This work provides novel mechanistic insights into how TopBP1 and the Rad9-Hus1-Rad1 (9-1-1) complex dock with one another at stalled replication forks. This step is necessary for the ATR-dependent activation of Chk1 during checkpoint responses.
Abstract
Rad17 is critical for the ATR-dependent activation of Chk1 during checkpoint responses. It is known that Rad17 loads the Rad9-Hus1-Rad1 (9-1-1) complex onto DNA. We show that Rad17 also mediates the interaction of 9-1-1 with the ATR-activating protein TopBP1 in Xenopus egg extracts. Studies with Rad17 mutants indicate that binding of ATP to Rad17 is essential for the association of 9-1-1 and TopBP1. Furthermore, hydrolysis of ATP by Rad17 is necessary for the loading of 9-1-1 onto DNA and the elevated, checkpoint-dependent accumulation of TopBP1 on chromatin. Significantly, a mutant 9-1-1 complex that cannot bind TopBP1 has a normal capacity to promote elevated accumulation of TopBP1 on chromatin. Taken together, we propose the following mechanism. First, Rad17 loads 9-1-1 onto DNA. Second, TopBP1 accumulates on chromatin in a manner that depends on both Rad17 and 9-1-1. Finally, 9-1-1 and TopBP1 dock in a Rad17-dependent manner before activation of Chk1.
INTRODUCTION
Coping with DNA lesions that arise because of cellular metabolism or external stress is a pivotal part of the cell cycle. Problematic DNA structures include double-stranded DNA breaks (DSBs) and stalled replication forks. Cells utilize checkpoint control systems in order to recognize aberrant DNA structures and halt further progression of the cell cycle until repair processes eliminate such defects. Failure in these responses leads to genomic instability, which can eventually result in the development of cancer.
Checkpoint systems function as signal transduction cascades that ultimately regulate key cell cycle control proteins such as Cdc25 and Wee1 (Sancar et al., 2004). At the apex of these pathways, sensor kinases such as ATM and ATR are involved in the initiation of signaling (Shiloh, 2006; Cimprich and Cortez, 2008). Various other sensors of DNA structures are also critical for the initial recognition of DNA lesions. These proteins include the Mre11-Rad50-Nbs1 (MRN) and the Rad9-Hus1-Rad1 (9-1-1) complexes (Parrilla-Castellar et al., 2004; Lee and Paull, 2007). The MRN complex recognizes DSBs and recruits ATM to these sites for activation. On the other hand, the 9-1-1 complex regulates activation of ATR by detecting recessed DNA ends at junctions between single-stranded and double-stranded DNA. These structures arise at both stalled replication forks and DSBs that have undergone resection.
ATR associates with DNA lesions independently of the 9-1-1 complex. In particular, ATR utilizes a binding partner called ATRIP to recognize RPA-coated, single-stranded regions at checkpoint-inducing DNA structures (Cortez et al., 2001; Zou and Elledge, 2003). Another group of checkpoint control proteins appears to possess functions as both sensors and mediators. Several BRCA1 carboxy-terminal (BRCT) domain-containing proteins such as 53BP1 and Mdc1 recognize DSBs by interacting with chromatin-bound proteins (Stewart et al., 2003; Huyen et al., 2004). On the other hand, Claspin monitors replication forks by interacting with replisome components (Lee et al., 2005). These proteins also associate with effector kinases such as Chk1 and Chk2 during checkpoint responses (Schwartz et al., 2002; Kumagai and Dunphy, 2003; Pellicioli and Foiani, 2005). ATM phosphorylates Chk2 in a manner that involves MRN, Mdc1, and 53BP1 (Lee and Paull, 2007; Wilson and Stern, 2008). ATR phosphorylates Chk1 in collaboration with Claspin (Kumagai et al., 2004). ATR also responds to resected DSBs by a mechanism that depends upon upstream activation of ATM (Cuadrado et al., 2006; Jazayeri et al., 2006; Yoo et al., 2007, 2009; Shiotani and Zou, 2009).
The ATR—ATRIP complex undergoes a dramatic increase in kinase activity upon association with TopBP1 (Kumagai et al., 2006; Mordes et al., 2008). TopBP1 and its yeast homologues (Dpb11/Cut5) are all required for both DNA replication and checkpoint responses (Navadgi-Patil and Burgers, 2009). The activation of ATR is mediated by the ATR-activating domain (AAD) of TopBP1, which is located in the C-terminal region of the protein outside of its conserved BRCT motifs. Because TopBP1 can activate ATR in vitro even in the absence of DNA, it was important to understand how DNA structures regulate the action of TopBP1 during checkpoint responses (Kumagai and Dunphy, 2006). Significantly, TopBP1 and Dpb11 display dynamic interactions with chromatin. Human TopBP1 does not colocalize with proliferating cell nuclear antigen (PCNA) during a normal cell cycle. However, it relocalizes to PCNA-containing foci upon inhibition of replication (Makiniemi et al., 2001). Dpb11 initially binds to origins of replication and then dissociates from the DNA during the elongation phase of replication. Dpb11 also associates with late origins upon stalling of replication forks (Masumoto et al., 2000). In Xenopus egg extracts, TopBP1 appears to have two modes for interacting with chromatin (Hashimoto and Takisawa, 2003; Hashimoto et al., 2006). Binding of TopBP1 to chromatin at low levels is sufficient for the initiation of DNA replication, but TopBP1 is not necessary for elongation (Van Hatten et al., 2002; Hashimoto and Takisawa, 2003). In the other mode of binding, TopBP1 accumulates to high levels at stalled replication forks (Hashimoto and Takisawa, 2003; Hashimoto et al., 2006). Collectively, these observations have suggested that dynamic association of TopBP1/Dpb11 with chromatin may be important for regulation of checkpoint responses.
At stalled replication forks, long stretches of single-stranded DNA arise from the uncoupling of helicase and polymerase activities (Byun et al., 2005). DNA polymerase α (Pol α) synthesizes short primers on the unwound single-stranded DNA. At this point, the 9-1-1 complex is deposited onto the 5′ ends of these primers (Parrilla-Castellar et al., 2004). The 9-1-1 complex shows structural homology with PCNA, the replicative sliding clamp. Moreover, by analogy with the loading of PCNA onto the 3′ ends of DNA primers by replication factor C (RFC), 9-1-1 is loaded onto the DNA with the assistance of Rad17. Rad17 forms a complex with the four small subunits of the replicative RFC, and this Rad17-RFC complex binds to chromatin in elevated amounts upon blockage of replication in Xenopus egg extracts (Jones et al., 2003; Lee et al., 2003). Rad17 and the RFC proteins belong to the AAA+-family of ATPases, which contain well-defined domains for binding and hydrolysis of ATP (Hanson and Whiteheart, 2005).
Recently, it was found that TopBP1 interacts with the Rad9 subunit of the 9-1-1 complex upon formation of DNA replication blockages (Delacroix et al., 2007; Lee et al., 2007). This interaction requires the phosphorylation of Ser373 in Xenopus Rad9 (Ser387 in human Rad9). Paradoxically, the phosphorylation of these residues is constitutive and thus apparently independent of cell cycle stage or checkpoint status (St. Onge et al., 2003; Lee et al., 2007). These observations suggest that there must be more regulatory steps that determine when TopBP1 would be able to activate ATR. In previous studies, we found that TopBP1 forms a complex not only with 9-1-1 but also with Rad17 (Lee et al., 2007). Here, we have further explored this observation to decipher the functional significance of the presence of Rad17 in this complex. We have found that Rad17 acts at multiple points to promote both the accumulation of TopBP1 on chromatin during genotoxic stress and the subsequent interaction of TopBP1 with the 9-1-1 complex.
MATERIALS AND METHODS
Xenopus Egg Extracts
Cell-free extracts from Xenopus eggs and demembranated sperm nuclei were prepared as described (Murray, 1991). The extracts were activated by addition of 0.4 mM CaCl2. To keep the extracts in interphase during incubations, 100 μg/ml cycloheximide was added. The DNA replication checkpoint response was triggered by addition of aphidicolin (50 μg/ml) to extracts containing sperm nuclei (4000/μl extract). Chromatin pellets were isolated according to the procedures described previously (Lee et al., 2003).
