Abstract
MicroRNAs (miRNAs) are small regulatory RNAs that participate in posttranscriptional gene regulation in a sequence-specific manner. However, little is understood about the role(s) of miRNAs in Alzheimer's disease (AD). We used miRNA expression microarrays on RNA extracted from human brain tissue from the University of Kentucky Alzheimer's Disease Center Brain Bank with near-optimal clinicopathological correlation. Cases were separated into four groups: elderly nondemented with negligible AD-type pathology, nondemented with incipient AD pathology, mild cognitive impairment (MCI) with moderate AD pathology, and AD. Among the AD-related miRNA expression changes, miR-107 was exceptional because miR-107 levels decreased significantly even in patients with the earliest stages of pathology. In situ hybridization with cross-comparison to neuropathology demonstrated that particular cerebral cortical laminas involved by AD pathology exhibit diminished neuronal miR-107 expression. Computational analysis predicted that the 3′-untranslated region (UTR) of β-site amyloid precursor protein-cleaving enzyme 1 (BACE1) mRNA is targeted multiply by miR-107. From the same RNA material analyzed on miRNA microarrays, mRNA expression profiling was performed using Affymetrix Exon Array microarrays on nondemented, MCI, and AD patients. BACE1 mRNA levels tended to increase as miR-107 levels decreased in the progression of AD. Cell culture reporter assays performed with a subset of the predicted miR-107 binding sites indicate the presence of at least one physiological miR-107 miRNA recognition sequence in the 3′-UTR of BACE1 mRNA. Together, the coordinated application of miRNA profiling, Affymetrix microarrays, new bioinformatics predictions, in situ hybridization, and biochemical validation indicate that miR-107 may be involved in accelerated disease progression through regulation of BACE1.
Keywords: Alzheimer's, miRNAs, microarray, in situ, BACE, noncoding
Introduction
MicroRNAs (miRNAs) are small regulatory RNAs with potent roles in metabolism, neurodevelopment, neuroplasticity, apoptosis, and other fundamental neurobiological processes (for review, see Nelson et al., 2003; Kosik and Krichevsky, 2005). miRNAs exert their function through base-pairing with specific “target” mRNAs with partially complementary sequences, resulting in decreased polypeptide formation from those mRNAs (Ambros, 2004). Depending on the algorithm used, bioinformatics predict that miRNAs regulate from 30% of human genes (Lewis et al., 2003, 2005) to as much as 92% (Miranda et al., 2006).
The study of miRNA biology is in its infancy, particularly in the human brain. Researchers have shown that RNA is pathologically altered in Alzheimer's disease (AD) brains (Ginsberg et al., 1997; Honda et al., 2005; Ding et al., 2006; Nelson and Keller, 2007). However, a role for miRNAs in the progression of AD has not been established firmly. Previous studies demonstrated that a subset of miRNAs show altered expression in advanced AD, and these changes may be stimulated by reactive metal elements in the brain (Lukiw, 2007; Lukiw and Pogue, 2007). These previous studies in AD brains were limited in the number of miRNAs that were evaluated.
Expression profiling has provided important insights into miRNA biology. Microarray studies have helped to describe which miRNAs are expressed during various normal and abnormal brain states, including in brain diseases (Babak et al., 2004; Ciafre et al., 2005; Nelson et al., 2006; Perkins et al., 2007). Although miRNA expression analyses can provide groundbreaking data for this novel research field, miRNA profiling benefits from the parallel use of more than a single technique. Previous work confirmed that understanding cellular level expression patterns is vitally important for brain miRNA studies (Nelson et al., 2006). In situ hybridization (ISH) pinpoints exactly where specific miRNAs are expressed: neurons, astrocytes, oligodendrocytes, microglia, and/or vasculature. Neurodegenerative diseases tend to affect distinct cell populations. Hence, knowing which miRNAs are expressed in which cell populations can facilitate the identification of disease-relevant miRNA genes and their mRNA targets.
β-Site amyloid precursor protein-cleaving enzyme 1 (BACE1) is a protein-coding gene known to be important in AD. BACE1 protein is an endopeptidase that cleaves the β-amyloid precursor protein to generate neurotoxic β-amyloid peptide Aβ1–42 (Vassar et al., 1999; Haniu et al., 2000). The dysregulation of BACE1 may play a major role in the pathogenesis of AD (Dominguez et al., 2004; Durham and Shepherd, 2006; Guo and Hobbs, 2006; John, 2006). BACE1 is enriched in neurons (Bigl et al., 2000; Fukumoto et al., 2002) and its expression is regulated posttranscriptionally (Marambaud et al., 1998; Rossner et al., 2006). However, BACE1 gene expression regulation is not completely understood. Here, we provide data indicating that miR-107 contributes to BACE1 posttranscriptional regulation and that this pathway is predicted to exacerbate pathology in AD brains.
Materials and Methods
RNA isolation from a human cerebral cortex.
RNA was extracted from snap-frozen brain tissue from the University of Kentucky Alzheimer's Disease Brain Bank under a University of Kentucky Institutional Review Board protocol. Premortem clinical evaluations were exhaustive, and the last cognitive evaluation was performed usually within the last year of life as described previously (Schmitt et al., 2000). Clinical diagnoses were established at Consensus Conference with University of Kentucky neuropsychologists, neurologists, and neuropathologists. The inclusion criteria that we applied were as follows: postmortem interval (PMI) <5 h, no argyrophilic grains, no cortical Lewy bodies, no evidence of frontotemporal dementia, no cancer in the brain parenchyma, and no infarctions within 3 cm of the brain tissue samples. These neuropathological confounds were assessed using standard neuropathological procedures as described in detail previously (Nelson et al., 2007). Demented controls were from the brains of patients with dementia with Lewy bodies (DLB), in comparison with “pure” AD that lacked Lewy bodies. Five DLB patients were of ages 68, 82, 85, 87, and 90 years. Braak stages for these patients were 2 or 3. Before RNA extraction, meninges and large vessels were removed; gray matter was dissected away from white matter, and only gray matter was used for these studies.
RNA was isolated from brain tissue cut from superior and middle temporal cortex and separately from motor cortex. Subjacent tissue was evaluated neuropathologically for all samples. Tissue (4–8 g) that had been snap-frozen in liquid nitrogen and then transferred to a −80°C freezer was thawed in isotonic lysis buffer with RNasin (160 U/ml; Promega, Madison, WI) and Complete protease inhibitor tablets (Roche, Indianapolis, IN). Tissue was homogenized for 5 min on ice using a glass and Teflon hand homogenizer followed by additional disruption with a Microson XL2000 hand sonicator (Fisher Scientific, Waltham, MA) (three times 10 s each on ice), and then the samples were centrifuged at 16,000 × g for 15 min at 4°C. The supernatant was used for RNA isolation. Trizol LS (Invitrogen, Carlsbad, CA) was used according to manufacturer's instructions, except for an additional overnight −20°C precipitation step during isopropanol precipitation. RNA quality was confirmed using A260/A280 readings as well as banding pattern and RNA Integrity Number using an Bioanalyzer (Agilent, Palo Alto, CA); degraded samples were discarded.
Microarray methods and analyses.
The miRCURY Labeling kit (Exiqon, Woburn, MA) was used for target labeling and hybridization to a conventional miRNA microarray. The oligonucleotide probe arrays were printed on Codelink substrates, processed, and scanned at the University of Pennyslvania Microarray Core Facility. Detailed methods with regard to miRNA labeling and hybridization are provided in supplemental file 1 (available at www.jneurosci.org as supplemental material).
cDNA microarrays: Affymetrix Exon Arrays.
RNA for the cDNA microarrays was from the same temporal isocortex samples as used for the miRNA microarray. Samples used were from AD (n = 3), mild cognitive impairment (MCI) (n = 4), and nondemented (n = 4) brains. The mRNA reverse transcription, labeling, hybridization, and other processing were performed exactly as written in the Affymetrix (Santa Clara, CA) technical manual. Exon Array data were also processed using the Affymetrix Expression Console software, and using that software, normalization was performed by using the gene level analysis, core annotation confidence, median polish summarization method, and global median normalization method.
Validation: in situ hybridization, Northern blots, and Western blots.
The seven different cases used for ISH were not the same as the cases used for any of the RNA expression profiling. For ISH, brain tissue from the following were used: AD case 1, 80-year-old female, Braak stage 6; AD case 2, 93-year-old female, Braak stage 5; AD case 3, 75-year-old male, Braak stage 6; nondemented case 1, 92-year-old male, Braak stage 0; nondemented case 2, 85-year-old female, Braak stage 0; nondemented case 3, 77-year-old male, Braak stage 4 (pathology “probable AD”); demented control with multiinfarct dementia related to cerebral amyloid angiopathy and an anomalous pattern of neurofibrillary pathology, 87-year-old male.
