Abstract
Brunfelsia calycina flowers change colour from purple to white due to anthocyanin degradation, parallel to an increase in fragrance and petal size. Here it was tested whether the production of the fragrant benzenoids is dependent on induction of the shikimate pathway, or if they are formed from the anthocyanin degradation products. An extensive characterization of the events taking place in Brunfelsia flowers is presented. Anthocyanin characterization was performed using ultraperfomance liquid chromatography–quadrupole time of flight–tandem mass specrometry (UPLC-QTOF-MS/MS). Volatiles emitted were identified by headspace solid phase microextraction–gas chromatography–mass spectrometry (HS-SPME-GC-MS). Accumulated proteins were identified by 2D gel electrophoresis. Transcription profiles were characterized by cross-species hybridization of Brunfelsia cDNAs to potato cDNA microarrays. Identification of accumulated metabolites was performed by UPLC-QTOF-MS non-targeted metabolite analysis. The results include characterization of the nine main anthocyanins in Brunfelsia flowers. In addition, 146 up-regulated genes, 19 volatiles, seven proteins, and 17 metabolites that increased during anthocyanin degradation were identified. A multilevel analysis suggests induction of the shikimate pathway. This pathway is the most probable source of the phenolic acids, which in turn are precursors of both the benzenoid and lignin production pathways. The knowledge obtained is valuable for future studies on degradation of anthocyanins, formation of volatiles, and the network of secondary metabolism in Brunfelsia and related species.
Keywords: Anthocyanin, benzenoid, Brunfelsia, lignin, secondary metabolism
Introduction
Brunfelsia calycina is a Solanaceae shrub, native to Brazil, with flowers that have a unique characteristic of changing colour from dark purple to white within 2–3 d after flower opening, and well before the onset of flower senescence (Heide, 1963). This whitening process is due to active in planta degradation of anthocyanins, dependent on de novo synthesis of mRNAs and proteins, after flower opening (Vaknin et al., 2005). Concomitant with the pigment degradation, Brunfelsia flowers undergo other changes such as fragrance emission and expansion of the petals. Based on an earlier study on B. australis (Bertrand et al., 2006), it is assumed that a major group of fragrant metabolites emitted from B. calycina are benzenoids. Benzenoids, anthocyanins, and cell wall components that may be involved in the petal expansion, such as lignin, are all derived from the phenylpropanoid pathway. In addition, the fact that the degradation of anthocyanins, increase in volatiles, and growth all occur at a short, well-defined stage of flower development makes Brunfelsia flowers a unique system for studying secondary metabolism and the possible inter-relations between the different processes.
The anthocyanin biosynthesis process ends in Brunfelsia by the day of flower opening, and no further accumulation was observed even when the degradation process was inhibited (Vaknin et al., 2005). Clearly, the two processes of anthocyanin synthesis and degradation are sequential and do not occur simultaneously in the flowers. Anthocyanin synthesis ends in most flowers between the end of cell division in the buds and before flower opening (Weiss, 2000). One example is petunia, in which the pigment concentration reaches a peak before petal unfurling and remains the same throughout the lifespan of the flowers (Jonsson et al., 1984).
Volatile benzenoids play an important role in determining the aroma of Brunfelsia flowers, and are the sole fragrant group in Petunia hybrida (Verdonk et al., 2005). Unlike the detailed knowledge on the biosynthesis and regulation of anthocyanins, the metabolism of the benzenoids is only partially understood, and little information is available on the regulation of this pathway (Schuurink et al., 2006). In petunia, the volatile benzenoid are derived from phenylalanine (Boatright et al., 2004). Overexpression of the Arabidopsis transcription factor PAP1, regulating the biosynthesis of anthocyanins in petunia flowers, caused a dramatic increase in both anthocyanins and volatiles derived from the phenypropanoid/benzenoid pathways (Ben Zvi et al., 2008). A study on the regulation of fragrance in petunia revealed a transcription factor (ODORANT1) that regulates the production of volatile benzenoids by activating the shikimate pathway, which precedes the formation of phenylalanine (Verdonk et al., 2005). Even though anthocyanin production is also dependent on this pathway, ODORANT1 had no effect on their regulation, probably since anthocyanin synthesis occurs earlier in flower development. This study suggests that the shikimate pathway is activated separately for anthocyanin production and later for benzenoid production in coloured fragrant flowers (Verdonk et al., 2005).
Since Brunfelsia is an attractive model system to study floral metabolic networking, transcript, protein, and metabolite databases have been generated, representing events taking place in petals following flower opening. In this study it was specifically investigated whether the production of volatiles in the open flower is driven by the degradation of the anthocyanins or, similarly to petunia, by the re-activation of the shikimate and phenylpropanoid pathways.
Materials and methods
Plant growth and sample collection
Brunfelsia calycina plants were grown in pots in a glass greenhouse under controlled conditions and induced for flowering according to Vaknin et al. (2005). Flowers were collected from a batch of 20 plants grown at 20 °C/12 °C day/night temperature conditions. For RNA, protein, and non-volatile metabolite characterization, flowers were detached from the plants on the day of flower opening (D0), transferred to a 2% sucrose solution, pH 5.5, and 80 mg ml−1 sodium-dichloro-isocyanurate (TOG-6), and sampled during the first 2 d after flower opening. The change in colour and petal growth is similar between flowers attached to the plant and detached flowers in the sucrose solution (Vaknin et al., 2005). Even though the increase in fragrance occurs in both attached and detached flowers as they whiten, the samples for characterization of volatile compounds were collected from flowers attached to the plant. Petunia flowers were obtained from Alexander Vainstein's laboratory (Spitzer et al., 2007).
Determination of anthocyanins by liquid chromatography–tandem mass specrtometry (LC-MS/MS)
Brunfelsia anthocyanins were extracted by grinding whole flowers (0.1–0.25 g) in liquid nitrogen and addition of extraction solution (70% methanol and 2% formic acid in H2O) in a ratio of 1 ml per 0.2 g followed by a 1 h incubation and 10 min centrifugation at 14 000 rpm at room temperature. Samples were filtered through a 0.22 μm PTFE membrane filter (Acrodisc® CR 13 mm, PALL) before injection into the LC-MS instrument. Petunia anthocyanins were extracted as described by Spitzer et al. (2007).