Antibodies and Immunodepletion
Rabbit polyclonal antisera against Xenopus Rad1 were raised against a bacterially expressed, His6-tagged fragment of the protein (amino acids 31-281) at a commercial facility (Covance Research Products, Madison, WI) and affinity-purified with this antigen. Antibodies against Xenopus Rad17, Hus1, TopBP1, ATR, RPA70, RFC40, and Orc2 were previously described (Guo et al., 2000; Lee et al., 2003, 2005, 2007; Kumagai et al., 2006). Antisera against Xenopus Rad9 were kindly provided by Dr. Howard Lindsay (Lancaster University, Lancaster, United Kingdom; Jones et al., 2003). Control IgG and anti-FLAG M2 agarose beads were purchased from Zymed Laboratories (South San Francisco, CA) and Sigma (St. Louis, MO), respectively. For immunodepletion of Rad17, 50 μg total of affinity-purified antibodies were bound to Affi-Prep protein A beads (Bio-Rad, Richmond, CA) and incubated with 100 μl of interphase egg extract in two successive rounds (each for 1 h) at 4°C. For immunodepletion of the 9-1-1 complex, purified antibodies against Rad1 (150 μg total) and Hus1 (50 μg total) were bound to beads as above and incubated with 100 μl of egg extract in three successive rounds (each 40 min) at 4°C. Immunodepletion of TopBP1 was performed as previously described (Kumagai et al., 2006).
Recombinant Proteins
35S-labeled Xenopus Chk1 and Rad17 were synthesized in vitro with TnT reticulocyte lysates (Promega, Madison, WI). Various deletion or amino acid substitution mutants of Rad17 were generated by PCR-based methods. The sequences of Rad17 and glutathione-S-transferase (GST) were subcloned into pFastBac-HTa to generate various His6-GST-Rad17 constructs. Clones of Xenopus Rad1 and Hus1 (gifts from Dr. Karlene Cimprich, Stanford University, CA), His6-Rad9, and various TopBP1 constructs were described previously (Lee et al., 2007). Recombinant baculoviruses were produced with Bac-to-Bac system (Invitrogen, Carlsbad, CA). For production of Rad17-RFC, Sf9 cells were coinfected with separate recombinant baculoviruses encoding the four small subunits of human RFC (RFC36, RFC37, RFC38, and RFC40; from Dr. Jerard Hurwitz, Memorial Sloan-Kettering Cancer Center, New York; Cai et al., 1996) and various forms of Xenopus Rad17. Expressed proteins were purified with nickel agarose beads. Purification of GST-p27 was previously described (Lee et al., 2005).
Protein-binding Assays
Up to 1 μg of His6-Rad9 protein purified from insect cells in a complex with Rad1 and Hus1 (r9-1-1) was added into 50 μl of interphase egg extract and incubated for 1 h at 23°C. Two volumes of buffer A (10 mM HEPES-KOH, pH 7.6, 150 mM NaCl, 5 mM EGTA, and 0.5% Triton X-100) and 10 μl of nickel agarose were added. After incubation for 1 h at 4°C, nickel beads were recovered, washed, and processed for SDS-PAGE. The FLAG-tagged BRCT I–II fragment of TopBP1 was used in pulldown assays as previously described (Lee et al., 2007). For immunoprecipitation assays, typically 2 μg of purified antibodies coupled to protein A beads were incubated in 100 μl of egg extract for 1 h.
Chromatin Transfer Experiments
Sperm chromatin (4000/μl) was incubated in interphase egg extracts containing 2 μM GST-p27 for 35 min. Mixtures were diluted in 10 volumes of buffer B (20 mM HEPES-KOH, pH 7.5, 80 mM KCl, 2.5 mM K-gluconate, and 10 mM Mg-gluconate). Diluted samples were overlaid onto 10 volumes of sucrose cushion (1 M sucrose in buffer B) and spun at 6100 × g for 5 min. After removal of the supernatant, mock-depleted or TopBP1-depleted extracts were added to the chromatin pellets in the absence or presence of aphidicolin for a further 75-min incubation.
RESULTS
The Rad17/9-1-1 Complex Binds to TopBP1 via Ser373 of Rad9
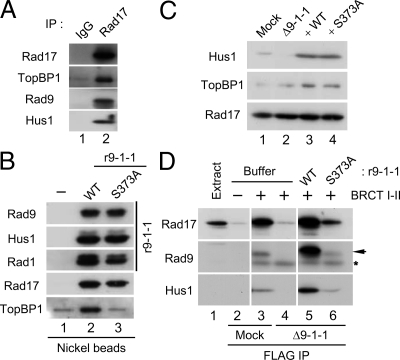
Binding of TopBP1 to the Rad9 subunit of the 9-1-1 complex is essential for activation of Chk1 (Delacroix et al., 2007; Lee et al., 2007). In Xenopus egg extracts, this binding involves interaction between the Ser373-phosphorylated form of the C-terminal domain of Rad9 and the region of TopBP1 containing its first two BRCT motifs (BRCT I–II). We observed that, in addition to the 9-1-1 complex, the isolated BRCT I–II fragment of TopBP1 also associates with Rad17 in egg extracts (Lee et al., 2007). To investigate this finding further, we immunoprecipitated the endogenous Rad17 from egg extracts with anti-Rad17 antibodies. As shown in Figure 1A, both TopBP1 and components of the 9-1-1 complex (Rad9 and Hus1) were coimmunoprecipitated with anti-Rad17 but not control antibodies. These observations establish that there is a tripartite complex composed of Rad17, the 9-1-1 complex, and TopBP1 in egg extracts.
Figure 1.
Binding of TopBP1 to the Rad17/9-1-1 complex is dependent on Ser373 of Rad9. (A) Control IgG (lane 1) and anti-Rad17 antibodies (lane 2) were used for immunoprecipitation of egg extracts. Associated proteins were examined by immunoblotting with indicated antibodies. (B) Recombinant 9-1-1 complexes containing WT His6-Rad9 (r9-1-1-WT, lane 2) or the S373A mutant form of His6-Rad9 (r9-1-1-S373A, lane 3) were incubated in interphase egg extracts. Control buffer was added in lane 1. Nickel agarose beads were incubated in the extracts, retrieved, and immunoblotted for the indicated proteins. (C) Mock-depleted (lane 1) and 9-1-1–depleted extracts supplemented with buffer alone (lane 2), r9-1-1-WT (lane 3), or r9-1-1-S373A (lane 4) were immunoblotted for Hus1 (to monitor the 9-1-1 complex), TopBP1, and Rad17. (D) Mock-depleted extracts (lanes 2 and 3) and 9-1-1–depleted extracts supplemented with buffer alone (lane 4), r9-1-1-WT (lane 5), or r9-1-1-S373A (lane 6) were prepared and incubated for 1 h with anti-FLAG antibody beads in the absence (lane 2) or presence of the FLAG-tagged BRCT I–II fragment of TopBP1 (lanes 3–6). The beads were retrieved, and associated proteins were examined by immunoblotting with indicated antibodies. Anti-Rad9 antibodies detected Rad9 (arrow) as well as the cross-reacting BRCT I–II band (asterisk). Lane 1 depicts 1.5 μl of initial egg extract.
We decided to investigate the topology of this tripartite complex in greater detail. To this end, we produced recombinant 9-1-1 complexes containing either wild-type (WT) His6-Rad9 (r9-1-1-WT) or a mutant version of His6-Rad9 in which Ser373 had been changed to Ala (r9-1-1-S373A). These complexes were incubated in egg extracts and recovered with nickel agarose beads (Figure 1B). We first observed that TopBP1 was copurified with the r9-1-1-WT complex but not the r9-1-1-S373A mutant complex. This observation demonstrates that TopBP1 cannot interact with any part of 9-1-1 independently of S373 on Rad9.
We also noted that Rad17 associated equally well with both the WT and mutant forms of the 9-1-1 complex. This finding suggests that Rad17 interacts with 9-1-1 independently of TopBP1. To test this idea, we examined whether Rad17 could bind to TopBP1 in the absence of 9-1-1 complex. We addressed this question by performing pulldown assays with the TopBP1 BRCT I–II fragment from egg extracts that had been depleted of the 9-1-1 complex. For these experiments, we used two antibodies against Rad1 and Hus1 to ensure complete depletion of the 9-1-1 complex from egg extracts (see Materials and Methods). Next, we added back r9-1-1-WT or r9-1-1-S373A to the depleted extracts (Figure 1C). Subsequently, we incubated the FLAG-tagged BRCT I–II fragment of TopBP1 in these extracts, recovered the fragment with anti-FLAG antibody beads, and examined the associated proteins (Figure 1D). Consistent with previous results, we observed that Rad17 as well as components of 9-1-1 associated with the BRCT I–II fragment in mock-depleted extracts. However, in extracts lacking 9-1-1, there was no binding of Rad17 to the TopBP1 fragment. Addition of r9-1-1-WT to the depleted extracts restored binding of Rad17 to the BRCT I–II fragment. By contrast, there was little restoration of binding in extracts containing the r9-1-1-S373A mutant complex. Taken together, these observations suggest that Rad17 does not interact with TopBP1 directly. Instead, 9-1-1 appears to bridge Rad17 and TopBP1 to form the tripartite complex.