ISH was performed as described previously (Nelson et al., 2006) with modifications. Tissue was obtained at autopsy, placed into 4% paraformaldehyde, and fixed for 24–36 h at 4°C. Tissue was then placed in 20% sucrose overnight, again at 4°C. The following day, tissue was cut on a freezing microtome at 25 μm thickness and mounted onto coated glass slides (VWR, West Chester, PA). After air-drying for 30 min, slides were stored at −80°C. For in situ hybridization, sections were first allowed to equilibrate to room temperature and then treated with Proteinase K (Roche; 40 μg/ml) for 20 min at room temperature, followed by quenching with glycine (Fisher Scientific; 1 mg/ml for 15 min). After PBS washes, slides were immersed in 10% formaldehyde (Fisher Scientific; 10 min), washed again twice in PBS, and treated with acetic anhydride/triethanolamine (Fisher Scientific; 10 min) at room temperature. Sections were then washed in PBS (four times, 10 min each) followed by 5× SSC (twice, 5 min each). Prehybridization was performed for 3 h at 37°C is in a buffer prepared thus: 650 ml of ultrapure formamide [EMD (Omnipur), Gibbstown, NJ]; 250 ml of 20× SSC; 1 ml of Tween 20; and 100 μg/ml purified yeast RNA (Roche). LNA probes (Exiqon) were diluted to 1.6 μm in prehybridization buffer. Care was taken not to refreeze diluted LNA probes. For both miR-107 and miR-124 probes, hybridization was performed at 37°C. After overnight hybridization, which was performed with mechanical agitation, slides were washed at 37°C (twice with 5× SSC for 5 min, and then three times with 2× SSC in 50% formamide for 20 min each), followed by three washes in PBS with 0.1% Triton X-100 (PBST) at room temperature. Nonspecific antigen blocking was performed at 4°C in PBST with 2% sheep serum (Jackson ImmunoResearch, West Grove, PA) and 2 mg/ml RNase-free BSA [EMD (Omnipur)] for 2 h, followed by the addition of 1:1000 diluted AP-conjugated anti-digoxigenin antibody (Roche) and incubation overnight at 4°C. The next day, tissue is washed in PBST (four times 10 min each), and then in staining solution (10 mm Tris-HCl, pH 9, 50 mm MgCl2, 100 mm NaCl, 0.1% Tween 20) twice for 5 min. Nitroblue tetrazolium and bromochloroindoyl phosphate (Roche) at 50 mg/ml each were added, and the staining reaction was at room temperature with gentle movement for 8–24 h. miR-24 and miR-204 were used in this application because the former has nearly identical Tm with miR-107, and miR-204 has low expression in the CNS and a low Tm.
Northern blots were performed as described previously (Nelson et al., 2004) with some modifications. The amount of RNA loaded in each well was normalized to the amount of total RNA and tRNA levels as described previously (Tang et al., 2007).
For Western blots, 20 μg of total protein was loaded onto each lane. BACE1 monoclonal antibody (R&D Systems, Minneapolis, MN) was used at 1:1000. β-Actin monoclonal antibody (Rockland, Gilbertsville, PA) was used at 1:1000.
Bioinformatics: rna22-based predictions of putative miR-107 miRNA targets prediction in BACE1 mRNA 3′-untranslated region.
To identify a list of putative gene targets for the microRNAs of interest, we used the rna22 algorithm described previously (Miranda et al., 2006). One strength of rna22 is its ability to identify putative microRNA binding sites along the length of the entire mRNA transcript [i.e., in 3′-untranslated regions (UTRs) as well as 5′-UTRs and CDSs]. Moreover, rna22 does not impose any cross-genome conservation constraints when determining the targets of a miRNA. For the analyses described below, we focused only on the 3′-UTR of BACE1. Our studies also considered predictions contained in other publicly available databases of predicted miRNA targets, as described in Results.
Validation: reporter vector constructs, transfection, and dual luciferase assays.
The evaluation of putative miRNA recognition elements (MREs) in the 3′-UTR of mRNAs was performed as described previously (Kiriakidou et al., 2004; Nelson et al., 2004) with modifications. To experimentally test two of the predicted miR-107 MREs in the 3′-UTR of BACE1, reporter vectors bearing these binding sites were subcloned into the mammalian expression vector pRL-TK (Promega) carrying the Renilla luciferase gene (Rluc) as a reporter. Predicted miR-107 binding sites, designated B1 (aatttaagtcgggaaattctgctgcttga) and B2 (attcccactgcacttggtgctgctttggc), which reside on BACE1 (NM_012104.3) 3′-UTR between 2208 and 2245 and between 3741 and 3774, respectively, were inserted into the 3′-UTR of Rluc gene at the XbaI and NotI restriction sites as described previously (Kiriakidou et al., 2004; Nelson et al., 2004). Mutated MREs (B1mut, aatttaagtcgggaaattcttggccttga; B2mut, attcccactgcacttgcgttgcgtttggc) containing sequence modifications predicted to affect the “seed region” targeted by miR-107 were used as negative controls such that the overall A/G/C/T nucleotide composition was not changed between the B1/B1mut or B2/B2mut. To monitor the reporter system, hsa-Let-7 MRE on LIN28 (LIN28), and its mutated sequence (M2) were used for positive and negative controls as described previously (Kiriakidou et al., 2004; Nelson et al., 2004). Correct construct sequences were confirmed by sequencing the 3′-UTR of Rluc gene on all of the pRL-TK vectors.
HeLa cells (American Type Culture Collection, Manassas, VA) were cultured under the vendor's recommended conditions. For the luciferase reporter assay, HeLa cells were plated at a density of 4 × 105 per well in a 24-well plate a day before transfection. Two separate transfections were performed, because otherwise the plasmid and short interfering RNA (siRNA) transfections interfered with each other (data not shown). For the first transfection, 0.4 μg of Rluc-encoding pRLTK reporter plasmid along with 0.2 μg Firefly luciferase (Fluc)-encoding PGL3 (Promega) control plasmid were transfected using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's instructions. Transfected cells were washed in antibiotic-free media after 6 h of transfection, and the cells were allowed to recover overnight. Twenty-four hours after the first transfection, cells were further transfected with 250 nm of either miR-107 siRNA duplex (sequences: agcagcauuguacagggcuauca and auagcccuguacaaugcugcguu), miR-320 siRNA duplex (sequences: aaaagcuggguugagagggcgaa and cgcccucucaacccagcuuuguu), or only with transfection reagent mixture, using siRMAX (Invitrogen) according to manufacturer's instructions. Luciferase reporter activity analysis was performed 24 h after the siRNA transfection using Dual Luciferase Assay System (Promega) and a Turner 20/20n luminometer (Fisher Scientific) according to the manufacturers' instructions. To control for transfection efficiency, relative luciferase activity was calculated by normalizing the RLuc (pRL-TK) to the Fluc (PGL-3) readouts. Each experimental condition was performed in triplicate within individual experiments, and the results shown represent two independent experiments.
Results
miRNA microarray results
RNA was isolated from patients with different clinical and neuropathological features as shown in Table 1. Note the short PMIs and the detailed correlation with neuropathology from subjacent tissue. Cases were separated into four groups: nondemented with no/negligible AD-type pathology, nondemented with incipient AD pathology, MCI with moderate AD pathology, and AD. The expression patterns for most miRNAs were stable across the different clinical states. Approximately 200 different miRNAs were expressed at levels above background on this array, but only ∼70 miRNAs were detected at a high enough level to merit comparative studies.
Table 1.
Cases used with neuropathological correlation
| Temporal cortex |
Pathology |
Counts |
Braak stage | PMI (h) | Age | ||
|---|---|---|---|---|---|---|---|
| Diffuse plaques | Neuritic plaques | NFT | |||||
| AD | Case 1 | 47 | 15 | 17 | 6 | 3.5 | 87 |
| Case 2 | 34 | 16 | 31 | 6 | 3 | 93 | |
| Case 3 | 50 | 30 | 31 | 6 | 3.33 | 84 | |
| Case 4 | 38 | 23 | 11 | 6 | 2.33 | 75 | |
| Case 5 | 18 | 22 | 35 | 6 | 4.5 | 84 | |
| Case 6 | 50 | 20 | 14 | 6 | 2.75 | 90 | |
| Average | 40 | 21 | 23 | 6.0 | 3.2 | 85.5 | |
| MCI | Case 1 | 7 | 9 | 1 | 3 | 2.45 | 97 |
| Case 2 | 47 | 12 | 0 | 3 | 5 | 91 | |
| Case 3 | 0 | 0 | 0 | 3 | 2.75 | 93 | |
| Case 4 | 50 | 9 | 1 | 4 | 3.5 | 87 | |
| Case 5 | 22 | 14 | 0 | 3 | 2.9 | 82 | |
| Case 6 | 50 | 5 | 5 | 5 | 2 | 99 | |
| Average | 29 | 8 | 1 | 3.5 | 3.1 | 91.5 | |
| Nondemented | Case 1 | 4 | 6 | 0 | 1 | 4 | 91 |
| with SMT | Case 2 | 3 | 0 | 0 | 0 | 3.25 | 92 |
| plaques | Case 3 | 37 | 9 | 0 | 2 | 2.75 | 93 |
| Case 4 | 17 | 2 | 0 | 2 | 2.4 | 87 | |
| Case 5 | 3 | 2 | 0 | 0 | 3.5 | 95 | |
| Average | 13 | 4 | 0 | 1.0 | 3.2 | 91.6 | |
| Nondemented | Case 1 | 0 | 0 | 0 | 2 | 1.66 | 85 |
| without SMT | Case 2 | 0 | 0 | 0 | 1 | 5 | 88 |
| plaques | Case 3 | 0 | 0 | 0 | 1 | 2.25 | 80 |
| Case 4 | 0 | 0 | 0 | 1 | 1.75 | 86 | |
| Case 5 | 0 | 0 | 0 | 2 | 4 | 90 | |
| Case 6 | 0 | 0 | 0 | 1 | 2 | 76 | |
| Average | 0 | 0 | 0 | 1.3 | 2.8 | 84.2 |
Cases used for miRNA microarray involved detailed evaluation, both prior and subsequent to patients' death. Brain samples were obtained immediately subjacent to tissue that was used for detailed neuropathological evaluation. “Pathology counts” represent a quantitation of neuropathological lesions as described in detail previously (Nelson et al., 2007). SMT signifies the superior and middle temporal gyri. Note that, in these areas, there is a graded increase in pathology between the four groups. NFT, Neurofibrillary tangle.
Among the few miRNAs that were consistently changed in AD versus nondemented brains was miR-107 (Fig. 1). One-way ANOVA shows that miR-107 shows a highly significant decrease across the four groups at p = 0.014. Of particular interest was the unique decrease between miR-107 levels in nondemented patients without pathology versus MCI (Student's t test, p = 0.008), suggesting that decreased miR-107 expression occurs in the earliest stages of AD.
Figure 1.
