Abstract
The formation of a metaphase spindle, a bipolar microtubule array with centrally aligned chromosomes, is a prerequisite for the faithful segregation of a cell’s genetic material. Using a full-genome RNA interference screen of Drosophila S2 cells, we identified about 200 genes that contribute to spindle assembly, more than half of which were unexpected. The screen, in combination with a variety of secondary assays, led to new insights into how spindle microtubules are generated; how centrosomes are positioned; and how centrioles, centrosomes, and kinetochores are assembled.
The diamond-shaped mitotic spindle has become one of the most widely recognized images in biology, emblematic of life’s propagation through cell division. In higher eukaryotes, the process of spindle formation begins after nuclear envelope breakdown (NEB) when microtubules (MTs), generated both from centrosomes and from the vicinity of chromatin, are organized into a bipolar array (1–5). Sister chromatids bind toMTs emanating from opposite poles, are aligned in the middle of the bipolar MT network, and then ultimately separate and move apart during anaphase. Failures early in mitosis result in the formation of an abnormal metaphase spindle, which can lead to mitotic delay and, potentially, chromosome missegregation during the ensuing anaphase.
To understand the mechanism of metaphase spindle assembly, it is critical to identify the proteins required for this process and then dissect how they function. Many mitotic proteins have been identified through genetic and RNAi screens (6–10), but the inventory is likely incomplete. Here, we present a genome-wide screen for mitotic spindle morphology in Drosophila S2 cells and the functional analysis of unexpected genes discovered through the screen.
Identification of genes involved in metaphase spindle formation by high-throughput microscopy
Because the percentage of S2 cells in mitosis is low (~1%), we conducted our RNAi screen in the presence of dsRNA (double-stranded RNA) to Cdc27 (a subunit of the anaphase-promoting complex) to delay anaphase and thereby increase the percentage of metaphase cells (~10% of the population). Thus, our screen was designed to investigate the assembly of the metaphase spindle, but not anaphase or cytokinesis. We also rescreened the final hits without Cdc27 RNAi–induced mitotic arrest. The majority (88%) showed identical phenotypes, although a few genes only manifest clear phenotypes under conditions of mitotic arrest (table S1).
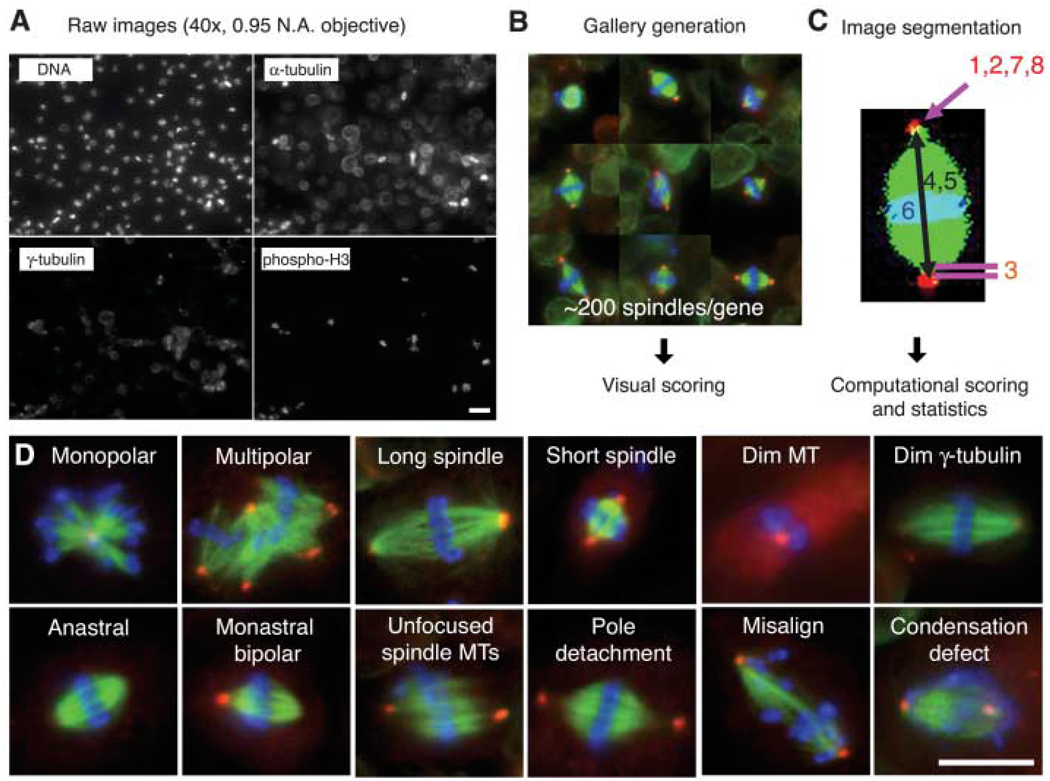
Using our custom, full-genome (14,425 genes) Drosophila RNAi library (11), we treated S2 cells with dsRNA for 4 days, conditions that generally reduce protein levels by >80% (12, 13). After dsRNA treatment, cells were fixed and stained for DNA, γ-tubulin, MT, and phosphohistone H3 (pH3) in 96 well plates, and about 40 sites per well were imaged by automated microscopy with a high numerical aperture air objective to obtain relatively high-resolution images of mitotic spindles (Fig. 1A). To reduce the complexity of this large amount of image data, a custom computer code was used to identify, crop, and arrange mitotic spindles into galleries, which were then blindly scored by an observer (Fig. 1B and fig. S1). In addition, computer algorithms measured eight parameters of spindle shape, as well as the intensity of γ-tubulin, cell number, and mitotic index (Fig. 1C) (11). More than 4,000,000 spindles were analyzed in this screen.