Rad17 Is Required for the Interaction between 9-1-1 and TopBP1
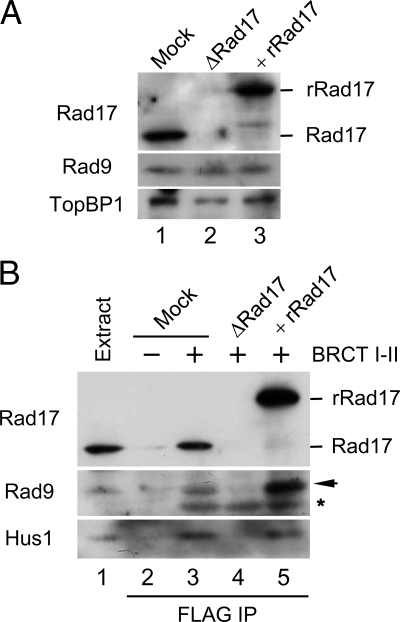
Next, we examined the role of Rad17 in the tripartite complex. To this end, we first depleted Rad17 from extracts with antibodies against Xenopus Rad17 (Figure 2A). Immunoblotting of the extracts indicated that the depletion procedure did not significantly reduce the amount of Rad9 or TopBP1 in extracts. Therefore, even though we can readily observe a tripartite complex of Rad17, 9-1-1, and TopBP1 in egg extracts, it appears that this complex must be very dynamic in nature. We also prepared a recombinant form of the Rad17 protein containing tandem tags of hexahistidine and GST on its N-terminal end (His6-GST-Rad17). We purified this protein from insect cells as a complex with recombinant forms of the four small subunits of human RFC (referred to as rRad17). This rRad17 was fully functional in supporting activation of Chk1 in egg extracts (see below), suggesting that depletion of Rad17 did not cause any additional perturbation of the extracts. We supplemented the depleted extracts with rRad17 and then performed pulldown assays with the BRCT I–II fragment of TopBP1 (Figure 2B). Significantly, we found that depletion of Rad17 completely disrupted the binding of 9-1-1 to the BRCT I–II fragment. Addition of rRad17 resulted in full restoration of the tripartite complex. Therefore, TopBP1 binds to 9-1-1 in a Rad17-dependent manner. Because Rad17 associates with 9-1-1 independently of TopBP1 (Figure 1), it appears that TopBP1 must recognize a pre-existing Rad17/9-1-1 bipartite complex.
Figure 2.
Rad17 regulates the interaction of 9-1-1 with TopBP1. (A) Mock-depleted (lane 1) and Rad17-depleted extracts supplemented with buffer (lane 2) or recombinant Rad17 (rRad17; lane 3) were immunoblotted for the indicated proteins. (B) The indicated extracts from A were incubated in the absence (lane 2) or presence of the FLAG-tagged BRCT I–II fragment of TopBP1 (lanes 3–5) for 1 h with anti-FLAG antibody beads. The beads were retrieved, and associated proteins were examined by immunoblotting with indicated antibodies. Rad9 (arrow) and the cross-reacting BRCT I–II band (asterisk) are marked. Lane 1 depicts 1.5 μl of initial egg extract.
The absolute requirement for Rad17 in this process suggests that association of Rad17 with 9-1-1 may cause structural changes within 9-1-1 that allow recognition of S373-phosphorylated Rad9 by TopBP1. We previously have shown that an isolated C-terminal fragment of Rad9 (residues 258-377, referred to as Rad9C) readily binds to both full-length TopBP1 and the BRCT I–II fragment (Lee et al., 2007). We also found that binding of Rad9C to TopBP1 occurred even in Rad17-depleted extracts (data not shown). This observation suggests that separation of Rad9C from the remainder of the 9-1-1 complex allows access to TopBP1, which implies that this region of Rad9 is normally inaccessible to TopBP1 without the assistance of Rad17. Overall, our work has revealed a novel role for Rad17 in regulating the binding between 9-1-1 and TopBP1.
Binding of ATP to Rad17 Is Required for Interaction with 9-1-1 and TopBP1
Thus far, our experiments have suggested that Rad17 and 9-1-1 form a bipartite complex first, and then TopBP1 recognizes Rad9 to form a tripartite complex. Therefore, it was important to ask how these steps relate to the sensing of DNA lesions by checkpoint-regulatory systems. We considered two possibilities. First, this tripartite complex may form in the cytosol and/or nucleus and then bind to damaged DNA. In this case, all three components of the complex would bind to DNA simultaneously. Alternatively, Rad17/9-1-1 and TopBP1 may separately recognize damaged DNA and then form a tripartite complex on the DNA.
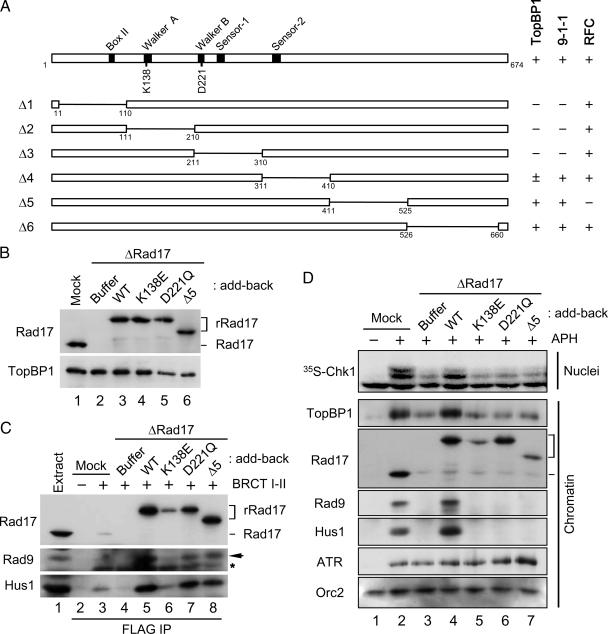
To distinguish between these possibilities, we screened for mutants of Rad17 that might help us to dissect the steps in the process. Rad17 belongs to the AAA+-family of ATPases. Members of this family possess an ATP-binding domain called the Walker-A motif and an ATP-hydrolysis domain containing the Walker-B, Sensor-1, and Sensor-2 motifs (Figure 3A; Hanson and Whiteheart, 2005). We generated six internal deletion mutants of Rad17 and synthesized them in vitro with rabbit reticulocyte lysates containing [35S]methionine. Next, we incubated these 35S-Rad17 polypeptides in egg extracts and performed immunoprecipitations with antibodies against Hus1 or RFC40. We also carried out pulldown assays with the BRCT I–II fragment of TopBP1. The results of these binding studies (see Supplemental Figure 1A) are summarized in Figure 3A. We found that C-terminal deletion mutants of Rad17 (Δ5 and Δ6) had essentially normal binding to 9-1-1 and TopBP1. By contrast, any deletion of the N-terminal half of Rad17 (Δ1, Δ2, and Δ3) resulted in greatly decreased binding of TopBP1 and 9-1-1, suggesting that the ATPase domain of Rad17 must be intact for these interactions. The Δ4 mutant, which is missing the Sensor-2 domain, showed intermediate defects. Finally, we observed that the Δ5 mutant, which lacks residues 411–525 of Rad17, was completely defective for binding of RFC40, a representative small RFC subunit.
Figure 3.
Rad17 regulates the chromatin accumulation of 9-1-1 and TopBP1 in an ATP-dependent manner. (A) Various constructs of Rad17 with internal deletions were generated and their binding to BRCT I–II (TopBP1), Hus1 (9-1-1), and RFC40 (RFC) was examined (see Supplemental Figure 1A). Binding of Rad17 to the indicated proteins is indicated as strong (+), weak (±), or negligible (−). (B) Mock-depleted (lane 1) and Rad17-depleted extracts supplemented with buffer alone (lane 2), rRad17-WT (lane 3), rRad17-K138E (lane 4), rRad17-D221Q (lane 5), or rRad17-Δ5 (lane 6) were immunoblotted for Rad17 and TopBP1. (C) The indicated extracts from B were incubated with anti-FLAG antibody beads in the absence (lane 2) or presence of the FLAG-tagged BRCT I–II fragment of TopBP1 (lanes 3–8). The beads were retrieved and immunoblotted for the indicated proteins. Lane 1 depicts 1.5 μl of initial egg extract. Rad9 (arrow) and the cross-reacting BRCT I–II band (asterisk) are marked. (D) The indicated extracts from B were incubated with sperm chromatin in the absence (lane 1) or presence of aphidicolin (lanes 2–7). Aliquots of the extracts were incubated with 35S-Chk1. Nuclear fractions from these incubations were processed for phosphorimaging (top panel). Other aliquots were processed for isolation of chromatin fractions and immunoblotting with indicated antibodies (bottom panels).