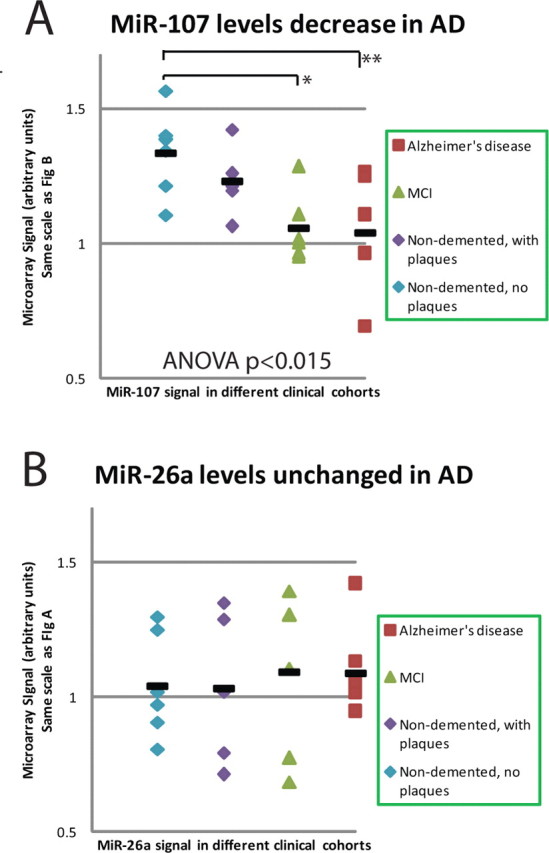
miRNA expression for a subset of miRNAs from temporal (superior and middle gyri) cortex. Note that miR-107 levels are decreased significantly already in MCI cases and trends toward a decrease in nondemented patients with incipient pathology. Although case-to-case variance is considerable, a trend is clearly discernible for miR-107. miR-26a was chosen for comparison because this miRNA has the closest expression to miR-107 on our microarray and was subject to very similar global normalization parameters. Hence, ordinate values are the same for A and B. The horizontal bars are the average for that condition. *p < 0.005; **p < 0.03, Student's one-tailed t tests.
Other miRNAs differed between nondemented and AD patients but did not appear changed in the subclinical stages of AD. miR-103 is a close paralog of miR-107, differing only at a single 3′ base. The expression of miR-107 was ∼20% higher than miR-103; however, as shown in Table 2, R2 (correlation coefficient) for miR-107 and miR-103 readout for the microarray was >0.91. Because the levels of miR-103 on the microarray closely mirrored miR-107, the miR-103 signal probably represents cross-hybridization as would be expected on a Northern blot. In addition to a decreased expression of miR-107 in temporal cortex, a trend for decrease in miR-107 was also seen in motor cortex in AD brains (data not shown) (one-way ANOVA for motor cortex is p = 0.050; for motor cortex, n = 19 different brains were assessed). These changes seem to indicate global change in brain miR-107 levels, even in areas less affected by AD pathology. Among other highly expressed miRNAs, ANOVA analyses indicate that miR-23b was also expressed at lower levels in AD brains than nondemented brains (p = 0.015).
Table 2.
Correlations between the expression of miRNAs and other genes
| Gene(s) assignment | R2 correlation coefficient versus miR-107 | R2 correlation coefficient versus miR-103 | R2 correlation: other intronic miRNAs versus mother mRNAs |
|---|---|---|---|
| miRNA microarray | |||
| miR-103 | (+) 0.912 | ||
| Exon array microarray | |||
| NM_148978//PANK1 | (+) 0.06 | (+) 0.14 | |
| NM_024960//PANK2 | (−) 0.071 | (−) 0.061 | |
| NM_024594//PANK3 | (+) 0.16 | (+) 0.236 | |
| miR-126 versus EGLF7 | (−) 0.11 | ||
| miR-128a versus R3HDM1 | (+) 0.005 | ||
| miR-342 versus EVL | (−) 0.006 |
The degree to which the expression of different miRNAs is correlated with expression of other genes is described for miR-103, miR-107, and other intronic miRNAs. The detected expressions of miR-103 and miR-107 are very highly correlated on our microarray, which may represent cross-hybridization. The other comparisons show the correlation between the expression of various miRNAs, using the miRNA microarray, versus the expression of genes in which those miRNAs are introns (n = 11 using a Affymetrix microarray as for Fig. 7). miR-107 resides in PANK1; paralogous miR-103 genes are present within PANK2 and PANK3; miR-126 is intronic within EGFL7, as are miR-128a and miR-342 within R3HDM1 and EVL, respectively. The (+) preceding the R2 numbers indicates a positive correlation, whereas (−) indicates a negative correlation. None of the correlations between intronic miRNAs and their mother mRNAs were statistically significant; an ANOVA to test the significance of the regression between miR-103 and PANK-3 mRNA levels was the lowest at 0.12.
We decided to focus on changes in miR-107 because these appeared to have the greatest statistical significance and emerged very early in the progression of the pathology. Demented controls are important to assess the specificity of the changes seen in AD. For these controls, isocortex from patients with DLB (n = 5) were used, in comparison with nondemented patients (n = 5) and those with pure AD (n = 2; positive control). A statistically significant drop in miR-107 was not seen in the DLB cases. A one-way Student's t test for miR-107 levels in DLB versus nondemented brains was p > 0.29.
Validation: Northern blots and in situ hybridization
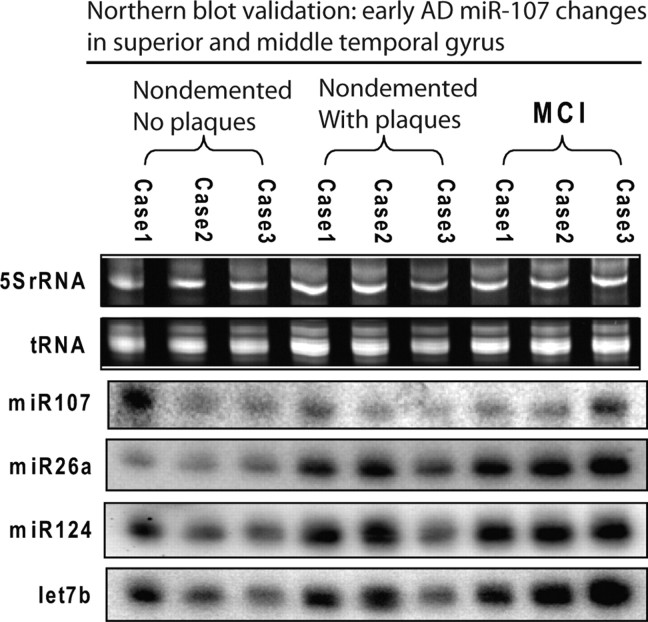
Microarray results were validated and extended using Northern blots and ISH. For initial post hoc validation Northern blotting (Fig. 2), we chose to evaluate the earlier stages of the disease (nondemented with no pathology, nondemented with mild or moderate pathology, and mildly cognitively impaired with moderate pathology) because this is where the miR-107 decrease was most significant and where the cellular populations are most comparable. The findings by Northern blot were consistent with those on the microarray; a number of miRNAs showed increased expression in the early progression of AD, but miR-107 levels decrease beginning early in the pathological evolution of the disease.
Figure 2.
Northern blots were used to validate the microarray data indicating that miRNA expression changes are seen in early AD pathogenesis. These results show that miR-107 levels are decreased early in the development of AD, whereas the expression of other miRNAs trend toward a slight increase. The miRNAs were chosen to represent a gamut of high and medium-expression level miRNAs.
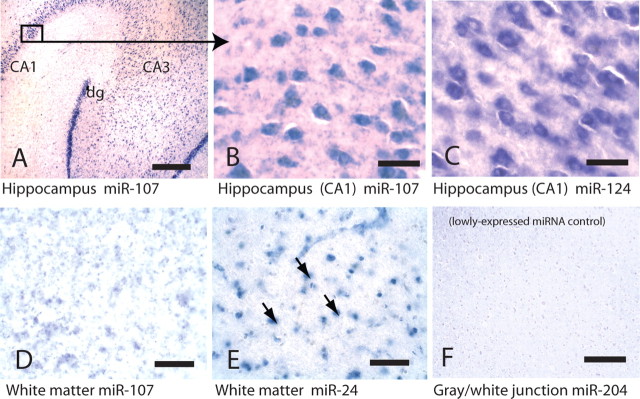
In situ hybridization (Figs. 3–5) revealed that miR-107 is expressed strongly in human neurons, as we have shown miR-124 to be previously (Nelson et al., 2006). In addition, the miR-107 probe labels some cells that by morphology appear to be astrocytes. However, other miRNAs such as miR-24 stain white matter, which includes glial cells with far fewer neurons, more strongly than miR-107 does (Fig. 3D,E). Different miRNAs appear to stain particular layering patterns in the hexalaminar isocortex of the superior and middle temporal (SMT) gyrus. Specifically, ISH with a miR-124 probe labels the superficial (layers I/II) and the deeper (layers IV/V) neuronal lamina. Even in relatively advanced AD, the pattern of miR-124 ISH is maintained. In a demented control brain with extensive cerebral amyloid angiopathy, miR-107 showed an identical staining pattern with distinct superficial and deep layer staining. In contrast, in brain sections with extensive AD pathology, miR-107 ISH still shows the superficial layers are stained; however, the deeper layers show a marked decrease in miR-107 hybridization (Fig. 4). This decrease of miR-107 in deep layers was observed also in a single nondemented case in which there was significant AD pathology (nondemented case 3 in Materials and Methods with pathology showing probable AD) (data not shown), an observation that correlates with the microarray data showing decreased miR-107 expression early in the pathology (Fig. 1). The cortical lamina with decreased miR-107 ISH signals coincide with cerebral cortical lamina where the neuropathological lesions of AD (thioflavine S-positive plaques and tangles) are seen (Fig. 4). In the perirhinal cortex, an isocortical (six-layered) region affected early by AD, ISH was performed on brain tissue from two elderly nondemented cases without appreciable pathology and compared with two AD cases (Fig. 5 shows representative photomicrographs from AD case 3 and nondemented case 1 described in Materials and Methods). In the AD cases, miR-124 remained strongly expressed in the surviving neurons, whereas miR-107 levels were decreased. In contrast, in the nondemented controls, miR-107 signal was present at a qualitatively higher level. These cases were different from the cases in Figure 1 or 5, thus providing cellular level insights, and independent replication.