Fig. 1.
An image-based, genome-wide RNAi screening ofmetaphase spindlemorphology. (A) Drosophila S2 cells were treated with dsRNAs of 14,425 genes, along with Cdc27/Apc3 dsRNA, to accumulate metaphase cells. After 4 days, cells were transferred and adhered onto ConA-coated glass-bottom plates. Cells were immunostained for DNA, α-tubulin, γ-tubulin and pH3 and imaged by high-throughput automated microscopy. Scale bar, 20 µm. (B) Mitotic cells were automatically selected by a computer algorithm that detects pH3 staining. The selected ~200 mitotic cells (mostly metaphase) were displayed in a gallery for an observer to score phenotypes. (C) Phenotypes were also analyzed by computer after image segmentation (1, monopolar; 2, multipolar; 3, pole detachment; 4, long spindle; 5, short spindle; 6, misalignment; 7, large γ-tubulin area; 8, dim γ-tubulin) (11). (D) Twelve major phenotypes identified in the screen. Scale bar, 10 µm.
Before beginning this screen, we annotated 49 genes that produce mitotic defects in S2 cells (table S2). Of these 49 genes, 45 were identified as hits in the primary screen, indicating a high success rate of identifying mitotic phenotypes. However, our final list of genes should not be considered as a complete or universal inventory, because genes can be missed (particularly those with subtle phenotypes), and some phenotypes (or lack thereof) may be specific to S2 cells. False positives by off-target effects of dsRNA can occur in RNAi screens (14, 15), so precautions were taken to minimize gene overlap in the dsRNA design, and all unexpected hits were confirmed with another dsRNA that had no overlap with the first dsRNA (11). To learn more about the functions of interesting genes, we determined protein localization by green fluorescence protein (GFP) tagging (38 genes, mostly tagged at both N and C termini to be certain of the localization pattern) (table S1 and figs. S3 to S7), analyzed RNAi phenotypes by live cell imaging, and/or examined the effect of gene knockdown on the localization of kinetochore or centrosome proteins.
Our screen identified ~150 unexpected or unknown genes that produced mitotic RNAi phenotypes, each of which was confirmed by 2 to 6 repeat experiments (Fig. 1D). In the following sections, we describe the roles of a subset of these genes in centrosome and γ-tubulin function, the shape and dynamics of the poles, and spindle size and chromosome alignment. The complete list of genes identified in the screen can be found in table S1 as well as our Web site, which also contains primary data on RNAi constructs, phenotypes, and protein localization (fig. S2 and http://rnai.ucsf.edu/mitospindlescreen).
Centrosomes and γ-tubulin localization
In preparation for mitosis, the centrioles duplicate, creating two γ-tubulin–containing centrosomes that nucleate MTs and ultimately become the poles of the mitotic spindle. However, centrioles are not needed for most cell divisions in flies, as MT nucleation around chromatin suffices for bipolar spindle formation (13, 16, 17). In our screen, depletion of proteins involved in centriole duplication would be expected to produce a mixture of anastral spindles (no γ-tubulin staining at the poles) and monastral bipolar spindles (only one pole with normal γ-tubulin staining), because centriole numbers only gradually diminished with dilution through successive cell cycles in a 4-day dsRNA treatment (Fig. 2A and fig. S3A).