We proceeded to introduce point mutations into the Walker-A (K138E) and Walker-B (D221Q) motifs, respectively. The K138E mutant (or its equivalents in other organisms) was previously reported to be defective in both binding to 9-1-1 and checkpoint activation in yeast, Xenopus, and humans (Jones et al., 2003; Majka et al., 2004; Wang et al., 2006). Moreover, the binding of this mutant to chromatin was shown to be impaired both in vivo and in vitro. Consistent with these reports, we observed that the 35S-Rad17-K138E (Walker A) mutant showed compromised binding to 9-1-1 and chromatin (Supplemental Figure 1, B and C). In addition, we observed that the 35S-Rad17-K138E protein could not bind to the BRCT I–II fragment of TopBP1. By contrast, the 35S-Rad17-D221Q (Walker B) mutant associated well with 9-1-1, the BRCT I–II fragment, and chromatin. Finally, we found that the 35S-Rad17-Δ5 mutant interacted well with 9-1-1 and BRCT I–II, but it could not associate with chromatin.
For further analysis of these mutants, we produced versions of rRad17 containing the WT, K138E, D221Q, and Δ5 forms of Rad17 in insect cells. We depleted the endogenous Rad17 from egg extracts, replaced it with these different versions of rRad17, and performed pulldown assays with the BRCT I–II fragment of TopBP1 (Figure 3, B and C). Consistent with above results, rRad17-K138E failed to support formation of the tripartite complex with 9-1-1 and TopBP1 in egg extracts. Thus, it appears that binding of ATP to Rad17 is required for interaction with 9-1-1 and TopBP1 in egg extracts. Intriguingly, both the D221Q and Δ5 mutants supported formation of the tripartite complex reasonably well. The D221Q (Walker B) mutant is expected to be defective for hydrolysis of ATP. Likewise, the Δ5 mutant, which cannot interact with the small RFC subunits, may have reduced ability to hydrolyze ATP relative to the intact Rad17-RFC complex. These observations suggest that hydrolysis of ATP is not required for the interaction of Rad17 with 9-1-1 and TopBP1. Moreover, our results indicate that the ability of Rad17 to interact with 9-1-1 (and thereby associate with TopBP1) is intrinsic to Rad17 and independent of the small RFC subunits.
Loading of the Rad17/9-1-1 Complex Leads to Accumulation of TopBP1 at Stalled Replication Forks
We were especially interested in the D221Q mutant because it still allowed formation of the tripartite complex in egg extracts and binding of Rad17 to chromatin. Therefore, we asked whether the ability to form the tripartite complex in egg extracts is sufficient for the three components of the complex to associate stably with chromatin and support the activation of Chk1. To pursue this issue, we examined the properties of the various Rad17 proteins in egg extracts containing reconstituted nuclei and aphidicolin (Figure 3D). As expected, depletion of Rad17 greatly compromised activation of Chk1 owing to the fact that there was no loading of 9-1-1 onto chromatin in these extracts. We observed rRad17-WT successfully restored the activation of Chk1 and loading of 9-1-1. However, all three mutants were unable to rescue this activation. This failure seemed to be primarily due to the inability of these proteins to load 9-1-1 stably onto chromatin.
Interestingly, we noticed that the amount of TopBP1 on chromatin correlated with binding of Rad17/9-1-1 to chromatin. For example, in the presence of aphidicolin, TopBP1 and Rad17/9-1-1 bound to chromatin in elevated amounts (cf. lanes 1 and 2 in Figure 3D). In the absence of Rad17, the accumulation of TopBP1 on chromatin was greatly reduced. Addition of rRad17-WT restored this accumulation, but all three mutants failed to rescue the defect. Importantly, even though rRad17-D221Q bound well to chromatin, it could not support the checkpoint-dependent accumulation of TopBP1 because this mutant does not mediate the loading of 9-1-1.
These studies rule out the possibility that formation of the tripartite complex composed of Rad17/9-1-1/TopBP1 in whole egg extracts is sufficient for recruitment of TopBP1 to chromatin. Instead, our studies suggest that Rad17/9-1-1 and TopBP1 associate with chromatin in a sequential manner. In other words, Rad17/9-1-1 appears to be required for the subsequent accumulation of TopBP1. The correlation between activation of Chk1 and the accumulation of Rad17, 9-1-1, and TopBP1 on chromatin also suggests that these proteins form a stable tripartite complex after association with stalled replication forks.
Phosphorylation of Ser373 of Rad9 Is Not Required for Accumulation of TopBP1 on Chromatin
The Ser373 site of Rad9 is essential for the interaction with TopBP1. Because the loading of 9-1-1 onto chromatin absolutely requires Rad17, it is important to answer the question of whether Rad9 is ultimately driving the accumulation of TopBP1 on chromatin. To this end, we immunodepleted the 9-1-1 complex from egg extracts and added back either r9-1-1-WT or r9-1-1-S373A (see Figure 1C). Next, we added sperm chromatin to the extracts and incubated them in the absence or presence of aphidicolin. Finally, we isolated chromatin fractions and analyzed bound proteins by immunoblotting (Figure 4). We found that binding of TopBP1 to chromatin was significantly reduced after depletion of 9-1-1 (lanes 2 and 3). Binding of RPA to chromatin was normal, which suggests that depletion of 9-1-1 does not perturb the integrity of replication forks. It was shown that RPA contributes to the accumulated binding of TopBP1 onto stalled replication forks (Hashimoto et al., 2006). Because 9-1-1 requires RPA for binding to primed DNA, our results suggest that the dependence of TopBP1 on RPA for its accumulation on chromatin may be explained by the lack of 9-1-1.
Figure 4.
Rad17/9-1-1 complex but not phospho-Ser373 of Rad9 is required for the checkpoint-related accumulation of TopBP1 at stalled replication forks. Mock-depleted extracts (lane 1 and 2) and 9-1-1–depleted extracts supplemented with buffer alone (lane 3), r9-1-1-WT (lane 4), or r9-1-1-S373A (lane 5) were incubated with sperm chromatin in the absence (lane 1) or presence of aphidicolin (lanes 2–5). Chromatin fractions were isolated and immunoblotted with the indicated antibodies.
Interestingly, binding of Rad17 to chromatin was also completely lost in the absence of the 9-1-1. Addition of either r9-1-1-WT or r9-1-1-S373A to the depleted extracts could restore the association of Rad17 with chromatin (lanes 4 and 5). This dependence of Rad17 on the 9-1-1 complex for retention on chromatin has not been described previously in any other system. The accumulation of TopBP1 on chromatin was also restored by addition of r9-1-1-WT. Surprisingly, we observed that addition of the r9-1-1-S373A mutant complex could also support the accumulation of TopBP1 equally well. We have previously reported that egg extracts containing the r9-1-1-S373A mutant complex are defective in the activation of Chk1 (Lee et al., 2007). Nonetheless, our current experiments demonstrate that r9-1-1-S373A is able to support the accumulation of TopBP1 on chromatin. These findings suggest that the presence of Rad17/9-1-1 on chromatin, but not the phosphorylation of Rad9 on Ser373, is necessary for this accumulation. Thus, it appears that TopBP1 does not need to associate with S373-phosphorylated Rad9 for accumulation on chromatin. These observations imply that docking between TopBP1 and S373-phosphorylated Rad9 occurs after both proteins have already associated with the DNA.
Elevated Accumulation of TopBP1 on Chromatin Is Required for Checkpoint Signaling
Even though our results suggest that Rad17/9-1-1 facilitates the accumulation of TopBP1 under checkpoint-inducing conditions, it is also possible that TopBP1 could stabilize the binding of Rad17/9-1-1 to chromatin. If this were the case, the binding of Rad17/9-1-1 and TopBP1 to chromatin would be mutually regulated. Recent studies have suggested that TopBP1 directly recruits 9-1-1 to stalled replication forks (Yan and Michael, 2009). Therefore, we asked whether the increased binding of TopBP1 to chromatin that we observe under checkpoint-inducing conditions is required for the binding of Rad17/9-1-1 to chromatin.
Because TopBP1 is essential for initiation of DNA replication, we cannot simply remove it from the extracts to investigate the DNA replication checkpoint. One widely used method to bypass this requirement involves incubating chromatin in egg extracts for a short period (in order to allow minimal binding of TopBP1 to a level that is sufficient to execute a step for initiation of replication), isolating the chromatin in a buffer containing moderately high salt (to strip TopBP1 from the chromatin), and finally transferring the chromatin to other egg extracts that had been depleted of TopBP1 (Van Hatten et al., 2002; Hashimoto and Takisawa, 2003; Hashimoto et al., 2006; Yan and Michael, 2009). However, we found that the salt conditions (250 mM KCl) used by Yan and Michael in their version of this approach stripped not only TopBP1 but also a large number of replication proteins off the DNA (Supplemental Figure 2A). Once this chromatin was mixed with a second extract, these proteins bound back to chromatin (Supplemental Figure 2B). These observations suggest that replication forks experience significant perturbation as a result of the salt-extraction procedure. We were therefore concerned that this relatively harsh method might lead to unpredictable results.