Figure 3.
ISH performed on human brains shows that miR-107 is enriched in neurons. A is a photomicrograph of a nondemented patient's hippocampus at low power probed for miR-107 and shows CA1 and CA3 as well as the dentate gyrus (dg). B and C show higher-power photomicrographs, from the area boxed in 3A, with miR-107 (B) and miR-124 (C), another neuron-enriched miRNA. miR-124 ISH was performed on a near-serial brain section. D, Unlike miR-124, miR-107 also stains white matter, albeit indistinctly in most cases. E, However, other miRNAs such as miR-24 show much more distinct glial profiles (arrows) in white matter. F shows a probe for the control miR-204, which was chosen because it has low CNS expression on microarray but has a low Tm. Scale bars: A, F, 1 mm; B–E, 100 μm.
Figure 4.
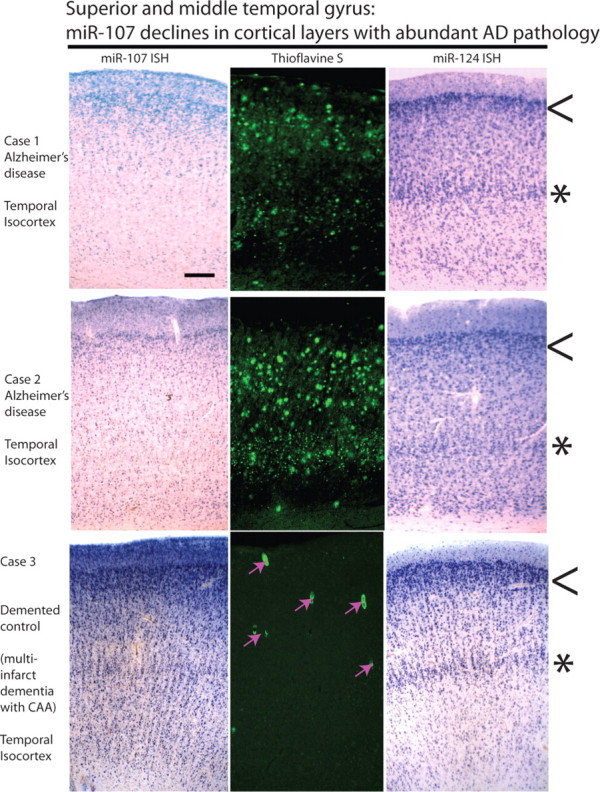
ISH-stained cells and AD-type neuropathology each show cerebral cortical lamina-specific staining patterns in human brains that differ between AD and control. These experiments were performed on superior and middle temporal gyri using sections cut at 25 μm. Photomicrographs depict near-serial sections through the middle temporal gyri of three human brains; pial surface is up. Thioflavine S shows green-fluorescent amyloid and neurofibrillary tangles. The top two cases were from AD brains, and the bottom was from a profoundly demented stroke patient (mini-mental status examination score, 0); demented control case, Thioflavine S shows cerebral amyloid angiopathy (CAA) with amyloid in small blood vessels (pink arrows). Note that the superficial layers (lamina II; arrowheads) appear relatively spared in AD brain; however, in AD brain the deeper layers (laminas IV/V; asterisks) are severely depleted for miR-107 (left) but not miR-124 (right). The deeper, miR-107-depleted laminas are vulnerable to AD pathology, compatible with the hypothesis that miR-107 depletion is associated with the development of AD pathology. Scale bar: Top left, 500 μm (for photomicrographs).
Figure 5.
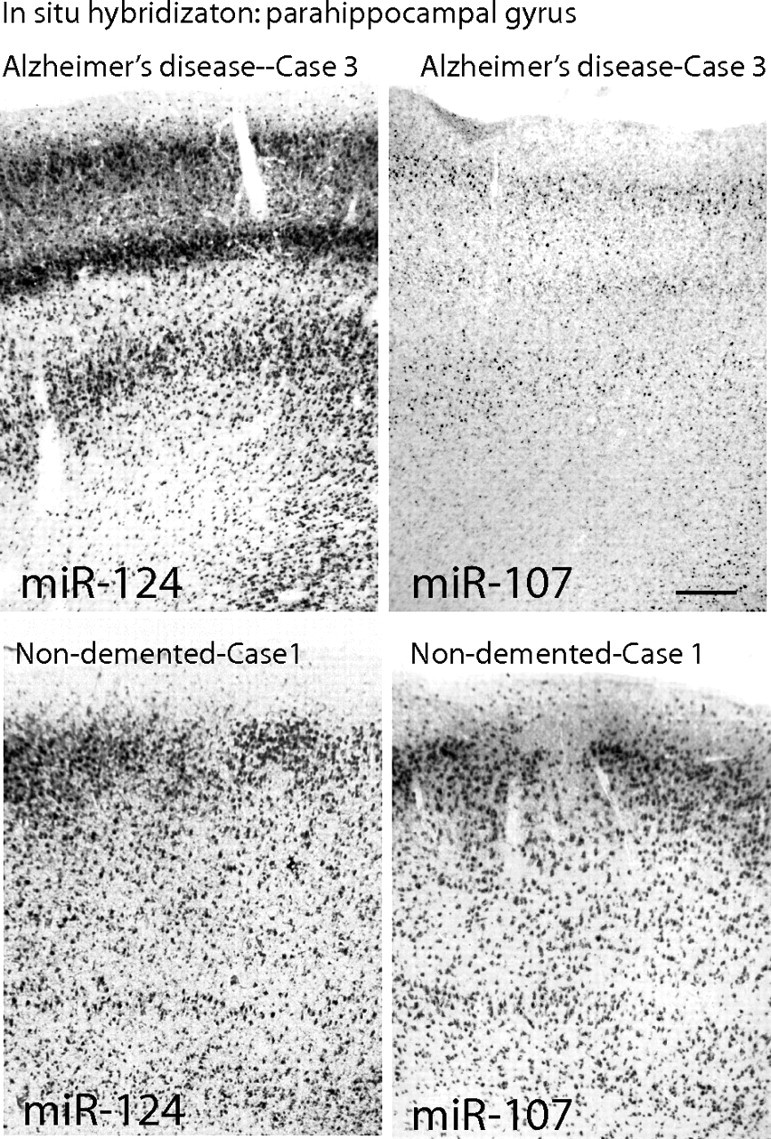
ISH on perirhinal gyrus comparing nondemented elderly control and AD on near-serial sections again shows that miR-107 levels are decreased in deep cortical lamina of AD brain. These are from different patients from those in Figure 4, and the result was replicated on other patients (see Results). Whereas miR-124 ISH signal remains strong in the neurons that survive AD neurodegeneration, miR-107 signal is qualitatively lower in neurons in AD brain. In contrast, in the nondemented elderly controls, both the neuronal miR-107 and miR-124 ISH signals are more robust. Scale bar, 500 μm (for all four photomicrographs).
miR-107 and BACE1: bioinformatic predictions
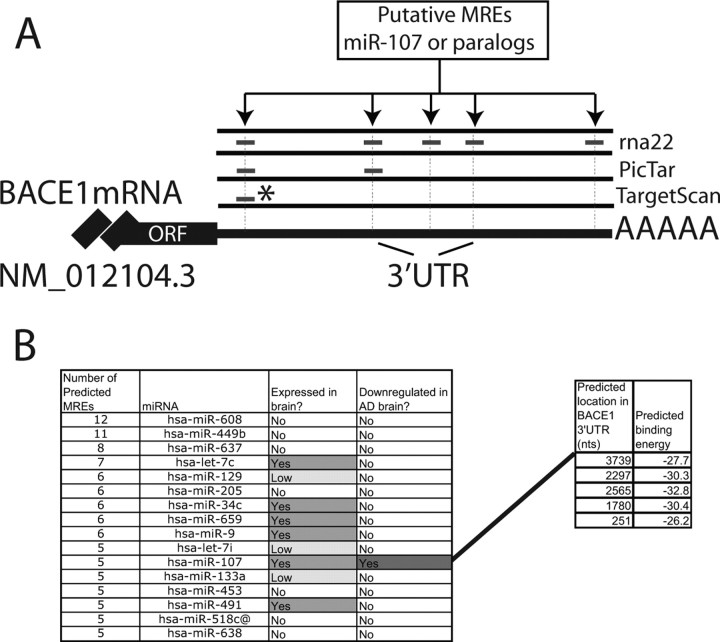
Having identified miR-107 as the miRNA with the most significant expression decrease early in the progression of AD, we sought to determine whether this change would have any impact on AD-related genes including BACE1, BACE2, APP, and SNCA, and others. We used the rna22 algorithm (Miranda et al., 2006) to examine the corresponding 3′-UTRs for potential miR-107 binding sites. This algorithm was chosen because rna22 does not rely on cross-genome sequence conservation to report putative binding sites for the microRNA under consideration. Using the default parameter settings (Miranda et al., 2006), rna22 identified five binding sites for miRNA miR-107 in the 3′-UTR of BACE1 mRNA (Fig. 6A). Taking into account additional experimental evidence (discussed below), which indicates that BACE1 is differentially expressed (upregulated) in AD, we also considered the possibility that miRNAs other than miR-107 may be involved in the regulation of this gene. Analysis of the 3′-UTR of BACE1 using rna22 indicates that at least 25 microRNAs from among those listed in Release 9 of miRBase (Griffiths-Jones, 2006) have three or more putative binding sites (Fig. 6B). More than one-half of the miRNAs that rna22 predicts target BACE1 are expressed in brain tissue; however, in view of our differential expression profiling studies, we decided to focus on the interactions between miR-107 and BACE1 for this first analysis. Detailed provision of the predicted miRNA:mRNA pairings are presented as supplemental file 2 (available at www.jneurosci.org as supplemental material). We also compared the predictions of rna22 to those made by Pictar (Krek et al., 2005) and TargetScan (Lewis et al., 2003). Across all the algorithms, we found that the 3′-UTR of most AD-related genes were not predicted to be targeted by miR-107 (data not shown). With regard to BACE1, one of the five sites predicted by rna22, BACE1 (NM_012104.3) 3′-UTR between mRNA nucleotides 2208 and 2245, was also reported by PicTar and TargetScan, although the latter two predicted the site would be recognized by the paralogous miRNAs including miR-16, which have the identical seed sequence as miR-107. The MREs predicted by the various programs and their relative locations are shown in Figure 6.