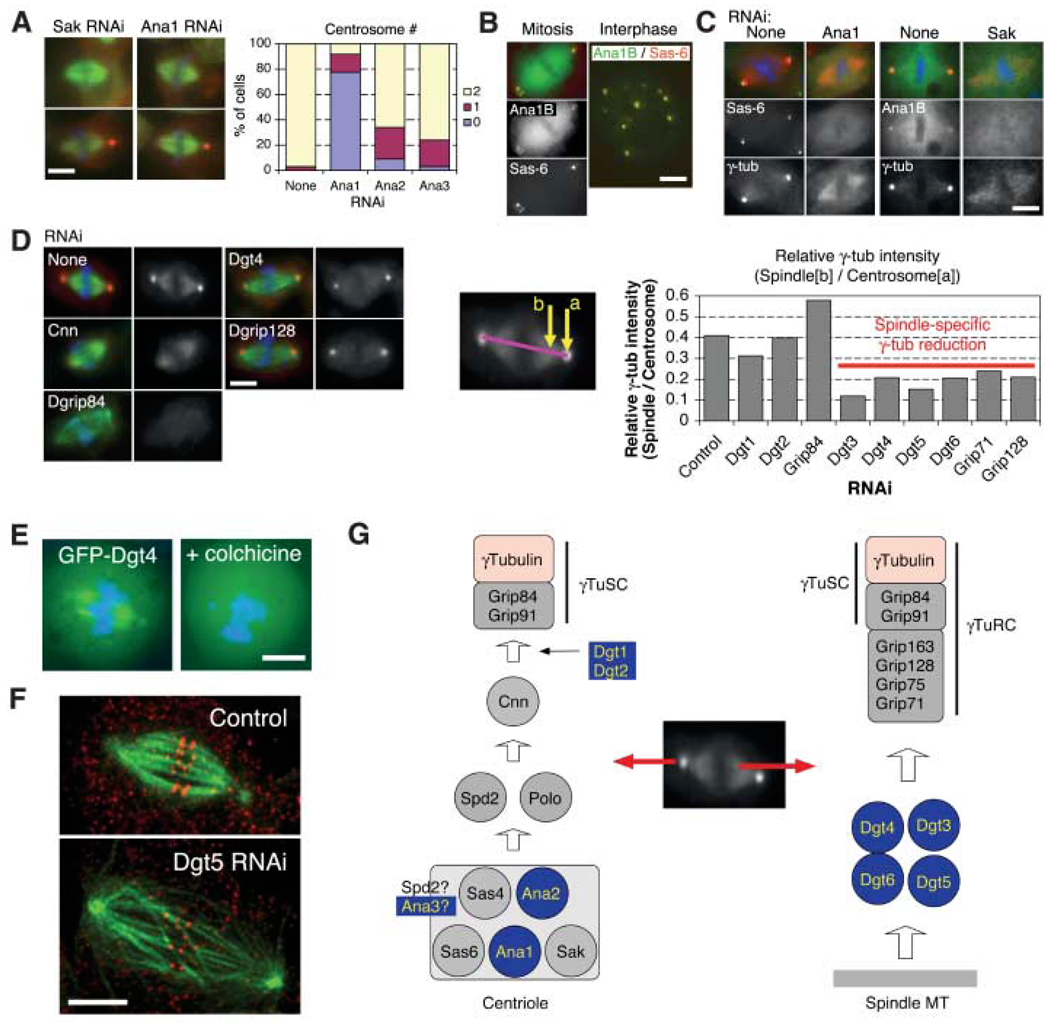
Fig. 2.
Genes required for localizing γ-tubulin to the centrosome and the spindle. (A) (Left) Anastral as well as monastral bipolar spindles were observed after RNAi to Sak or three previously unknown Ana genes (the latter two shown in fig. S3). (Right) Centrosome number was counted for bipolar spindles after four rounds of RNAi treatment. (B) Ana1 (isoform B) was colocalized with the centriole marker mRFP-Sas-6. (C) Ana1 RNAi caused the loss of GFP-Sas-6 from the pole region of anastral spindles, whereas SAK RNAi interfered with GFP-Ana1B localization to poles. (D) (Left) Various dim γ-tubulin RNAi phenotypes. The γ-tubulin signal is reduced at the centrosome alone (Cnn), at both the centrosome and spindle (Dgrip84), or only at the spindle (Dgt4 and Dgrip128). (Right) Intensity at centrosome (a) and spindle region (b) was measured, and the relative intensity was calculated (11). (E) Spindle localization of GFP-Dgt4 is lost after colchicine-induced MT depolymerization. (F) RNAi of Dgt5 (as well as other genes required for spindle localization of γ-tubulin) produces long spindles with low MT density and misaligned chromosomes. Shown is a maximum intensity projection of four optical slices obtained at 0.25-µm intervals by spinning-disk confocal microscope. Red, antibody to CENP-A; green, MT. Scale bar, 5 µm. (G) Model for recruitment pathway of γ-tubulin to the centrosome and spindle MTs. Previously unknown or unexpected genes are highlighted in blue in this and other figures.
Our screen identified several known proteins (Sak kinase, DSas-4, and Sas-6), as well as three previously unknown genes [anastral spindle phenotype (Ana)]. Consistent with their RNAi phenotype, GFP-Ana1 and -Ana2 colocalized during interphase and mitosis with the centriolar markers mRFP-Sas-6 (Fig. 2B and fig. S3C) and Sak and DSas-4 (fig. S3D). GFP-Ana1 was not detected at anastral spindle poles after Sak RNAi, and RNAi depletion of Ana1 resulted in a substantial decrease in GFP-Sas6 fluorescence from spindle poles, suggesting an important role in centriole formation (Fig. 2C). Thus, Ana1 and Ana2 (and possibly Ana3) may be core components of the centriole that are necessary for centriole duplication.