To circumvent this problem, we utilized a procedure to allow limited binding of TopBP1 to chromatin (in amounts that would be sufficient for DNA replication). To this end, we first incubated sperm chromatin in interphase egg extracts containing p27 for 35 min. p27, an inhibitor of Cdk2-cyclin E, blocks initiation of DNA replication in egg extracts by preventing the binding of Cdc45 to replication origins. Incubation with p27 allows only limited binding of TopBP1 to chromatin (Hashimoto and Takisawa, 2003). However, this amount is sufficient to support the initiation of DNA replication.
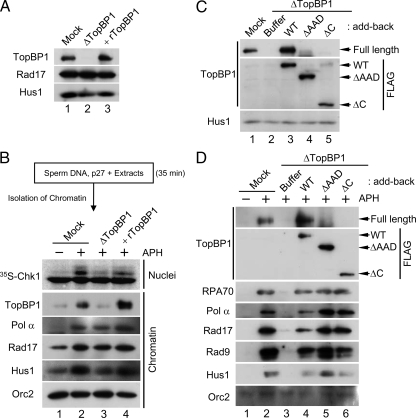
Next, we isolated chromatin from these extracts with a low-salt buffer (containing 80 mM KCl) that does not alter the profile of chromatin-binding proteins significantly. Subsequently, we transferred the p27-treated chromatin to mock-depleted or TopBP1-depleted extracts and assayed responses to aphidicolin (Figure 5, A and B). In the mock-depleted recipient extracts, activation of Chk1 occurred normally (Figure 5B, lanes 1 and 2). Moreover, TopBP1, Rad17, and Hus1 displayed elevated binding to aphidicolin-treated chromatin in these extracts.
Figure 5.
Initial binding of the N-terminal half of TopBP1 to chromatin is sufficient for the formation of replication forks. (A) Mock-depleted extracts (lane 1) and TopBP1-depleted extracts supplemented with buffer alone (lane 2) or rTopBP1 (lane 3) were immunoblotted for the indicated proteins. (B) Sperm chromatin was isolated after a 35-min incubation in egg extracts in the presence of 2 μM p27. Next, the p27-treated chromatin was transferred to mock-depleted extracts (lanes 1 and 2) or TopBP1-depleted extracts supplemented with buffer (lane 3) or rTopBP1 (lane 4). Extracts were incubated in the absence (lane 1) or presence of aphidicolin (lanes 2–4). Aliquots of the extracts were incubated with 35S-Chk1. Nuclear fractions from these incubations were processed for phosphorimaging (top panel). Other aliquots were processed for isolation of chromatin fractions and immunoblotting with indicated antibodies (bottom panels). (C) Mock-depleted extracts (lane 1) and TopBP1-depleted extracts supplemented with buffer (lanes 2) or the indicated versions of recombinant TopBP1 (lanes 3–5) were immunoblotted with anti-TopBP1 (top), anti-FLAG (middle), and anti-Hus1 antibodies (bottom). (D) Chromatin fractions from the extracts in C were immunoblotted with anti-TopBP1 antibodies (top panel), anti-FLAG antibodies (second panel from top), and the additional indicated antibodies (bottom panels).
By contrast, in the TopBP1-depleted recipient extracts (which contained only the low amount of TopBP1 on chromatin that had bound during the initial incubation in p27-treated extracts), there was little or no activation of Chk1 (lane 3). This observation is consistent with the previous findings of Hashimoto et al. (2006). We could restore activation of Chk1 by addition of rTopBP1 to the TopBP1-depleted extracts (lane 4). Significantly, despite the low amount of TopBP1 on chromatin in these TopBP1-depleted extracts, Rad17 and Hus1 bound in normal amounts (lane 3). We observed that the levels of Pol α on chromatin were also normal. Hashimoto et al. (2006) have shown that, under the similar conditions, ATR, Claspin, and many DNA replication proteins display normal binding to chromatin. Addition of rTopBP1 led to increased accumulation of TopBP1 on chromatin, but there was no effect on Rad17, Hus1, and Pol α. Taken together, these observations suggest that activation of Chk1 requires accumulation of TopBP1 above the level that is minimally necessary for replication. Importantly, the binding of Rad17/9-1-1 to chromatin is independent of this checkpoint-related accumulation of TopBP1. We conclude that only a low amount of TopBP1 on chromatin is sufficient for the binding of Pol α, Rad17, and 9-1-1. Furthermore, the presence of Rad17/9-1-1 on chromatin is necessary for the accumulation of TopBP1 in amounts sufficient for activation of ATR.
The N-terminal Region of TopBP1 is Sufficient for Accumulation of Pol α, Rad17, and 9-1-1 on Chromatin
It has been shown that roughly the N-terminal half of TopBP1 (approximately residues 1-760) is sufficient for supporting full genomic replication in Xenopus egg extracts (Hashimoto et al., 2006; Yan et al., 2006). However, it was recently suggested that some portions of the C-terminal half of TopBP1, including the AAD, are required for the elevated binding of Pol α and Rad9 to chromatin (Yan and Michael, 2009). To assess these findings, we used TopBP1 proteins lacking either the AAD (ΔAAD) or most of the C-terminal half of the protein (ΔC; Lee et al., 2007). We immunodepleted TopBP1 from egg extracts, replaced it with either WT TopBP1 or these mutants, and then incubated the extracts with sperm nuclei in the absence or presence of aphidicolin (Figure 5C). As expected, depletion of TopBP1 blocked the recruitment of both RPA and Pol α to chromatin because there was no DNA replication (Figure 5D). Consequently, there was also complete loss of binding of Rad17 and 9-1-1, owing to the fact that there is no DNA priming under these conditions. We found that both ΔAAD and ΔC versions of TopBP1 bound to chromatin as efficiently as the WT protein. WT TopBP1 restored the normal binding of Pol α, Rad17, and the 9-1-1. In contrast to the observations of Yan and Michael (2009), both ΔAAD and ΔC TopBP1 proteins also restored the normal binding of Rad17 and 9-1-1. Furthermore, we observed higher accumulation of Pol α onto chromatin in the presence of the two mutant proteins. This effect is most likely due to the fact that these mutants are defective for activation of ATR. It is well established that inhibition of ATR relieves an inhibitory constraint on initiation of replication (Lee et al., 2003; Luciani et al., 2004; Shechter et al., 2004). In summary, we conclude that once the N-terminal domain of TopBP1 fulfills its role in initiation of replication, Pol α is able to synthesize the primers that serve as the platform for loading of Rad17/9-1-1.
DISCUSSION
In this study, we have explored the role of Rad17 in establishing an ATR-activating complex at stalled replication forks. We have found that Rad17 is essential for mediating the interaction between Rad9 of 9-1-1 and TopBP1 in egg extracts. This interaction is crucial for the activation of ATR. We have also investigated the dynamic nature of the binding of TopBP1 to stalled replication forks. We found that elevated binding of TopBP1 to stalled replication forks is necessary for activation of Chk1. Moreover, we have shown that this elevated binding of TopBP1 requires the prior association of both Rad17 and 9-1-1 with chromatin. Interestingly, however, phosphorylation of Ser373 on Rad9 is not necessary for this accumulation of TopBP1.
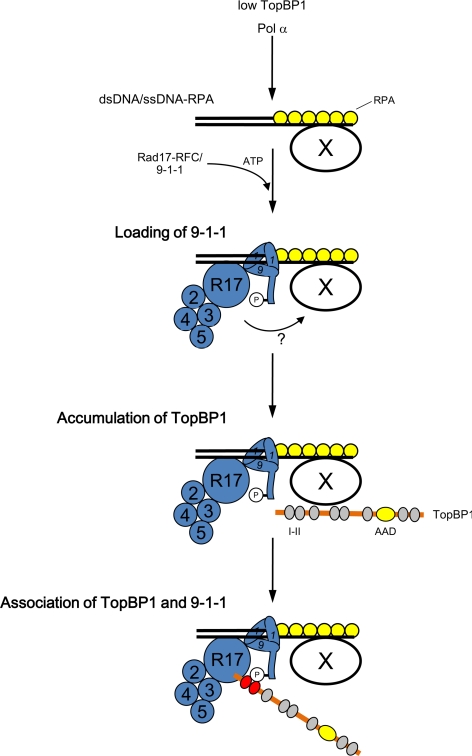
On the basis of these observations, we propose the following model (see Figure 6). First, TopBP1 binds to chromatin at a low level to promote the initiation of replication. In the event of replication-fork stalling, Pol α is thought to synthesize multiple primers along the unwound DNA (Byun et al., 2005). These primers serve as good substrates for the binding of Rad17/9-1-1. Rad17 can form a complex with 9-1-1, provided that there is an adequate supply of ATP. On hydrolysis of ATP by Rad17-RFC, 9-1-1 becomes loaded onto the primer-template junctions. This binding of Rad17/9-1-1 onto chromatin seems to induce either a structural change within chromatin or modulation of some unknown factor so that TopBP1 may begin to accumulate on chromatin in the vicinity of 9-1-1. Finally, the accumulated TopBP1 now recognizes pSer373 on Rad9 through its BRCT I–II domain. This binding facilitates the interaction of ATR—ATRIP with the AAD of TopBP1.