Figure 6.
A, A graphic depiction of BACE1 3′-UTR sequence showing the location of the MREs that were predicted by rna22 and several other public-access miRNA target prediction databases. The asterisk denotes sequences that were predicted to be recognized by homologous miRs (miR-15 family) that share the same seed sequence as miR-107. B, A partial list of microRNAs that rna22 predicts target the BACE1 3′-UTR; note that the list includes only microRNAs that are predicted to target the BACE1 3′-UTR at three or more sites. Also shown are the starting locations for the five miR-107 binding sites that are predicted by rna22.
Correlating miRNA expression with mRNA expression profiling microarrays and Western blot results
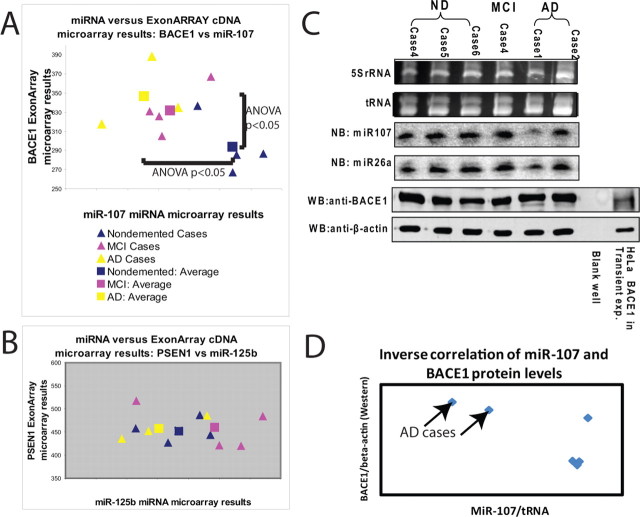
Because numerous recent studies have shown that changes in miRNA levels lead to altered levels of mRNA targets (Lim et al., 2005; Wang and Wang, 2006; Grimson et al., 2007), we performed correlative mRNA studies on a subset of the cases of nondemented (n = 4), MCI (n = 4), and AD (n = 3) brains using the same RNA extracted from temporal cortex and used in the miRNA microarray studies. The results of these experiments with regard to BACE1 and miR-107 expression are shown in Figure 7. These experiments show a clear trend: cases with lower miR-107 (which tend to have more AD pathology) tend to have higher BACE1 mRNA than cases with higher miR-107 and vice versa. Separate, independent one-way ANOVA results indicate that both the increase in BACE1 mRNA as well as the decrease in miR-107 levels were statistically significant (p < 0.05). For comparison purposes, we show analogous results for presenilin (PSEN1) and for miR-125b in which such trends were absent (Fig. 7B).
Figure 7.
At both the mRNA (A, B) and protein (C, D) levels, decreased miR-107 levels correlate with increased BACE1 levels in AD pathological progression. Cross-correlation was performed between cDNA microarrays (Affymetrix Exon Array) and the miRNA microarray to assess the correlation between changes in miR-107 and in BACE1 mRNA in individual cases. RNA was isolated from temporal cortex and processed in parallel using the two expression arrays. Results indicate that, in the progression of AD pathology, miR-107 levels tend to decrease and BACE1 mRNA levels increase such that AD cases (yellow; n = 3) have high BACE1 mRNA and low miR-107 levels, nondemented cases (blue; n = 4) have low BACE1 mRNA levels and high miR-107 levels, and MCI cases (pink; n = 4) are in the intermediate. For the sake of comparison, miR-125b and PSEN1 are chosen; miR-125b was the highest-expressing miRNA in this group, and PSEN1 is known to be related to AD pathology. The triangles indicate individual cases, and the squares are the average for a condition. Separately, six different human cases were compared for miR-107 [Northern blots (NB)] and BACE1 protein [Western blots (WB)]. These results are shown in C. The miR-107 signals were normalized to tRNA and the BACE1 Western blot results normalized to β-actin, and these results are shown in D. Note that the cases with a high relative amount of BACE1 and low miR-107 are AD cases.
Having performed miRNA- and mRNA-related profiling on the same 11 total RNA samples, we were able to also test the correlation between the levels of intronic miRNAs (such as miR-107 and the paralogous miR-103s) and those of the mRNAs of their “mother” genes. These results are shown in Table 2. The correlations of expression were highest for both miR-107 and miR-103 to PANK3 > PANK1 > PANK2. This was surprising because miR-107 is present in PANK1. Nonetheless, as with the correlations for other intronic miRNAs and their mother genes, as shown in Table 2, none was statistically significant.
mRNA and protein levels are frequently discrepant (Nelson and Keller, 2007). Hence, we performed post hoc studies of six additional aged human patients that were diagnosed as nondemented, MCI, or AD, to correlate the expression of miR-107 and BACE1 protein as detected by Northern and Western blots, respectively. These results are shown in Figure 7, C and D. These results show that, at the protein level, as well as the mRNA level, a decrease of miR-107 and an increase in BACE1 expression tend to be seen in AD brains.
Validation: tissue culture miRNA recognition element studies
Guided by the computational predictions presented above, we focused on the two MREs that were consistently predicted by at least the rna22 and PicTar methods. Their sequences are designated B1 and B2, respectively (Fig. 8). Each of the two MREs was subcloned separately into pRL-TK expression plasmids and assessed in HeLa cells to determine its physiological relevance. These experiments depend partly on endogenous miRNAs, and miR-107 was originally described in a study of HeLa cells (Mourelatos et al., 2002). Figure 8 shows the two sites B1 and B2 highlighted and also shows the sequences used and the results of two different experiments (n = 3 replicates each). A third experiment (n = 3 replicates) showed the same result (data not shown). To establish valid negative controls for particular MRE sequences, we scrambled the seed region sequences of the B1 and B2 MREs, without changing the A/G/C/T content of the plasmid. The highly similar control sequences were subcloned into separate pRLTK plasmids. Compared with the control sequences, the reporter assays showed that Rluc activity of the reporter carrying B1 MRE was clearly reduced as the result of endogenous miRNAs (Student's t test, p < 0.0001). This reduction was further augmented by the addition of miR107 siRNAs, indicating the specificity of miR-107 action on this MRE. To confirm that siR107 action is specific, the unrelated siR320 was included in the experiment. Addition of siR320 did not further decrease Rluc activity as observed with siR107, but increased luciferase activity relative to control, which may indicate a separate regulatory mechanism or a possible compromise of miRNA function. Unlike MRE B1, the subcloning of MRE B2 into the 3′-UTR of pRLTK did not have any effect on Rluc activity, suggesting that B2 putative MRE is probably not an miRNA recognition element or that it acts as a MRE only in a specific context. In these experiments, the very strong lin-28 MRE recognized by endogenous let-7 miRNAs and its scrambled M2 were used as controls to monitor the reporter assay system. The activity of the luciferase reporter bearing lin-28 MRE was dramatically reduced as result of endogenous let-7 expression in HeLa cells as seen previously (Kiriakidou et al., 2004). Note that neither siR107 nor siR320 had any effect on the luciferase activity of lin28 reporter.
Figure 8.
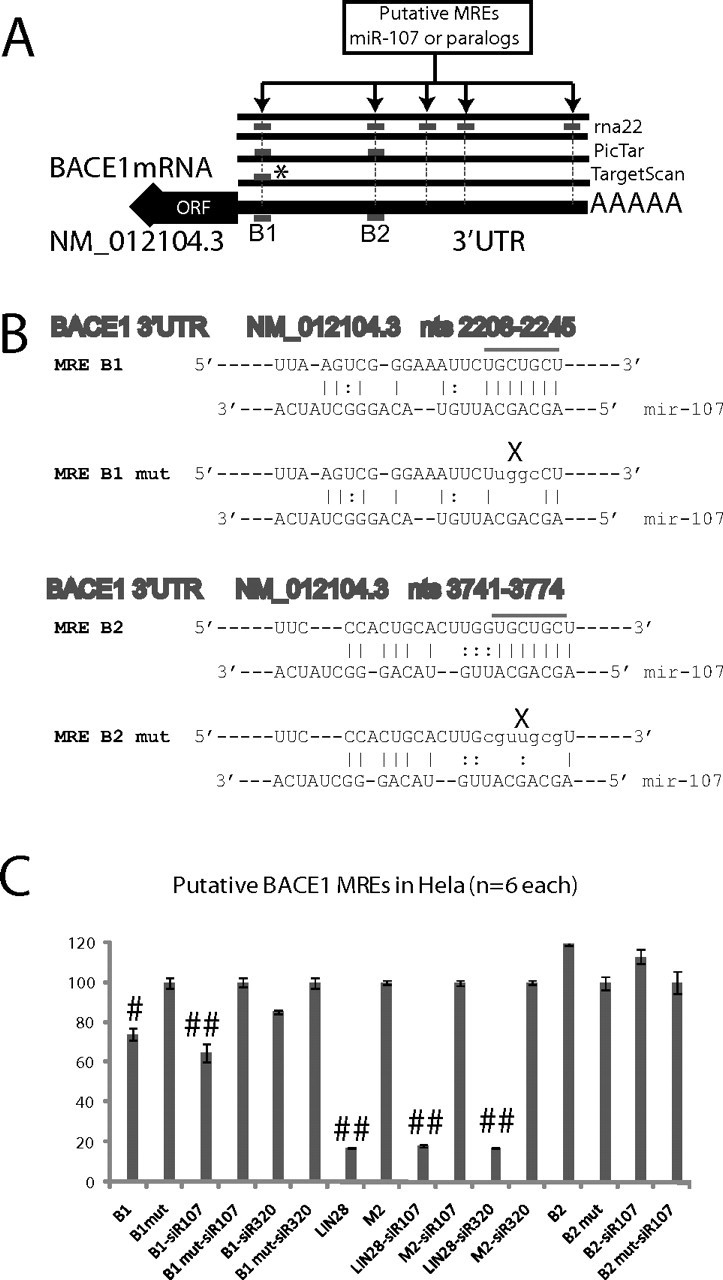
A, Tissue culture validation was performed using HeLa cells to demonstrate whether two of the putative MRE sequences predicted by multiple algorithms on BACE1 are recognized physiologically by miR-107. These results indicate that, of the two tested putative MREs, one (B1) is physiologically relevant but not the other (B2). B, For this experiment, six different MREs were subcloned into pRLTK expression plasmids, along with PGL-3 control plasmids, to understand whether the MREs (designated B1 and B2) are changed as a result of endogenous miRNAs. Furthermore, siRNAs were transfected that mimic miRNAs. B1 and B2 each were compared versus appropriate controls that were matched except for a small shifting of nucleotides at the predicted seed-pair region. In addition, known positive and negative controls were used, namely LIN-28 and M2, the former a very strong MRE recognized by endogenous let-7 miRNAs as described previously (Kiriakidou et al., 2004, 2005). Luciferase experiment results are presented in C. The bar signals shown to the right are the result of six different experiments (n = 3 performed twice); the signals represent the Renilla luciferase signal that has been normalized using the Firefly luciferase as a transfection efficiency control. Note that miR-107 siRNAs cause a statistically significant additional drop in the reporter construct expression, but miR-320 actually caused an apparent lack of inhibition on the B1 MRE. For C, Student's t tests were performed. ##p < 0.0001; #p < 0.01. For the siR-107 experiments, # refers to the statistical significance of the difference in the overall reporter decrease with, versus without, siR-107 transfection.