Mass spectral analyses were carried out by the ultraperformance liquid chromatography–quadrupole time of flight (UPLC-QTOF) instrument (Waters Premier QTOF, Milford, MA, USA), with the UPLC column connected on-line to a UV detector (measuring at 530 nm; Waters Acquity), and then to the MS detector equipped with an electrospray ion (ESI) source (performed in ESI-positive mode). Separation of metabolites was performed on the 100×2.1 mm id, 1.7 μm UPLC BEH C18 column (Waters Acquity). The chromatographic and MS parameters were as described previously by Mintz-Oron et al. (2008). A mixture of 15 standard compounds (Supplementary Table S1 available at JXB online), injected after each batch of 10 Brunfelsia samples, was used for instrument quality control. The UV spectra (200–600 nm) were acquired on a UPLC instrument (Waters, Acquity) equipped with an Acquity 2996 PDA under LC conditions as described above for the UPLC-QTOF analysis.
Metabolomics of Brunfelsia flowers
For non-targeted metabolomics analysis of semi-polar compounds, whole Brunfelsia white and purple flowers (n=6 each) were extracted as described above and analysed by UPLC-QTOF-MS, essentially as described above for the analysis of anthocyanins.Acquisition was performed in both the ESI-positive and ESI-negative modes. For the UPLC-MS/MS run, collision energies of 10 eV and 25 eV and of 15 eV and 30 eV for the positive and negative modes, respectively, were used. Metabolites were identified using standard compounds by comparison of their retention times, UV spectra, and MS/MS fragments. In cases where the corresponding standards were not available, compounds were putatively identified applying several steps. First, the elemental composition was selected according to the accurate masses and the isotopic pattern using the MassLynx software. Then the elemental composition obtained was searched against the KNApSAcK metabolite database for petunia flowers (http://prime.psc.riken.jp/KNApSAcK; (Shinbo et al., 2006) and the Dictionary of Natural Products (Version 16:2, Chapman & Hall/CRC). When a suitable candidate was not found, more comprehensive chemical databases were searched using the SciFinder tool (SciFinder Scholar 2007). Predicted Log D values for pH 3 (pH of the UPLC mobile phase), found by the SciFinder tool, were utilized for the retention time prediction in order to narrow down the number of proposed structures. The interpretation of the observed UV and MS/MS spectra in comparison with those found in the literature (when possible) was the main tool for putative identification of metabolites.
Metabolomics raw data analysis
Peak picking and data processing were performed by the MarkerLynx 4.1 software (Waters Inc.) with the following parameters: mass tolerance, 0.03 Da; peak width, 5%; height, 30 s; peak-to-peak baseline noise, 60; intensity threshold, 50 counts; mass window, 0.02 Da; retention time (RT) window, 0.3 min; noise elimination level, 4. The automatic chromatogram smoothing was applied. The data from the beginning of the chromatogram (the first 1.05 min), representing the column void volume, and the end of the chromatogram (starting from 23 min), corresponding to the column washing and equilibration, were removed from the analysis. Since injections of samples in the positive and negative ionization modes were performed in separate injection sets, MarkerLynx pre-processing was done for each ionization mode independently and the mass signals lists with RT, m/z, and peak area intensities were further used for post-processing and statistical analysis using Matlab v. 7.3 (The MathWorks Inc.) as follows: (i) spurious zeros produced by MarkerLynx which hamper statistical analyses of the data were either replaced by a low threshold or removed from further analyses (a full description of the procedure is presented in the Supplementary Materials and methods S1 at JXB online); and (ii) a two-sample t-test was carried out on the data with replaced zeros. Differential markers were determined by applying the false discovery rate (FDR) procedure on the t-test results with FDR (q-value) set to 5%. Differential masses which are higher in white flowers were clustered to find masses belonging to the same metabolite. A custom computer program implemented in MATLAB was used for this purpose. The program accepted as an input the differential mass signals in positive and negative ionization modes separately. Using a greedy clustering process, the mass signals were grouped according to the similarity in their abundance profiles across different samples and according to the proximity in their retention times. Pearson correlation was used as the distance measure.
Headspace collection of flower volatiles
Individual flowers collected from day 0 to day 3 were placed in a 1.0 l glass sealed with a ‘cooky bag’ and incubated under ambient conditions. The volatile metabolites were analysed by headspace solid phase microextraction–gas chromatography–mass spectrometry (HS-SPME-GC-MS). The volatile metabolites were adsorbed for 30 min by manual HS-SPME at ambient temperature by 65 μm polydimethylsiloxane/divinylbenzene (PDMS/DVB) fiber (Supelco, PA, USA). The fiber was inserted into the injection port of the GC-MS for 10 min (splitless) for desorption of the volatiles.
Gas chromatography–mass spectrometry
GC-MS analysis was carried out on Agilent GC-MSD system (CA, USA) equipped with an Rtx-5 SIL MS column (30 m length×0.25 mm id, 0.25 μm film thickness, stationary phase 95% dimethyl–5% diphenyl polysiloxane). Oven temperature was set at an initial temperature of 50 °C for 1 min, increased to 200 °C with 5 °C min−1 increments, followed by a ramp of 15 °C min−1 to 230 °C min−1, and an additional 4 min at the same temperature. The inlet temperature was 250 °C and the transfer line temperature was 280 °C. The carrier gas was helium at 0.8 ml min−1. A quadruple mass detector with electron ionization at 70 eV was used to obtain the MS data in the range of 41–350 m/z. A mixture of straight-chain alkanes (C6–C16) was injected into the column under the above-mentioned conditions to determine the retention indices. Identification of the volatile metabolites was done by matching the retention indices with those of the authentic standards and by comparison of the spectral data with the NIST98 GC-MS library.
Protein extraction and 2D gel separation
Total proteins were extracted according to Hurkman and Tanaka (1986). For initial separation, the protein samples were loaded on 13 cm IEF (isoelectric focusing) dry strips having pH 3–10 (Amersham Biosciences) and run according to Berkelman and Stenstedt (1998). For the second dimension runs, the strips were loaded on a 12% polyacrylamide gel.