Figure 6.
Summary of Rad17-mediated interaction between 9-1-1 and TopBP1 at stalled replication forks. See text for detailed discussion.
Our findings differ in several respects with those in a recent report suggesting that TopBP1 directly recruits the 9-1-1 complex to stalled replication forks in Xenopus egg extracts (Yan and Michael, 2009). In our work, we have observed dynamic binding of TopBP1 to chromatin, which was originally described by the Takisawa laboratory (Hashimoto and Takisawa, 2003; Hashimoto et al., 2006). In particular, TopBP1 binds to chromatin initially at a low level near the commencement of replication. Subsequently, the binding of TopBP1 increases significantly in a manner that depends on the pre-RC and Cdk2. By the end of replication, the level of TopBP1 on chromatin decreases to a low level again. Moreover, TopBP1 displays increased binding to chromatin containing replication blockages, which is a common behavior for numerous replication and checkpoint proteins. However, Yan and Michael (2009) did not observe such dynamic binding. The reason for this discrepancy is unclear. However, we have further shown that this elevated binding is necessary for activation of Chk1 (Figure 5B).
Accordingly, we have focused on the issue of what causes the accumulation of TopBP1 and how it relates to Rad17 and 9-1-1. As discussed above, Rad17/9-1-1 is necessary for the accumulation of TopBP1. Conversely, we also examined whether TopBP1 would be necessary for the accumulation of Rad17 and 9-1-1 under our experimental conditions. Because TopBP1 is essential for initiation of replication, we designed our experiments to bypass this early requirement of TopBP1. Instead of transferring chromatin through a buffer containing high salt, we used a less disruptive approach in which we incubated chromatin in egg extracts containing p27, washed this chromatin in a buffer containing 80 mM KCl (which does not remove TopBP1), and then transferred the chromatin to TopBP1-depleted extracts. Under these conditions, replication occurs normally in the TopBP1-depleted extract because of the presence of the small amount of TopBP1 that had bound to chromatin in the first p27-treated extract. We found that the 9-1-1 complex could still bind normally to stalled replication forks in these TopBP1-depleted extracts. Importantly, activation of Chk1 does not occur in these extracts because TopBP1 does not accumulate to the elevated level that is necessary for this activation. These results clearly demonstrate that TopBP1 is not driving the accumulation of 9-1-1 on chromatin for activation of Chk1. To rule out the possibility that the small amount of TopBP1 on p27-treated chromatin could promote binding of 9-1-1 to chromatin (albeit under conditions that are inadequate for activation of Chk1), we also performed chromatin-transfer experiments in which we stripped TopBP1 off chromatin with salt solutions containing up to 250 mM KCl before addition to TopBP1-depleted extracts. With this protocol, we observed that 9-1-1 bound to stalled replication forks in undiminished amounts in the absence of TopBP1 (Supplemental Figure 2B). Because these conditions were essentially identical to those of Yan and Michael, it is unclear why we obtained different results.
It has also been reported that TopBP1 is necessary for the recruitment of Rad1 to etoposide-damaged chromatin (Parrilla-Castellar and Karnitz, 2003). However, etoposide generates DSBs, and the role of TopBP1 in response to DSBs appears to bear significant differences from its role at stalled replication forks (Cuadrado et al., 2006; Jazayeri et al., 2006; Yoo et al., 2007; Shiotani and Zou, 2009; Yoo et al., 2009). In particular, DSBs must undergo resection before activation of ATR. This process leads to the generation of single-stranded DNA and recessed DNA ends, both of which are necessary for this activation. Interestingly, TopBP1 also interacts with the MRN complex (Shiotani and Zou, 2009; Yoo et al., 2009), a major regulator of resection in yeast and vertebrates (Lee and Paull, 2007; Williams et al., 2007; Rupnik et al., 2008). Therefore, it is possible that TopBP1 might be required in some way for appropriate processing of DSBs so that Rad1 can be recruited to recessed DNA ends.
Our studies raise the question of what causes the elevated accumulation of TopBP1 on chromatin during a checkpoint response. On damaged DNA, 9-1-1 functions as a recruiting center for multiple proteins involved in DNA repair and replication processing (Parrilla-Castellar et al., 2004; Helt et al., 2005). These proteins include Pol κ and Pol ζ (DNA translesion synthesis), Myh (base excision repair), and Fen1 and DNA ligase I (Okazaki fragment processing). Also, our laboratory has observed that phosphorylation of the helicase BLM depends upon Rad17 (Li et al., 2004). Interestingly, BLM forms a complex with proteins such as BRCA1, MRN, ATM, and RFC (Wang et al., 2000), a number of which have also been implicated in the regulation of TopBP1. Another possibility would be histone variants such as phosphorylated H2AX. For example, once Rad17 loads 9-1-1 onto chromatin, these proteins may promote exposure of γ-H2AX for recruitment of TopBP1. Because ATR also phosphorylates H2AX upon stalling of replication (Ward and Chen, 2001; Syljuasen et al., 2005), this initial modification may increase the accumulation of TopBP1 for further activation of ATR and thus create a positive feedback loop. Indeed, it was shown in human cells that TopBP1 accumulates in γ-H2AX–containing foci at the sites of DSBs in a Rad9-dependent manner (Greer et al., 2003). Therefore, it seems interesting to pursue the idea that 9-1-1–mediated modification on chromatin components may direct accumulation of TopBP1 at sites of damaged DNA.
Thus far, Rad17 has been known as a checkpoint clamp loader that deposits the checkpoint clamp 9-1-1 onto DNA. Studies on the replicative RFC and PCNA suggest that RFC undergoes conformational change upon binding to ATP, which is essential for association with PCNA (Bowman et al., 2004; Indiani and O'Donnell, 2006; Bloom, 2009). Binding to DNA results in activation of the ATPase of RFC, and the resulting hydrolysis of ATP powers the opening of the PCNA ring. After loading of PCNA onto DNA, RFC dissociates from PCNA in order to catalyze another cycle of loading. By contrast, our work suggests that Rad17 needs to remain associated with 9-1-1 on DNA. In particular, Rad17 is required for the interaction between Rad9 and TopBP1. Therefore, unlike RFC-PCNA, Rad17 seems to be in contact with 9-1-1 even after loading onto DNA. Consistent with this idea, we have not observed dissociation of Rad17 from chromatin as long as 9-1-1 remains on the DNA. Moreover, 9-1-1 is likewise critical for the retention of Rad17 on chromatin (Figure 4). Collectively, these observations reveal a novel role of Rad17 in establishing and maintaining an active checkpoint complex.
This regulatory role of Rad17 sheds light on some puzzling issues regarding the phosphorylation of Ser373 in Rad9. We find that, even though this site is constitutively phosphorylated, TopBP1 can associate efficiently with Rad9 only when Rad17 is associated with the 9-1-1 complex. This finding implies Rad17 may elicit some structural change that allows TopBP1 to dock with Rad9 (Figure 2). We can observe a soluble tripartite complex of Rad17, 9-1-1, and TopBP1. However, this soluble complex appears to be relatively unstable. Instead, our results suggest that this complex is more stable once all three components have associated with chromatin. Moreover, it appears that the Rad17-RFC complex must hydrolyze ATP in the process of loading 9-1-1 onto DNA in order for Rad17-RFC, 9-1-1, and TopBP1 to form this final assemblage on chromatin. Significantly, this mechanism would render the interaction between 9-1-1 and TopBP1 dependent on the presence of a checkpoint-inducing DNA template.