Discussion
miR-107 levels are decreased in human cerebral cortical gray matter early in the pathological progression of AD, as shown by expression profiling using microarrays and Northern blots. In situ hybridization on human cerebral cortex demonstrated that miR-107 is strongly expressed in neurons with a characteristic laminar expression pattern. Bioinformatics approaches predict that as many as five different sites on the 3′-UTR of BACE1 mRNA may be targeted by miR-107. Luciferase reporter assays directly support the hypothesis that miR-107 regulates BACE1 through at least one discrete 3′-UTR MRE. Together, these data indicate that the change in neuronal miR-107 expression contributes to the pathogenesis of AD.
There are several limitations to our data. The microarray we used was unable to differentiate specifically between the paralogs miR-103 and miR-107, which differ by a single nucleotide near their 3′ ends. Nonetheless, the signal levels referent to miR-107 spots were consistently higher than miR-103 on the arrays, suggesting that miR-107 (and not miR-103) was likely the predominant miRNA detected. In any case, because of their extreme sequence identity and the location of their single-nucleotide difference, the target prediction algorithms would have generated the same list of putative binding sites. Previous studies have indicated that miRNA hybridization is not specific to the level of a single-nucleotide difference at that part of the molecule (Kiriakidou et al., 2004; Goff et al., 2005). However, this issue will be addressed in the immediate future. We also were surprised to find a very low detection of miR-9 on this microarray platform. We found discrepancies previously in miR-9 detection, which were not linked to our microarray platform, and may indicate inherent difficulty in phosphorylating or ligating to the UCUUU 5′ sequence of miR-9 (data not shown). Notwithstanding this technical shortcoming, the total number of miRNAs expressed above background is quite compatible with the previous findings of other brain miRNA microarray studies (Baskerville and Bartel, 2005; Gaur et al., 2007; Tang et al., 2007).
In addition to potential cross-reaction on this specific microarray platform, there are other caveats that pertain to most human studies. When performing human brain studies, controlling for all relevant variables is virtually impossible. In the current study, we included important non-AD dementia controls, namely DLB. Nonetheless, it is possible that different patient groups will be treated subtly differently, or manifest distinct comorbidities and/or behaviors that cause different environmental conditions (such as circadian rhythms and agonal states/medicines) that might induce bias in miRNA expression patterns.
Our results indicate a regulatory mechanism involving molecules, miR-107 and BACE1, heretofore not known to interact. Between those molecules, far less is known about miR-107. The nearly identical paralogs miR-107 and miR-103 (Mourelatos et al., 2002) (miR-103/7) have been shown to be expressed in many mammalian organs with highest concentration in brain tissue (Babak et al., 2004; Baskerville and Bartel, 2005; Wang and Wang, 2006). miR-103/7 are differentially expressed in a number of contexts, including during development (Esau et al., 2004; Miska et al., 2004), neoplastic transformation (Roldo et al., 2006), hypoxia (Kulshreshtha et al., 2007), and both cold and heat stress (Marsit et al., 2006; van Rooij et al., 2006).
Intriguingly, miR-107 expression may relate to the coordinated regulation of a pathway related to lipid metabolism (Wilfred et al., 2007). The DNA templates for miR-107 and miR-103 each exist within introns in the genes that encode the pantothenate kinase (PANK) enzymes. In humans, there are at least three highly homologous PANK genes. miR-103(1), miR-103(2), and miR-107 reside within PANK3, PANK2, and PANK1 genes, respectively (Ying and Lin, 2005). The basic arrangement of these miRNAs in PANK genes remains constant through 500 million years of vertebrate evolution (Wilfred et al., 2007). PANK enzymes serve a universal catalytic function, namely, the phosphorylation of pantothenate (vitamin B5) (Tahiliani and Beinlich, 1991). This is the rate-limiting step in generating coenzyme A (CoA), and PANKs are central enzymes in the regulation of cellular CoA levels (Leonardi et al., 2005). CoA is a necessary cofactor in ∼4% of known enzymes, including >100 reactions involved in metabolism (Leonardi et al., 2005).
The location of miR-103/7 genes within introns of the PANK genes may be physiologically relevant. Expression levels of intronic miRNAs and their host genes are in some but not all circumstances well correlated, presumably because they are cotranscribed by RNA polymerase II (Baskerville and Bartel, 2005; Ying and Lin, 2005, 2006); however, we find that in our comparison of miRNA and mRNA microarrays, the correlations for intronic miRNAs and mother mRNAs was (from a statistical standpoint) nonexistent. We speculate that the downstream miRNA processing is differentially regulated, or, conceivably, these intronic miRNAs may be transcribed separately from the mRNAs. Additional work remains to be performed in terms of the regulation of processing in the context of intronic miRNAs.
BACE1 cleaves the β-amyloid precursor protein creating the neurotoxic β-amyloid peptide Aβ1–42 (Vassar et al., 1999; Haniu et al., 2000). The dysregulation of BACE1 appears to directly contribute to the pathogenesis of AD (Dominguez et al., 2004; Durham and Shepherd, 2006; Guo and Hobbs, 2006; John, 2006). Our ISH show that miR-107, like BACE1 (Bigl et al., 2000; Fukumoto et al., 2002), is enriched in neurons. A striking laminar staining pattern is observed for miRNAs miR-107, miR-124, and AD neuropathology alike. Laminas with evident decreased miR-107 relative to miR-124 signal are vulnerable to developing AD pathology. This correlation is by no means proof of the hypothesis that decreases in miR-107 levels cause increased vulnerability to AD. Nonetheless, it is suggestive and demonstrates the importance of complementing tissue-level profiling modalities with ISH.
Our computational analyses indicate that miR-107 may target BACE1 mRNA posttranscriptionally through as many as five distinct binding sites in the 3′-UTR. One of these five sites was common across three different target prediction algorithms. However, it is entirely likely that the action of miR-107 on BACE1 is also effected by one or more of the other predicted sites. Indeed, experimental evidence, using reporter constructs with subcloned MREs in HeLa cells, corroborates the computational predictions that endogenous miR-107 participates in the regulation of BACE1. These reporter constructs have been useful in previous description of miRNA:mRNA interactions (Kiriakidou et al., 2004, 2005, 2007; Nelson et al., 2004). Our data suggest that at least one of the putative miR-107 MREs is physiologically relevant, albeit less potent than the very strongly inhibitory let-7 MRE. Previous studies have supported a role for miRNAs in biologically important partial, as opposed to complete, suppression of target mRNAs (Sevignani et al., 2006; Ying et al., 2006; Li et al., 2007).
BACE1 genetic regulation is complex (Preece et al., 2003; Lahiri et al., 2006; Li et al., 2006; Rossner et al., 2006; Mihailovich et al., 2007; Stockley and O'Neill, 2007; Tesco et al., 2007) and has been shown to occur partly posttranscriptionally (Marambaud et al., 1998; Rossner et al., 2006). Different regulatory mechanisms probably overlap in time and in cytoplasmic domains with multiple transcriptional, translational, and posttranslational processes involved in BACE1 regulation. Because a pharmacological regulation of BACE1 endopeptidase activity at its active site has proved a challenge (Beher and Graham, 2005; Hull et al., 2006; Ohno, 2006), each of these regulatory mechanisms represent an alternative potential “handle” for altering BACE1 expression pharmacologically.
Previous studies have suggested roles for miRNAs in neurodegenerative disease. Recent reviews have summarized these data (Hebert and De Strooper, 2007; Nelson and Keller, 2007; Singh, 2007). One previous study has shown that particular miRNAs are altered in CA1 hippocampal cells of AD; miR-124, for example, is increased (Lukiw, 2007). Our findings show a trend toward increases in miR-124 levels in AD, but this trend is not statistically significant. Another previous study has shown that a miRNA may be relevant in Parkinson's disease and to normal dopaminergic function (Kim et al., 2007). Furthermore, studies in mammals and in invertebrates have suggested that miRNAs may play a role in neuroprotection (Bilen et al., 2006a,b). However, there have been as yet no data suggesting a direct genetic link between miRNAs or miRNA recognition elements and neurodegenerative disease. Together, these observations may indicate that neurodegenerative diseases like AD, which takes place over multiple decades rather than just years (Scarmeas et al., 2005), tend to reflect a “downward spiral” effect of multiple processes, and miRNAs may play a contributory role.