Gel analysis and protein identification (sequencing)
Protein gels were stained with 0.1% Coomassie Brilliant Blue and scanned using an Image-Scanner (Amersham Biosciences). Intensity differences of the protein spots were compared using Z3 software version 1.5 (Compugen, Tel Aviv, Israel). Individual protein spots were manually excised from the 2D gels and in-gel digested with trypsin based on Shevchenko et al. (1996). The MS analysis was carried out in the interdepartmental Equipment Unit, School of Medicine at the Hebrew University of Jerusalem. Analysis of the digest was performed by LC-MS/MS on the QTOF II (Micromass, UK) mass spectrometer with online nano-ESI connected to Waters CapLC system. Spectra results were analysed using MassLynx software and submitted to the MASCOT server for protein identification. Results were compared with the general database of Swiss-Prot and TrEMBL via the Expasy proteomics tool set.
RNA extraction
Total RNA was extracted from flowers at different stages of development (days 0.5, 0, 1, and 2) using the RNeasy kit (Qiagen, Valencia, CA, USA), according to the manufacturer's instructions for plant tissue material.
Experimental design for microarray
Cross-species hybridization of Brunfelsia RNA to a potato microarray was performed. The potato microarray was printed by TIGR as a cDNA spotted array from a set of 15 264 non-redundant cDNA clones from the TIGR Potato Gene Index. Two time points were analysed by a direct comparison of RNA sample design (Churchill, 2002). In this experimental design, RNA test samples (i.e. RNA samples extracted from the flower tissue at specific time points) from two time points were co-hybridized to the cDNA microarray. Comparison between the two time points was facilitated by bioinformatics means (see below). Three biological independent replicates and two dye-swap (Cy3-dUTP/Cy5-dUTP) technical replicates were performed.
Microarray hybridizations
Pre-hybridizations and hybridizations were performed as described in http://132.183.243.28/assets/pdf/protocols_niddk_oligo_cdna_microarray.pdf, supplied by the Keck Biotechnology Resource Laboratory (Yale University, CT, USA) with modifications, as described in Bar Or et al. (2005). Initial image analysis was performed by using the histogram method of QuantArray version 3 software (Packard BioScience).
Microarray data analysis
Data normalization was performed by applying per-spot and per-chip normalization (GeneSpring 5.1, Silicon Genetics, Redwood City, CA, USA) for each two co-hybridized samples. The following are the five filters used for the data for picking candidate genes: (i) the signal to noise ratio for each microarray spot was checked to exceed 2.0 for all five replicates; (ii) to reduce cross-species hybridization effects (Bar-Or et al., 2007) the data were filtered for spots representing spot diameter >75 and <100; (iii) unchanged genes (i.e. clones with values of >0.66 and <1.5 over all five replicates) were removed from the data; (iv) clones representing t-test P-values <0.05 for the five replicates that passed Benjamini and Hochberg multiple testing correction (FDR=0.05) were retained; and (v) clones showing up-regulation (gene expression change >2) were singled out.
Isolation of Brunfelsia genes and quantitative validation of Brunfelsia gene induction during anthocyanin degradation by quantitative real-time PCR (RT-PCR)
For isolation of Brunfelsia genes that have sequence similarity to those of potato as represented in the potato microarray (and that were selected as significantly and differentially induced, i.e. target genes), primers were designed from Solanaceae conserved target gene regions by BLAST-based alignment (http://blast.ncbi.nlm.nih.gov/Blast.cgi). cDNA preparation was performed as described (Gal et al., 2006) and PCR was done using the ReddyMix kit (ABgene). Amplification conditions were as follows: 5 min at 95 °C, 35 cycles of 1 min at 93 °C, 30 s at 45 °C, and 1 min at 72 °C. PCR products were separated by electrophoresis and gel-isolated PCR fragments were purified using the PCR DNA Extraction kit (RBC), according to the manufacturer's instructions, and sequenced. Annotation of the isolated gene fragments was done based on homology searches using BLAST. Gene-specific primers were designed for PCR fragments that had sequence similarity to the target genes, and quantitative PCR was performed as described before (Bar-Or et al., 2005), using ABGene's ABsolute qPCR SYBR Green ROX Mix (ABGene, Espom, UK), and an ABI PRISM 7700 Real-Time PCR machine (Applied Biosystems). To minimize mRNA quantification errors and to correct for intersample variations, the 18S ribosomal Brunfelsia gene (accession no. L49274) was used as an internal control using specific forward and reverse primers. The level of expression of target genes was calculated relative to that of the reference (18S ribosomal) mRNA; the relative efficiency of the target and reference was validated to be approximately equal. Three technical replicates and three independent biological replicates were performed for each examined time point. Means were calculated for all replicates of a time point. Statistical analysis and determination of significance of changes in the level of transcripts in D1 versus that of D0 were done by one-way analysis of variance (ANOVA; general linear model) using The Statistical Discovery Software Institute (SAS JMP™); P ≤0.05.
Results
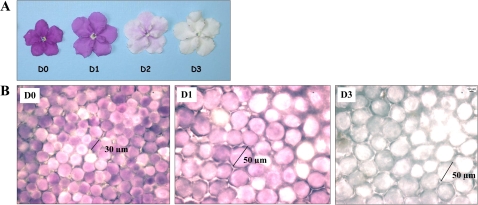
The anthocyanin concentration in Brunfelsia flowers decreases to ∼10% of its initial concentration (Vaknin et al., 2005) while the flowers grow and become fragrant. To investigate whether the production of benzenoids is dependent on the induction of the shikimate pathway, or on anthocyanin degradation, and to further develop Brunfelsia as a model plant for future metabolic studies, several profiling approaches were employed. The developmental stages examined in this study were the following: D0, the day of petal unfurling before anther opening and pollen release; D1, D2, and D3, 1, 2, and 3 d, respectively, after flower opening and expansion of the petal cells (Fig. 1).
Fig. 1.