The requirement for Rad17 in this process might have been missed in previous studies for various reasons. For example, the human Rad9 C-terminal tail fused to PCNA or histone H2B could activate the replication checkpoint even in cells lacking Rad17 (Delacroix et al., 2007). However, we have observed that a similar fragment from Xenopus Rad9 (Rad9C) can bind to TopBP1 without Rad17. In budding yeast, forced colocalization of overexpressed Ddc1 (homolog of Rad9) and Ddc2 (homolog of ATRIP) on chromatin could elicit activation of Rad53 (homolog of Chk2) in cells lacking Rad24 (homolog of Rad17) (Bonilla et al., 2008). Based on this observation, it was proposed that the sole role of Rad24-RFC would be the loading of 9-1-1 onto damaged DNA (Bonilla et al., 2008). This proposal is most likely correct under these conditions of overexpression because Ddc1 (Rad9) itself appears to be able to activate the Mec1-Ddc2 kinase directly (Majka et al., 2006). However, it is still possible that the overexpressed, isolated Ddc1 subunit may be able to bypass the requirement of Rad24-RFC for the interaction with Dpb11 (TopBP1 homolog).
In summary, we have provided data suggesting that Rad17 regulates the interaction between 9-1-1 and TopBP1. Moreover, both Rad17 and 9-1-1 promote the initial accumulation of TopBP1 at DNA lesions before the ultimate docking of TopBP1 with 9-1-1. Overall, our data suggest that Rad17 acts at multiple points to promote TopBP1-dependent activation of ATR.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Drs. Karlene Cimprich (Stanford University), Jerard Hurwitz (Memorial Sloan-Kettering Cancer Center), and Howard Lindsay (Lancaster University, Lancaster, United Kingdom) for kindly supplying key reagents. We also thank other members of the laboratory for fruitful comments on this manuscript. This work was supported by grants from the National Institutes of Health (GM043974 and GM070891) to W.G.D.
Abbreviations used:
- ATM
ataxia-telangiectasia mutated
- ATR
ATM- and Rad3-related
- BRCT
BRCA1 carboxy-terminal
- DSB
double-stranded DNA break
- MRN
Mre11-Rad50-Nbs1
- TopBP1
topoisomerase II–binding protein 1
- 9-1-1
Rad9-Hus1-Rad1.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-11-0958) on January 28, 2010.
REFERENCES
- Bloom L. B. Loading clamps for DNA replication and repair. DNA Repair. 2009;8:570–578. doi: 10.1016/j.dnarep.2008.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonilla C. Y., Melo J. A., Toczyski D. P. Colocalization of sensors is sufficient to activate the DNA damage checkpoint in the absence of damage. Mol. Cell. 2008;30:267–276. doi: 10.1016/j.molcel.2008.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman G. D., O'Donnell M., Kuriyan J. Structural analysis of a eukaryotic sliding DNA clamp-clamp loader complex. Nature. 2004;429:724–730. doi: 10.1038/nature02585. [DOI] [PubMed] [Google Scholar]
- Byun T. S., Pacek M., Yee M. C., Walter J. C., Cimprich K. A. Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev. 2005;19:1040–1052. doi: 10.1101/gad.1301205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai J., Uhlmann F., Gibbs E., Flores-Rozas H., Lee C. G., Phillips B., Finkelstein J., Yao N., O'Donnell M., Hurwitz J. Reconstitution of human replication factor C from its five subunits in baculovirus-infected insect cells. Proc. Natl. Acad. Sci. USA. 1996;93:12896–12901. doi: 10.1073/pnas.93.23.12896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimprich K. A., Cortez D. ATR: an essential regulator of genome integrity. Nat. Rev. Mol. Cell Biol. 2008;9:616–627. doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez D., Guntuku S., Qin J., Elledge S. J. ATR and ATRIP: partners in checkpoint signaling. Science. 2001;294:1713–1716. doi: 10.1126/science.1065521. [DOI] [PubMed] [Google Scholar]
- Cuadrado M., Martinez-Pastor B., Murga M., Toledo L. I., Gutierrez-Martinez P., Lopez E., Fernandez-Capetillo O. ATM regulates ATR chromatin loading in response to DNA double-strand breaks. J. Exp. Med. 2006;203:297–303. doi: 10.1084/jem.20051923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delacroix S., Wagner J. M., Kobayashi M., Yamamoto K., Karnitz L. M. The Rad9-Hus1-Rad1 (9-1-1) clamp activates checkpoint signaling via TopBP1. Genes Dev. 2007;21:1472–1477. doi: 10.1101/gad.1547007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer D. A., Besley B. D., Kennedy K. B., Davey S. hRad9 rapidly binds DNA containing double-strand breaks and is required for damage-dependent topoisomerase II beta binding protein 1 focus formation. Cancer Res. 2003;63:4829–4835. [PubMed] [Google Scholar]
- Guo Z., Kumagai A., Wang S. X., Dunphy W. G. Requirement for Atr in phosphorylation of Chk1 and cell cycle regulation in response to DNA replication blocks and UV-damaged DNA in Xenopus egg extracts. Genes Dev. 2000;14:2745–2756. doi: 10.1101/gad.842500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson P. I., Whiteheart S. W. AAA+ proteins: have engine, will work. Nat. Rev. Mol. Cell Biol. 2005;6:519–529. doi: 10.1038/nrm1684. [DOI] [PubMed] [Google Scholar]
- Hashimoto Y., Takisawa H. Xenopus Cut5 is essential for a CDK-dependent process in the initiation of DNA replication. EMBO J. 2003;22:2526–2535. doi: 10.1093/emboj/cdg238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto Y., Tsujimura T., Sugino A., Takisawa H. The phosphorylated C-terminal domain of Xenopus Cut5 directly mediates ATR-dependent activation of Chk1. Genes Cells. 2006;11:993–1007. doi: 10.1111/j.1365-2443.2006.00998.x. [DOI] [PubMed] [Google Scholar]
- Helt C. E., Wang W., Keng P. C., Bambara R. A. Evidence that DNA damage detection machinery participates in DNA repair. Cell Cycle. 2005;4:529–532. doi: 10.4161/cc.4.4.1598. [DOI] [PubMed] [Google Scholar]
- Huyen Y., Zgheib O., Ditullio R. A., Jr., Gorgoulis V. G., Zacharatos P., Petty T. J., Sheston E. A., Mellert H. S., Stavridi E. S., Halazonetis T. D. Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature. 2004;432:406–411. doi: 10.1038/nature03114. [DOI] [PubMed] [Google Scholar]
- Indiani C., O'Donnell M. The replication clamp-loading machine at work in the three domains of life. Nat. Rev. Mol. Cell Biol. 2006;7:751–761. doi: 10.1038/nrm2022. [DOI] [PubMed] [Google Scholar]
- Jazayeri A., Falck J., Lukas C., Bartek J., Smith G. C., Lukas J., Jackson S. P. ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat. Cell Biol. 2006;8:37–45. doi: 10.1038/ncb1337. [DOI] [PubMed] [Google Scholar]
- Jones R. E., Chapman J. R., Puligilla C., Murray J. M., Car A. M., Ford C. C., Lindsay H. D. XRad17 is required for the activation of XChk1 but not XCds1 during checkpoint signaling in Xenopus. Mol. Biol. Cell. 2003;14:3898–3910. doi: 10.1091/mbc.E03-03-0138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai A., Dunphy W. G. Repeated phosphopeptide motifs in Claspin mediate the regulated binding of Chk1. Nat. Cell Biol. 2003;5:161–165. doi: 10.1038/ncb921. [DOI] [PubMed] [Google Scholar]
- Kumagai A., Dunphy W. G. How cells activate ATR. Cell Cycle. 2006;5:1265–1268. doi: 10.4161/cc.5.12.2834. [DOI] [PubMed] [Google Scholar]
- Kumagai A., Kim S. M., Dunphy W. G. Claspin and the activated form of ATR-ATRIP collaborate in the activation of Chk1. J. Biol. Chem. 2004;279:49599–49608. doi: 10.1074/jbc.M408353200. [DOI] [PubMed] [Google Scholar]
- Kumagai A., Lee J., Yoo H. Y., Dunphy W. G. TopBP1 activates the ATR-ATRIP complex. Cell. 2006;124:943–955. doi: 10.1016/j.cell.2005.12.041. [DOI] [PubMed] [Google Scholar]
- Lee J., Gold D. A., Shevchenko A., Shevchenko A., Dunphy W. G. Roles of replication fork-interacting and Chk1-activating domains from Claspin in a DNA replication checkpoint response. Mol. Biol. Cell. 2005;16:5269–5282. doi: 10.1091/mbc.E05-07-0671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Kumagai A., Dunphy W. G. Claspin, a Chk1-regulatory protein, monitors DNA replication on chromatin independently of RPA, ATR, and Rad17. Mol. Cell. 2003;11:329–340. doi: 10.1016/s1097-2765(03)00045-5. [DOI] [PubMed] [Google Scholar]