RNAi of the known genes Spd-2, Polo, centrosomin, Dgrip84, and Dgrip91 [these Dgrips make up the stable core (γTuSC) of the γ-tubulin ring complex (γTuRC)] decreased γ-tubulin staining at the pole without interfering with centriole marker localization (fig. S3, F and G). By examining the effects of RNAi of these genes on centrosomin and γ-tubulin localization, we suggest a molecular pathway leading to γ-tubulin localization at the spindle pole (Fig. 2G). We also found two previously unknown genes, Dgt1 and Dgt2 (dim γ-tubulin), that decreased spindle pole γ-tubulin staining without affecting centrosomin [a known γ-tubulin localization factor (18)]. RNAi of these genes also produced long spindles, a phenotype characteristic of γ-tubulin RNAi, thus further suggesting a role in γ-tubulin function.
In addition to centrosome localization, a subset of γ-tubulin localizes to the spindle (16, 19), where it might contribute to MT nucleation within the spindle (16). Phosphorylation of a γTuRC subunit is required for spindle localization of γ-tubulin in mammalian cells (19), but otherwise, little is known about this population of spindle-localized γ-tubulin. In our screen, we identified genes that are needed to localize γ-tubulin to the spindle but not the pole (Fig. 2D and fig. S4A) (11). Among these are components of the γTuRC (Dgrip71, Dgrip75, Dgrip128, and Dgrip163). RNAi of several previously unknown genes (Dgt3 to Dgt6) also diminished γ-tubulin selectively within the spindle compared with the pole. Consistent with a role in targeting γ-tubulin to the spindle, GFP-tagged Dgt4, Dgt5, and Dgt6 localized uniformly on spindle MTs, with no enrichment at the centrosome; the spindle staining was lost upon MT depolymerization and was cell cycle dependent (noMT localization in interphase) (Fig. 2E and fig. S4B). High-throughput, automated imaging of living cells expressing mCherry-tubulin and H2B-GFP as well as high-resolution confocal imaging of MTs further revealed that RNAi of these Dgts reducedMT density inside the spindle, increased monopolar spindle formation, and caused chromosome/kinetochore misalignment and mitotic delay (Fig. 2F; and movie S1 of Dgt5 RNAi). Recently, γTuRC was implicated in the spindle-assembly checkpoint (SAC) signaling through binding Cdc20 and BubR1 (20), but our results suggest that the loss of proper kinetochore-MT interactions after removal of spindle-localized γTuRC may constitute the primary reason for failure to satisfy the SAC.
This study suggests two pathways for γ-tubulin localization in mitosis (Fig. 2G). At the spindle pole, a core set of proteins build centrioles (Ana1, Ana2, Sak, DSas-4, and Sas-6), which provide scaffolds for Spd-2, polo kinase, and centrosomin to recruit γ-tubulin through the γTuSC subunits. However, this pathway is dispensable for cell division in S2 cells. A second pathway involving a new set of proteins (Dgt3–6) and the outer γTuRC subunits recruit γ-tubulin to spindle MTs. Surprisingly, this site of γ-tubulin function is more important than the centrosome in building a normal-length bipolar spindle with properly aligned chromosomes.
The shape and dynamics of the poles
Mitotic spindles normally have two well-focused poles. In our screen, we identified a series of genes that affect the number or organization of spindle poles (table S1 and fig. S5). Unfocused kinetochore fibers (K-fibers), a rare phenotype, were obtained for RNAi of calmodulin (CaM), a broadly functioning calcium effector (Fig. 3A). By time-lapse imaging, CaM RNAi caused K-fiber detachment from centrosomes, a finding that mimicked observations for RNAi of Abnormal Spindle protein (Asp) (movie S2) (21). Like Asp, CaM localized at the minus ends of K fibers, even when these ends were disconnected from the centrosome byNcd/Dhc RNAi (Fig. 3B). Furthermore, CaM and Asp mutually depended upon one another for localization to the minus end of K fibers (fig S5F). Thus, Asp, which has multiple, predicted CaM-binding IQ motifs (22), requires CaM for its localization and function. Surprisingly, the specificity of the CaM RNAi phenotype suggests that CaM’s main function during mitosis is serving as a cofactor in Asp-mediated pole-focusing.
