et al. The organization and evolution of the dipteran and hymenopteran Down syndrome cell adhesion molecule (Dscam) genes. RNA. 2004;10:1499–1506. doi: 10.1261/rna.7105504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Watson FL, et al. Extensive diversity of Ig-superfamily proteins in the immune system of insects. Science. 2005;309:1874–1878. doi: 10.1126/science.1116887. [DOI] [PubMed] [Google Scholar]
- 22.Grillenzoni N, van Helden J, Dambly-Chaudiere C, Ghysen A. The iroquois complex controls the somatotopy of Drosophila notum mechanosensory projections. Development. 1998;125:3563–3569. doi: 10.1242/dev.125.18.3563. [DOI] [PubMed] [Google Scholar]
- 23.Meijers R, et al. Structural basis of Dscam isoform specificity. Nature. 2007;449:487–491. doi: 10.1038/nature06147. [DOI] [PubMed] [Google Scholar]
- 24.Sawaya MR, et al. A double S shape provides the structural basis for the extraordinary binding specificity of Dscam isoforms. Cell. 2008;134:1007–1018. doi: 10.1016/j.cell.2008.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adams MD, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- 26.Kaufman L, Rousseeuw PJ. Finding Groups in Data: an Introduction to Cluster Analysis. Wiley; 1990. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.