Brunfelsia flowers during anthocyanin degradation. D0 is the day of flower opening and D1–3 are consecutive days after flower opening. Flowers were detached at D0 and grown in sucrose media as described in Materials and methods. (A) Detached flowers at D0–D3. (B) A photomicrograph of petal tissues at D0, D1, and D3 (×40 enlargement).
Characterization of anthocyanins in Brunfelsia flowers on the day of flower opening
A detailed molecular characterization of the anthocyanin molecules in Brunfelsia flowers at D0 was performed using UPLC-QTOF-MS/MS. The analysis revealed nine distinct anthocyanins as described in Fig. 2 and Supplementary Table S1 (at JXB online). Different anthocyanins, putatively assigned on the basis of their ESI MS/MS fragmentation spectra, appeared to be acyl and glucose derivatives of malvidin, petunidin, and delphinidin. The use of a high resolution TOF mass analyser allowed a distinction to be made between acid and sugar substituents with closely related masses (e.g. hexose and caffeic acid). The most abundant anthocyanin in Brunfelsia flowers was malvidin-O-coumaroylrutinoside-O-glucoside. Since Brunfelsia and Petunia are related species, we compared the composition of anthocyanins in these two species. Most anthocyanins found in Brunfelsia were identified based on petunia MS/MS data from our laboratory or from the literature (Alcalde-Eon et al., 2004; Ando et al., 1999).
Fig. 2.
Characterization of the main anthocyanins in Brunfelsia flower, using UPLC-QTOF-MS/MS. (A) A UV chromatogram at 530 nm of the different anthocyanins in Brunfelsia flower petals at D0. (B) The main anthocyanins detected in Brunfelsia flowers.
Volatile compounds emitted following Brunfelsia flower opening
The volatile metabolites emitted by Brunfelsia flowers were characterized on D0, D1, and D2–3 by HS-SPME/GC-MS (Table 1). The volatiles were collected from flowers attached to the shrub, and the time after flower opening was determined according to the developmental stage of the flower. Nineteen different volatiles were identified, all of which were present in the flowers at D0 and in most cases increased in quantity during flower de-pigmentation, up to D2–3. The set of volatile compounds identified could be divided into terpenoids and phenol derivatives. Most of the identified phenolics were benzenoids, originating from the phenylpropanoid pathway that also leads to anthocyanin biosynthesis.
Table 1.
Relative concentration of volatile compounds during flower opening based on HS-SPME-GC-MS measurements
| Pathway | Peak area×106 | |||
| Compound name | Day 0 | Day 1 | Day 2–3 | |
| Terpenoids | E-β-Ocimene | 235 | 385 | 462 |
| (E)-4,8-Dimethyl-1,3,7-nonatriene | 6 | 14 | 25 | |
| Linalool | 31 | 64 | 109 | |
| z-Linalool oxide (furanoid) | 9 | 5 | 18 | |
| E-Nerolidol | 4 | 6 | 10 | |
| Geranyl acetone | 4 | 2 | 9 | |
| Phenyl propanoids | Cinnamyl acetate | 4 | 16 | 39 |
| Methyl benzoate | 11 | 38 | 37 | |
| Benzyl benzoate | 2 | 11 | 17 | |
| Methyl salicylate | 5 | 6 | 12 | |
| Benzyl acetate | 1 | 2 | 4 | |
| Methyl anthranilate | 5 | 13 | 18 | |
| Benzyl tiglate | 2 | 6 | 9 | |
| Isoamyl benzoate | 2 | 11 | 7 | |
| Eugenol. | 1 | 8 | 30 | |
| p-Cresol | 4 | 22 | 30 | |
| Benzyl alcohol | 3 | 8 | 27 | |
| Cinnamyl alcohol | 1 | 5 | 12 | |
| Nitrogen containing | Indole | 3 | 11 | 17 |
The results are an average of two biological replicates.
Proteomic analysis of Brunfelsia flowers
In a search for proteins that may be involved in the secondary metabolic processes occurring simultaneously, total soluble protein extracts were prepared from Brunfelsia flowers at D0 and D2. The extracts were separated by 2D gel electrophoresis and proteins that increased in intensity by >2-fold were selected from the D2 sample (see Supplementary Fig. S1 at JXB online) and partially sequenced for identification (Table 2). Since no proteomic or genomic data on Brunfelsia are available, the selected proteins were identified by comparison with sequence databases of plants belonging mainly to the Solanaceae family. Seven proteins were annotated from the peptide sequences (Table 2); three of them represented three different enzymes that play a major part in the biosynthesis of lignins [catechol O-methyl transferase (COMT), caffeoyl-CoA O-methyl transferase (CCoA-OMT), and cinnamyl-alcohol dehydrogenase (CAD)]. The levels of an additional protein, 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase (DAHP), increased between D0 and D2. DAHP is a key enzyme in the shikimate pathway, leading to biosynthesis of the aromatic amino acids phenylalanine, tyrosine, and tryptophan. The activity of this enzyme might be related to the supply of precursors to benzenoids as well as to lignin biosynthesis in expanding Brunfelsia flowers.
Table 2.
Proteomic analysis of Brunfelsia flowers
| No.a | Identity | Accession no.b | Source | Function |
| 1 | Caffeoyl-CoA O-methyl transferase 6 | Q42945 | Nicotiana tabacum | Lignification |
| 2 | Catechol O-methyl transferase | CAA52462 | Nicotiana tabacum | Lignification |
| 3 | Cinnamyl-alcohol dehydrogenase | CAA44217 | Nicotiana tabacum | Lignification |
| 4 | Cinnamyl-alcohol dehydrogenase | CAA44217 | Nicotiana tabacum | Lignification |
| 5 | ATPase β-subunit | CAA43612 | Nicotiana tabacum | Proton transport through membranes |
| 6 | 3-Deoxy-D-arabino-heptulosonate 7-phosphate synthase | CAA75092 | Morinda citrifolia | Formation of aromatic amino acids |
| 7 | 3-Ketoacyl-CoA thiolase; acetyl-CoA acyltransferase | Q87ZB3 | Tomato | Oxidation of fatty acids |
All seven proteins were partially sequenced and identified using sequence homologues of other plants.