Footnotes
This work was supported by National Institutes of Health (NIH) Grants K08 NS050110, P20 RR16481, and P30 AGO 28383. We thank members of the Sanders-Brown Center on Aging, and we are deeply grateful to all of the participants in our longitudinal aging study and to the patients in the University of Kentucky Alzheimer's Disease Center's research clinic. We also thank Ela Patel and Ann Tudor for technical support and Paul Thomason for help with preparation of this manuscript. We thank Dr. Kuey Chen for assistance with the Affymetrix Exon Arrays, and Dr. Don Baldwin and Robert Benjamin Isett for assistance in miRNA microarrays. We thank Dr. M. Paul Murphy for advice about BACE1 biochemistry. Finally, we are grateful to Dr. William Markesbery for his support in this project.
References
- Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- Babak T, Zhang W, Morris Q, Blencowe BJ, Hughes TR. Probing microRNAs with microarrays: tissue specificity and functional inference. RNA. 2004;10:1813–1819. doi: 10.1261/rna.7119904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskerville S, Bartel DP. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA. 2005;11:241–247. doi: 10.1261/rna.7240905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beher D, Graham SL. Protease inhibitors as potential disease-modifying therapeutics for Alzheimer's disease. Expert Opin Investig Drugs. 2005;14:1385–1409. doi: 10.1517/13543784.14.11.1385. [DOI] [PubMed] [Google Scholar]
- Bigl M, Apelt J, Luschekina EA, Lange-Dohna C, Rossner S, Schliebs R. Expression of beta-secretase mRNA in transgenic Tg2576 mouse brain with Alzheimer plaque pathology. Neurosci Lett. 2000;292:107–110. doi: 10.1016/s0304-3940(00)01452-x. [DOI] [PubMed] [Google Scholar]
- Bilen J, Liu N, Burnett BG, Pittman RN, Bonini NM. MicroRNA pathways modulate polyglutamine-induced neurodegeneration. Mol Cell. 2006a;24:157–163. doi: 10.1016/j.molcel.2006.07.030. [DOI] [PubMed] [Google Scholar]
- Bilen J, Liu N, Bonini NM. A new role for microRNA pathways: modulation of degeneration induced by pathogenic human disease proteins. Cell Cycle. 2006b;5:2835–2838. doi: 10.4161/cc.5.24.3579. [DOI] [PubMed] [Google Scholar]
- Ciafre SA, Galardi S, Mangiola A, Ferracin M, Liu CG, Sabatino G, Negrini M, Maira G, Croce CM, Farace MG. Extensive modulation of a set of microRNAs in primary glioblastoma. Biochem Biophys Res Commun. 2005;334:1351–1358. doi: 10.1016/j.bbrc.2005.07.030. [DOI] [PubMed] [Google Scholar]
- Ding Q, Markesbery WR, Cecarini V, Keller JN. Decreased RNA, and increased rna oxidation, in ribosomes from early Alzheimer's disease. Neurochem Res. 2006;31:705–710. doi: 10.1007/s11064-006-9071-5. [DOI] [PubMed] [Google Scholar]
- Dominguez DI, Hartmann D, De Strooper B. BACE1 and presenilin: two unusual aspartyl proteases involved in Alzheimer's disease. Neurodegener Dis. 2004;1:168–174. doi: 10.1159/000080982. [DOI] [PubMed] [Google Scholar]
- Durham TB, Shepherd TA. Progress toward the discovery and development of efficacious BACE inhibitors. Curr Opin Drug Discov Devel. 2006;9:776–791. [PubMed] [Google Scholar]
- Esau C, Kang X, Peralta E, Hanson E, Marcusson EG, Ravichandran LV, Sun Y, Koo S, Perera RJ, Jain R, Dean NM, Freier SM, Bennett CF, Lollo B, Griffey R. MicroRNA-143 regulates adipocyte differentiation. J Biol Chem. 2004;279:52361–52365. doi: 10.1074/jbc.C400438200. [DOI] [PubMed] [Google Scholar]
- Fukumoto H, Cheung BS, Hyman BT, Irizarry MC. Beta-secretase protein and activity are increased in the neocortex in Alzheimer disease. Arch Neurol. 2002;59:1381–1389. doi: 10.1001/archneur.59.9.1381. [DOI] [PubMed] [Google Scholar]
- Gaur A, Jewell DA, Liang Y, Ridzon D, Moore JH, Chen C, Ambros VR, Israel MA. Characterization of microRNA expression levels and their biological correlates in human cancer cell lines. Cancer Res. 2007;67:2456–2468. doi: 10.1158/0008-5472.CAN-06-2698. [DOI] [PubMed] [Google Scholar]
- Ginsberg SD, Crino PB, Lee VM, Eberwine JH, Trojanowski JQ. Sequestration of RNA in Alzheimer's disease neurofibrillary tangles and senile plaques. Ann Neurol. 1997;41:200–209. doi: 10.1002/ana.410410211. [DOI] [PubMed] [Google Scholar]
- Goff LA, Yang M, Bowers J, Getts RC, Padgett RW, Hart RP. Rational probe optimization and enhanced detection strategy for microRNAs using microarrays. RNA Biol. 2005;2:93–100. doi: 10.4161/rna.2.3.2059. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S. miRBase: the microRNA sequence database. Methods Mol Biol. 2006;342:129–138. doi: 10.1385/1-59745-123-1:129. [DOI] [PubMed] [Google Scholar]
- Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo T, Hobbs DW. Development of BACE1 inhibitors for Alzheimer's disease. Curr Med Chem. 2006;13:1811–1829. doi: 10.2174/092986706777452489. [DOI] [PubMed] [Google Scholar]
- Haniu M, Denis P, Young Y, Mendiaz EA, Fuller J, Hui JO, Bennett BD, Kahn S, Ross S, Burgess T, Katta V, Rogers G, Vassar R, Citron M. Characterization of Alzheimer's beta-secretase protein BACE. A pepsin family member with unusual properties. J Biol Chem. 2000;275:21099–21106. doi: 10.1074/jbc.M002095200. [DOI] [PubMed] [Google Scholar]
- Hebert SS, De Strooper B. Molecular biology. miRNAs in neurodegeneration. Science. 2007;317:1179–1180. doi: 10.1126/science.1148530. [DOI] [PubMed] [Google Scholar]
- Honda K, Smith MA, Zhu X, Baus D, Merrick WC, Tartakoff AM, Hattier T, Harris PL, Siedlak SL, Fujioka H, Liu Q, Moreira PI, Miller FP, Nunomura A, Shimohama S, Perry G. Ribosomal RNA in Alzheimer disease is oxidized by bound redox-active iron. J Biol Chem. 2005;280:20978–20986. doi: 10.1074/jbc.M500526200. [DOI] [PubMed] [Google Scholar]
- Hull M, Berger M, Heneka M. Disease-modifying therapies in Alzheimer's disease: how far have we come? Drugs. 2006;66:2075–2093. doi: 10.2165/00003495-200666160-00004. [DOI] [PubMed] [Google Scholar]
- John V. Human beta-secretase (BACE) and BACE inhibitors: progress report. Curr Top Med Chem. 2006;6:569–578. doi: 10.2174/156802606776743084. [DOI] [PubMed] [Google Scholar]
- Kim J, Inoue K, Ishii J, Vanti WB, Voronov SV, Murchison E, Hannon G, Abeliovich A. A microRNA feedback circuit in midbrain dopamine neurons. Science. 2007;317:1220–1224. doi: 10.1126/science.1140481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiriakidou M, Nelson PT, Kouranov A, Fitziev P, Bouyioukos C, Mourelatos Z, Hatzigeorgiou A. A combined computational-experimental approach predicts human microRNA targets. Genes Dev. 2004;18:1165–1178. doi: 10.1101/gad.1184704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiriakidou M, Nelson P, Lamprinaki S, Sharma A, Mourelatos Z. Detection of microRNAs and assays to monitor microRNA activities in vivo and in vitro. Methods Mol Biol. 2005;309:295–310. doi: 10.1385/1-59259-935-4:295. [DOI] [PubMed] [Google Scholar]
- Kiriakidou M, Tan GS, Lamprinakin S, De Plannell-Saguer M, Nelson PT, Mourelatos Z. An mRNA m7G cap binding-like motif within human Ago2 represses translation. Cell. 2007;129:1141–1151. doi: 10.1016/j.cell.2007.05.016. [DOI] [PubMed] [Google Scholar]
- Kosik KS, Krichevsky AM. The elegance of the microRNAs: a neuronal perspective. Neuron. 2005;47:779–782. doi: 10.1016/j.neuron.2005.08.019. [DOI] [PubMed] [Google Scholar]
- Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, Rajewsky N. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- Kulshreshtha R, Ferracin M, Wojcik SE, Garzon R, Alder H, Agosto-Perez FJ, Davuluri R, Liu CG, Croce CM, Negrini M, Calin GA, Ivan M. A microRNA signature of hypoxia. Mol Cell Biol. 2007;27:1859–1867. doi: 10.1128/MCB.01395-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahiri DK, Maloney B, Ge YW. BACE1 gene promoter is differentially regulated: detection of a novel promoter region for its cell type-specific regulation. J Mol Neurosci. 2006;28:193–210. doi: 10.1385/JMN:28:2:193. [DOI] [PubMed] [Google Scholar]
- Leonardi R, Zhang YM, Rock CO, Jackowski S. Coenzyme A: back in action. Prog Lipid Res. 2005;44:125–153. doi: 10.1016/j.plipres.2005.04.001. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Li QJ, Chau J, Ebert PJ, Sylvester G, Min H, Liu G, Braich R, Manoharan M, Soutschek J, Skare P, Klein LO, Davis MM, Chen CZ. miR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell. 2007;129:147–161. doi: 10.1016/j.cell.2007.03.008. [DOI] [PubMed] [Google Scholar]