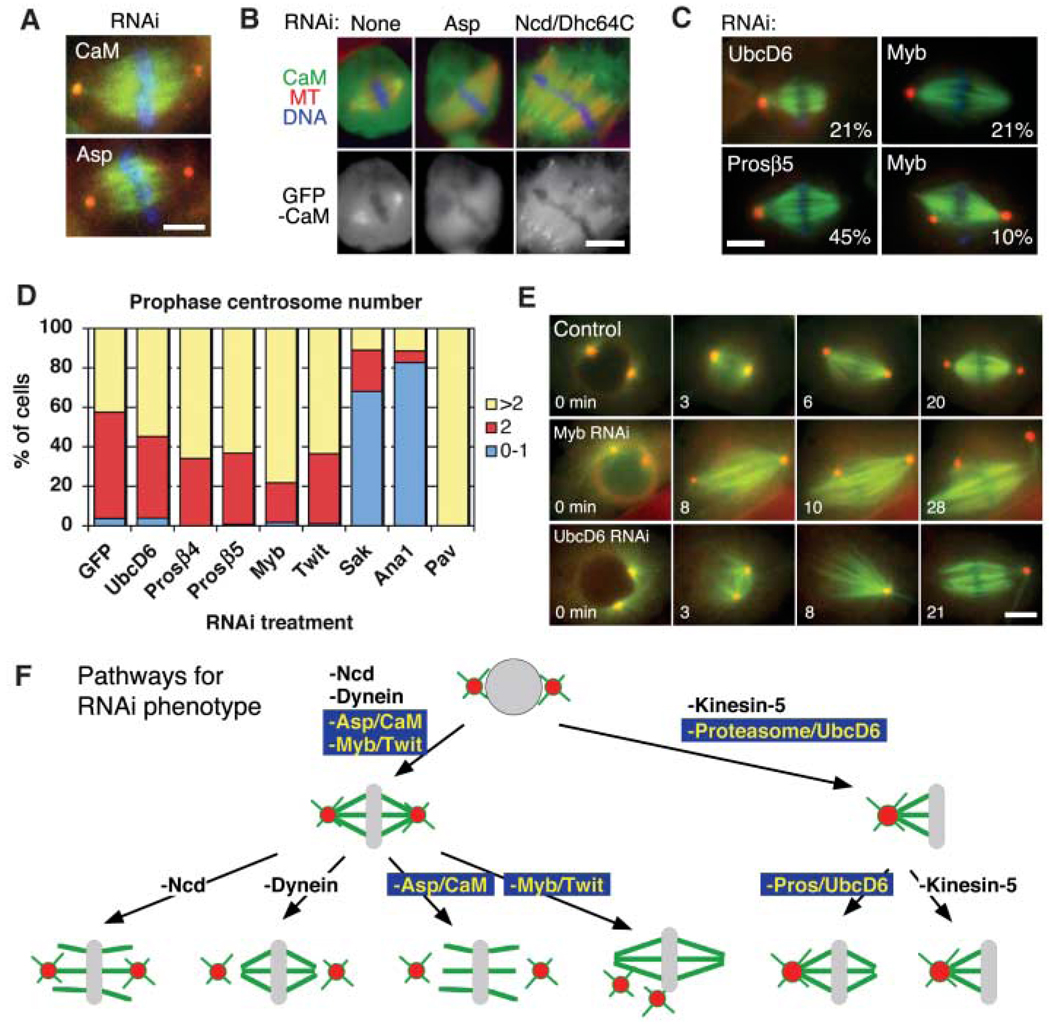
Fig. 3.
Genes required for centrosome separation and association to spindle poles. (A) Identical kinetochore-fiber unfocusing and centrosome-detachment phenotypes were observed after Asp and calmodulin (CaM) RNAi. See movie S2. (B) GFP-CaM was localized at spindle poles in an Asp-dependent manner, but was still detected at unfocused poles induced by Ncd/Dhc64C RNAi. (C) Monastral bipolar spindles produced by UbcD6, proteasome subunit β5, or Myb RNAi (percentage of cells with phenotype indicated; 3% to 8% for untreated cells). Centrosome detachment also was detected after Myb RNAi. (D) At prophase, most proteosome (Prosβ4 and Prosβ5), UbcD6, Myb, and Twit RNAi cells (n > 65) had >1 centrosome; Sak, Ana1 (centriole duplication), and Pav (cytokinesis) served as control RNAis that decrease or increase centrosome numbers. (E) Time-lapse imaging of γ-tubulin-GFP (red) and mCherry-tubulin (green) from prophase to metaphase shows bipolar spindle formation, and then centrosome detachment after Myb RNAi (80%) (n = 20), and immediate centrosome fusion after NEB for UbcD6 RNAi (25%) (n = 20). See movie S3. Scale bars, 5 µm. (F) Models for various spindle-pole phenotypes in response to various protein depletions.
An unexpected phenotype was increased numbers of monastral bipolar (but not anastral) spindles after RNAi of multiple proteasome subunits, the E2 ubiquitin conjugating enzyme UbcD6, the transcription factor Myb, and a Myb-interacting protein (Twit) (Fig. 3C). Proteasome RNAi showed other phenotypes, such as severe proliferation defects, but the monastral bipolar spindle phenotype was not characteristic of other RNAi treatments that reduced cell numbers. Moreover, RNAi of UbcD6 did not inhibit growth.
To better understand this phenotype, we scored the number of γ-tubulin–staining centrosomes in prophase to determine whether centrosome numbers were reduced (Fig. 3D). However, comparable (proteasome and UbcD6 RNAi) or higher [Myb and twit RNAi; see also (23)] numbers of prophase centrosomes were observed compared with control cells, suggesting that these RNAi treatments caused centrosome fusion after NEB to generate monastral bipolar spindles. To test this idea, we performed automated, live imaging of cells coexpressing γ-tubulin-GFP and mCherry-tubulin (Fig. 3E and movie S3). Myb RNAi cells initially formed normal bipolar (or occasionally multipolar) spindles after NEB. However, subsequently, one or more of the centrosomes detached from the poles and wandered toward the center [in contrast to dynein RNAi, which caused centrosome to detach and move away from the spindle (24)]. In some instances, centrosome fusion was observed, creating a monastral bipolar spindle as seen in fixed cells. This phenotype may reflect a direct action of Myb at the centrosome or could be mediated indirectly through its role in gene transcription. In contrast, RNAi of UbcD6 induced centrosome fusion shortly after NEB, generating monopolar spindles, which then converted into monastral bipolar spindles by chromatin-dependent MT generation.