The numbers refer to spot numbers as given in the Supplementary data.
The accession numbers represents protein entries if available (until December 2008).
Transcriptome analysis of Brunfelsia flowers
In search for genes activated during the de-pigmentation process, gene expression arrays were used to compare the transcription profile of D0 and D1 flowers. Since Brunfelsia microarrays are unavailable, cross-species hybridization of Brunfelsia cDNAs to potato cDNA microarrays was performed. Out of 15 266 clones present on the array (representing 11 412 verified cDNA clones), 1823 were selected as differentially expressed between the two developmental stages sampled (see Materials and methods.) Among the 1823 differential genes, 146 and 98 were significantly up- or down-regulated at the D1 stage by at least 2-fold, respectively (see Supplementary Table S2 at JXB online). In this study the focus was on the up-regulated genes, and a list of these genes with their suggested functions is presented in Supplementary Table S3 at JXB online. Out of the 146 up-regulated genes, 21 were related to secondary metabolism (Table 3), and among those there were 10 with a suggested function related to the biosynthesis of lignins and suberins and six putatively related to the biosynthesis of either terpenoid or flavonoid pathway-derived volatiles. RT-PCR on four selected genes, namely copper amine oxidase, COMT, CCoA-OMT, and CAD, confirmed the up-regulation of these genes at D1 (Table 4). The increase in levels of three of these genes was further evidenced by the accumulation of their gene products detected by proteomics analysis (Table 4).
Table 3.
Secondary metabolism genes up-regulated in Brunfelsia flowers after opening
| Systematic name | Average fold change (range) | Annotation | Function |
| STMHW02 | 5.39 (3.246–10.18) | (P15004) Suberization-associated anionic peroxidase 2 precursor (TMP2) | Suberanization |
| STMCL66 | 5.271 (4.096–7.023) | (Q9M527) Phenylcoumaran benzylic ether reductase homologue Fi2 | Lignification |
| STMDQ29 | 4.608 (3.016–9.118) | (Q41242) Alcohol dehydrogenase ADH | |
| STMCC79 | 3.475 (2.195–4.905) | (Q42958) Catechol O-methyltransferase (EC 2.1.1.6) | Lignification |
| STMJL95 | 3.387 (1.797–5.991) | (Q8H9B6) Caffeoyl-CoA O-methyltransferase (trans-caffeoyl-CoA 3-O-methyltransferase) (CCoAMT) | Lignification |
| STMIC60 | 3.361 (2.379–4.822) | (Q42945) Caffeoyl-CoA O-methyltransferase 6 (trans-caffeoyl-CoA 3-O-methyltransferase 6) (CCoAMT-6) | Lignification |
| STMCL31 | 3.037 (0.936–5.418) | (Q9AVG9) S-adenosyl-L-methionine:salicylic acid carboxyl methyltransferase | Scent/enzymatic activity that catalyses the formation of methylsalicylate from salicylic acid |
| STMER77 | 3.023 (1.58–7.432) | (P15004) Suberization-associated anionic peroxidase 2 precursor (TMP2) | Suberanization |
| STMCK29 | 2.867 (2.037–4.372) | (Q9XH50) 1-D-Deoxyxylulose 5-phosphate synthase | Terpenoids/IPP synthesis |
| STMCY65 | 2.745 (1.409–5.622) | (O82676) 1-Deoxyxylulose 5-phosphate synthase | Terpenoids/IPP synthesis |
| STMHZ50 | 2.586 (1.723–3.886) | (Q8H9B6) Caffeoyl-CoA O-methyltransferase (trans-caffeoyl-CoA 3-O-methyltransferase) (CCoAMT) | Lignification |
| STMEM71 | 2.4 (0.927–5.05) | (Q9MB73) Limonoid UDP-glucosyltransferase (limonoid glucosyltransferase) (limonoid GTase) (LGTase) | Terpenoids/glucosylation of limonoids |
| STMIN59 | 2.38 (1.399–4.517) | (Q8GVE3) Flavonoid 1–2 rhamnosyltransferase | Biosynthesis of flavonoids |
| STMEU71 | 2.365 (1.076–3.65) | (68418.m05207) O-Methyltransferase N-terminus domain-containing protein contains Pfam profile PF02409: | Lignification |
| STMIX91 | 2.322 (0.126–4.496) | (P30359) Cinnamyl-alcohol dehydrogenase (CAD) (EC 1.1.1.195) | Lignification |
| STMIJ60 | 2.267 (1.662–3.439) | (Q9SXJ0) Transcription factor Ntlim1 | Lignification/zinc binding |
| STMHR20 | 2.151 (1.657–3.4) | Adenosine monophosphate-binding protein 7 (AMPBP7_) | Acyl Co-A synthase/secondary metabolite biosynthesis |
| STMGM34 | 2.113 (1.5–3.42) | (Q42698) Geranylgeranyl pyrophosphate synthetase chloroplast precursor (GGPP synthetase) (GGPS) | Carotenoids |
| STMEP23 | 2.056 (1.16–3.063) | (Q41437) 3-Hydroxy-3-methylglutaryl-coenzyme A reductase 2 (HMG-CoA reductase 2) (HMG2.2) | Isoprenoid synthesis/mevalonate synthesis |
| STMCB66 | 2.054 (1.652–2.731) | (68417.m04881) LytB family protein contains Pfam profile: PF02401 LytB protein | Isoprenoid synthesis/enzyme |
| STMIV38 | 2.037 (0.684–5.894) | (P28554) Phytoene dehydrogenase chloroplast precursor (phytoene desaturase) | Carotenoids |
Genes were identified by heterologous hybridization of RNA from Brunfelsia flowers from D1 and D0 onto potato micrroarray, with a suggested function in the category of secondary metabolism. Genes were identified as significantly regulated (P = 0.05) at least one of the time points (D0 or D1) and have a >2-fold increase in D1 versus D0.
Table 4.