- Li Y, Zhou W, Tong Y, He G, Song W. Control of APP processing and Abeta generation level by BACE1 enzymatic activity and transcription. FASEB J. 2006;20:285–292. doi: 10.1096/fj.05-4986com. [DOI] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Lukiw WJ. Micro-RNA speciation in fetal, adult and Alzheimer's disease hippocampus. NeuroReport. 2007;18:297–300. doi: 10.1097/WNR.0b013e3280148e8b. [DOI] [PubMed] [Google Scholar]
- Lukiw WJ, Pogue AI. Induction of specific micro RNA (miRNA) species by ROS-generating metal sulfates in primary human brain cells. J Inorg Biochem. 2007;101:1265–1269. doi: 10.1016/j.jinorgbio.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marambaud P, Chevallier N, Ancolio K, Checler F. Post-transcriptional contribution of a cAMP-dependent pathway to the formation of alpha- and beta/gamma-secretases-derived products of beta APP maturation in human cells expressing wild-type and Swedish mutated beta APP. Mol Med. 1998;4:715–723. [PMC free article] [PubMed] [Google Scholar]
- Marsit CJ, Eddy K, Kelsey KT. MicroRNA responses to cellular stress. Cancer Res. 2006;66:10843–10848. doi: 10.1158/0008-5472.CAN-06-1894. [DOI] [PubMed] [Google Scholar]
- Mihailovich M, Thermann R, Grohovaz F, Hentze MW, Zacchetti D. Complex translational regulation of BACE1 involves upstream AUGs and stimulatory elements within the 5′ untranslated region. Nucleic Acids Res. 2007;35:2975–2985. doi: 10.1093/nar/gkm191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda KC, Huynh T, Tay Y, Ang YS, Tam WL, Thomson AM, Lim B, Rigoutsos I. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126:1203–1217. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- Miska EA, Alvarez-Saavedra E, Townsend M, Yoshii A, Sestan N, Rakic P, Constantine-Paton M, Horvitz HR. Microarray analysis of microRNA expression in the developing mammalian brain. Genome Biol. 2004;5:R68. doi: 10.1186/gb-2004-5-9-r68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourelatos Z, Dostie J, Paushkin S, Sharma A, Charroux B, Abel L, Rappsilber J, Mann M, Dreyfuss G. miRNPs: a novel class of ribonucleoproteins containing numerous microRNAs. Genes Dev. 2002;16:720–728. doi: 10.1101/gad.974702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson P, Kiriakidou M, Sharma A, Maniataki E, Mourelatos Z. The microRNA world: small is mighty. Trends Biochem Sci. 2003;28:534–540. doi: 10.1016/j.tibs.2003.08.005. [DOI] [PubMed] [Google Scholar]
- Nelson PT, Keller JN. RNA in brain disease: no longer just “the messenger in the middle.”. J Neuropathol Exp Neurol. 2007;66:461–468. doi: 10.1097/01.jnen.0000240474.27791.f3. [DOI] [PubMed] [Google Scholar]
- Nelson PT, Hatzigeorgiou AG, Mourelatos Z. miRNP:mRNA association in polyribosomes in a human neuronal cell line. RNA. 2004;10:387–394. doi: 10.1261/rna.5181104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson PT, Baldwin DA, Kloosterman WP, Kauppinen S, Plasterk RH, Mourelatos Z. RAKE and LNA-ISH reveal microRNA expression and localization in archival human brain. RNA. 2006;12:187–191. doi: 10.1261/rna.2258506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson PT, Jicha GA, Schmitt FA, Liu HL, Davis DG, Mendiondo MS, Abner E, Markesbery WR. Clinicopathological correlations in a large Alzheimer's disease center autopsy cohort: neuritic plaques and neurofibrillary tangles “do count” when staging disease severity. J Neuropathol Exp Neurol. 2007;66:1136–1146. doi: 10.1097/nen.0b013e31815c5efb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno M. Genetic and pharmacological basis for therapeutic inhibition of beta- and gamma-secretases in mouse models of Alzheimer's memory deficits. Rev Neurosci. 2006;17:429–454. doi: 10.1515/revneuro.2006.17.4.429. [DOI] [PubMed] [Google Scholar]
- Perkins DO, Jeffries CD, Jarskog LF, Thomson JM, Woods K, Newman MA, Parker JS, Jin J, Hammond SM. microRNA expression in the prefrontal cortex of individuals with schizophrenia and schizoaffective disorder. Genome Biol. 2007;8:R27. doi: 10.1186/gb-2007-8-2-r27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preece P, Virley DJ, Costandi M, Coombes R, Moss SJ, Mudge AW, Jazin E, Cairns NJ. Beta-secretase (BACE) and GSK-3 mRNA levels in Alzheimer's disease. Brain Res Mol Brain Res. 2003;116:155–158. doi: 10.1016/s0169-328x(03)00233-x. [DOI] [PubMed] [Google Scholar]
- Roldo C, Missiaglia E, Hagan JP, Falconi M, Capelli P, Bersani S, Calin GA, Volinia S, Liu CG, Scarpa A, Croce CM. MicroRNA expression abnormalities in pancreatic endocrine and acinar tumors are associated with distinctive pathologic features and clinical behavior. J Clin Oncol. 2006;24:4677–4684. doi: 10.1200/JCO.2005.05.5194. [DOI] [PubMed] [Google Scholar]
- Rossner S, Sastre M, Bourne K, Lichtenthaler SF. Transcriptional and translational regulation of BACE1 expression—implications for Alzheimer's disease. Prog Neurobiol. 2006;79:95–111. doi: 10.1016/j.pneurobio.2006.06.001. [DOI] [PubMed] [Google Scholar]
- Scarmeas N, Habeck CG, Hilton J, Anderson KE, Flynn J, Park A, Stern Y. APOE related alterations in cerebral activation even at college age. J Neurol Neurosurg Psychiatry. 2005;76:1440–1444. doi: 10.1136/jnnp.2004.053645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt FA, Davis DG, Wekstein DR, Smith CD, Ashford JW, Markesbery WR. “Preclinical” AD revisited: neuropathology of cognitively normal older adults. Neurology. 2000;55:370–376. doi: 10.1212/wnl.55.3.370. [DOI] [PubMed] [Google Scholar]
- Sevignani C, Calin GA, Siracusa LD, Croce CM. Mammalian microRNAs: a small world for fine-tuning gene expression. Mamm Genome. 2006;17:189–202. doi: 10.1007/s00335-005-0066-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SK. miRNAs: from neurogeneration to neurodegeneration. Pharmacogenomics. 2007;8:971–978. doi: 10.2217/14622416.8.8.971. [DOI] [PubMed] [Google Scholar]
- Stockley JH, O'Neill C. The proteins BACE1 and BACE2 and beta-secretase activity in normal and Alzheimer's disease brain. Biochem Soc Trans. 2007;35:574–576. doi: 10.1042/BST0350574. [DOI] [PubMed] [Google Scholar]
- Tahiliani AG, Beinlich CJ. Pantothenic acid in health and disease. Vitam Horm. 1991;46:165–228. doi: 10.1016/s0083-6729(08)60684-6. [DOI] [PubMed] [Google Scholar]
- Tang X, Gal J, Zhuang X, Wang W, Zhu H, Tang G. A simple array platform for microRNA analysis and its application in mouse tissues. RNA. 2007;13:1803–1822. doi: 10.1261/rna.498607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesco G, Koh YH, Kang EL, Cameron AN, Das S, Sena-Esteves M, Hiltunen M, Yang SH, Zhong Z, Shen Y, Simpkins JW, Tanzi RE. Depletion of GGA3 stabilizes BACE and enhances beta-secretase activity. Neuron. 2007;54:721–737. doi: 10.1016/j.neuron.2007.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rooij E, Sutherland LB, Liu N, Williams AH, McAnally J, Gerard RD, Richardson JA, Olson EN. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc Natl Acad Sci USA. 2006;103:18255–18260. doi: 10.1073/pnas.0608791103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassar R, Bennett BD, Babu-Khan S, Kahn S, Mendiaz EA, Denis P, Teplow DB, Ross S, Amarante P, Loeloff R, Luo Y, Fisher S, Fuller J, Edenson S, Lile J, Jarosinski MA, Biere AL, Curran E, Burgess T, Louis JC, et al. Beta-secretase cleavage of Alzheimer's amyloid precursor protein by the transmembrane aspartic protease BACE. Science. 1999;286:735–741. doi: 10.1126/science.286.5440.735. [DOI] [PubMed] [Google Scholar]
- Wang X, Wang X. Systematic identification of microRNA functions by combining target prediction and expression profiling. Nucleic Acids Res. 2006;34:1646–1652. doi: 10.1093/nar/gkl068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilfred BR, Wang WX, Nelson PT. Energizing miRNA research: a review of the role of miRNAs in lipid metabolism, with a prediction that miR-103/107 regulates human metabolic pathways. Mol Genet Metab. 2007;91:209–217. doi: 10.1016/j.ymgme.2007.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying SY, Lin SL. Intronic microRNAs. Biochem Biophys Res Commun. 2005;326:515–520. doi: 10.1016/j.bbrc.2004.10.215. [DOI] [PubMed] [Google Scholar]
- Ying SY, Lin SL. Current perspectives in intronic micro RNAs (miRNAs) J Biomed Sci. 2006;13:5–15. doi: 10.1007/s11373-005-9036-8. [DOI] [PubMed] [Google Scholar]
- Ying SY, Chang DC, Miller JD, Lin SL. The microRNA: overview of the RNA gene that modulates gene functions. Methods Mol Biol. 2006;342:1–18. doi: 10.1385/1-59745-123-1:1. [DOI] [PubMed] [Google Scholar]