- Lee J., Kumagai A., Dunphy W. G. The Rad9-Hus1-Rad1 checkpoint clamp regulates interaction of TopBP1 with ATR. J. Biol. Chem. 2007;282:28036–28044. doi: 10.1074/jbc.M704635200. [DOI] [PubMed] [Google Scholar]
- Lee J. H., Paull T. T. Activation and regulation of ATM kinase activity in response to DNA double-strand breaks. Oncogene. 2007;26:7741–7748. doi: 10.1038/sj.onc.1210872. [DOI] [PubMed] [Google Scholar]
- Li W., Kim S. M., Lee J., Dunphy W. G. Absence of BLM leads to accumulation of chromosomal DNA breaks during both unperturbed and disrupted S phases. J. Cell Biol. 2004;165:801–812. doi: 10.1083/jcb.200402095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luciani M. G., Oehlmann M., Blow J. J. Characterization of a novel ATR-dependent, Chk1-independent, intra-S-phase checkpoint that suppresses initiation of replication in Xenopus. J. Cell Sci. 2004;117:6019–6030. doi: 10.1242/jcs.01400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majka J., Chung B. Y., Burgers P. M. Requirement for ATP by the DNA damage checkpoint clamp loader. J. Biol. Chem. 2004;279:20921–20926. doi: 10.1074/jbc.M400898200. [DOI] [PubMed] [Google Scholar]
- Majka J., Niedziela-Majka A., Burgers P. M. The checkpoint clamp activates Mec1 kinase during initiation of the DNA damage checkpoint. Mol. Cell. 2006;24:891–901. doi: 10.1016/j.molcel.2006.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makiniemi M., et al. BRCT domain-containing protein TopBP1 functions in DNA replication and damage response. J. Biol. Chem. 2001;276:30399–30406. doi: 10.1074/jbc.M102245200. [DOI] [PubMed] [Google Scholar]
- Masumoto H., Sugino A., Araki H. Dpb11 controls the association between DNA polymerases alpha and epsilon and the autonomously replicating sequence region of budding yeast. Mol. Cell. Biol. 2000;20:2809–2817. doi: 10.1128/mcb.20.8.2809-2817.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mordes D. A., Glick G. G., Zhao R., Cortez D. TopBP1 activates ATR through ATRIP and a PIKK regulatory domain. Genes Dev. 2008;22:1478–1489. doi: 10.1101/gad.1666208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray A. W. Cell cycle extracts. Methods Cell Biol. 1991;36:581–605. [PubMed] [Google Scholar]
- Navadgi-Patil V.M., Burgers P.M. A tale of two tails: activation of DNA damage checkpoint kinase Mec1/ATR by the 9-1-1 clamp and by Dpb11/TopBP1. DNA Repair. 2009;8:996–1003. doi: 10.1016/j.dnarep.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrilla-Castellar E. R., Arlander S. J., Karnitz L. Dial 9-1-1 for DNA damage: the Rad9-Hus1-Rad1 (9-1-1) clamp complex. DNA Repair. 2004;3:1009–1014. doi: 10.1016/j.dnarep.2004.03.032. [DOI] [PubMed] [Google Scholar]
- Parrilla-Castellar E. R., Karnitz L. M. Cut5 is required for the binding of Atr and DNA polymerase alpha to genotoxin-damaged chromatin. J. Biol. Chem. 2003;278:45507–45511. doi: 10.1074/jbc.C300418200. [DOI] [PubMed] [Google Scholar]
- Pellicioli A., Foiani M. Signal transduction: how rad53 kinase is activated. Curr. Biol. 2005;15:R769–771. doi: 10.1016/j.cub.2005.08.057. [DOI] [PubMed] [Google Scholar]
- Rupnik A., Grenon M., Lowndes N. The MRN complex. Curr. Biol. 2008;18:R455–457. doi: 10.1016/j.cub.2008.03.040. [DOI] [PubMed] [Google Scholar]
- Sancar A., Lindsey-Boltz L. A., Unsal-Kacmaz K., Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- Schwartz M. F., Duong J. K., Sun Z., Morrow J. S., Pradhan D., Stern D. F. Rad9 phosphorylation sites couple Rad53 to the Saccharomyces cerevisiae DNA damage checkpoint. Mol. Cell. 2002;9:1055–1065. doi: 10.1016/s1097-2765(02)00532-4. [DOI] [PubMed] [Google Scholar]
- Shechter D., Costanzo V., Gautier J. ATR and ATM regulate the timing of DNA replication origin firing. Nat. Cell Biol. 2004;6:648–655. doi: 10.1038/ncb1145. [DOI] [PubMed] [Google Scholar]
- Shiloh Y. The ATM-mediated DNA-damage response: taking shape. Trends Biochem. Sci. 2006;31:402–410. doi: 10.1016/j.tibs.2006.05.004. [DOI] [PubMed] [Google Scholar]
- Shiotani B., Zou L. Single-stranded DNA orchestrates an ATM-to-ATR switch at DNA breaks. Mol. Cell. 2009;33:547–558. doi: 10.1016/j.molcel.2009.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Onge R. P., Besley B. D., Pelley J. L., Davey S. A role for the phosphorylation of hRad9 in checkpoint signaling. J. Biol. Chem. 2003;278:26620–26628. doi: 10.1074/jbc.M303134200. [DOI] [PubMed] [Google Scholar]
- Stewart G. S., Wang B., Bignell C. R., Taylor A. M., Elledge S. J. MDC1 is a mediator of the mammalian DNA damage checkpoint. Nature. 2003;421:961–966. doi: 10.1038/nature01446. [DOI] [PubMed] [Google Scholar]
- Syljuasen R. G., Sorensen C. S., Hansen L. T., Fugger K., Lundin C., Johansson F., Helleday T., Sehested M., Lukas J., Bartek J. Inhibition of human Chk1 causes increased initiation of DNA replication, phosphorylation of ATR targets, and DNA breakage. Mol. Cell. Biol. 2005;25:3553–3562. doi: 10.1128/MCB.25.9.3553-3562.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hatten R. A., Tutter A. V., Holway A. H., Khederian A. M., Walter J. C., Michael W. M. The Xenopus Xmus101 protein is required for the recruitment of Cdc45 to origins of DNA replication. J. Cell Biol. 2002;159:541–547. doi: 10.1083/jcb.200207090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Zou L., Lu T., Bao S., Hurov K. E., Hittelman W. N., Elledge S. J., Li L. Rad17 phosphorylation is required for claspin recruitment and Chk1 activation in response to replication stress. Mol. Cell. 2006;23:331–341. doi: 10.1016/j.molcel.2006.06.022. [DOI] [PubMed] [Google Scholar]
- Wang Y., Cortez D., Yazdi P., Neff N., Elledge S. J., Qin J. BASC, a super complex of BRCA1-associated proteins involved in the recognition and repair of aberrant DNA structures. Genes Dev. 2000;14:927–939. [PMC free article] [PubMed] [Google Scholar]
- Ward I. M., Chen J. Histone H2AX is phosphorylated in an ATR-dependent manner in response to replicational stress. J. Biol. Chem. 2001;276:47759–47762. doi: 10.1074/jbc.C100569200. [DOI] [PubMed] [Google Scholar]
- Williams R. S., Williams J. S., Tainer J. A. Mre11-Rad50-Nbs1 is a keystone complex connecting DNA repair machinery, double-strand break signaling, and the chromatin template. Biochem. Cell Biol. 2007;85:509–520. doi: 10.1139/O07-069. [DOI] [PubMed] [Google Scholar]
- Wilson K. A., Stern D. F. NFBD1/MDC1, 53BP1 and BRCA1 have both redundant and unique roles in the ATM pathway. Cell Cycle. 2008;7:3584–3594. doi: 10.4161/cc.7.22.7102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan S., Lindsay H. D., Michael W. M. Direct requirement for Xmus101 in ATR-mediated phosphorylation of Claspin bound Chk1 during checkpoint signaling. J. Cell Biol. 2006;173:181–186. doi: 10.1083/jcb.200601076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan S., Michael W. M. TopBP1 and DNA polymerase-alpha directly recruit the 9-1-1 complex to stalled DNA replication forks. J. Cell Biol. 2009;184:793–804. doi: 10.1083/jcb.200810185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo H. Y., Kumagai A., Shevchenko A., Shevchenko A., Dunphy W. G. Ataxia-telangiectasia mutated (ATM)-dependent activation of ATR occurs through phosphorylation of TopBP1 by ATM. J. Biol. Chem. 2007;282:17501–17506. doi: 10.1074/jbc.M701770200. [DOI] [PubMed] [Google Scholar]
- Yoo H. Y., Kumagai A., Shevchenko A., Shevchenko A., Dunphy W. G. The Mre11-Rad50-Nbs1 complex mediates activation of TopBP1 by ATM. Mol. Biol. Cell. 2009;20:2351–2360. doi: 10.1091/mbc.E08-12-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L., Elledge S. J. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.