Thus, we observed two previously unreported phenotypes concerning centrosome dynamics. Loss of a ubiquitin-dependent proteolysis pathway induced excess centrosome fusion, and Myb complex depletion caused centrosomes to reposition toward the spindle interior (Fig. 3F).
Spindle size and chromosome alignment
In addition to identifying known regulators of S2 cell spindle length (25), the screen also identified several previously unknown genes that produced short-spindle RNAi phenotypes [short spindle (Ssp) genes], and three of these localized to mitotic spindles (Fig. 4A, table S1, fig. S6). Depletion of one of these proteins [CG33130 (termed Ssp4)] caused pronounced MT severing in interphase cells (Fig. 4B and movie S4), a surprising effect because severing is rarely observed in untreated cells. The severed MTs often continued to grow at their plus ends and depolymerize at their minus ends, causing them to treadmill through the cytoplasm (26). Thus, Ssp4 regulates MT severing, and the shorter-spindle phenotype might be due to enhanced MT severing and depolymerization at the poles.
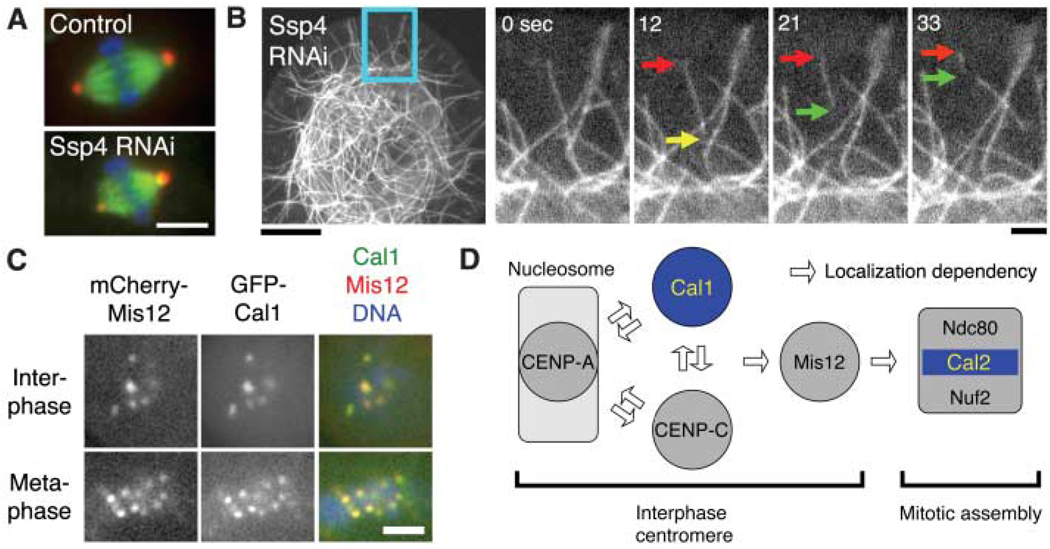
Fig. 4.
Regulation of spindle length and chromosome alignment. (A) Spindle length was altered after RNAi depletion of the novel protein Ssp4. Scale bar, 5 µm. (B) MT severing (yellow arrow) frequently occurred after Ssp4 RNAi. Severed MTs often showed treadmilling behavior (red and green arrows) and then disappeared. Scale bars, 10 µm (left), 2 µm (right). See also movie S4. (C) Previously unknown Cal1 protein localizes to the centromere (marked by mCherry-Mis12). (Localization data for other proteins are in fig. S7). Scale bar, 2 µm. (D) Model for kinetochore assembly in S2 cells based on protein localization and RNAi. (Data are in fig. S7, D to F).
Our screen also identified genes involved in chromosome alignment (table S1, fig. S7, and movie S5). Chromosome misalignment frequently coincided with an increase in spindle length (table S1), consistent with a model in which defective chromosome-MT interactions result in an imbalance of forces acting upon the spindle (25). Five genes with severe-misalignment RNAi phenotypes encode proteins that localized at the kinetochore, and two (CG18156 and CG5148) also localized at centromeres in interphase nuclei (Fig. 4C and fig. S7, B and C). These five genes were unannotated at the time of our screen, but sequence alignments performed by our group and others (27) identified CG9938, CG8902, and CG18156 as fly homologs of Ndc80/Hec1, Nuf2, and Mis12, respectively.