The up-regulation of selected transcripts and their products after flower opening CCoA-OMT
| Systematic name | Annotation | Microarray (D1-D0) | RT-PCRa (D1–D0) | 2D gel (D2–D0) |
| STMIV90 | Copper amine oxidase | 2.47 | 3.65 | – |
| STMCC79 | Catechol O-methyltransferase | 3.47 | 5.95 | >2 |
| STMIC60 | Caffeoyl-CoA O-methyltransferase | 3.36 | 3.94 | >2 |
| STMIX91 | Cinnamyl-alcohol dehydrogenase | 2.32 | 6.39 | >2 |
The numbers are average fold changes, as measured by the microarray analysis, real-time PCR (4–5 biological repeats) and 2D gel analysis.
The RT-PCR results were statistically significant (P <0.05) in all genes except catechol O-methyltransferase.
Non-targeted metabolomic analysis of Brunfelsia flowers
Further understanding of processes occurring in Brunfelsia flowers during the first 2 d after flower opening was achieved by the putative identification of >12 accumulated metabolites. Non-targeted metabolite analysis performed by UPLC-QTOF-MS resulted in the detection of 2522 and 1348 mass signals in positive and negative ionization modes, respectively. To estimate the number of metabolites up-regulated, statistical filtering was applied to the mass signal data. Totals of 353 and 501 mass signals in the positive and negative ionization modes, respectively, were at least 2-fold more abundant in white versus purple flowers. This set of differential mass signals was analysed in order to cluster together masses belonging to the same metabolite (see Materials and methods for details). After clustering of differential masses, 118 and 188 groups were formed in the positive and negative ionization modes, respectively. It was possible to assign 17 putative Brunfelsia metabolites that accumulated in the flowers between D0 and D2 on the basis of accurate mass measurements, information available from the literature, and MS/MS analyses (Table 5, see Supplementary Table S4 in JXB online). The metabolites found belong to amino acid, phenolic acid, benzoic acid, phenylpropanoid, flavonoid, and alkaloid classes.
Table 5.
Putative metabolite accumulating in Brunfelsi a between D0 and D2, identified by UPLC-QTOF-MS
| Putative metabolite | Compound type | Retention time (min) | Molecular formula | Molecular weight | Level of confidencea |
| Tyrosine | Amino acid | 1.17 | C9H11NO3 | 181.0738 | A |
| Tryptophan | Amino acid | 3.86 | C11H12N2O2 | 204.0898 | A |
| Phenylalanine | Amino acid | 2.18 | C9H11NO2 | 165.0788 | A |
| Hydroxybenzoic acid hexose | Benzoic acids and esters | 1.72 | C13H16O8 | 300.0838 | B |
| Caffeic acid hexose-I | Phenolic acid | 2.83 | C15H18O9 | 342.0938 | B |
| Caffeic acid hexose-II | Phenolic acid | 3.45 | C15H18O9 | 342.0948 | B |
| Benzyl alcohol dihexose | Phenolic acid | 5.06 | C19H28O11 | 432.1638 | B |
| Ferulic acid hexose | Phenolic acid | 6.72 | C16H20O9 | 356.1098 | B |
| Caffeic acid di-glucoside | Phenolic acid | 2.52 | C21H28O14 | 504.1478 | B |
| Dihydromyricetin+glucose | Dihydroflavonols | 4.05 | C21H22O13 | 482.1058 | B |
| Dicaffeoylspermidine | Polyamine alkaloids | 7.55 | C25H31N3O6 | 469.2212 | B |
| Caffeoylputrescine | Polyamine alkaloids | 2.18 | C13H18N2O3 | 250.1318 | B |
| 4-Coumaroylputrescine | Polyamine alkaloids | 3.30 | C13H18N2O2 | 234.1358 | B |
| Esculetin (6,7-dihydroxycoumarin) or caffeoquinone | Phenylpropanoids | 2.31 | C9H6O4 | 178.0261 | C |
| Cinnamic acid di-glucoside or coumaric acid rutinoside | Phenolic acid | 8.54 | C21H28O12 | 472.1582 | C |
| Caffeic acid rutinoside | Phenolic acid | 4.62 | C21H28O13 | 488.1508 | C |
| trans-Caffeoyl ethyl ester (ethyl 3,4-dihydroxycinnamate) | Phenylpropanoids | 9.82 | C11H12O4 | 208.0742 | C |
The metabolites were characterized in negative ion mode.
Confidence level of the identification of metabolites: A, identified compounds (identified by a standard); B, putatively annotated compounds (identified by MS/MS fragments, comparing with the literature); C, putatively characterized compound (based upon Solanaceae-specific compounds from the ‘Dictionary of natural products’ and identified partly by MS/MS fragments or a similar standard).
Discussion
The profiling of secondary metabolism taking place in Brunfelsia flowers after opening reveals induction of metabolic pathways preceding and branching from the phenylpropanoid pathway, as well as a severe reduction in the activity of the flavonoid and anthocyanin pathways (Fig. 3).
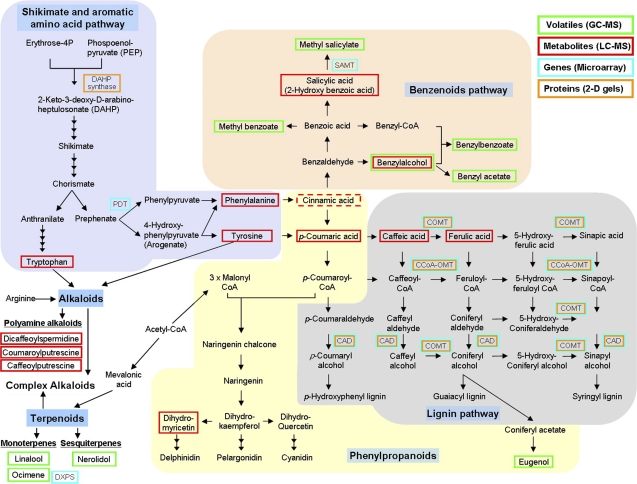
Fig. 3.