Two genes, CG5148 [chromosome alignment defect (Cal1)] and CG7242 (Cal2) did not display sequence similarity to known kinetochore proteins. To understand how these proteins are integrated and function in the molecular assembly pathway of the Drosophila kinetochore, we investigated the localization dependency of the kinetochore proteins using GFP fusion proteins and RNAi. RNAi of Cal2 affected mitotic localization of Ndc80 and Nuf2, but not the constitutive centromere proteins Cal1, Mis12, and CENP-A/Cid, similar to results for Spc25 RNAi in HeLa cells (28). Thus, Cal2 [a 26-kD protein with known two-hybrid interaction with Ndc80 (29)] may be Drosophila Spc25. RNAi of Cal1, on the other hand, affected localization of all markers tested (Fig. 4D; and fig. S7, D to F). Different from other systems (30), centromere localizations of CENP-A, CENP-C, and Cal1 were mutually dependent, because RNAi depletion of any single protein disrupted or diminished the localization of the other two. Thus, S2 cell inner kinetochore formation may involve a coassembly process of CENP-A, -C, and the previously unknown protein Cal1 (Fig. 4D). Once these core proteins are assembled, Mis12 and the Ndc80 outer kinetochore complex are recruited in a linear pathway, similar to that described for C. elegans embryos (31).
Several known genes, including transcription factors (Spt), chromatin-binding proteins (Dmt/Dalmatian), and signaling proteins [target of rapamycin (TOR)–associated protein (Raptor) and the GTPase RheB], also produced unexpected RNAi chromosome misalignment phenotypes (table S1). The misalignment effects of Raptor and RheB RNAi, as well as those produced by the TOR inhibitor rapamycin (fig. S7H), were only observed with Cdc27 arrest. RNAi of numerous spliceosome components also caused chromosome misalignment. Although this finding is largely unexpected, mutations in splicing factors cause missegregation of mini-chromosomes in yeast (32). RNA splicing might regulate proteins involved in kinetochore structure or generate RNAs that have structural roles within the spindle (33). Thus, chromatin structure, RNA, and signaling pathways can influence chromosome alignment, although the mechanisms of these effects remain to be understood at a molecular level.
Conclusion
This morphological screen of the Drosophila mitotic spindle was made possible by computer-assisted identification of mitotic cells for visual scoring and quantitative computational image analysis. However, the resultant ~200-gene RNAi “hit” list, by itself, was insufficient to gain new insight into spindle formation. A suite of secondary assays was needed to decipher the site of action and mechanism of previously unknown proteins, as well as to develop an integrated understanding of how these proteins work together.
In addition to implicating many unexpected genes, this study has revealed unanticipated processes involved in spindle assembly. For example, the identification of Dgt proteins led to the finding that the activity of γ-tubulin within the spindle is more critical for spindle architecture and chromosome alignment than its better-known function at the centrosome. We also uncovered unanticipated RNAi phenotypes, such as excessive centrosome fusion (UbcD6 RNAi), centrosome detachment and motion within the spindle (Myb RNAi), and activation of MT severing (Ssp4 RNAi). Interfering with general cellular machines also gave rise to distinct spindle defects such as those seen with the proteasome (monastral bipolar spindles), RNA polymerase II (long spindles), and the spliceosome (chromosome misalignment). This work also provides a systematic analysis of the assembly of Drosophila centrosomes and inner kinetochores, ordering several proteins into these pathways. Many of the molecules and pathways described in this study are likely to be conserved in human cells. Therefore, the alterations in centrosome function and chromosome alignment observed in this screen may provide insight into how these commonly observed defects arise in human cancers (34–36).
Supplementary Material
Footnotes
Supporting Online Material
www.sciencemag.org/cgi/content/full/1141314/DC1
Materials and Methods
Figs. S1 to S7
Tables S1 and S2
References
Movies S1 to S5
Web site S1
References and Notes
- 1.Mitchison TJ, Salmon ED. Nat. Cell Biol. 2001;3:E17. doi: 10.1038/35050656. [DOI] [PubMed] [Google Scholar]
- 2.McIntosh JR, Grishchuk EL, West RR. Annu. Rev. Cell Dev. Biol. 2002;18:193. doi: 10.1146/annurev.cellbio.18.032002.132412. [DOI] [PubMed] [Google Scholar]
- 3.Rieder CL, Khodjakov A. Science. 2003;300:91. doi: 10.1126/science.1082177. [DOI] [PubMed] [Google Scholar]
- 4.Gadde S, Heald R. Curr. Biol. 2004;14:R797. doi: 10.1016/j.cub.2004.09.021. [DOI] [PubMed] [Google Scholar]
- 5.Scholey JM, Brust-Mascher I, Mogilner A. Nature. 2003;422:746. doi: 10.1038/nature01599. [DOI] [PubMed] [Google Scholar]
- 6.Yanagida M. Trends Cell Biol. 1998;8:144. doi: 10.1016/s0962-8924(98)01236-7. [DOI] [PubMed] [Google Scholar]
- 7.Doheny KF, et al. Cell. 1993;73:761. doi: 10.1016/0092-8674(93)90255-O. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nasmyth K. Science. 2002;297:559. doi: 10.1126/science.1074757. [DOI] [PubMed] [Google Scholar]
- 9.Bettencourt-Dias M, et al. Nature. 2004;432:980. doi: 10.1038/nature03160. [DOI] [PubMed] [Google Scholar]
- 10.Sonnichsen B, et al. Nature. 2005;434:462. doi: 10.1038/nature03353. [DOI] [PubMed] [Google Scholar]
- 11.Materials and methods are available as supporting material on Science Online.