Multilevel analysis reveals metabolic processes occurring in Brunfelsia flowers during the first 2 d after opening and during the degradation of anthocyanins. The diagram shows the different pathways. Genes, proteins, and metabolites that have increased significantly are boxed. The diagram summarizes the results of GC-MS, LC-MS, microarray, and 2D gel analyses. The colour of the box indicates the method by which they were identified. The dashed line box indicates a lower level of confidence of metabolite identification. Enzymes: SAMT, salicylic acid carboxyl methyltransferase; COMT, catechol O-methyl transferase; CCoA-OMT, caffeoyl-CoA O-methyl transferase; CAD, cinnamyl-alcohol dehydrogenase; DAHP synthase, 3-deoxy-D-arabino-heptulosonate 7-phosphate; PDT, prephenate dehydratase; DXPS, 1-deoxyxylulose 5-phosphate synthase.
The shikimate pathway, linking carbohydrate metabolism to the synthesis of aromatic amino acids which serve as precursors for the synthesis of anthocyanins, benzenoids, and lignin, is induced in the flowers at these later stages (Fig. 3). The increased concentration of tryptophan, tyrosine, and phenylalanine, the end-products of the shikimate and aromatic amino acid pathways, the increase in the DAHP synthase levels, and up-regulation of PDT gene expression (see Supplementary Table S3 general metabolism category at JXB online, Fig. 3) provide evidence for this induction. Furthermore, the increase in phenylalanine and tyrosine is accompanied by an increase in their products, cinnamic and coumaric acids, which are early precursors of the phenylpropanoid pathway. Further indication for the increase in phenolic acids is the increase in the polyamine alkaloids containing coumaric and caffeic moieties (Table 5, Fig. 3.). Thus, there are probably two events of induction of these metabolic pathways: first at the early stages of flower development for the production of anthocyanins; and then, following flower opening, for the production of volatile compounds and probably also lignin.
The metabolism of the volatile benzenoid is induced in Brunfelsia after flower opening and results in a significant increase in a group of floral fragrance components that is similar to the profile emitted by Brunfelsia australis flowers (Bertrand et al., 2006) (Table 1, Fig. 3). Additional indications of the induction of the benzenoid pathway are the increase in its precursors, namely cinnamic and salicylic acid, and the gene encoding salicylic acid carboxyl methyltransferase (SAMT), which may possibly transform salicylic acid to the volatile methyl salicylate (Fig. 3). Two additional benzenoid derivatives, benzyl alcohol dihexose and the volatile benzyl alcohol, were identified among the compounds elevated after flower opening (Tables 2, 5, Fig. 3).
Lignin biosynthesis is the second metabolic pathway branching from the phenylpropanoid pathway that is induced in Brunfelsia flowers after opening and during petal expansion (Fig. 3). The genes putatively encoding the three major enzymes in the lignin biosynthesis pathway, COMT, CCoA-OMT, and CAD (Rastogi and Dwivedi, 2008), were induced during the first day after flower opening and their products increased in concentration during the first 2 d. The induction of this pathway may be essential for the expansion of the petals and the accumulation of lignin along the petal veins. The three enzymes, COMT, CCoA-OMT, and CAD, are also involved in the synthesis of one of the volatile compounds in Brunfelsia flowers, eugenol (Koeduka et al., 2006).
The biosynthesis of anthocyanins in Brunfelsia ends before flower opening (Vaknin et al., 2005) and therefore it is not surprising that apart from an increase in dihydromyricetin, no increase in metabolite levels or protein or gene expression related to the flavonoid or anthocyanin pathway could be detected (Fig. 3). Moreover, the transcriptomic analysis suggests down-regulation of chalcone synthase, the first committed enzyme for flavonoid and anthocyanin biosynthesis (Supplementary Table S2 at JXB online).
The three phenolic acids, coumaric, caffeic, and ferulic, that have increased in concentration (Fig. 3) are constituents of the Brunfelsia anthocyanin molecules (Fig. 2), and therefore may be products of the pigment degradation. These products, however, are probably not precursors for the production of phenolic acids, which in turn are precursors of both the benzenoid and lignin. In order for the phenolic acids originating from the degraded anthocyanins to serve as precursors for benzenoid and lignin, they would have to be transported out of the vacuole. The export of anthocyanin breakdown products, such as the phenolic acids, from the vacuoles has not yet been demonstrated. In addition, it is not clear whether the biosynthetic enzymes producing benzenoids in Brunfelsia are expressed in the epidermal cells of the flowers, where anthocyanins accumulate, as has been shown in snapdragon and Clarkia brewei (Kolosova et al., 2001). Future studies, in which labelled anthocyanin precursors are fed to detached Brunfelsia flowers, may clarify the destiny of the degradation products.
Here, for the first time, extensive molecular and biochemical data from Brunfelsia flowers are presented that provide a knowledge base regarding changes that occur during the de-pigmentation period. The knowledge obtained in this study is very valuable for future studies regarding the process of active in planta degradation of anthocyanins, formation of volatiles, and the network of secondary metabolism in flowers of Brunfelsia and related species, such as petunia.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. 2D gels of total protein extracts from Brunfelsia flowers at D0 (left) and D2 (right) after opening.
Table S1. Characterization of the main anthocyanins in Brunfelsia flower, using UPLC-QTOF-MS and MS/MS analyses.
Table S2. A list of up- and down-regulated genes between D1 and D0.
Table S3. A list of up-regulated genes between D1 and D0 with their suggested function.
Table S4. A list of putative metabolites that have accumulated in Brunfelsia flowers between D0 and D2 and identified by UPLC-QTOF-MS and MS/MS analyses.
Materials and methods S1. MarkerLynx data treatment.
Supplementary Material
Acknowledgments
We are grateful to Ilya Venger for his help with LC-MS data analysis and Chanita Zemah for help with lignin staining. AA is the incumbent of the Adolpho and Evelyn Blum Career Development Chair of Cancer Research. The work in the Aharoni laboratory was supported by Mrs Louise Gartner, Dallas, TX, USA and Mr and Mrs Mordechai Segal, Israel.