- 12.Rogers SL, Wiedemann U, Stuurman N, Vale RD. J. Cell Biol. 2003;162:1079. doi: 10.1083/jcb.200303023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goshima G, Vale RD. J. Cell Biol. 2003;162:1003. doi: 10.1083/jcb.200303022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma Y, Creanga A, Lum L, Beachy PA. Nature. 2006;443:359. doi: 10.1038/nature05179. [DOI] [PubMed] [Google Scholar]
- 15.Kulkarni MM, et al. Nat. Methods. 2006;3:833. doi: 10.1038/nmeth935. [DOI] [PubMed] [Google Scholar]
- 16.Mahoney NM, Goshima G, Douglass AD, Vale RD. Curr. Biol. 2006;16:564. doi: 10.1016/j.cub.2006.01.053. [DOI] [PubMed] [Google Scholar]
- 17.Basto R, et al. Cell. 2006;125:1375. doi: 10.1016/j.cell.2006.05.025. [DOI] [PubMed] [Google Scholar]
- 18.Li K, Kaufman TC. Cell. 1996;85:585. doi: 10.1016/s0092-8674(00)81258-1. [DOI] [PubMed] [Google Scholar]
- 19.Luders J, Patel UK, Stearns T. Nat. Cell Biol. 2006;8:137. doi: 10.1038/ncb1349. [DOI] [PubMed] [Google Scholar]
- 20.Muller H, Fogeron ML, Lehmann V, Lehrach H, Lange BM. Science. 2006;314:654. doi: 10.1126/science.1132834. [DOI] [PubMed] [Google Scholar]
- 21.Morales-Mulia S, Scholey JM. Mol. Biol. Cell. 2005;16:3176. doi: 10.1091/mbc.E04-12-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saunders RD, Avides MC, Howard T, Gonzalez C, Glover DM. J. Cell Biol. 1997;137:881. doi: 10.1083/jcb.137.4.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fung SM, Ramsay G, Katzen AL. Development. 2002;129:347. doi: 10.1242/dev.129.2.347. [DOI] [PubMed] [Google Scholar]
- 24.Maiato H, Rieder CL, Khodjakov A. J. Cell Biol. 2004;167:831. doi: 10.1083/jcb.200407090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goshima G, Wollman R, Stuurman N, Scholey JM, Vale RD. Curr. Biol. 2005;15:1979. doi: 10.1016/j.cub.2005.09.054. [DOI] [PubMed] [Google Scholar]
- 26.Rodionov VI, Borisy GG. Science. 1997;275:215. doi: 10.1126/science.275.5297.215. [DOI] [PubMed] [Google Scholar]
- 27.Meraldi P, McAinsh AD, Rheinbay E, Sorger PK. Genome Biol. 2006;7:R23. doi: 10.1186/gb-2006-7-3-r23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bharadwaj R, Qi W, Yu H. J. Biol. Chem. 2004;279:13076. doi: 10.1074/jbc.M310224200. [DOI] [PubMed] [Google Scholar]
- 29.Giot L, et al. Science. 2003;302:1727. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- 30.Chan GK, Liu ST, Yen TJ. Trends Cell Biol. 2005;15:589. doi: 10.1016/j.tcb.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 31.Desai A, et al. Genes Dev. 2003;17:2421. doi: 10.1101/gad.1126303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takahashi K, Yamada H, Yanagida M. Mol. Biol. Cell. 1994;5:1145. doi: 10.1091/mbc.5.10.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blower MD, Nachury M, Heald R, Weis K. Cell. 2005;121:223. doi: 10.1016/j.cell.2005.02.016. [DOI] [PubMed] [Google Scholar]
- 34.Storchova Z, Pellman D. Nat. Rev. Mol. Cell Biol. 2004;5:45. doi: 10.1038/nrm1276. [DOI] [PubMed] [Google Scholar]
- 35.Nigg EA. Nat. Rev. Cancer. 2002;2:815. doi: 10.1038/nrc924. [DOI] [PubMed] [Google Scholar]
- 36.Yuen KW, Montpetit B, Hieter P. Curr. Opin. Cell Biol. 2005;17:576. doi: 10.1016/j.ceb.2005.09.012. [DOI] [PubMed] [Google Scholar]
- 37.We thank K. Pollard for valuable suggestions on statistics; Y. Guo, I. Vasenkova, and R. De Breuil for help in dsRNA synthesis at Open Biosystems Inc.; I. Cheeseman for advice on homology searches; A. Straight for sharing data in advance of publication; and S. Henikoff, M. Bettencourt-Dias, D. Glover, T. Kaufman, D. Agard, R. Tsien, and E. Griffis for reagents. G.G. received support from the Human Frontier Science Program and R.W. from a University of California GREAT Training Grant. We thank the Sandler Foundation (UCSF) for support.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.