References
- Alcalde-Eon C, Saavedra G, de Pascual-Teresa S, Rivas-Gonzalo JC. Identification of anthocyanins of pinta boca (Solanum stenotomum) tubers. Food Chemistry. 2004;86:441–448. [Google Scholar]
- Ando T, Saito N, Tatsuzawa F, et al. Floral anthocyanins in wild taxa of Petunia (Solanaceae) Biochemical Systematics and Ecology. 1999;27:623–650. [Google Scholar]
- Bar-Or C, Kapulnik Y, Koltai H. A broad characterization of the transcriptional profile of the compatible tomato response to the plant parasitic root knot nematode Meloidogyne javanica. European Journal of Plant Pathology. 2005;111:181–192. [Google Scholar]
- Bar-Or C, Novikov E, Reiner A, Czosnek H, Koltai H. Utilizing microarray spot characteristics to improve cross-species hybridization results. Genomics. 2007;90:636–645. doi: 10.1016/j.ygeno.2007.06.008. [DOI] [PubMed] [Google Scholar]
- Ben Zvi MM, Florence NZ, Masci T, Ovadis M, Shklarman E, Ben-Meir H, Tzfira T, Dudareva N, Vainstein A. Interlinking showy traits: co-engineering of scent and colour biosynthesis in flowers. Plant Biotechnology Journal. 2008;6:403–415. doi: 10.1111/j.1467-7652.2008.00329.x. [DOI] [PubMed] [Google Scholar]
- Berkelman T, Stenstedt T. 2-D Electrophoresis: principles and methods. Amsterdam: Amersham Bioscience; 1998. [Google Scholar]
- Bertrand C, Comte G, Piola F. Solid-phase microextraction of volatile compounds from flowers of two Brunfelsia species. Biochemical Systematics and Ecology. 2006;34:371–375. [Google Scholar]
- Boatright J, Negre F, Chen XL, Kish CM, Wood B, Peel G, Orlova I, Gang D, Rhodes D, Dudareva N. Understanding in vivo benzenoid metabolism in petunia petal tissue. Plant Physiology. 2004;135:1993–2011. doi: 10.1104/pp.104.045468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill GA. Fundamentals of experimental design for cDNA microarrays. Nature Genetics. 2002;32:490–495. doi: 10.1038/ng1031. [DOI] [PubMed] [Google Scholar]
- Gal TZ, Aussenberg ER, Burdman S, Kapulnik Y, Koltai H. Expression of a plant expansin is involved in the establishment of root knot nematode parasitism in tomato. Planta. 2006;224:155–162. doi: 10.1007/s00425-005-0204-x. [DOI] [PubMed] [Google Scholar]
- Heide OM. Effect of temperature and day-length on flower initiation of Brunfelsia calycina (Hook) benth. Physiologia Plantarum. 1963;16:104–109. [Google Scholar]
- Hurkman WJ, Tanaka CK. Solubilization of plant membrane proteins for analysis by two-dimensional gel electrophoresis. Plant Physiology. 1986;81:802–806. doi: 10.1104/pp.81.3.802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson LMV, Donker Koopman WE, Schram AW. Turnover of anthocyanins and tissue compartmentation of anthocyanin biosynthesis in flowers of Petunia hybrida. Journal of Plant Physiology. 1984;115:29–37. doi: 10.1016/S0176-1617(84)80048-6. [DOI] [PubMed] [Google Scholar]
- Koeduka T, Fridman E, Gang DR, et al. Eugenol and isoeugenol, characteristic aromatic constituents of spices, are biosynthesized via reduction of a coniferyl alcohol ester. Proceedings of the National Academy of Sciences, USA. 2006;103:10128–10133. doi: 10.1073/pnas.0603732103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolosova N, Sherman D, Karlson D, Dudareva N. Cellular and subcellular localization of S-adenosyl-l-methionine: benzoic acid carboxyl methyltransferase, the enzyme responsible for biosynthesis of the volatile ester methylbenzoate in snapdragon flowers. Plant Physiology. 2001;126:956–964. doi: 10.1104/pp.126.3.956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mintz-Oron S, Mandel T, Rogachev I, Feldberg L, Lotan O, Yativ M, Wang Z, Jetter R, Venger I, Adato A, Aharoni A. Gene expression and metabolism in tomato fruit surface tissues. Plant Physiology. 2008;147:823–851. doi: 10.1104/pp.108.116004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastogi S, Dwivedi UN. Manipulation of lignin in plants with special reference to O-methyltransferase. Plant Science. 2008;174:264–277. [Google Scholar]
- Schuurink RC, Haring MA, Clark DG. Regulation of volatile benzenoid biosynthesis in petunia flowers. Trends in Plant Science. 2006;11:20–25. doi: 10.1016/j.tplants.2005.09.009. [DOI] [PubMed] [Google Scholar]
- Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins from silver stained polyacrylamide gels. Analytical Chemistry. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- Shinbo Y, Nakamura Y, Altaf-Ul-Amin M, Asahi H, Kurokawa K, Arita M, Saito K, Ohta D, Shibata D, Kanaya S. KNApSAcK: a comprehensive species–metabolite relationship database. In: Saito K, Dixon RA, Willmitzer L, editors. Plant metabolomics. Biotechnology in agriculture and forestry. Vol. 57. Berlin: Springer; 2006. pp. 165–181. [Google Scholar]
- Spitzer B, Ben Zvi MM, Ovadis M, et al. Reverse genetics of floral scent: application of tobacco rattle virus-based gene silencing in petunia. Plant Physiology. 2007;145:1241–1250. doi: 10.1104/pp.107.105916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaknin H, Bar-Akiva A, Ovadia R, Nissim-Levi A, Forer I, Weiss D, Oren-Shamir M. Active anthocyanin degradation in Brunfelsia calycina (yesterday-today-tomorrow) flowers. Planta. 2005;222:19–26. doi: 10.1007/s00425-005-1509-5. [DOI] [PubMed] [Google Scholar]
- Verdonk JC, Haring MA, van Tunen AJ, Schuurink RC. ODORANT1 regulates fragrance biosynthesis in petunia flowers. The Plant Cell. 2005;17:1612–1624. doi: 10.1105/tpc.104.028837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss D. Regulation of flower pigmentation and growth: multiple signaling pathways control anthocyanin synthesis in expanding petals. Physiologia Plantarum. 2000;110:152–157. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.